ABSTRACT
Cellular processes requiring access to the DNA genome are regulated by an overlay of epigenetic modifications, including histone modification and chromatin remodeling. Similar to the cellular host, many nuclear DNA viruses that depend upon the host cell’s transcriptional machinery are also subject to the regulatory impact of chromatin assembly and modification. Infection of cells with alphaherpesviruses (herpes simplex virus [HSV] and varicella-zoster virus [VZV]) results in the deposition of nucleosomes bearing repressive histone H3K9 methylation on the viral genome. This repressive state is modulated by the recruitment of a cellular coactivator complex containing the histone H3K9 demethylase LSD1 to the viral immediate-early (IE) gene promoters. Inhibition of the activity of this enzyme results in increased repressive chromatin assembly and suppression of viral gene expression during lytic infection as well as reactivation from latency in a mouse ganglion explant model. However, available small-molecule LSD1 inhibitors are not originally designed to inhibit LSD1, but rather monoamine oxidases (MAO) in general. Thus, their specificity for and potency to LSD1 is low. In this study, a novel specific LSD1 inhibitor was identified that potently repressed HSV IE gene expression, genome replication, and reactivation from latency. Importantly, the inhibitor also suppressed primary infection of HSV in vivo in a mouse model. Based on common control of a number of DNA viruses by epigenetic modulation, it was also demonstrated that this LSD1 inhibitor blocks initial gene expression of the human cytomegalovirus and adenovirus type 5.
IMPORTANCE Epigenetic mechanisms, including histone modification and chromatin remodeling, play important regulatory roles in all cellular processes requiring access to the genome. These mechanisms are often altered in disease conditions, including various cancers, and thus represent novel targets for drugs. Similarly, many viral pathogens are regulated by an epigenetic overlay that determines the outcome of infection. Therefore, these epigenetic targets also represent novel antiviral targets. Here, a novel inhibitor was identified with high specificity and potency for the histone demethylase LSD1, a critical component of the herpes simplex virus (HSV) gene expression paradigm. This inhibitor was demonstrated to have potent antiviral potential in both cultured cells and animal models. Thus, in addition to clearly demonstrating the critical role of LSD1 in regulation of HSV infection, as well as other DNA viruses, the data extends the therapeutic potential of chromatin modulation inhibitors from the focused field of oncology to the arena of antiviral agents.
IMPORTANCE
Epigenetic mechanisms, including histone modification and chromatin remodeling, play important regulatory roles in all cellular processes requiring access to the genome. These mechanisms are often altered in disease conditions, including various cancers, and thus represent novel targets for drugs. Similarly, many viral pathogens are regulated by an epigenetic overlay that determines the outcome of infection. Therefore, these epigenetic targets also represent novel antiviral targets. Here, a novel inhibitor was identified with high specificity and potency for the histone demethylase LSD1, a critical component of the herpes simplex virus (HSV) gene expression paradigm. This inhibitor was demonstrated to have potent antiviral potential in both cultured cells and animal models. Thus, in addition to clearly demonstrating the critical role of LSD1 in regulation of HSV infection, as well as other DNA viruses, the data extends the therapeutic potential of chromatin modulation inhibitors from the focused field of oncology to the arena of antiviral agents.
Introduction
Similar to their cellular host, many DNA viruses that replicate in the nucleus are also subject to chromatin-mediated regulation of gene transcription and DNA replication (1, 2). In addition to the direct DNA binding factors that specify activation or repression of transcription, the complex overlay of nucleosome assembly, modification, and remodeling plays a critical role in determining the progression of the lytic replication cycle. For pathogens such as herpesviruses, chromatin modulation is also a regulatory component of the viral latency and reactivation cycles (3–9).
