ABSTRACT
Pyridoxal 5′-phosphate (PLP) is a coenzyme synthesized by all forms of life. Relevant to the work reported here is the mechanism of the PLP-dependent threonine/serine dehydratases, which generate reactive enamine/imine intermediates that are converted to keto acids by members of the RidA family of enzymes. The RidA protein of Salmonella enterica serovar Typhimurium LT2 is the founding member of this broadly conserved family of proteins (formerly known as YjgF/YER057c/UK114). RidA proteins were recently shown to be enamine deaminases. Here we demonstrate the damaging potential of enamines in the absence of RidA proteins. Notably, S. enterica strains lacking RidA have decreased activity of the PLP-dependent transaminase B enzyme IlvE, an enzyme involved in branched-chain amino acid biosynthesis. We reconstituted the threonine/serine dehydratase (IlvA)-dependent inhibition of IlvE in vitro, show that the in vitro system reflects the mechanism of RidA function in vivo, and show that IlvE inhibition is prevented by RidA proteins from all domains of life. We conclude that 2-aminoacrylate (2AA) inhibition represents a new type of metabolic damage, and this finding provides an important physiological context for the role of the ubiquitous RidA family of enamine deaminases in preventing damage by 2AA.
IMPORTANCE
External stresses that disrupt metabolic components can perturb cellular functions and affect growth. A similar consequence is expected if endogenously generated metabolites are reactive and persist in the cellular environment. Here we show that the metabolic intermediate 2-aminoacrylate (2AA) causes significant cellular damage if allowed to accumulate aberrantly. Furthermore, we show that the widely conserved protein RidA prevents this accumulation by facilitating conversion of 2AA to a stable metabolite. This work demonstrates that the reactive metabolite 2AA, previously considered innocuous in the cell due to a short half-life in aqueous solution, can survive in the cellular environment long enough to cause damage. This work provides insights into the roles and persistence of reactive metabolites in vivo and shows that the RidA family of proteins is able to prevent damage caused by a reactive intermediate that is created as a consequence of PLP-dependent chemistry.
Introduction
The classic view of metabolism holds that it is comprised of discrete but integrated biochemical pathways that generate the building blocks of life. A complex repertoire of regulatory systems maintain metabolic homeostasis at the transcriptional, translational, and posttranslational level, in addition to ensuring the availability of metals and cofactors that are required for enzyme function. Disruption of any of these systems has deleterious consequences on the growth and/or behavior of an organism. Metabolic imbalance can also be caused by reactive metabolites generated nonenzymatically or as a part of normal metabolic pathways. For instance, if left uncontrolled, reactive oxygen species (ROS) generated during aerobic growth can damage DNA, proteins, lipids, and cofactors. To meet the ROS challenge, organisms have evolved peroxidases, catalases, and dismutases which prevent cell death (1). Similarly, intermediates of other pathways can be detrimental if they accumulate. For example, 2-keto-3-deoxygluconate 6-phosphate (2KDGP), an intermediate in the Entner-Doudoroff pathway, and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), an intermediate in purine biosynthesis, can inhibit central metabolic enzymes if they accumulate (2–6). Here, we describe cellular damage caused by a reactive enamine, which is generated as an unavoidable consequence of a reaction mechanism common to all organisms, and we demonstrate an important role for the RidA protein in preventing cellular damage by this reactive enamine.
Pyridoxal 5′-phosphate (PLP)-dependent threonine/serine dehydratases (ammonia lyases; EC 4.3.1.19 and 4.3.1.17) are found in all domains of life. These well-characterized enzymes dehydrate threonine or serine to form the corresponding enamine intermediates. The enamines subsequently tautomerize to imines, which are then hydrolyzed nonenzymatically to form the final keto acid products (7, 8). The bacterium Salmonella enterica has two PLP-dependent threonine/serine dehydratases, the biosynthetic IlvA enzyme, involved in isoleucine biosynthesis, and the catabolic TdcB enzyme, involved in the anaerobic catabolism of threonine. IlvA and TdcB have the same biochemical activity and catalytic mechanism, but IlvA has a regulatory domain that responds to isoleucine, while TdcB is not allosterically regulated (9, 10).
RidA is the founding member of a broadly conserved family of proteins (formerly known as YjgF/YER057c/UK114) found in all domains of life (11–14). We recently showed that RidA homologs increase the rate of hydrolysis of the enamine/imine products generated by PLP-dependent dehydratases (15). Prior to the above report, it was generally accepted that the hydrolysis of these intermediates into keto acid products was nonenzymatic in vivo. Importantly, the previous mechanistic studies were performed under conditions in which the half-lives of the enamine/imine intermediates were relatively short (3 to 4 min for the threonine-derived intermediates [16, 17] and 1.5 s for the serine-derived intermediates [18]). However, multiple metabolic phenotypes of an S. enterica ridA mutant (11, 12, 19–21) suggested that the reactive intermediate substrates of RidA accumulated in vivo to levels that affected other areas of metabolism.
Notably, the activity of isoleucine transaminase B was decreased in strains lacking RidA in both S. enterica and Saccharomyces cerevisiae (12, 22). In S. enterica, the decrease in the specific activity of transaminase B (IlvE) depended on the activity of IlvA (12) but not its normal substrate, threonine (23). Thus, the possibility that the IlvA-dependent inhibition of IlvE could be exerted when serine was the substrate was considered. There was precedent for the inhibition of IlvE by 2-aminoacrylate (2AA), the serine-derived enamine that could also be derived from the serine analog and suicide inhibitor 3-chloro-l-alanine (3CA) (24). Although the mechanism of IlvE inhibition by 3CA is unknown, some insights were obtained from studies of other PLP-dependent enzymes inhibited by 3CA (25–28) and other serine analogs (29–34). In each of the above-mentioned studies, a 2AA intermediate was generated in the active site of the PLP-dependent enzyme, which led to the formation of a 2AA-PLP adduct covalently bound to the enzyme (27, 32, 34–36). These studies suggested a possible mechanism underpinning the phenotypes of S. enterica ridA strains.
This study was initiated to probe the deaminase activity of RidA in the context of cellular metabolism. Specifically, we sought to define the mechanism responsible for the decreased IlvE activity in a ridA strain and to address how this mechanism was related to the characterized in vitro activity of RidA. Data reported here support a model in which the dehydration of serine by IlvA generates 2AA, which in turn modifies and inhibits cellular targets in vivo in the absence of RidA. We suggest that the role for RidA in cell physiology is to prevent metabolic damage caused by reactive enamines and that the presence of such reactive molecules in all forms of life could be a major selective pressure for the evolution and conservation of RidA proteins.
RESULTS
Increased levels of threonine dehydratase TdcB diminish IlvE activity.
