Abstract
We investigated the role of variation in putative psychosis genes coding for elements of the white matter system by examining the contribution of genotypic variation in three single-nucleotide polymorphisms (SNPs) neuregulin 1 (NRG1) SNP8NRG221533, myelin oligodendrocytes glycoprotein (MOG) rs2857766 and CNP (rs2070106) and one haplotype HAPICE (deCODE) to white matter volume in patients with psychotic disorder and their unaffected relatives. Structural magnetic resonance imaging and blood samples for genotyping were collected on 189 participants including patients with schizophrenia (SZ) or bipolar I disorder (BDI), unaffected first-degree relatives of these patients and healthy volunteers. The association of genotypic variation with white matter volume was assessed using voxel-based morphometry in SPM5. The NRG1 SNP and the HAPICE haplotype were associated with abnormal white matter volume in the BDI group in the fornix, cingulum and parahippocampal gyrus circuit. In SZ the NRG1 SNP risk allele was associated with lower white matter volume in the uncinate fasciculus (UF), right inferior longitudinal fasciculus and the anterior limb of the internal capsule. Healthy G-homozygotes of the MOG SNP had greater white matter volume in areas of the brainstem and cerebellum; this relationship was absent in those with a psychotic disorder and the unaffected relatives groups. The CNP SNP did not contribute to white matter volume variation in the diagnostic groups studied. Variation in the genes coding for structural and protective components of myelin are implicated in abnormal white matter volume in the emotion circuitry of the cingulum, fornix, parahippocampal gyrus and UF in psychotic disorders.
Keywords: CNP, HAPICE, MOG, NRG1, voxel-based morphometry, white matter
Introduction
Bipolar I disorder (BDI) and schizophrenia (SZ) share a number of common genetic risk loci and susceptibility genes.1 Several of these converge on pathways that regulate oligodendrocytes proliferation, assembly, protection and degeneration. These include the genes coding for neuregulin 1 (NRG1)2 and myelin oligodendrocytes glycoprotein (MOG)3, 4 and in SZ the 2′,3′-cyclic nucleotide 3′ phosphodiesterase enzyme (E.C. 3.1.4.37, CNPase) genes.5 Postmortem gene expression and mRNA levels support a regional specificity for the effects of these risk alleles in SZ and to some extent in BD. In particular, differential expression for several genes involved in myelination has been reported in the dorsolateral prefrontal cortex of postmortem brains from individuals with an ante-mortem diagnosis of SZ.6 However, brain-wide analyses of such relationships are not yet available.
Non-invasive in vivo structural magnetic resonance imaging (MRI) provides a means by which we can examine the regional effects of these risk polymorphisms that code for structural and protective elements of white matter, on the volume of white matter throughout the brain. Moreover, incorporating groups of affected and related individuals who are unaffected provides the potential to implicate genetic markers potentially responsible for white matter dysfunction that contribute to the development of SZ and BD, and those that confer endophenotypic risk.
NRG1 single-nucleotide polymorphism (SNP)8NRG221533 and the HAPICE (deCODE) haplotype
The NRG1 gene is located on chromosome 8p13 within the SZ susceptibility loci identified at 8p22–p12. The 8p region7, 8, 9 and several loci in the NRG1 gene2 confer risk for SZ. In particular, the C-allele of the NRG1 SNP, SNP8NRG221533 (rs35753505) located in the noncoding 5′-flanking region of the NRG1 gene, was implicated as giving the best uncorrected single marker association in the Icelandic study and is part of a core haplotype consisting of five SNPs and two microsatellites termed hereafter the HAPICE haplotype.10 NRG1 SNP8NRG221533 has been associated with SZ in Icelandic, Scottish, British/Irish, Dutch, African American, South African and Han Chinese populations10, 11, 12, 13, 14 but not in one Finnish15 and one Irish population,16 and in a meta-analysis.17
The NRG1 gene codes for six types of neuronal NRG1, of which types 1 and 4 are implicated in SZ. NRG1 binds the epidermal growth factor receptor family and receptor tyrosine-protein kinase erbB-3 and is involved in axonal myelination,18, 19, 20 and NMDA receptor signaling21 among a number of other functions.2 In the prefrontal cortex in SZ and BD, altered expression and protein levels of the NRG1-ErbB signaling system have been detected but are not consistent. These reports include increased expression of ErbB4,22 reduced expression of ErbB3, the alpha isoform of NRG1 (which is increased in hippocampus23) and NRG1 mRNA,24 and no change in expression or protein levels of NRG1.25, 26, 27 However, these studies may be confounded by the effects of antipsychotics medication.28 Finally, in animal studies, NRG1 heterozygous mice have a behavioral phenotype that overlaps with the signs and symptoms of SZ,11 and in NRG1 hypomorphs these are partially reversed by clozapine administration.11, 29, 30 The functional implications of altered expression, protein levels or signaling via NRG1-ErbB include abnormal neuronal growth, neurodegeneration and reduced glutamatergic signaling among other effects.2, 27, 31, 32
In vivo imaging in the human brain has provided further evidence for the role of the NRG1 SNP8NRG221533 in white matter pathophysiology in SZ. C-carriers with SZ had reduced microstructural uniformity in the organization (fractional anisotropy (FA)) of their white matter on the left anterior thalamic radiation33 and the subcortical medial frontal region34 relative to the TT-genotype group. However, a contrasting study reported reduced FA in the anterior cingulum in the T-carriers relative to the CC-genotype.35 The risk genotype (TT) of a NRG1 promoter region SNP8NRG243177 has been associated with reduced FA and white matter volume in the anterior limb of the internal capsule (ALIC)36 and in the anterior thalamic radiation33 relative to the C-carriers. Using functional MRI, the latter risk genotype has additionally been associated with reduced medial prefrontal (BA9) and temporo–occipital junction (BA39 and BA19) activation while performing a sentence completion task, and the development of psychotic symptoms, as well as a lower premorbid IQ.37 This relationship was not present with the SNP examined in this study, SNP8NRG221533.
MOG rs2857766
Oligodendroglia abnormalities have been observed in both SZ and BD38 with increasing frequency in recent years. In SZ a structural component of myelin, MOG has been found to be differentially expressed in the dorsolateral prefrontal cortex,39 thalamus40 and anterior temporal lobe,41 and MOG mRNA levels were lower in several regions examined.42 However, MOG mRNA levels did not differ in one study in individuals with BD.42 Negative findings for association of MOG gene variation and SZ also exist: four polymorphisms in MOG ((CA)n, (TAAA)n, C1334T and C10991T) were examined in a family study of 111 probands and transmission frequency was not significantly associated with disease.43 Weak but positive associations with SZ have been detected for three MOG microsatellites in a Han Chinese population.44 Despite these inconclusive data, myelin as a structural axonal component restricting diffusion, have been indirectly implicated in studies detecting reduced diffusion parameters that represent the orientation and microstructural organization of white matter in SZ and BD.45, 46 In the human MOG gene (6p21.3) a missense variation in a SNP rs2857766 (511G>C, V142L, constitutively spliced exon-3 coding for a transmembrane segment of the MOG protein47) has been implicated as an independent MS susceptibility-modulating factor in the histocompatibility complex class I region, suggesting a possible role in structural degradation of myelin.48 However, rs2857766 has not previously been examined for association with SZ or BD or with white matter abnormalities in psychosis.