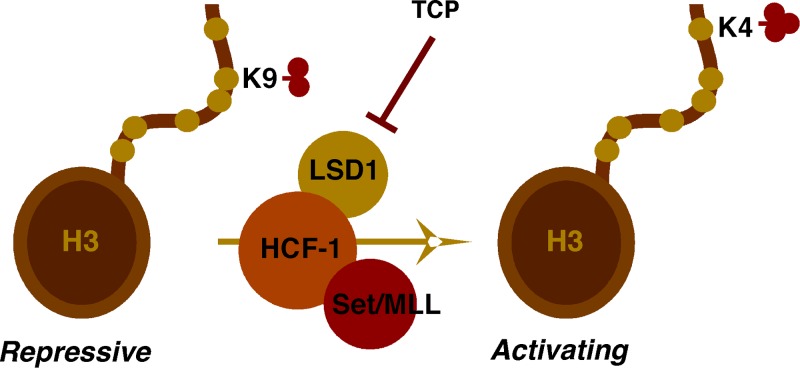
The additional layer of regulatory complexity mediated by assembled nucleosomes represents a dynamic interplay between the host cell chromatin modulation machinery and the pathogen. For the alphaherpesviruses, herpes simplex virus (HSV) and varicella-zoster virus (VZV), infection of host cells results in rapid deposition of nucleosomes (10) bearing repressive histone marks (11, 12). Expression of the viral lytic immediate-early (IE) genes and progression of lytic infection depend upon modulating these modifications to counter the accumulation of repressive marks. To do this, both viruses utilize transcriptional activators packaged in the viral particle to recruit the HCF-1 cellular coactivator complex containing both histone H3K9 demethylases (LSD1 [lysine-specific demethylase 1]) and histone H3K4 methyltransferases (Set1 or MLL family members) to the viral IE gene promoters (11, 13, 14). Recruitment of this coactivator complex thus contains the required enzymatic activities to limit or reverse the accumulation of the repressive H3K9 methylation while promoting the accumulation of the activating H3K4 methylation.
With respect to the significance of the chromatin modulation components of the HCF-1 complex, it has been clearly demonstrated that small interfering RNA (siRNA)-mediated depletion of the H3K9 demethylase LSD1 or inhibition of its activity with monoamine oxidase inhibitors (MAOIs) results in the accumulation of repressive H3K9 methylation across the viral IE gene promoters and suppression of viral IE gene expression (Fig. 1). Significantly, these inhibitors also block the reactivation of HSV from latency in sensory neurons, indicating that LSD1-mediated reversal of repressive H3K9 methylation is a crucial component of the reactivation mechanism (11). These studies demonstrate that histone modification activities such as LSD1 may represent targets for the development of novel antiviral therapeutics to control herpesvirus infection and related diseases.
FIG 1 .
Inhibition of LSD1 by monoamine oxidase inhibitors (MAOIs). Upon infection, the HSV or VZV IE gene activator recruits the HCF-1 coactivator complex to viral IE gene promoters to limit the accumulation of repressive histone H3K9 methylation and promote the installation of activating marks such as histone H3K4 methylation. The HCF-1 coactivator complex consists of a set of histone modification components, including the histone H3K9 demethylase LSD1 and the H3K4 methyltransferase Set1 or MLL. MAOIs such as tranylcypromine (TCP) block LSD1 demethylation activity, resulting in stable or increased levels of repressive marks on viral IE gene promoters and suppression of IE gene transcription.
Importantly, MAOIs such as tranylcypromine (TCP) and pargyline are not specific to LSD1 (15–17) but rather were originally designed to inhibit MAO-A and/or MAO-B for the treatment of psychiatric illnesses (i.e., severe depression). Thus, two limiting factors for the use of these compounds to inhibit alphaherpesvirus infection and reactivation are (i) the high 50% inhibitory concentration (IC50) for LSD1 relative to the MAO-A/B targets (15, 16) and (ii) the broad-spectrum (i.e., non-LSD1) activity.
Therefore, in this study, a series of novel selective LSD1 inhibitors were screened for efficacy in suppressing HSV IE gene expression in vitro. Of these inhibitors, compound OG-L002 demonstrated significantly enhanced potency both in inhibition of IE gene expression and in suppression of viral reactivation from latency in a mouse ganglion explant model. Additionally, OG-L002 substantially suppressed HSV primary lytic infection of mice as determined by reduced viral loads in sensory ganglia. Interestingly, OG-L002 also suppressed the expression of the initial class of human cytomegalovirus (hCMV) and adenovirus genes in vitro, suggesting that chromatin modulation components such as LSD1 may represent common elements of multiple viral pathogens. The results, therefore, indicate that inhibition of the expression of the first class of viral genes via targeting of histone modification and chromatin-remodeling activities could hold significant potential for the control of a class of viruses and related diseases.
RESULTS
Identification of the novel LSD1 inhibitor OG-L002 as a potent inhibitor of HSV IE gene expression in cell culture.