The decreased activity of isoleucine transaminase B (IlvE) in a ridA strain depended on the activity of the biosynthetic threonine dehydratase (IlvA), a PLP-dependent enzyme (12). S. enterica has a second PLP-dependent threonine/serine dehydratase, the catabolic enzyme TdcB, whose expression is inhibited in the presence of molecular oxygen and in the presence of glucose (10, 37). To determine the basis of the IlvA requirement, tdcB was provided ectopically under the control of a constitutive lac promoter in ridA+ and ridA strains, allowing for expression of TdcB under the growth conditions used here. Isoleucine was added to the growth medium to allosterically inhibit IlvA and thus differentiate between the effects of the isozymes TdcB and IlvA. IlvE activity was assayed in crude extracts of relevant strains, and the data are shown in Table 1.
TABLE 1.
IlvE activity is decreased in a ridA strain by serine/threonine dehydratasesa
| Relevant genotype | IlvE sp act (nmol 2KMV ⋅ min−1 mg−1 protein) in indicated medium |
|
|---|---|---|
| Minimal | Minimal + isoleucine | |
| ridA+ pSU18 (empty) | 73 ± 9 | 52 ± 2 |
| ridA pSU18 (empty) | 18 ± 5 | 51 ± 11 |
| ridA+ pSU18-tdcB+ | 72 ± 8 | 77 ± 6 |
| ridA pSU18-tdcB+ | 2 ± 4 | 6 ± 3 |
Cells were grown in the indicated media, and IlvE activity was measured in permeabilized cells as described in Materials and Methods. IlvE specific activity is expressed in nmol 2KMV ⋅ min−1 mg−1 protein; shown are averages and standard deviations of results from three biological replicates.
In a ridA+ strain, the ectopic expression of tdcB had no effect on IlvE activity in the presence or absence of isoleucine. In contrast, in a ridA strain, ectopic expression of tdcB decreased the activity of IlvE to 3% (>30-fold) compared to that of the ridA+ strain (2 versus 72 nmol 2-keto-3-methylvalerate [2KMV] ⋅ min−1 mg−1 of protein). Unlike the inhibition resulting from IlvA activity, TdcB-dependent inhibition was unaffected by the addition of isoleucine. In the vector control experiments, IlvE activity in the ridA strain was decreased to 25% (4-fold) of that in the ridA+ strain (18 versus 73 nmol 2KMV ⋅ min−1 mg−1). This decrease was eliminated by the presence of isoleucine in the medium due to allosteric inhibition of IlvA (12). In total, these data indicated that in the absence of RidA in vivo, IlvE could be inhibited by two distinct PLP-dependent threonine dehydratases.
PLP-dependent threonine/serine dehydratases inhibit IlvE activity in vitro.
The threonine/serine dehydratase-dependent inhibition of IlvE was reconstituted in vitro. Based on previous in vivo data, serine was expected to be the relevant substrate for IlvA in the context of inhibiting IlvE (23). To test whether IlvA and serine could produce an intermediate that would inhibit IlvE, purified IlvE, IlvA, and l-serine were incubated together. Control reaction mixtures lacked either IlvA or serine, and alternatively, serine was replaced with l-threonine in the reaction mixture. The reaction mixtures were preincubated at pH 9.0 to maximize the half-lives of the intermediate metabolites formed in the dehydration reaction (15, 17, 38) at 37°C for 2 h (to allow ample time for the intermediate to affect IlvE). After the preincubation period, IlvE activity was assayed from each reaction by quantifying the conversion of [14C]isoleucine to [14C]2KMV (Fig. 1).
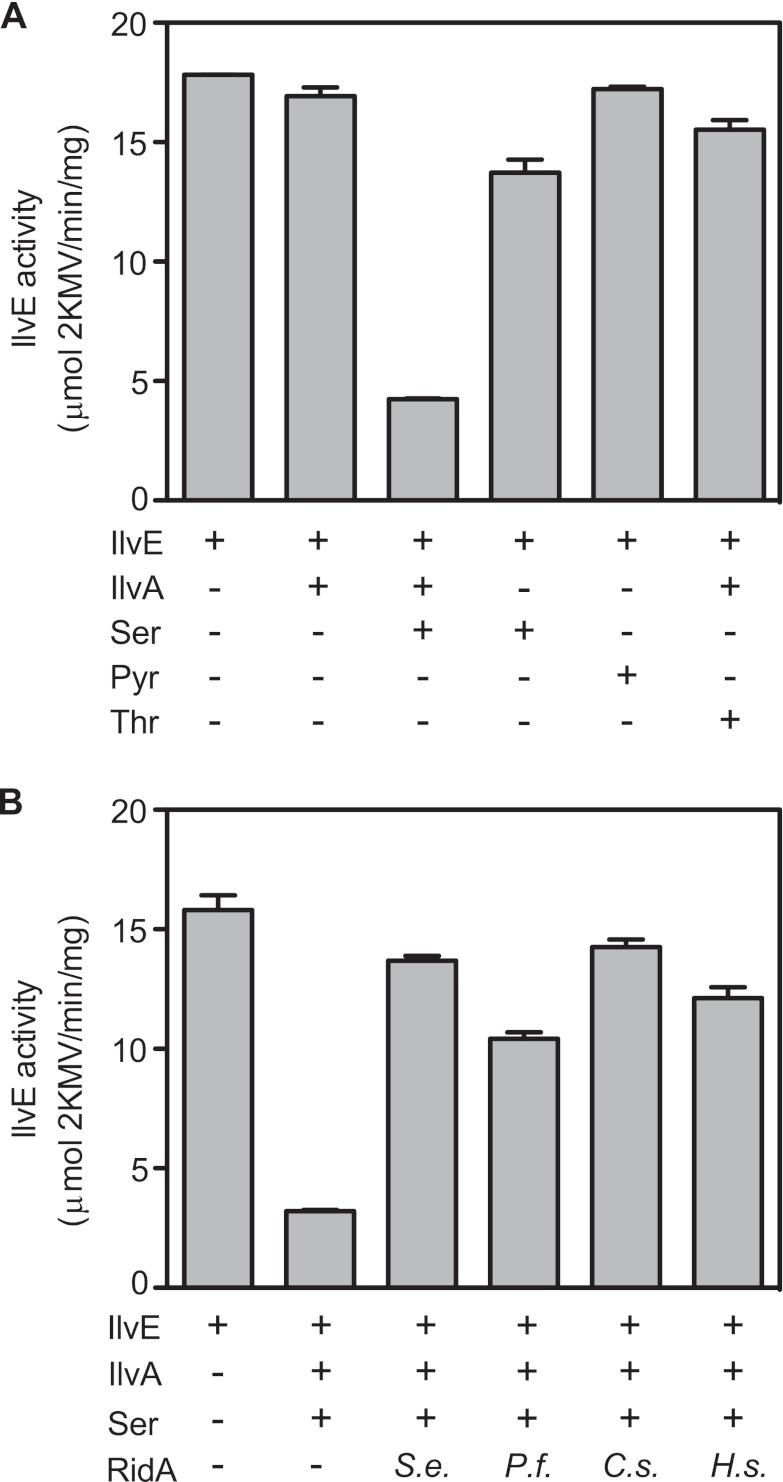
FIG 1 .