CNP rs2070106
The 2′,3′-cyclic nucleotide 3′ phosphodiesterase enzyme (E.C. 3.1.4.37, CNPase) resides in the oligodendrocytes membrane sheath, catalyzes 2′,3′-nucleotides hydrolysis to form 2′-nucleotides and shows high activity in myelinated regions, reduced levels around plaques in the brain and elevated levels in cerebrospinal fluid during worsening periods in MS sufferers (for historical review see Vogel et al.49). It is involved in a signaling cascade responsible for an increase in the number and size of microtubule/CNPase-like structures among other changes and thereby the elongation of oligodendrocytes processes, and expansion of membrane sheaths and may thereby ultimately have a role in the assembly and/or maintenance of myelin.50, 51 In mice, the absence of CNPase results in axonal degradation but myelin assembly or the physical stability of myelin appears to be intact. These mice extend smaller outgrowths with less arborized processes52 and such oligodendrocyte dysfunction may be sufficient to result in secondary axonal loss.53 Reductions in CNPase levels in animal models have been associated with reduced learning ability.49
CNPase and in particular the SNP rs2071006 has been implicated in SZ by a number of types of evidence including case–control54 and family55 association, and postmortem gene expression studies but not consistently in all populations56, 57, 58 and not by genome-wide analyses.59, 60, 61, 62 In addition, the risk-conferring allele is in question and this ‘flip-flop' phenomenon63 may be the result of another functionally linked locus acting together to contribute to disease, differences in linkage disequilibrium between the two populations studied or a false-positive in one or other study.55 The CNPase gene SNP rs2071016 has been shown to be functional, affecting the expression of CNPase in a transcript-specific manner,5 with the presence of the polymorphism (A-allele) predicting low expression of the gene64 without resulting in an amino acid change in CNPase.54 Accordingly, reduced CNPase gene expression is associated with the A-allele of rs2070106 in SZ (the more common allele in affected individuals54) in the dorsolateral prefrontal cortex,6, 39 anterior temporal lobe41 and anterior cingulate cortex65 but not in all studies.64 In major depressive disorder (MDD), CNPase expression was reduced in the temporal lobes compared with controls.66 In concordance with reduced CNPase expression, lower mRNA levels were reported in SZ, but not BD or MDD, compare with controls along with mRNA for GALC, MAG and MOG.42 CNPase dysfunction appears to be regionally specific and certainly the anterior cingulate cortex is repeatedly implicated where both mRNA and protein expression have been reported to be reduced in contrast to the putamen where neither was altered in an elderly schizophrenic group.67
Given the evidence implicating each of these myelination genes in psychotic illness, this study aims to explore whether genotypic variation potentially conferring risk for psychosis was associated in vivo with white matter volume deviation in three groups of individuals at varying levels of genetic risk for psychosis: in those individuals who actually developed psychotic illness, in their unaffected first-degree relatives who are likely to be carrying susceptibility genes for illness and in healthy volunteers.
Materials and methods
Participants
Patients affected with SZ or BDI and unaffected first-degree relatives of these individuals were recruited through voluntary support groups or through direct psychiatric referral. All members of the BDI group had additionally experienced at least one psychotic episode involving delusions and hallucination as described previously.68 In addition, all patients had at least one first- or second-degree relative affected with a psychotic disorder while none of the healthy volunteers had a personal or family history of psychosis or as personal history of any other psychiatric disorder. Related pairs of unaffected SZ or BD relative were included to preserve power and findings qualified by examining the effect of removing related subjects post hoc. The healthy volunteers were recruited from the community through advertisements in local newspapers or from staff. All of the 189 subjects (Table 1) were aged 16–69 years, and their first language was English. The participants were excluded if they had experienced organic brain disease, head trauma resulting in loss of consciousness for >5 min, or DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition) substance or alcohol dependence in the 12 months before assessment. The study was approved by the relevant Local Ethics Committees and informed written consent obtained. Structural MRI brain scans were obtained for 189 subjects (Table 1). The recruitment and assessment of these participants has previously been described in detail.68, 69 There was no overlap between the subjects included in this study and those included in our previous region-of-interest studies of families affected with SZ.70, 71
Table 1. Demographic description of the participants for each pair of genotype groups compared.
| HC | SZ | SZrel | BD | BDrel | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NRG rs35753505 | C-carriers | TT | C-carriers | TT | C-carriers | TT | C-carriers | TT | C-carriers | TT |
| n | 27 | 10 | 25 | 8 | 27 | 9 | 18 | 9 | 22 | 17 |
| % Female (n) | 41 (11) | 80 (8) | 20 (5) | 13 (1) | 48 (13) | 56 (5) | 61 (11) | 67 (6) | 55 (12) | 41 (7) |
| Mean age±s.d. | 40±15 | 44±15 | 37±9 | 33±7 | 50±14 | 51±12 | 42±11 | 37±11 | 41±17 | 46±15 |
| PANSS total (mean±s.d.) | 0±0.7 | 0 | 25±12 | 28±9 | 1±2 | 1±2 | 5±5 | 5±5 | 1±1 | 1±2 |
| Current exposurea | ||||||||||
| Medication free | 27 | 10 | 0 | 0 | 27 | 9 | 3 | 0 | 21 | 16 |
| Atypical APs | 0 | 0 | 18 | 6 | 0 | 0 | 3 | 2 | 0 | 0 |
| Other APs | 0 | 0 | 5 | 3 | 0 | 0 | 1 | 0 | 0 | 0 |
| Mood stabilizers | 0 | 0 | 2 | 1 | 0 | 0 | 14 | 8 | 0 | 0 |
| SSRIs | 0 | 0 | 6 | 0 | 0 | 0 | 4 | 4 | 1 | 0 |
| Other Ads | 0 | 0 | 4 | 3 | 0 | 0 | 0 | 1 | 0 | 1 |
| HAPICE haplotype | Arh0 | Arh1 | Arh0 | Arh1 | Arh0 | Arh1 | Arh0 | Arh1 | Arh0 | Arh1 |
| n | 26 | 11 | 22 | 11 | 24 | 10 | 23 | 8 | 30 | 10 |
| % Female (n) | 58 (15) | 36 (4) | 23 (5) | 18 (2) | 50 (12) | 70 (7) | 61 (14) | 63 (5) | 47 (14) | 60 (6) |
| Mean age±s.d. | 43±16 | 36±13 | 35±9 | 36±8 | 50±13 | 52±16 | 41±12 | 44±11 | 42±16 | 48±17 |