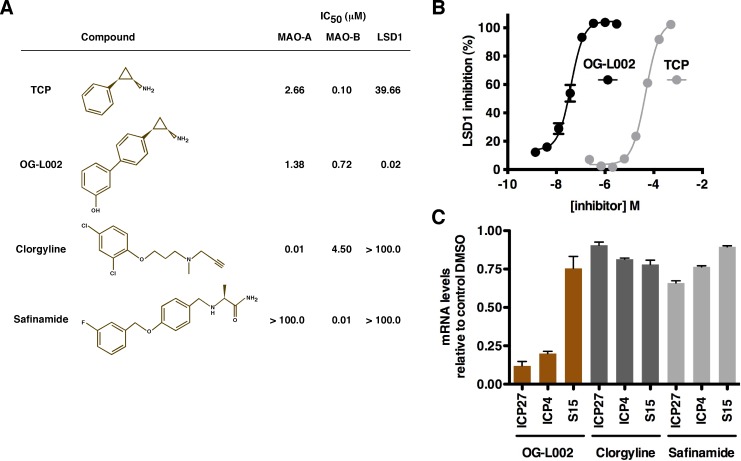
As noted, MAOIs such as tranylcypromine (TCP) were originally designed to target the MAO-A/B. While these compounds also inhibit LSD1, they have a significantly higher IC50 for LSD1 than for their intended targets (Fig. 2). Therefore, to identify compounds with reduced IC50s for LSD1, a series of lead compounds were screened in an in vitro LSD1 demethylation assay using purified LSD1 protein. The IC50s for each compound were determined for LSD1 and MAO-A/B as an indication of specificity (data not shown).
FIG 2 .
Screening novel LSD1 inhibitors identifies lead compounds with improved potency in the repression of HSV-1 IE gene expression. (A) Structures of TCP, OG-L002, clorgyline (CLG), and safinamide (SAF). IC50s were determined in vitro for monoamine oxidase-A/B (MAO-A/B), monoamine oxidase-B (MAO-B), and LSD1. (B) Inhibition of LSD1 in vitro by TCP and OG-L002 is graphically represented. (C) HeLa cells were pretreated for 5 h with DMSO control or the indicated compound (50 µM) followed by infection with HSV-1 (0.1 PFU/cell) for 2 h. The levels of viral immediate-early (ICP4 and ICP27) and cellular control (S15) mRNAs were determined by qRT-PCR and are expressed as ratios to the levels in control DMSO-treated cells. OG-L002 and clorgyline were selected for further comparative analyses.
Compounds with low IC50s for LSD1 relative to MAO-A/B were selected and tested for their efficacy in suppression of HSV IE gene expression. HeLa cells were pretreated for 5 h with the dimethyl sulfoxide (DMSO) control or 50 µM of each compound, followed by infection with HSV for 2 h. The levels of viral immediate-early (ICP4 and ICP27) and cellular control (S15) mRNAs were quantitated by quantitative reverse transcription-PCR (qRT-PCR). As shown in Fig. 2, compound OG-L002, which is highly specific to LSD1 (IC50, ~0.02 µM), potently inhibited the expression of viral IE (ICP27 and ICP4) genes, but not cellular control genes. In contrast, control inhibitors that are more specific to MAO-A (clorgyline) or MAO-B (safinamide) did not show substantial inhibition of viral IE gene expression. The results further support the model that LSD1 is an integral component of the HSV IE gene regulatory paradigm and that the previously described impact of MAOI treatment on viral IE gene expression is not due to inhibition of MAO-A/B.
OG-L002 potently inhibits HSV IE gene expression and the production of progeny virus in both HeLa and HFF cells.