IlvE activity is inhibited by serine and IlvA. IlvE assays were performed using a radioactive assay as described in Materials and Methods. (A) IlvE assays were performed in the presence of IlvA, serine (Ser), pyruvate (Pyr), or threonine (Thr), as indicated. (B) RidA homologs were added to IlvE that had been preincubated with IlvA and serine (Ser), as indicated. RidA homologs tested were from S. enterica (S.e.), P. furiosus (P.f.), C. sativus (C.s.), and H. sapiens (H.s.). Shown are averages and ranges from two replicates for each condition (ranges, ≤1 in all cases). Each assay was performed independently at least twice.
Preincubation of IlvE with IlvA and serine decreased the activity of the enzyme to 24% of that of the IlvE protein incubated in the absence of IlvA and serine (4 versus 18 µmol 2KMV ⋅ min−1 mg−1) (Fig. 1A). Significantly, both serine and IlvA were required to decrease IlvE activity. Further, IlvE was not inhibited by incubation with pyruvate, a product of serine deamination, or when threonine was substituted for serine. Collectively, these data suggested that IlvE inhibition was caused by an intermediate metabolite derived from serine by IlvA activity. Providing TdcB instead of IlvA produced a similar result, with IlvE activity decreasing to 14% of that found in the reaction mixtures lacking TdcB. Taken together, the above data suggested that 2AA was likely the reactive species generated by IlvA and TdcB and that 2AA inactivated IlvE.
RidA prevents IlvA-dependent inhibition of IlvE.
The system described above reconstituted one phenotype of a ridA strain, namely, the serine/threonine dehydratase-dependent inhibition of IlvE. Purified S. enterica RidA was added to the in vitro assay to define the physiological relevance of this system. The presence of RidA in the reaction mixture dramatically decreased the negative effect of serine and IlvA on IlvE activity (Fig. 1B). The presence of RidA in the reaction mixture restored IlvE activity from 20% to 87% of those of the uninhibited control reaction mixtures (3 versus 14 µmol KMV min−1 mg−1, respectively). RidA homologs from the archaeon Pyrococcus furiosus (PF0668), cucumber plant Cucumis sativus (ChrD), and Homo sapiens (UK114) were all proficient at preventing the dehydratase-dependent inhibition of IlvE (Fig. 1B). These results showed that, like other activities of RidA (15), the function of RidA that protected IlvE was conserved across all domains of life.
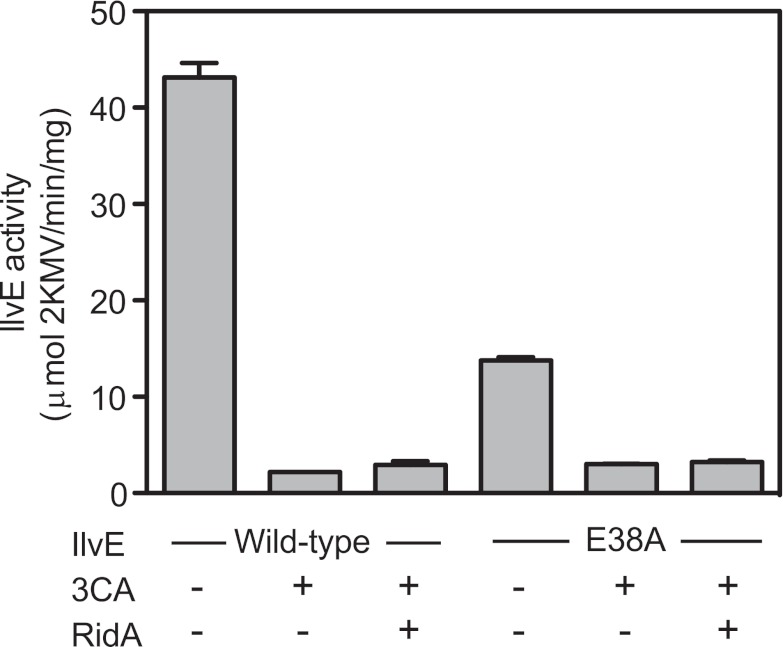
The suicide inhibitor 3CA has been shown to work via a 2AA attack on the active site of PLP enzymes (27, 32, 34–36). The presence of 3CA decreased IlvE activity to 5% of that of the uninhibited control (Fig. 2). While the reactive molecule derived from both 3CA and IlvA/serine is expected to be 2AA, RidA had no effect on the inhibition of IlvE by 3CA (Fig. 2). These data appeared to distinguish the two mechanisms of inhibition. Consistently with previous reports (25–28), these data suggested that the 3CA-derived 2AA was generated within the same active site that it targeted; however, IlvA/serine-derived 2AA was generated in the active site of the IlvA dehydratase, was released, and targeted the active site of a different protein (IlvE). Together, these data supported the conclusion that RidA acted on free 2AA.
FIG 2 .
3CA inhibits both wild-type IlvE and IlvEE38A activity. IlvE assays were performed with pure protein using DNPH derivatization and subsequent organic extraction to quantify the product 2KMV as described in Materials and Methods. Wild-type IlvE and IlvEE38A were assayed in the absence and presence of the inhibitor 3CA and RidA as indicated. Shown are averages and ranges from two replicates for each condition (ranges, ≤1 in all cases). Each assay was performed independently at least twice.
Modification with 3CA or IlvA and serine leads to a 317-Da adduct on IlvE.
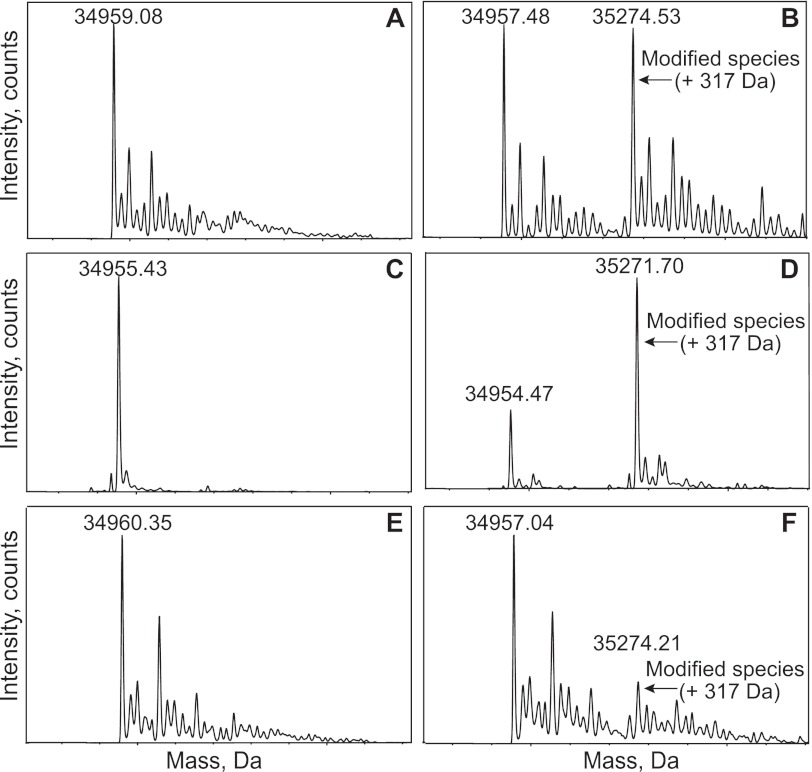
A mass spectrometry approach was used to characterize the protein modification generated by 2AA. Purified IlvE protein was either left untreated or incubated with 3CA or IlvA and serine. The resulting samples were subjected to intact-protein electrospray ionization-time of flight mass spectrometry (ESI-TOF MS) analysis (Fig. 3). In all cases, a peak close to the expected mass for IlvE (35,072 Da) was observed. Significantly, the IlvE protein that was incubated with 3CA had a species with a higher mass (an additional 317 Da) than the expected IlvE peak (Fig. 3A and B). This species was not present in the IlvE sample from a control reaction mixture lacking serine. The sample incubated with IlvA/serine also had a peak with a mass of 317 mass units more than the parental IlvE peak (Fig. 3C and D). When IlvE lacking PLP was treated with 3CA, only a single peak with the IlvE mass was visible (data not shown), demonstrating that PLP was required for the modification. Together, these data suggested that the same protein modification was generated on IlvE by 3CA or IlvA/serine. These data were also consistent with IlvE being inhibited by a mechanism involving a covalent adduct of 2AA and PLP similar to those implicated in the suicide inhibition of other PLP-dependent enzymes (27, 32, 34–36).