| PANSS total (mean±s.d.) | 0±0.5 | 0±0.6 | 26±9 | 26±15 | 1±2 | 1±2 | 5±4 | 7±7 | 1±2 | 1±2 |
| Current exposurea | ||||||||||
| Medication free | 26 | 11 | 0 | 0 | 24 | 10 | 3 | 0 | 27 | 9 |
| Atypical APs | 0 | 0 | 15 | 9 | 0 | 0 | 4 | 2 | 0 | 0 |
| Other APs | 0 | 0 | 7 | 1 | 0 | 0 | 4 | 3 | 0 | 0 |
| Mood stabilizers | 0 | 0 | 2 | 1 | 0 | 0 | 18 | 7 | 0 | 0 |
| SSRIs | 0 | 0 | 3 | 4 | 0 | 0 | 7 | 3 | 1 | 0 |
| Other Ads | 0 | 0 | 6 | 1 | 0 | 0 | 4 | 0 | 1 | 1 |
| MOG rs2857766 | GG | C-carriers | GG | C-carriers | GG | C-carriers | GG | C-carriers | GG | C-carriers |
| n | 21 | 15 | 21 | 12 | 20 | 10 | 19 | 11 | 28 | 11 |
| % Female (n) | 48 (10) | 47 (7) | 29 (6) | 17 (2) | 65 (13) | 40 (4) | 68 (13) | 55 (6) | 61 (17) | 27 (3) |
| Mean age±s.d. | 45±12 | 34±16 | 35±9 | 36±9 | 51±13 | 43±17 | 41±12 | 40±10 | 46±15 | 38±18 |
| PANSS total (mean±s.d.) | 0±0.7 | 0±0.5 | 27±10 | 21±14 | 2±3 | 0±0.3 | 6±5 | 6±6 | 1±2 | 1±1 |
| Current exposurea | ||||||||||
| Medication free | 21 | 15 | 0 | 0 | 20 | 10 | 4 | 0 | 25 | 11 |
| Atypical APs | 0 | 0 | 13 | 11 | 0 | 0 | 5 | 3 | 0 | 0 |
| Other APs | 0 | 0 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mood stabilizers | 0 | 0 | 3 | 0 | 0 | 0 | 15 | 10 | 0 | 0 |
| SSRIs | 0 | 0 | 1 | 5 | 0 | 0 | 6 | 3 | 1 | 0 |
| Other Ads | 0 | 0 | 6 | 1 | 0 | 0 | 2 | 1 | 2 | 0 |
| CNP rs2070106 | GG | A-carriers | GG | A-carriers | GG | A-carriers | GG | A-carriers | GG | A-carriers |
| n | 21 | 16 | 7 | 21 | 9 | 24 | 16 | 14 | 12 | 24 |
| % Female (n) | 43 (9) | 50 (8) | 29 (2) | 24 (5) | 89 (8) | 46 (11) | 63 (10) | 64 (9) | 75 (9) | 38 (9) |
| Mean age±s.d. | 39±14 | 45±17 | 42±8 | 35±9 | 53±14 | 49±14 | 40±12 | 41±12 | 42±17 | 44±16 |
| PANSS total (mean±s.d.) | 0±0.5 | 0±0.9 | 25±6 | 24±13 | 1±2 | 1±2 | 7±6 | 4±4 | 1±3 | 1±1 |
| Current exposurea | ||||||||||
| Medication free | 21 | 16 | 0 | 0 | 9 | 24 | 1 | 3 | 12 | 21 |
| Atypical APs | 0 | 0 | 3 | 17 | 0 | 0 | 5 | 1 | 0 | 0 |
| Other APs | 0 | 0 | 3 | 3 | 0 | 0 | 1 | 0 | 0 | 0 |
| Mood stabilizers | 0 | 0 | 1 | 2 | 0 | 0 | 14 | 11 | 0 | 0 |
| SSRIs | 0 | 0 | 2 | 4 | 0 | 0 | 5 | 4 | 0 | 1 |
| Other Ads | 0 | 0 | 1 | 5 | 0 | 0 | 2 | 0 | 0 | 2 |
Abbreviations: AD, antidepressant; AP, antipsychotic; LSD, lysergic acid diethylamide; MOG, myelin oligodendrocytes glycoprotein; PANSS, Positive and Negative Syndrome Scale; SSRI, selective serotonin reuptake inhibitor.
Dependence constituted an exclusion criteria for the study, this item refers to regular use at some point in their lives meeting criteria for abuse.
Clinical assessments
Structured diagnostic interviews were performed on all subjects with the Schedule for Affective Disorders and SZ—Lifetime Version,72 and additional clinical information was collected to enable lifetime DSM-IV diagnoses to be made. For relatives not assessed directly, information regarding psychiatric diagnoses was obtained from the most reliable informants with the Family Interview for Genetic Studies73 and supplemented by medical notes where available. The patients were also assessed with the Positive and Negative Syndrome Scale (PANSS).74 the schedule for schizotypal personalities75 was used to assess nonpsychotic relatives and comparison subjects for schizotypal traits. Three unaffected SZ relatives and no other unaffected subjects reached criteria for schizotypal personality disorder.
MRI acquisition and analyses
Coronal 1.5-mm-thick contiguous T1-weighted MRI images of the entire brain were obtained by using a three-dimensional spoiled gradient recall echo sequence on a 1.5-T GE N/Vi Signa System scanner (General Electric, Milwaukee, WI, USA) and the following protocol: TR=13.1, TI=450 TE=5.8 ms, number of excitations=1, flip angle=20° and acquisition matrix=256 × 256 × 128. Images were processed for voxel-by-voxel-based analysis including reorientation to align images along the anterior commissure–posterior commissure axis in the sagittal plane and along the interhemispheric fissure in the coronal and axial planes using an automated reorientation matlab script by Carlton Chu and SPM5.76 Tissue class segmentation and smoothing to a full-width half-maximum kernel of 8 mm3 as performed using SPM 5.76 The resulting white matter images were grouped into two genotype-based groups, (Table 1) and voxel-based analysis using the flexible factorial design and a single covariate to account for age-based changes in white matter was performed. Analyses involved a relative threshold that discarded the lower 10% of voxel intensities and a classical model. Modulation was performed and age covaried for, therefore the outcome parameter used refers to relative white matter volume changes that cannot be explained by age.77, 78
Genotyping
The SNPs, MOG rs2857766 and CNP rs2070106, were genotyped using KBiosciences (http://www.kbioscience.co.uk), with a competitive allele-specific PCR system. As described in Williams et al.,79 SNP8NRG221533 was genotyped using the primer extension SNuPe and the genotyping platform Megabace (Amersham Bioscience, Buckinghamshire, UK), and the microsatellites were genotyped using a fluorescently labeled primer PCR assay, and were analyzed by the ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA). As defined by Stefansson et al.,10, 11 the core NRG1 at-risk haplotype (the HAPICE haplotype) consists of one SNP marker (SNP8NGR221533) and two microsatellites (478 B14-848 and 420 M9-1395).
Statistics
The Hardy–Weinberg equilibrium was calculated using Haploview version 4.180 (Table 2). Analyses of the results of the voxel-based analysis included using T-contrasts between the two allele frequency-based genotype groups (Table 2) within each diagnostic group covarying for age. A height threshold of T=2.35 (0.05/5, P<0.01) was used for each T-contrast to correct for that comparison within each of five diagnostic groups in addition to voxel-based corrections for multiple comparisons using random field theory.81 Clusters with a corrected P-value <0.0125 (0.05/4) were reported to correct for comparison across four genotypes. Pearson's χ2 test was performed to examine the frequency of distribution of gender across genotype groups. Non-parametric Mann–Whitney U-tests were used to confirm that PANSS total score and the age at symptoms onset did not differ between genotype groups within each diagnostic group in these three significant findings (Table 3). Statistical tests were carried out using SPSS version 15 (http://www.spss.com).