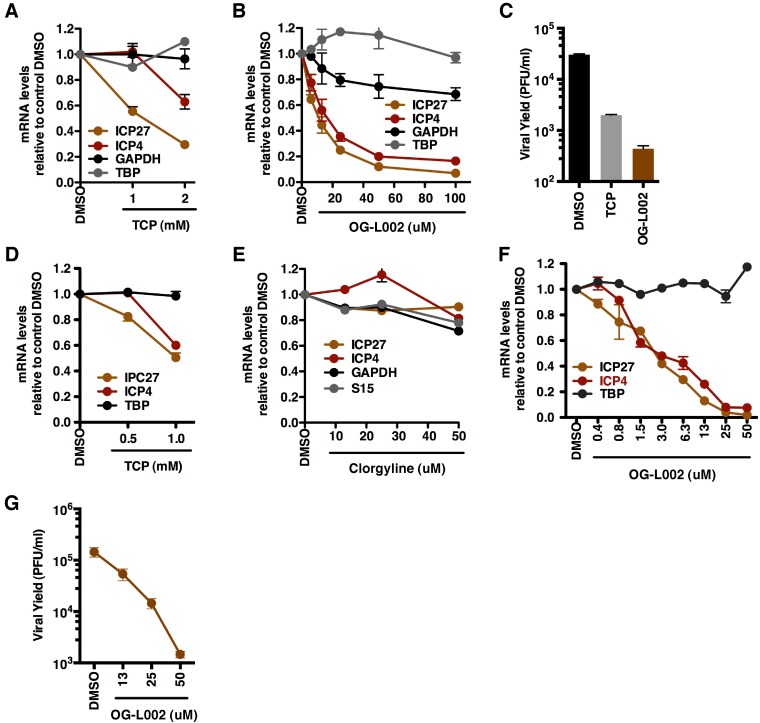
Given the impact of the novel LSD1 inhibitor OG-L002 on HSV IE gene expression, the antiviral potential was further characterized in HeLa (Fig. 3A to C) or human foreskin fibroblast (HFF) (Fig. 3D to G) cells. Following pretreatment with the indicated concentrations of control MAOI TCP, OG-L002, MAO-A inhibitor CLG, or DMSO for 4 h, the cells were infected with HSV for 2 h (mRNA levels [Fig. 3A and B and D to F]) or 24 h (viral yields [Fig. 3C and G]). As shown, OG-L002 inhibited viral IE gene expression in both cells with a significantly reduced IC50 (IC50, ~10 µM in HeLa cells; IC50, ~3 µM in HFF cells) relative to the control MAOI TCP (IC50, ~1 mM). In addition, OG-L002 potently reduced viral yields (~100-fold) in comparison to control DMSO treatment. As shown in Fig. S1 in the supplemental material, no significant toxicity was evident in HeLa or HFF cells from treatment with 50 µM OG-L002.
FIG 3 .
OG-L002 potently inhibits HSV-1 IE gene expression and viral yield in both HeLa and HFF cells. HeLa (A to C) or HFF (D to G) cells were treated with the indicated concentrations (millimolar or micromolar) of TCP, OG-L002, clorgyline, or DMSO control for 4 h, followed by infection with HSV-1 (0.1 PFU/cell) (A and B and D to F). mRNA levels of viral and cellular genes were measured by qRT-PCR at 2 h postinfection and expressed as ratios to the levels of the DMSO control (C and G). Viral yields were determined at 24 h postinfection. In panel C, 1 mM TCP and 50 µM OG-L002 were used.
Specific inhibition of LSD1 by OG-L002 but not treatment with an MAO inhibitor results in accumulation of repressive chromatin on viral IE gene promoters.
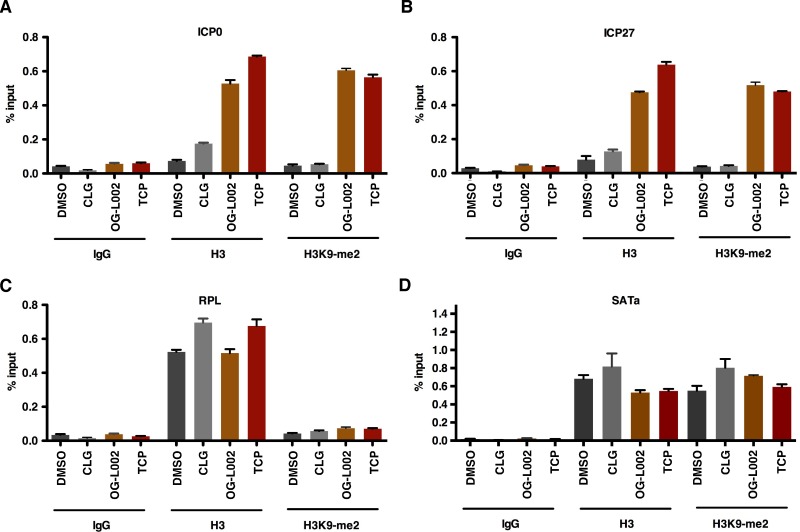
As previously shown, treatment of cells with the MAOI TCP resulted in accumulation of chromatin bearing repressive H3K9 methylation on the HSV IE gene promoters and suppression of viral IE gene expression upon infection (11). To confirm that the novel LSD1 inhibitor OG-L002 also results in accumulation of repressive chromatin marks, HeLa cells were pretreated with DMSO, OG-L002, MAO-A inhibitor clorgyline, or TCP for 4 h, followed by infection with HSV. The levels of dimethyl-H3K9 (H3K9-me2) and total histone H3 associated with viral IE promoters (ICP0 and ICP27) and cellular controls (RPL and SATa) were determined by chromatin immunoprecipitation (ChIP) assays. As shown in Fig. 4, the transcriptionally repressed cellular SATa region exhibits a high level of repressive H3K9-me2 compared to the transcriptionally active cellular RPL gene. In contrast, the total histone H3 levels associated with these regions are comparable, demonstrating the specificity of the H3K9-me2 ChIP assays (Fig. 4C and D). Notably, treatment of cells with TCP and OG-L002, but not clorgyline (CLG), substantially increased the levels of total histone H3 and H3K9-me2 (20- to 30-fold) associated with viral IE promoters. The data are consistent with increased accumulation of repressive chromatin at the IE gene promoters that results in decreased viral IE gene expression.