FIG 3 .
IlvE proteins are modified by 2AA. IlvE protein samples were subjected to the relevant treatment prior to MS analysis as described in Materials and Methods. (A) Untreated; (B) incubated with 3CA; (C) untreated; (D) incubated with IlvA/serine; (E) IlvE purified from the ridA+ strain; (F) IlvE purified from a ridA strain. Samples in panels A, B, E, and F were precipitated with acetone and resuspended in acid before mass spectral analysis, while samples in panels C and D were separated by LC before mass analysis. In samples A, C, and E, no species with an additional 317 amu was visible.
A 317-Da modification is also found on protein purified from a ridA strain.
To confirm that the in vitro reconstitution of IlvA-dependent inhibition of IlvE was connected to the phenotypes of ridA strains, IlvE protein was isolated from both ridA+ and ridA strains. To maximize detection of any modification, TdcB was overproduced in the strains from which IlvE was purified. After purification, the IlvE protein isolated from the ridA strain had 80% the specific activity of the IlvE protein isolated from the ridA+ strain. The reduced magnitude of the inhibition was due to the expression of IlvE above chromosomal levels to facilitate purification and was consistent with previous results that showed that the deleterious effect on IlvE was titratable (12). Both IlvE protein preparations were analyzed by mass spectrometry. In both samples a peak of the appropriate size for the intact IlvE protein was detected (~34,957 Da). However, in the sample isolated from the ridA strain, a protein species that was 317-Da larger (35,274 Da) was also detected (Fig. 3E and F). Significantly, this peak was not present in the protein sample that was purified from the ridA+ strain. These two protein preparations were also analyzed by nanoESI ultra-high-resolution quadrupole TOF MS (QTOF MS), and the presence of the minor 317-Da peak was confirmed in the IlvE purified from the ridA strain and was not found in the IlvE purified from the ridA+ strain. These results supported the conclusion that in strains lacking RidA, the 2AA generated by IlvA accumulated to levels that were sufficiently high to modify IlvE by the mechanism observed in vitro.
Properties of the posttranslational modification suggest a 2AA-PLP adduct.
Based on the 317-Da mass difference between the modified and unmodified IlvE enzymes, and considering previously described modifications of PLP-dependent enzymes, we concluded that the modification on IlvE was a 2AA-PLP adduct. The mechanisms described for the inactivation of PLP enzymes by 3CA gave two possible scenarios for a 2AA-PLP adduct on IlvE. In the first scenario, characterized with the PLP-dependent enzyme alanine racemase, 2AA attacks the PLP bound to the active-site lysine residue, generating a pyruvate-PLP adduct covalently bound to the active-site lysine and preventing further chemistry by the enzyme (27, 30, 31, 39). This mechanism resulted in an adduct that added 319 Da to the mass of the unmodified protein and could be removed intact from the protein with base treatment (27). Here, IlvE protein that had been incubated with either 3CA or serine/IlvA was treated with base, but it failed to release a stable product (data not shown).
Alternatively, a 2AA-PLP adduct could be formed as an external aldimine in the same way that PLP reacts with other nitrogen-containing substrates. The resulting 2AA-PLP can be attacked by a nonlysine nucleophilic residue in the active site, resulting in attachment of the adduct to this residue (Fig. 4A) (26, 32–36). This second mechanism would yield a modification with a mass of 317 Da when fully protonated for mass spectral analysis and allow for the loss of a proton from the enzyme during the nucleophilic attack. As shown in Fig. 3, whole IlvE protein treated with IlvA/serine or 3CA resulted in a product with an additional 317 Da, but efforts using standard methods of trypsin or endoproteinase Glu-C digestion failed to detect the modified fragment. These results suggested that the relevant adduct was not stable in one or more steps of the digestion process.
FIG 4 .
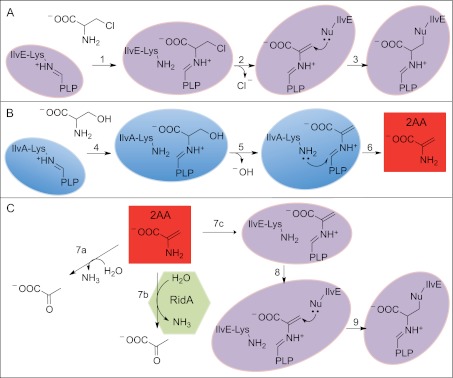
Distally generated 2AA modifies PLP enzymes unless it was removed by RidA. Both the inhibition of a PLP enzyme by 3CA and the generation of 2AA and its three fates in vivo are schematically represented. (A) Inhibition of PLP-dependent enzyme IlvE by the suicide substrate 3CA. In step 1, IlvE forms an internal aldimine between PLP and the suicide substrate 3CA. In step 2 the 3CA is dechlorinated, forming a 2AA-PLP intermediate that is attacked by a nucleophilic residue (Nu) in the IlvE active site. Step 3 results in an inactive IlvE enzyme, with 2AA-PLP attached to a nucleophilic residue. (B) Schematic of the PLP-dependent mechanism of serine dehydration by IlvA that results in the formation of 2AA. In step 4, IlvA forms an internal aldimine between PLP and serine. In step 5, the serine is dehydrated, generating 2AA in the enzyme active site. In step 6, the 2AA is released from the PLP into solution. (C) Three fates of 2AA in vivo. Step 7a reflects the hydrolysis of 2AA that can occur on the enzyme or in solution. Step 7b depicts the RidA-assisted hydrolysis of 2AA. Step 7c depicts the attack of the IlvE active site by 2AA and the resulting modification, which is also generated by 3CA attack in panel A. Purple shading, IlvE enzyme; blue shading, IlvA enzyme; green shading, RidA enzyme; red shading, reactive metabolite 2-aminoacrylate (2AA).