Table 2. Sample size for each genotype or haplotype investigated.
| Genotype/haplotype sample size | HC | SZ | SZrel | BD | BDrel | Total | χ2 (P) | HWp |
|---|---|---|---|---|---|---|---|---|
| NRG1 rs35753505 | ||||||||
| C-carriers | 27 | 25 | 27 | 18 | 22 | 119 | ||
| TT | 10 | 8 | 9 | 9 | 17 | 53 | 4.5 (0.34) | 0.24 |
| HAPICE haplotype | ||||||||
| Arh0 | 26 | 22 | 24 | 23 | 30 | 125 | ||
| Arh1 | 11 | 11 | 10 | 8 | 10 | 50 | 0.77 (0.94) | NA |
| MOG rs2857766 | ||||||||
| GG | 21 | 21 | 20 | 19 | 28 | 109 | ||
| C-carriers | 15 | 12 | 10 | 11 | 11 | 59 | 1.59 (0.81) | 0.26 |
| CNP rs2070106 | ||||||||
| GG | 21 | 7 | 9 | 16 | 12 | 65 | ||
| A-carriers | 16 | 21 | 24 | 14 | 24 | 99 | 12.1 (0.017) | 1.00 |
| Total number of subjects | 39 | 37 | 40 | 33 | 40 | 189 | ||
Abbreviations: MOG, myelin oligodendrocytes glycoprotein; NA, not applicable; NRG, neuregulin.
Table 3. White matter variation across genotype or haplotype detected by voxel-based analysis.
| Genotype, group and direction | Maxima (MNI, T) | Cluster KE (P) |
|---|---|---|
| NRG SNP8NRG221533 | ||
| BDI c-carriers>tt | ||
| Cingulum/PHG, splenium and anterior CC | −16 −12 −28 (4.66), 18 −42 14 (4.34), 20 26 36 (4.32) | 17862 (0.0001) |
| SZ c-carriers<tt | ||
| Right UF, right ILF, right ALIC (atr, cpt) | 12 14 −16 (3.61), 38 −16 −2 (3.54), 22 4 −10 (3.52) | 5004 (0.0001) |
| HAPICE haplotype | ||
| Unaffected individuals with a relative with SZ arh0>arh1 | ||
| SCP and fornix | −2 −34 −4 (3.48), −12 −48 −18 (3.46), 6 −28 14 (3.26) | 903 (0.006) |
| BDI arh0<arh1 | ||
| Fornix, caudate, posterior cingulum bundle | 24 −42 8 (4.34), −8 6 12 (4.23), 28 −40 −10 (3.14) | 1625 (0.0001) |
| MOG rs2857766 | ||
| Healthy controls gg>c-carriers | ||
| Corticopontine/spinal tract, MCP, ICP | 6 −28 −40 (3.46), 12 −44 −50 (3.34), 6 −52 −52 (3.25) | 1305 (0.001) |
Abbreviations: ALIC, anterior limb of the internal capsule; BDI, bipolar I disorder; ICP, inferior cerebellar peduncle; ILF, inferior longitudinal fasciculus; MCP, middle cerebellar peduncle; NRG1, neuregulin 1; PHG, parahippocampal gyrus; SCP, superior cerebellar peduncle; SZ, schizophrenia; UF, uncinate fasciculus.
KE is the cluster size (number of voxels) and the P-value refers to the cluster level-corrected P value (cutoff P<0.0125).
Results
In total, 70 individuals with a psychotic disorder (37 SZ and 33 BDI) and 119 unaffected subjects (39 HC, 40 with a relative with SZ, 40 with a relative with BDI) were studied. Of the 37 individuals in the SZ group, 3 had schizoaffective disorder, 16 were undifferentiated, 17 of the paranoid and 1 of the disorganized subtype and none were related to any member of the BD group. There was no significant difference in gender distribution between the genotype or haplotype groups within any of the diagnostic groups with three exceptions. In NRG8SNP221533, the HC group has a lower proportion of males in the TT-group (n=2). In MOG rs2857766, the unaffected group with a BD relative had a lower proportion of females among the C-carriers group (n=3) compared with the GG-genotype group. In the CNP rs2070106, GG-genotype had a lower proportion of males (n=1 and n=3) among the unaffected individuals with a SZ relative and those with a BD relative, respectively, compared with the A-carriers. The SZ and BD groups had significantly greater total PANSS scores than the three unaffected groups. At time of scanning a number of members of the SZ and BD groups were receiving medication (Table 1).
NRG1 rs35753505 (NRG8SNP221533)
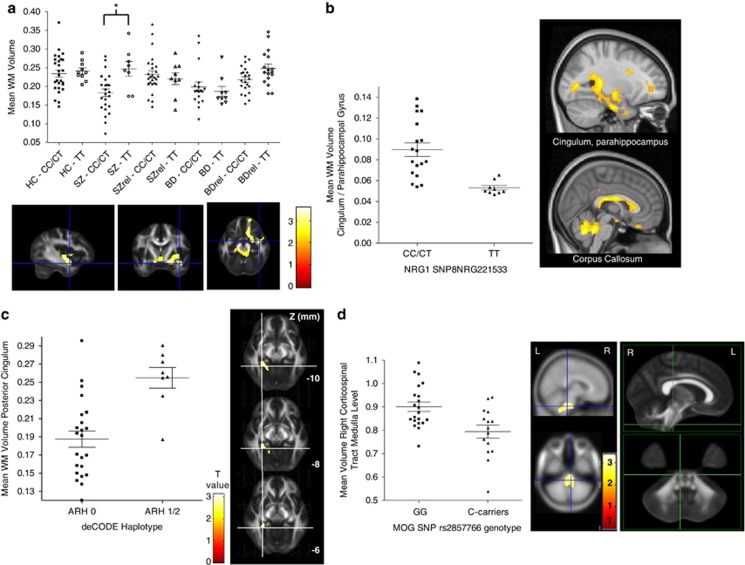
No effect of the NRG8SNP221533 genotype was detected in the HC group, nor among unaffected individuals with a relative with a psychotic disorder on white matter volume. In the SZ group, the NRG8SNP221533 risk allele, C-carrier genotype was associated with reduced white matter volume relative to the TT-genotype group in the region of the right inferior longitudinal fasciculus, ALIC (partially overlapping the anterior thalamic radiations and corticopontine tracts) and right uncinate fasciculus (Figure 1). In the BDI-group, the risk allele C-carriers was associated with greater white matter relative to those of the TT-genotype in several regions including the cingulum/parahippocampal gyrus (Figure 1b) and the callosal body. The PANSS total, the age at symptom onset nor the presence of atypical antipsychotic medication in the SZ group differed significantly between C-carriers and the T-homozygotes. Similarly, in the BDI group C-carriers and T-homozygotes did not differ significantly on PANSS total, the age at symptom onset not the presence of mood stabilizing medication.
Figure 1.
The association of white matter (WM) volume in psychotic disorders with genotypic variation in neuregulin 1 (NRG1) and myelin oligodendrocytes glycoprotein (MOG). (a) C-allele carrier genotype of the NRG1 single-nucleotide polymorphism (SNP), SNP8NRG221533 is associated with lower white matter volume in the region of the uncinate fasciculus (UF) in patients with schizophrenia (SZ). Scatter plot of mean (±s.e.m.) white matter volume across the TT versus C-carriers genotype groups in all five diagnostic categories in a sphere of 2 mm radius centered on the cluster peak (12 14 −16) implicating the UF in SZ. Beneath is the cluster T-map overlaid on sections of the MNI152 T1 template. (b). The risk genotype (TT) of the NRG1 SNP8NRG221533 and bipolar I disorder (BD) is associated with reduced white matter volume in the posterior inferior portion of the cingulum, parahippocampal gyrus and in the callosal body. Scatter plot of mean (±s.e.m.) white matter volume in a sphere of 2 mm radius centered on the posterior cingulum/parahippocampal gyrus cluster peak (−16 −12 −28). Top right illustrates the cluster extent and magnitude on a T-map, bottom right, the callosal body cluster effect for the finding in the same group and direction. (c). BD and having the HAPICE haplotype is associated with greater white matter volume in the posterior portion of the cingulum bundle. Scatter plot of mean (±s.e.m.) white matter volume in a sphere of 2 mm radius centered on the cluster peak (28 −40 −10) and adjacent, the cluster T-map overlaid on slices (Z −10 to −6 mm) of the functional MRI (fMRI)B58 FA map. (d). Psychiatrically healthy controls who are G-homozygotes of the MOG SNP rs2857766 have greater corticospinal tract white matter volume at the level of the medulla relative to C-carriers. Scatter plot of mean (±s.e.m.) white matter volume in a sphere of 2 mm radius centered on the cluster peak (6 −28 −40), middle panel illustrated the cluster on a statistical T-map. The cluster peak is at the level of the medulla in the right corticopontine/corticospinal tract and this is indicated by the crosshairs on sagittal and axial views of the fMRIBs mean FA map (FMRIB58_FA). L, left; R, right.
HAPICE haplotype
There were no significant differences in white matter volume between groups possessing one or two copies of the HAPICE haplotype versus having no copies in the HC or SZ groups or in the unaffected group with a BD relative. Those unaffected individuals with a relative with SZ and one or two copies showed a trend toward lower white matter volume (29–33%) in the superior cerebellar peduncle and the fornix relative to those with no copies. Removing all related individuals (25–46% lower) or removing one of each pair of related participants (preferring first the minor allele and then the first recruited subject, 22–28% lower) did not change the magnitude or direction of this finding. In the BDI group those carrying one or two copies of the haplotype had greater white matter than those carrying none in the fornix, caudate and cingulum (Figure 1c). The PANSS total, the age at symptom onset nor the presence of mood stabilizing or antidepressant medication in the BDI-group differed significantly between those having no copies versus those with one or two copies of the arh haplotype.
MOG rs2857766
The genotypic frequency for rs2857766 among the sample of 168 Europeans was 0.65 for G-homozygotes and 0.33 for C-carriers. Consistent with the frequency previously reported for C-homozygotes (0.086), we detected a total of three C-homozygotes (0.018). Among Europeans, the genotypic frequency is reported to be 0.483, 0.431 and 0.086 for the G-homozygotes, heterozygotes and C-homozygotes, respectively. Healthy controls who were G-homozygotes of the MOG SNP rs2857766 had greater white matter volume in the middle and inferior cerebellar peduncles and in medulla-level corticopontine/corticospinal tracts relative to C-carriers (Figure 1d). In contrast, the BD or SZ group, or in the unaffected individuals with a SZ or BD relative, no white matter differences were detected between the G-homozygotes and C-carriers of the MOG SNP rs2857766.
CNP rs2070106
None of the groups showed any significant differences in white matter across the CNP rs2070106 genotype.
Discussion
The major finding from this study is the association between genotypic variation in NRG, previously linked to psychotic illness, and white matter abnormalities in psychotic patients and their relatives. Individuals with SZ and carrying the risk allele C, of the NRG1 SNP8NRG221533, had lower white matter volume in the regions of the right uncinate fasciculus, right inferior longitudinal fasciculus and in the ALIC (overlapping portions of the anterior thalamic radiations and the corticopontine tracts). Unaffected relatives of patients with SZ possessing at least one copy of the HAPICE haplotype had less white matter volume in the fornix and superior cerebellar peduncle compared with those with no copies. In contrast, those BD1 patients carrying the NRG risk allele C of SNP8NRG221533 had greater white matter volume in the posterior cingulum/parahippocampal gyrus and a number of regions of the corpus callosum. Similarly, individuals with BDI and at least one copy of the HAPICE haplotype, which includes the risk allele of the NRG1 SNP8NRG221533, possessed greater white matter volume in the cingulum and fornix.
There was no association between white matter volume and genotypic variation in SNP8NRG221533 in healthy volunteers, consistent with the study by Winterer et al 34 using a similar voxel-based approach (optimized VBM and SPM5). However, Winterer et al.34 did detect reduced medial frontal FA using diffusion tensor imaging in C-carriers of the SNP8NRG221533. McIntosh et al.36 have additionally reported reduced white matter density and FA in the ALIC associated with the TT-genotype of another NRG1 SNP in the promoter region of the gene (SNP8NRG243177). Finally, the NRG1 gene SNP and exact population studied herein has been examined previously for contribution to variance in the volume of the lateral ventricles or hippocampus and no relationship was detected.82 The present finding implicates the NRG1 gene SNP, SNP8NRG221533 in white matter-related abnormalities of the emotion circuitry in BD,83 and in previously implicated regions in SZ including the ALIC. It is not clear why in contrast to the SZ group, the SZ risk genotype is associated with increased white matter in the cingulum and corpus callosum in the BDI group. These data suggest the possibility that another currently unknown factor associated with having BDI may be involved in modulating the relationship between the NRG1 SNP genotype and white matter volume. In addition, long-term antipsychotic medication exposure may mediate structural changes as has been recently demonstrated.84 Thus, it remains possible that in the SZ group, an as yet unidentified, interaction between genotype and medication response may contribute to the present findings.
Several models of the neurocircuitry underlying psychiatric disorders have been developed many based on structural and functional evidence from in vivo neuroimaging studies. These and similar recent studies33, 36, 37, 85, 86, 87 substantiate a growing body of evidence implicating genetic susceptibility to developing abnormalities within and between (white matter volume and microstructural organization) emotion-related structures. These studies collectively contribute to progress toward the inevitable future clinical application of such in vivo knowledge, which may include earlier detection of susceptibility, monitoring progression, presaging treatment response and potentially diagnosing based classification.88
It is not known whether MOG rs2857766 confers risk for SZ or BD and, in our study we found no relationship between genotype and white matter volume in the brain in these groups. However, we provide preliminary evidence suggesting a possible relationship between G-homozygosity (the ancestral allele) in MOG rs2857766 and greater brainstem level white matter volume in psychiatrically healthy individuals that was not detected in psychotic patients or their unaffected relatives. The uniformity of microstructural organization of white matter in this region, the middle cerebellar peduncle has been reported to be disrupted in SZ using diffusion-weighted imaging.89 However, there is insufficient evidence to directly implicate MOG rs2857766 in disruption of the middle cerebellar peduncle.
Despite previous reports of the involvement of CNPase SNP rs2070106 in SZ and BD, this study did not detect any relationship between white matter volume and genetic variation in this SNP in any patient or relative group examined or among the healthy controls.