Inspection of the crystal structure of the E. coli IlvE enzyme (1A3G [40; http://www.pdb.org/pdb/home/home.do.]) showed that the closest nucleophilic residue to the active portion of the PLP molecule was Glu38. This residue was changed to an alanine by site-directed mutagenesis, generating variant protein IlvEE38A. The IlvEE38A variant was purified from a ridA+ strain and incubated with 3CA. If Glu38 were the site of the 2AA-PLP modification, the IlvEE38A variant would be resistant to 3CA inhibition. Though the IlvEE38A variant had a reduced specific activity, the activity was still eliminated by incubation with 3CA (Fig. 2), indicating that Glu38 was not a functionally significant site of modification. Consistently, when the untreated and 3CA-exposed IlvEE38A variant were subjected to mass spectral analysis, the variant exposed to 3CA had a species that was 317-Da larger than the untreated protein (see Fig. S1 in the supplemental material). The activity of the IlvEE38A variant was also inhibited by incubation with IlvA and serine (data not shown), indicating that the Glu38 residue was not the site of the modification generated by IlvA/serine. Other nucleophilic residues in the active site are implicated in binding the PLP cofactor, and these were not substituted due to their expected requirement for activity (41).
DISCUSSION
The mechanism of PLP-dependent serine/threonine dehydratases involves the formation of reactive enamine/imine intermediates that are hydrolyzed to keto acid products by water. To our knowledge, an impact of these metabolites in vivo has not been reported. A role for reactive enamines like 2AA in the cell may have been overlooked because these metabolites are easily hydrolyzed in vitro and thus not expected to persist in an aqueous solution. Here, we showed that enamine/imine metabolites attacked the active site of an enzyme distinct from the dehydratase that formed them, both in vitro and in vivo. The ability of RidA proteins to prevent this damage supported the conclusion that this family of proteins has an important role in preventing damage from enamines. Evidence that isoleucine transaminase B (IlvE) was damaged by 2AA in strains lacking RidA (Fig. 3) validated the physiological significance of the data from in vitro experiments (Fig. 1) and emphasized the significance of this finding to our understanding of both mechanistic biochemistry and cellular physiology.
The data presented herein warrant a paradigm shift in our understanding of the persistence of enamine species in vivo (Fig. 4). Previous reports described how 3CA and related compounds damage PLP-dependent enzymes in vitro via the formation of 2AA (25–34). This scenario as it relates to IlvE (Fig. 4A) leads to a 2AA-PLP adduct attached to a nucleophilic residue in the IlvE active site. In contrast to this situation, our data showed that when the PLP-dependent threonine dehydratase IlvA dehydrated serine (Fig. 4B), the resulting 2AA product had three potential fates (Fig. 4C). First, the canonical attack on 2AA by water results in the formation of pyruvate. Second, 2AA released into solution can be deaminated by the RidA enzyme (15). Third, in the absence of RidA, the free 2AA is able to modify and thus inactivate IlvE. To our knowledge, this is the first evidence of 2AA inactivating a site distinct from the enzymatic active site that generated it. Previously, we showed that the reduced specific activity of IlvE that is found in a ridA strain required the activity of IlvA (12). Those data, in combination with the results here, demonstrated that endogenously generated 2AA inactivates enzymes in vivo. These findings demonstrate that the reactive enamine intermediates formed as part of PLP-dependent chemistry can persist in the cellular environment long enough to damage target enzymes. Although it was formally possible that the 2AA was channeled to the IlvE active site and not present in the cellular milieu, the finding that other PLP-dependent enzymes, including alanine racemase (Alr), are similarly damaged by 2AA minimizes this possibility (J. M. Flynn and D. M. Downs, unpublished data). Thus, in total, the in vivo and in vitro data showed that the presence of RidA prevented damage by free 2AA. These results allow the conclusion that the enamine deaminase activity of RidA has a critical role in cell physiology.
We suggest that the RidA family of proteins comprises a damage prevention system that intercepts reactive metabolites to preempt damage to cellular proteins. The RidA protein family is broadly conserved and consists of both high- and low-identity members (42). High-identity members from all domains are functionally interchangeable in vivo and in vitro (15, 21). We suggest that the high-identity RidA proteins have a housekeeping role in preventing damage from enamines generated by PLP-dependent enzymes in central metabolism. The conservation of RidA suggests that damage from 2AA may be a universal challenge that organisms must manage. IlvA is not the only enzyme whose mechanism could produce free 2AA in vivo. Organisms have other PLP enzymes whose reaction mechanisms may involve 2AA derived from serine, cysteine, tryptophan, or other metabolites (43–47). PLP-dependent enzymes are ubiquitous, and the need for this cofactor is a selective pressure for the evolution of RidA enzymes to avert damage caused by enamine/imine intermediates.
There are examples of high-identity RidA homologs in operons with biosynthetic genes that include a PLP-dependent serine/threonine dehydratase (48, 49) and low-identity RidA homologs that exist in pathways generating reactive enamine intermediates distinct from 2AA (50–56). These findings suggest that some RidA homologs may have a different function and are thus clustered with genes involved in pathways that produce specific enamines or other reactive metabolites.
The results of in vivo genetic analyses, conservation of the protein family, and a preliminary bioinformatic analysis emphasized a role for the RidA protein family beyond protecting only IlvE. Phenotypes of a ridA mutant of S. enterica include sensitivity to serine and cysteine, the inability to use pyruvate or glycerol as a carbon source, decreased activity of IlvE, decreased activity of Alr, and the ability to synthesize the thiamine precursor phosphoribosyl amine (PRA) by a novel mechanism (11, 12, 19, 20; J.M. Flynn and D.M. Downs, unpublished data). PRA formation required the 2-aminocrotonate intermediate of threonine dehydration, while the decrease in both IlvE and Alr required the 2AA intermediate of serine dehydration (15, 21, 57). The consequences of the accumulation of these reactive metabolites were apparent only in the absence of RidA. It is not yet clear how many of the metabolic phenotypes of a ridA strain require the activity of a serine/threonine dehydratase or how many target proteins of 2AA modification exist. Insights generated by the current study will facilitate our ability to define additional metabolic phenotypes of a ridA strain on a mechanistic level.
MATERIALS AND METHODS
Strain construction.
The gene encoding IlvE was cloned into plasmid pET28b(+) (Novagen) at NcoI and XhoI sites, creating a pET28-ilvE+ construct that directed the synthesis of IlvE with a hexahistidine tag fused to its C terminus (IlvE-H6). Strains used for the overexpression of ilvE were derived from SB300A1 (58), a derivative of S. enterica that carries a chromosomally borne gene for T7 polymerase under the control of the arabinose-inducible PBAD promoter (aadA::araC-PBAD T7 pol+). Strain SB300A1carrying plasmid pET28-ilvE+ was named DM13730. The pET28-ilvE+ plasmid was also moved into ridA+ and ridA1::Tn10d (tet+) derivatives of strain SB300A1 that harbored plasmid pSU18-tdcB+. The resulting strains were named DM13914 (ridA+) and DM13915 (ridA), respectively.