Individual SNPs are unlikely to confer more than a relatively minor proportion of variation in white matter volume, however, such associations may serve to highlight biological processes that are involved in dysfunctional white matter circuits and point to other genetic variants or functional units that are tightly linked to such pathophysiology and that warrant further exploration. A limitation of this study is the small number of subjects in some of the genotype groups and it is possible we were underpowered to detect relationships with white matter that may be detected in the future with more sensitive technology, analysis methods and larger cohorts of subjects. In addition, the inclusion of a related pairs of individuals within the unaffected SZ and BD relative groups to preserve power represents a limitation of this study. Despite their inclusion, however, no positive finding was detected for any genotype examined in the unaffected BD group or for three of the four genotypes examined in the case of the unaffected SZ group. In the unaffected SZ group, those having one or two copies of the HAPICE haplotype showed a trend toward lower white matter volume in the superior cerebellar peduncle and the fornix relative to those with no copies. Removing all related individuals from this group did not significantly alter the magnitude or direction of this finding. A particular limitation of the voxel-based approach to image analysis is the possibility of errors due to registration and segmentation steps.77 To minimize these possible sources of error, registration and segmentation performance were visually assessed on an individual basis. A particular strength of this study was the inclusion of subjects who were unaffected but had a relative with a psychotic disorder. In the case of the MOG SNP examined, the absence in all affected groups and their relatives of the relationship detected in healthy controls suggests a possible pathological role for this SNP in psychotic disorders. Future linkage, case–control, family and genome-wide studies may further elucidate the role of this MOG SNP in psychosis.
In summary, these findings provide support for the theory that genotypic variation in neuregulin may confer risk for psychosis by influencing white matter in areas known to underlie the emotional circuitry of the human brain.83 The deficits in white matter volume detected in this study have a number of possible underlying explanations including reduced axonal projections or a reduction in the volume of non-axonal white matter components including glia. An optimum ratio of axonal diameter to myelin sheath thickness termed the g-ratio has been described in terms of efficiency of conductivity.90, 91 It is plausible that regional perturbations of this ratio may manifest as signs and symptoms associated with psychotic disorders.92 Volume changes in white matter are inherently limited in forming the nature or identity of molecular factors involved in abnormalities detected, and future studies examining the uniformity of microstructural organization of white matter using diffusion-weighted MR imaging with genotype among other approaches will aid in the identification of the mechanism by which the NRG risk allele contributes to the signs and symptoms of psychotic disorders.
Acknowledgments
We gratefully acknowledge the support of the Wellcome Trust, all the families for participating in the study, the National SZ Fellowship (Rethink) and the Manic Depressive Fellowship for help with recruitment. We also thank the NUI Galway School of Medicine students Cliodhna NiFoghlu, David Cormican, Kealan Westby, Larissa Higgins, Marie K. Ryan, Sharon Holland, Zhu Lim, Fiona Fonseca and Marc Curtin for their contribution during the neuroimaging special study option.
The authors declare no conflict of interest.
References
- Craddock N, O'Donovan MC, Owen MJ. Psychosis genetics: modeling the relationship between schizophrenia, bipolar disorder, and mixed (or "schizoaffective") psychoses. Schizophr Bull. 2009;35:482–490. doi: 10.1093/schbul/sbp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Law AJ. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol Psychiatry. 2006;60:132–140. doi: 10.1016/j.biopsych.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Walker RM, Christoforou A, Thomson PA, McGhee KA, Maclean A, Muhleisen TW, et al. Association analysis of Neuregulin 1 candidate regions in schizophrenia and bipolar disorder. Neurosci Lett. 2010;478:9–13. doi: 10.1016/j.neulet.2010.04.056. [DOI] [PubMed] [Google Scholar]
- Sokolov BP. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse: comorbidity, shared traits, or molecular phenocopies. Int J Neuropsychopharmacol. 2007;10:547–555. doi: 10.1017/S1461145706007322. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Ueda J, Bundo M, Nakano Y, Kato T. Effect of a functional single nucleotide polymorphism in the 2′,3′-cyclic nucleotide 3′-phosphodiesterase gene on the expression of oligodendrocyte-related genes in schizophrenia. Psychiatry Clin Neurosci. 2008;62:103–108. doi: 10.1111/j.1440-1819.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, et al. Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet. 1999;65:1096–1103. doi: 10.1086/302579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JZ, Si TM, Ruan Y, Ling YS, Han YH, Wang XL, et al. Association study of neuregulin 1 gene with schizophrenia. Mol Psychiatry. 2003;8:706–709. doi: 10.1038/sj.mp.4001377. [DOI] [PubMed] [Google Scholar]
- Hall D, Gogos JA, Karayiorgou M. The contribution of three strong candidate schizophrenia susceptibility genes in demographically distinct populations. Genes Brain Behav. 2004;3:240–248. doi: 10.1111/j.1601-183X.2004.00078.x. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Stopkova P, Rafael MA, Saito T. Association of schizophrenia in African Americans to polymorphism in synapsin III gene. Psychiatr Genet. 2005;15:127–132. doi: 10.1097/00041444-200506000-00009. [DOI] [PubMed] [Google Scholar]
- Kampman O, Anttila S, Illi A, Saarela M, Rontu R, Mattila KM, et al. Neuregulin genotype and medication response in Finnish patients with schizophrenia. Neuroreport. 2004;15:2517–2520. doi: 10.1097/00001756-200411150-00017. [DOI] [PubMed] [Google Scholar]
- Thiselton DL, Webb BT, Neale BM, Ribble RC, O'Neill FA, Walsh D, et al. No evidence for linkage or association of neuregulin-1 (NRG1) with disease in the Irish study of high-density schizophrenia families (ISHDSF) Mol Psychiatry. 2004;9:777–783. doi: 10.1038/sj.mp.4001530. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11:539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- Corfas G, Roy K, Buxbaum JD. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nat Neurosci. 2004;7:575–580. doi: 10.1038/nn1258. [DOI] [PubMed] [Google Scholar]
- Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, et al. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Gu Z, Jiang Q, Fu AK, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25:4974–4984. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno A, Fischbach GD. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol. 2001;11:287–296. doi: 10.1016/s0959-4388(00)00210-5. [DOI] [PubMed] [Google Scholar]
- Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5' SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Bertram I, Bernstein HG, Lendeckel U, Bukowska A, Dobrowolny H, Keilhoff G, et al. Immunohistochemical evidence for impaired neuregulin-1 signaling in the prefrontal cortex in schizophrenia and in unipolar depression. Ann NY Acad Sci. 2007;1096:147–156. doi: 10.1196/annals.1397.080. [DOI] [PubMed] [Google Scholar]
- Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- Wang XD, Su YA, Guo CM, Yang Y, Si TM. Chronic antipsychotic drug administration alters the expression of neuregulin 1beta, ErbB2, ErbB3, and ErbB4 in the rat prefrontal cortex and hippocampus. Int J Neuropsychopharmacol. 2008;11:553–561. doi: 10.1017/S1461145707008371. [DOI] [PubMed] [Google Scholar]
- Rimer M, Barrett DW, Maldonado MA, Vock VM, Gonzalez-Lima F. Neuregulin-1 immunoglobulin-like domain mutant mice: clozapine sensitivity and impaired latent inhibition. Neuroreport. 2005;16:271–275. doi: 10.1097/00001756-200502280-00014. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Pisacane P, Erickson S. Heregulin but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav Brain Res. 2000;109:219–227. doi: 10.1016/s0166-4328(99)00175-8. [DOI] [PubMed] [Google Scholar]
- Cannella B, Pitt D, Marchionni M, Raine CS. Neuregulin and erbB receptor expression in normal and diseased human white matter. J Neuroimmunol. 1999;100:233–242. doi: 10.1016/s0165-5728(99)00201-5. [DOI] [PubMed] [Google Scholar]
- Barros CS, Calabrese B, Chamero P, Roberts AJ, Korzus E, Lloyd K, et al. Impaired maturation of dendritic spines without disorganization of cortical cell layers in mice lacking NRG1/ErbB signaling in the central nervous system. Proc Natl Acad Sci USA. 2009;106:4507–4512. doi: 10.1073/pnas.0900355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprooten E, Lymer GK, Maniega SM, McKirdy J, Clayden JD, Bastin ME, et al. The relationship of anterior thalamic radiation integrity to psychosis risk associated neuregulin-1 variants. Mol Psychiatry. 2009;14:237–238. doi: 10.1038/mp.2008.136. [DOI] [PubMed] [Google Scholar]
- Winterer G, Konrad A, Vucurevic G, Musso F, Stoeter P, Dahmen N. Association of 5′ end neuregulin-1 (NRG1) gene variation with subcortical medial frontal microstructure in humans. Neuroimage. 2008;40:712–718. doi: 10.1016/j.neuroimage.2007.12.041. [DOI] [PubMed] [Google Scholar]
- Wang F, Jiang T, Sun Z, Teng SL, Luo X, Zhu Z, et al. Neuregulin 1 genetic variation and anterior cingulum integrity in patients with schizophrenia and healthy controls. J Psychiatry Neurosci. 2009;34:181–186. [PMC free article] [PubMed] [Google Scholar]
- McIntosh AM, Moorhead TW, Job D, Lymer GK, Munoz Maniega S, McKirdy J, et al. The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry. 2008;13:1054–1059. doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- Hall J, Whalley HC, Job DE, Baig BJ, McIntosh AM, Evans KL, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Miguel-Hidalgo JJ. Gliogenesis and glial pathology in depression. CNS Neurol Disord Drug Targets. 2007;6:219–233. doi: 10.2174/187152707780619326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Maccarrone G, Hunyadi-Gulyas E, Eberlin MN, et al. Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophrenia. J Psychiatr Res. 2009;43:978–986. doi: 10.1016/j.jpsychires.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Maccarrone G, Wobrock T, Zerr I, Gormanns P, Reckow S, et al. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. J Psychiatr Res. 2010;44:1176–1189. doi: 10.1016/j.jpsychires.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Rewerts C, Marangoni S, Novello JC, et al. Alterations in oligodendrocyte proteins, calcium homeostasis and new potential markers in schizophrenia anterior temporal lobe are revealed by shotgun proteome analysis. J Neural Transm. 2009;116:275–289. doi: 10.1007/s00702-008-0156-y. [DOI] [PubMed] [Google Scholar]
- Barley K, Dracheva S, Byne W. Subcortical oligodendrocyte- and astrocyte-associated gene expression in subjects with schizophrenia, major depression and bipolar disorder. Schizophr Res. 2009;112:54–64. doi: 10.1016/j.schres.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Zai G, King N, Wigg K, Couto J, Wong GW, Honer WG, et al. Genetic study of the myelin oligodendrocyte glycoprotein (MOG) gene in schizophrenia. Genes Brain Behav. 2005;4:2–9. doi: 10.1111/j.1601-183X.2004.00089.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Qin W, He G, Yang Y, Chen Q, Zhou J, et al. A family-based association study of the MOG gene with schizophrenia in the Chinese population. Schizophr Res. 2005;73:275–280. doi: 10.1016/j.schres.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Bora E, Fornito A, Radua J, Walterfang M, Seal M, Wood SJ, et al. Neuroanatomical abnormalities in schizophrenia: a multimodal voxelwise meta-analysis and meta-regression analysis. Schizophr Res. 2011;127:46–57. doi: 10.1016/j.schres.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Ellison-Wright I, Bullmore E. Meta-analysis of diffusion tensor imaging studies in schizophrenia. Schizophr Res. 2009;108:3–10. doi: 10.1016/j.schres.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Jensen CJ, Stankovich J, Butzkueven H, Oldfield BJ, Rubio JP. Common variation in the MOG gene influences transcript splicing in humans. J Neuroimmunol. 2010;229:225–231. doi: 10.1016/j.jneuroim.2010.07.027. [DOI] [PubMed] [Google Scholar]
- D'Alfonso S, Bolognesi E, Guerini FR, Barizzone N, Bocca S, Ferrante D, et al. A sequence variation in the MOG gene is involved in multiple sclerosis susceptibility in Italy. Genes Immun. 2008;9:7–15. doi: 10.1038/sj.gene.6364437. [DOI] [PubMed] [Google Scholar]
- Vogel US, Thompson RJ. Molecular structure, localization, and possible functions of the myelin-associated enzyme 2′,3′-cyclic nucleotide 3′-phosphodiesterase. J Neurochem. 1988;50:1667–1677. doi: 10.1111/j.1471-4159.1988.tb02461.x. [DOI] [PubMed] [Google Scholar]
- Lintner RN, Dyer CA. Redistribution of cholesterol in oligodendrocyte membrane sheets after activation of distinct signal transduction pathways. J Neurosci Res. 2000;60:437–449. doi: 10.1002/(SICI)1097-4547(20000515)60:4<437::AID-JNR2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Dyer CA, Matthieu JM. Antibodies to myelin/oligodendrocyte-specific protein and myelin/oligodendrocyte glycoprotein signal distinct changes in the organization of cultured oligodendroglial membrane sheets. J Neurochem. 1994;62:777–787. doi: 10.1046/j.1471-4159.1994.62020777.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Gravel M, Zhang R, Thibault P, Braun PE. Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol. 2005;170:661–673. doi: 10.1083/jcb.200411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Peirce TR, Bray NJ, Williams NM, Norton N, Moskvina V, Preece A, et al. Convergent evidence for 2′,3′-cyclic nucleotide 3′-phosphodiesterase as a possible susceptibility gene for schizophrenia. Arch Gen Psychiatry. 2006;63:18–24. doi: 10.1001/archpsyc.63.1.18. [DOI] [PubMed] [Google Scholar]
- Voineskos AN, de Luca V, Bulgin NL, van Adrichem Q, Shaikh S, Lang DJ, et al. A family-based association study of the myelin-associated glycoprotein and 2′,3′-cyclic nucleotide 3′-phosphodiesterase genes with schizophrenia. Psychiatr Genet. 2008;18:143–146. doi: 10.1097/YPG.0b013e3282fa1874. [DOI] [PubMed] [Google Scholar]