To generate the IlvEE38A variant, site-directed mutagenesis of pET28-ilvE+ was performed using the QuikChange Lightening (Agilent) kit according to the manufacturer’s protocol. The resulting plasmid was introduced into the aadA::araC-PBAD T7 pol+ strain SB300A1, generating strain DM13974.
Protein purification.
The purifications of the S. enterica IlvA and RidA proteins and RidA homologs were described elsewhere (15, 21). To purify wild-type IlvE and the IlvEE38A proteins, strains DM13730 and DM13974 were grown in 3 liters of superbroth (32 g tryptone, 20 g yeast extract, 5 g sodium chloride, and 1 ml 5 N sodium hydroxide per liter) containing kanamycin (50 mg/liter). The cultures were incubated at 37°C with shaking until they reached an optical density (650 nm) of 0.35, at which time ilvE+ expression was induced with 0.2% (wt/vol) l-(+)-arabinose (herein arabinose) and 0.2 mM isopropyl-β-d-thiogalactoside (IPTG). Cells were grown for 3 more hours and then harvested by centrifugation. Alternatively, strains DM13914 and DM13915 were grown in 4.5 liters of minimal no-carbon E medium (59) containing glucose (22 mM), MgSO4 (1 mM), trace minerals (adapted from reference 60), histidine (100 µM), pantothenate (100 µM), kanamycin (50 mg/liter), and chloramphenicol (5 mg/liter). In this case, there was no addition of arabinose or IPTG and ilvE expression was at a basal level from the T7 promoter. These cultures were grown for 16 h at 37°C, and then the cells were harvested by centrifugation.
Cell pellets were resuspended in potassium phosphate buffer (50 mM, pH 7.7) containing NaCl (100 mM), imidazole (5 mM), and glycerol (10%, vol/vol) and disrupted using a French press. Cell-free extracts were clarified by centrifugation (48,000 × g for 45 min) and applied to nickel-nitrilotriacetic (Ni-NTA) Superflow resin. The relevant protein was purified according to the manufacturer’s protocol (Qiagen) using column chromatography and an imidazole gradient. The purified protein was concentrated ~5-fold and desalted into potassium phosphate buffer (50 mM, pH 7.7) containing NaCl (100 mM) and glycerol (10%, vol/vol) using a PD-10 Sephadex G-25M column developed by gravity (GE Healthcare).
IlvE enzyme activity assays. (i) Nonradioactive assays.
Assays of IlvE activity were performed with permeabilized cells as described previously (12, 61). Briefly, cells were grown in minimal medium with glucose (11 mM) as the carbon source, pelleted, and resuspended in potassium phosphate buffer (pH 8). An aliquot of cells was permeabilized by 10% PopCulture (Novagen) in the presence of the assay reagents potassium phosphate buffer (100 mM, pH 8), PLP (50 µM), and 2-ketoglutarate (10 mM). The substrate l-isoleucine (20 mM) was added to initiate reactions. Alternatively, 0.5 µg of purified IlvE protein was used in lieu of permeabilized cells when assays were performed in the presence of the inhibitor 3CA. The formation of product 2-keto-3-methylvalerate (2KMV) was measured after 20 min by 2,4-dinitrophenylhydrazine (DNPH) derivatization and subsequent organic extraction. Protein from permeabilized cells was quantified by a bicinchoninic acid assay kit (Pierce). IlvE activity was reported as nmol 2KMV ⋅ min−1 mg−1 protein.
(ii) Radioactive assays.
Reaction mixtures containing potassium phosphate (50 mM, pH 9.0), PLP (50 µM), 2-ketoglutarate (12.5 mM), IlvE (0.75 µg), IlvA (10 µg), serine (50 mM), and RidA (16 µg) as indicated in the figures, were preincubated at 37°C for 2 h to allow for IlvE inhibition. l-[U-14C]isoleucine (American Radiolabeled Chemicals) was added to a final concentration of 5 mM (20 nCi) to initiate IlvE reactions. Reaction mixtures were incubated for 15 min at 37°C, and reactions were stopped by spotting 5 µl (0.5 nCi) onto a prewashed polyethyleneimine cellulose thin-layer chromatography plate. The plate was developed using a butanol-acetic acid-water (4:2:1) mobile phase to separate [14C]isoleucine from [14C]2KMV. The relative mobilities of radioactive spots were detected using a Cyclone Storage Phosphor Screen (Packard Instrument Company) and a Typhoon FLA 9000 scanner (GE Healthcare). The concentration of [14C]2KMV was quantified, and specific activity is reported as µmol of 2KMV ⋅ min−1 mg−1 IlvE.
Mass spectrometry analyses.
Reaction mixtures containing potassium phosphate buffer (50 mM, pH 9.0), PLP (33 µM), IlvE (20 µg), and IlvA (1 µg), with or without l-serine (100 mM), were incubated at 37°C for 4 h. Alternatively, reaction mixtures containing potassium phosphate buffer (50 mM, pH 9.0), PLP (33 µM), IlvE (20 µg), and 3CA (1 mM) were incubated at 37°C for 3 h. Subsequent mass spectrometry (MS) analyses were performed at the University of Wisconsin Biotechnology Center. For IlvE proteins isolated from the ridA+ and ridA strains, 100 µg of treated protein was precipitated with acetone, and then samples were resolubilized in acid before direct MS analysis. The 3CA-modified sample and unmodified control were also treated with acetone and acid before MS analysis. The serine-treated sample and control were chromatographically resolved with a Jupiter C4 300Å 50- by 2-mm column (Phenomenex) on an Agilent 1200 high-performance liquid chromatograph (HPLC), using a linear gradient of 0.1% acetic acid in water and 0.1% acetic acid in acetonitrile over 30 min. The mass spectrometer used was an Agilent LC/TOF MS with electrospray ionization (ESI) used in positive-ion mode. Acquired data were processed using Analyst QS 1.1 (build 9865) software (Agilent) to monitor masses observed in the range of 100 to 3,200 amu. Intact protein species deconvolution was carried out using the ProteinApp module of Agilent BioConfirm software. An independent verification of intact protein masses was carried out at the University of Wisconsin—Madison School of Pharmacy Analytical Instrumentation Center. Treated and untreated IlvE samples were dialyzed against ammonium bicarbonate buffer (20 mM). Protein was then diluted 1:10 into 20% acetonitrile with 0.1% formic acid and analyzed directly on a nanoESI ultra-high-resolution QTOF MS (MaXis 4G) in positive-ion mode.