- Che R, Tang W, Zhang J, Wei Z, Zhang Z, Huang K, et al. No relationship between 2′,3′-cyclic nucleotide 3′-phosphodiesterase and schizophrenia in the Chinese Han population: an expression study and meta-analysis. BMC Med Genet. 2009;10:31. doi: 10.1186/1471-2350-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Qu M, Wang L, Ruan Y, Lu T, Zhang H, et al. Case-control association study of the 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP) gene and schizophrenia in the Han Chinese population. Neurosci Lett. 2007;416:113–116. doi: 10.1016/j.neulet.2007.01.054. [DOI] [PubMed] [Google Scholar]
- Usui H, Takahashi N, Saito S, Ishihara R, Aoyama N, Ikeda M, et al. The 2′,3′-cyclic nucleotide 3′-phosphodiesterase and oligodendrocyte lineage transcription factor 2 genes do not appear to be associated with schizophrenia in the Japanese population. Schizophr Res. 2006;88:245–250. doi: 10.1016/j.schres.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, et al. Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet. 2000;9:1049–1057. doi: 10.1093/hmg/9.7.1049. [DOI] [PubMed] [Google Scholar]
- Bailer U, Leisch F, Meszaros K, Lenzinger E, Willinger U, Strobl R, et al. Genome scan for susceptibility loci for schizophrenia. Neuropsychobiology. 2000;42:175–182. doi: 10.1159/000026690. [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, et al. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Kirby A, et al. Genome scan of schizophrenia. Am J Psychiatry. 1998;155:741–750. doi: 10.1176/ajp.155.6.741. [DOI] [PubMed] [Google Scholar]
- Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkus SN, Hyde TM, Vakkalanka R, Kolachana B, Weinberger DR, Kleinman JE, et al. Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res. 2008;98:129–138. doi: 10.1016/j.schres.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullumsmith RE, Gupta D, Beneyto M, Kreger E, Haroutunian V, Davis KL, et al. Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia. Schizophr Res. 2007;90:15–27. doi: 10.1016/j.schres.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Dracheva S, Davis KL, Chin B, Woo DA, Schmeidler J, Haroutunian V. Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol Dis. 2006;21:531–540. doi: 10.1016/j.nbd.2005.08.012. [DOI] [PubMed] [Google Scholar]
- McDonald C, Marshall N, Sham PC, Bullmore ET, Schulze K, Chapple B, et al. Regional brain morphometry in patients with schizophrenia or bipolar disorder and their unaffected relatives. Am J Psychiatry. 2006;163:478–487. doi: 10.1176/appi.ajp.163.3.478. [DOI] [PubMed] [Google Scholar]
- McDonald C, Bullmore ET, Sham PC, Chitnis X, Wickham H, Bramon E, et al. Association of genetic risks for schizophrenia and bipolar disorder with specific and generic brain structural endophenotypes. Arch Gen Psychiatry. 2004;61:974–984. doi: 10.1001/archpsyc.61.10.974. [DOI] [PubMed] [Google Scholar]
- McDonald C, Grech A, Toulopoulou T, Schulze K, Chapple B, Sham P, et al. Brain volumes in familial and non-familial schizophrenic probands and their unaffected relatives. Am J Med Genet. 2002;114:616–625. doi: 10.1002/ajmg.10604. [DOI] [PubMed] [Google Scholar]
- Schulze K, McDonald C, Frangou S, Sham P, Grech A, Toulopoulou T, et al. Hippocampal volume in familial and nonfamilial schizophrenic probands and their unaffected relatives. Biol Psychiatry. 2003;53:562–570. doi: 10.1016/s0006-3223(02)01910-8. [DOI] [PubMed] [Google Scholar]
- Spitzer R, Endicott J. Schedule for Affective Disorders and Schizophrenia-Lifetime Version. New York State Psychiatric Institute, Biometrics Research: New York; 1978. [Google Scholar]
- Maxwell M. Family Interview for Genetic Studies. National Institute of Mental Health, Intramural Research Program, Clinical Neurogenetics Branch: Rockville, MD; 1992. [Google Scholar]
- Kay SR, Opler LA, Lindenmayer JP. The Positive and Negative Syndrome Scale (PANSS): rationale and standardisation. Br J Psychiatry Suppl. 1989;7:59–67. [PubMed] [Google Scholar]
- Baron M, Asnis L, Gruen R. The schedule for Schizotypal Personalities (SSP): a diagnostic interview for schizotypal features. Psychiatry Res. 1981;4:213–228. doi: 10.1016/0165-1781(81)90024-x. [DOI] [PubMed] [Google Scholar]
- FIL Statistical Parametric Mapping The FIL methods group; 1994 ; spm@fil.ion.ucl.ac.uk . [Google Scholar]
- Mechelli A, Price C, Friston K, Ashburner J. Voxel-based morphometry of the human brain: methods and applications. Curr Med Imaging Rev. 2005;1:105–113. [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11 (Part 1:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Williams NM, Preece A, Spurlock G, Norton N, Williams HJ, Zammit S, et al. Support for genetic variation in neuregulin 1 and susceptibility to schizophrenia. Mol Psychiatry. 2003;8:485–487. doi: 10.1038/sj.mp.4001348. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage. 2004;22:676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Dutt A, McDonald C, Dempster E, Prata D, Shaikh M, Williams I, et al. The effect of COMT, BDNF, 5-HTT, NRG1 and DTNBP1 genes on hippocampal and lateral ventricular volume in psychosis. Psychol Med. 2009;39:1783–1797. doi: 10.1017/S0033291709990316. [DOI] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct. 2008;213:93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Andreasen NC, Ziebell S, Pierson R, Magnotta V. Long-term antipsychotic treatment and brain volumes: a longitudinal study of first-episode schizophrenia. Arch Gen Psychiatry. 2011;68:128–137. doi: 10.1001/archgenpsychiatry.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminowicz DA, Mayberg HS, McIntosh AR, Goldapple K, Kennedy S, Segal Z, et al. Limbic-frontal circuitry in major depression: a path modeling metanalysis. Neuroimage. 2004;22:409–418. doi: 10.1016/j.neuroimage.2004.01.015. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Maniega SM, Lymer GK, McKirdy J, Hall J, Sussmann JE, et al. White matter tractography in bipolar disorder and schizophrenia. Biol Psychiatry. 2008;64:1088–1092. doi: 10.1016/j.biopsych.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Cannon DM, Klaver JK, Gandhi SK, Solorio G, Peck SA, Erickson K, et al. Genetic variation in cholinergic muscarinic-2 receptor gene modulates M2 receptor binding in vivo and accounts for reduced binding in bipolar disorder. Mol Psychiatry. 2011;16:407–418. doi: 10.1038/mp.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardekani BA, Tabesh A, Sevy S, Robinson DG, Bilder RM, Szeszko PR. Diffusion tensor imaging reliably differentiates patients with schizophrenia from healthy volunteers. Hum Brain Mapp. 2011;32:1–9. doi: 10.1002/hbm.20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okugawa G, Nobuhara K, Minami T, Tamagaki C, Takase K, Sugimoto T, et al. Subtle disruption of the middle cerebellar peduncles in patients with schizophrenia. Neuropsychobiology. 2004;50:119–123. doi: 10.1159/000079101. [DOI] [PubMed] [Google Scholar]
- Chomiak T, Hu B. What is the optimal value of the g-ratio for myelinated fibers in the rat CNS? A theoretical approach. PLoS One. 2009;4:e7754. doi: 10.1371/journal.pone.0007754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraher J, Dockery P. A strong myelin thickness-axon size correlation emerges in developing nerves despite independent growth of both parameters. J Anat. 1998;193:195–201. doi: 10.1046/j.1469-7580.1998.19320195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T, Keshavan M, Giedd JN. Why do many psychiatric disorders emerge during adolescence. Nat. Rev. 2008;9:947–957. doi: 10.1038/nrn2513. [DOI] [PMC free article] [PubMed] [Google Scholar]