SUPPLEMENTAL MATERIAL
IlvEE38A is modified with 3CA.The variant protein IlvEE38A was purified and analyzed by mass spectroscopy as described in Materials and Methods. Sample A was untreated, and sample B was incubated with 3CA. Download
ACKNOWLEDGMENTS
We thank Grzegorz Sabat and Cameron Scarlett for performing mass spectral procedures and assisting with data analysis, and we thank Jorge Escalante-Semerena for critical reading of the manuscript.
This work was supported by NIH competitive grant R01 GM095837 (D.M.D.), NIH training grant T32 GM07215 (J.A.L.), and a University of Wisconsin Department of Bacteriology Predoctoral Fellowship (J.A.L.).
Footnotes
Citation Lambrecht JA, Schmitz GE, Downs DM. 2013. RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. mBio 4(1):e00033-13. doi:10.1128/mBio.00033-13.
REFERENCES
- 1. Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Faik P, Kornberg HL, McEvoy-Bowe E. 1971. Isolation and properties of Escherichia coli mutants defective in 2-keto 3-deoxy 6-phosphogluconate aldolase activity. FEBS Lett. 19:225–228 [DOI] [PubMed] [Google Scholar]
- 3. Fradkin JE, Fraenkel DG. 1971. 2-Keto-3-deoxygluconate 6-phosphate aldolase mutants of Escherichia coli. J. Bacteriol. 108:1277–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuhrman LK, Wanken A, Nickerson KW, Conway T. 1998. Rapid accumulation of intracellular 2-keto-3-deoxy-6-phosphogluconate in an Entner-Doudoroff aldolase mutant results in bacteriostasis. FEMS Microbiol. Lett. 159:261–266 [DOI] [PubMed] [Google Scholar]
- 5. Baggott JE, Vaughn WH, Hudson BB. 1986. Inhibition of 5-aminoimidazole-4-carboxamide ribotide transformylase, adenosine deaminase and 5′-adenylate deaminase by polyglutamates of methotrexate and oxidized folates and by 5-aminoimidazole-4-carboxamide riboside and ribotide. Biochem. J. 236:193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan ES, Cronstein BN. 2002. Molecular action of methotrexate in inflammatory diseases. Arthritis Res. 4:266–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chargaff E, Sprinson DB. 1943. Studies on the mechanism of deamination of serine and threonine in biological systems. J. Biol. Chem. 151:273–280 [Google Scholar]
- 8. Phillips AT, Wood WA. 1965. The mechanism of action of 5′-adenylic acid-activated threonine dehydrase. J. Biol. Chem. 240:4703–4709 [PubMed] [Google Scholar]
- 9. Umbarger HE, Brown B. 1957. Threonine deamination in Escherichia coli. II. Evidence for two l-threonine deaminases. J. Bacteriol. 73:105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luginbuhl GH, Hofler JG, Decedue CJ, Burns RO. 1974. Biodegradative l-threonine deaminase of Salmonella typhimurium. J. Bacteriol. 120:559–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enos-Berlage JL, Langendorf MJ, Downs DM. 1998. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J. Bacteriol. 180:6519–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmitz G, Downs DM. 2004. Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:803–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leitner-Dagan Y, Ovadis M, Zuker A, Shklarman E, Ohad I, Tzfira T, Vainstein A. 2006. CHRD, a plant member of the evolutionarily conserved YjgF family, influences photosynthesis and chromoplastogenesis. Planta 225:89–102 [DOI] [PubMed] [Google Scholar]
- 14. Oxelmark E, Marchini A, Malanchi I, Magherini F, Jaquet L, Hajibagheri MA, Blight KJ, Jauniaux JC, Tommasino M. 2000. Mmf1p, a novel yeast mitochondrial protein conserved throughout evolution and involved in maintenance of the mitochondrial genome. Mol. Cell. Biol. 20:7784–7797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lambrecht JA, Flynn JM, Downs DM. 2012. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J. Biol. Chem. 287:3454–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flavin M, Slaughter C. 1964. An intermediate trapped by maleimides in a pyridoxal-phosphate potentiated enzymatic elimination reaction. Biochemistry 3:885–893 [DOI] [PubMed] [Google Scholar]
- 17. Datta P, Bhadra R. 1978. Biodegradative threonine dehydratase. Reduction of ferricyanide by an intermediate of the enzyme-catalyzed reaction. Eur. J. Biochem. 91:527–532 [DOI] [PubMed] [Google Scholar]
- 18. Hillebrand GG, Dye JL, Suelter CH. 1979. Formation of an intermediate and its rate of conversion to pyruvate during the tryptophanase-catalyzed degradation of S-o-nitrophenyl-l-cysteine. Biochemistry 18:1751–1755 [DOI] [PubMed] [Google Scholar]
- 19. Browne BA, Ramos AI, Downs DM. 2006. PurF-independent phosphoribosyl amine formation in yjgF mutants of Salmonella enterica utilizes the tryptophan biosynthetic enzyme complex anthranilate synthase-phosphoribosyltransferase. J. Bacteriol. 188:6786–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Christopherson MR, Schmitz GE, Downs DM. 2008. YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J. Bacteriol. 190:3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambrecht JA, Browne BA, Downs DM. 2010. Members of the YjgF/YER057c/UK114 family of proteins inhibit phosphoribosylamine synthesis in vitro. J. Biol. Chem. 285:34401–34407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim JM, Yoshikawa H, Shirahige K. 2001. A member of the YER057c/YjgF/UK114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells 6:507–517 [DOI] [PubMed] [Google Scholar]
- 23. Christopherson MR, Lambrecht JA, Downs D, Downs DM. 2012. Suppressor analyses identify threonine as a modulator of ridA mutant phenotypes in Salmonella enterica. PLoS One 7:e43082 http://dx.doi.org/10.1371/journal.pone.0043082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arfin SM, Koziell DA. 1971. Inhibition of growth of Salmonella typhimurium and of threonine deaminase and transaminase B by beta-chloroalanine. J. Bacteriol. 105:519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tate SS, Relyea NM, Meister A. 1969. Interaction of l-aspartate beta-decarboxylase with beta-chloro-l-alanine. Beta-elimination reaction and active-site labeling. Biochemistry 8:5016–5021 [DOI] [PubMed] [Google Scholar]
- 26. Kishore GM. 1984. Mechanism-based inactivation of bacterial kynureninase by beta-substituted amino acids. J. Biol. Chem. 259:10669–10674 [PubMed] [Google Scholar]
- 27. Badet B, Roise D, Walsh CT. 1984. Inactivation of the dadB Salmonella typhimurium alanine racemase by D and L isomers of beta-substituted alanines: kinetics, stoichiometry, active site peptide sequencing, and reaction mechanism. Biochemistry 23:5188–5194 [DOI] [PubMed] [Google Scholar]
- 28. Whalen WA, Wang MD, Berg CM. 1985. Beta-chloro-l-alanine inhibition of the Escherichia coli alanine-valine transaminase. J. Bacteriol. 164:1350–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. John RA, Fasella P. 1969. The reaction of l-serine O-sulfate with aspartate aminotransferase. Biochemistry 8:4477–4482 [DOI] [PubMed] [Google Scholar]
- 30. Ueno H, Likos JJ, Metzler DE. 1982. Chemistry of the inactivation of cytosolic aspartate aminotransferase by serine O-sulfate. Biochemistry 21:4387–4393 [DOI] [PubMed] [Google Scholar]
- 31. Likos JJ, Ueno H, Feldhaus RW, Metzler DE. 1982. A novel reaction of the coenzyme of glutamate decarboxylase with l-serine O-sulfate. Biochemistry 21:4377–4386 [DOI] [PubMed] [Google Scholar]
- 32. Wang EA, Kallen R, Walsh C. 1981. Mechanism-based inactivation of serine transhydroxymethylases by d-fluoroalanine and related amino acids. J. Biol. Chem. 256:6917–6926 [PubMed] [Google Scholar]
- 33. Silverman RB, Abeles RH. 1976. Inactivation of pyridoxal phosphate dependent enzymes by mono- and polyhaloalanines. Biochemistry 15:4718–4723 [DOI] [PubMed] [Google Scholar]
- 34. Rando RR. 1974. Irreversible inhibition of aspartate aminotransferase by 2-amino-3-butenoic acid. Biochemistry 13:3859–3863 [DOI] [PubMed] [Google Scholar]
- 35. Walsh C. 1982. Suicide substrates: mechanism-based enzyme inactivators. Tetrahedron 38:871–909 [DOI] [PubMed] [Google Scholar]
- 36. Relyea NM, Tate SS, Meister A. 1974. Affinity labeling of the active center of l-aspartate-beta-decarboxylase with beta-chloro-l-alanine. J. Biol. Chem. 249:1519–1524 [PubMed] [Google Scholar]
- 37. Shizuta Y, Hayaishi O. 1970. Regulation of biodegradative threonine deaminase synthesis in Escherichia coli by cyclic adenosine 3',5'-monophosphate. J Biol Chem 245:5416–5423 [PubMed] [Google Scholar]
- 38. Feldberg RS, Datta P. 1971. l-Threonine deaminase of Rhodospirillum rubrum. Purification and characterization. Eur. J. Biochem. 21:438–446 [DOI] [PubMed] [Google Scholar]
- 39. Roise D, Soda K, Yagi T, Walsh CT. 1984. Inactivation of the Pseudomonas striata broad specificity amino acid racemase by D and L isomers of beta-substituted alanines: kinetics, stoichiometry, active site peptide, and mechanistic studies. Biochemistry 23:5195–5201 [DOI] [PubMed] [Google Scholar]
- 40. Okada K, Hirotsu K, Sato M, Hayashi H, Kagamiyama H. 1997. Three-dimensional structure of Escherichia coli branched-chain amino acid aminotransferase at 2.5 A resolution. J. Biochem. 121:637–641 [DOI] [PubMed] [Google Scholar]
- 41. Okada K, Hirotsu K, Hayashi H, Kagamiyama H. 2001. Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: induced fit and substrate recognition of the enzyme. Biochemistry 40:7453–7463 [DOI] [PubMed] [Google Scholar]
- 42. Parsons L, Bonander N, Eisenstein E, Gilson M, Kairys V, Orban J. 2003. Solution structure and functional ligand screening of HI0719, a highly conserved protein from bacteria to humans in the YjgF/YER057c/UK114 family. Biochemistry 42:80–89 [DOI] [PubMed] [Google Scholar]
- 43. Newton WA, Snell EE. 1964. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl. Acad. Sci. U. S. A. 51:382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cook PF, Wedding RT. 1976. A reaction mechanism from steady state kinetic studies for O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2. J. Biol. Chem. 251:2023–2029 [PubMed] [Google Scholar]
- 45. Schnackerz KD, Ehrlich JH, Giesemann W, Reed TA. 1979. Mechanism of action of d-serine dehydratase. Identification of a transient intermediate. Biochemistry 18:3557–3563 [DOI] [PubMed] [Google Scholar]
- 46. Awano N, Wada M, Mori H, Nakamori S, Takagi H. 2005. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl. Environ. Microbiol. 71:4149–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bisht S, Rajaram V, Bharath SR, Kalyani JN, Khan F, Rao AN, Savithri HS, Murthy MR. 2012. Crystal structure of Escherichia coli diaminopropionate ammonia-lyase reveals mechanism of enzyme activation and catalysis. J. Biol. Chem. 287:20369–20381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sawers G. 1998. The anaerobic degradation of l-serine and l-threonine in enterobacteria: networks of pathways and regulatory signals. Arch. Microbiol. 171:1–5 [DOI] [PubMed] [Google Scholar]
- 49. Zelyas NJ, Cai H, Kwong T, Jensen SE. 2008. Alanylclavam biosynthetic genes are clustered together with one group of clavulanic acid biosynthetic genes in Streptomyces clavuligerus. J. Bacteriol. 190:7957–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He Z, Spain JC. 1998. A novel 2-aminomuconate deaminase in the nitrobenzene degradation pathway of Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 180:2502–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park HS, Kim HS. 2000. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takenaka S, Murakami S, Kim YJ, Aoki K. 2000. Complete nucleotide sequence and functional analysis of the genes for 2-aminophenol metabolism from Pseudomonas sp. AP-3. Arch. Microbiol. 174:265–272 [DOI] [PubMed] [Google Scholar]
- 53. Colabroy KL, Begley TP. 2005. Tryptophan catabolism: identification and characterization of a new degradative pathway. J. Bacteriol. 187:7866–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Orii C, Takenaka S, Murakami S, Aoki K. 2004. A novel coupled enzyme assay reveals an enzyme responsible for the deamination of a chemically unstable intermediate in the metabolic pathway of 4-amino-3-hydroxybenzoic acid in Bordetella sp. strain 10d. Eur. J. Biochem. 271:3248–3254 [DOI] [PubMed] [Google Scholar]
- 55. Takenaka S, Sato T, Koshiya J, Murakami S, Aoki K. 2009. Gene cloning and characterization of a deaminase from the 4-amino-3-hydroxybenzoate-assimilating Bordetella sp. strain 10d. FEMS Microbiol. Lett. 298:93–98 [DOI] [PubMed] [Google Scholar]
- 56. Kim KS, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE. 2010. The rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J. Bacteriol. 192:4089–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lambrecht JA, Downs DM. 2012. Anthranilate phosphoribosyl transferase (TrpD) generates phosphoribosylamine for thiamine synthesis from enamines and non-enzymatic chemistry. ACS Chem. Biol. 8:242-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McKinney J, Guerrier-Takada C, Galán J, Altman S. 2002. Tightly regulated gene expression system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:6056–6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97–106 [PubMed] [Google Scholar]
- 60. Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Duggan DE, Wechsler JA. 1973. An assay for transaminase B enzyme activity in Escherichia coli K-12. Anal. Biochem. 51:67–79 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IlvEE38A is modified with 3CA.The variant protein IlvEE38A was purified and analyzed by mass spectroscopy as described in Materials and Methods. Sample A was untreated, and sample B was incubated with 3CA. Download