FIG 4 .
Specific inhibition of LSD1 by OG-L002 but not treatment with an MAO inhibitor results in accumulation of repressive chromatin on viral IE gene promoters. HeLa cells were pretreated with DMSO, OG-L002 (50 µM), clorgyline (50 µM), or TCP (2 mM) for 4 h, followed by infection with HSV-1 (0.1 PFU/cell) for 3 h. ChIP assays were performed using control IgG, H3K9-me2, or histone H3 antibodies. The levels of H3K9-me2 and histone H3 associated with viral IE promoters (ICP0 and ICP27) and cellular controls (RPL and SATa) are expressed as the percentage of input. RPL is a transcriptionally active cellular gene, and SATa is a transcriptionally repressed cellular region. The data shown are representative of two independent experiments.
OG-L002 represses HSV primary infection in vivo.
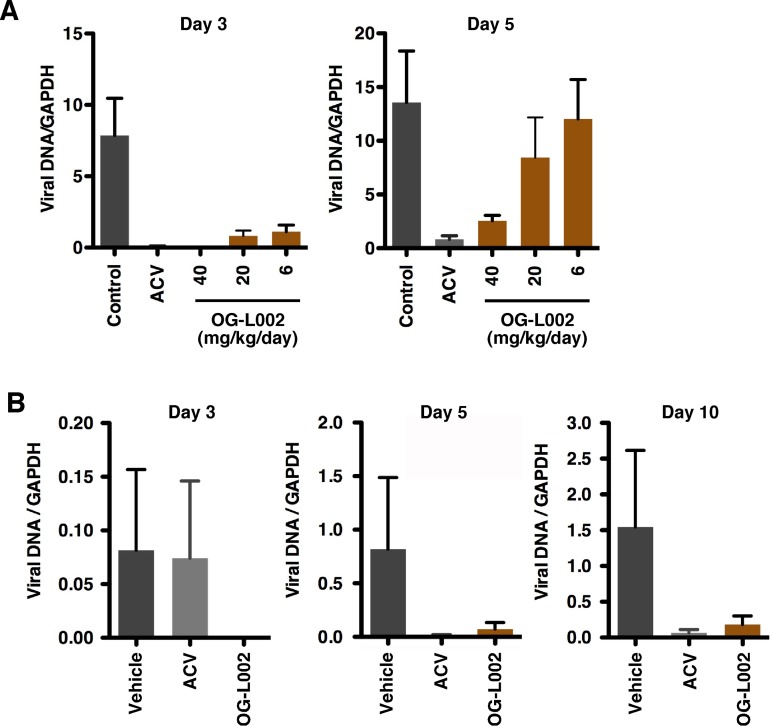
The data derived from cell culture system assays support the role of LSD1 in HSV lytic infection in vitro and suggest that inhibition of LSD1 activity would suppress a primary infection in vivo in a mouse model system. To assess this, BALB/c mice were pretreated with vehicle or OG-L002 (6 to 40 mg/kg of body weight/day) for 7 days followed by infection with a lethal dose of HSV (90% lethal dose [LD90]). As an additional control, infected mice were treated with the viral DNA replication inhibitor acyclovir (ACV) (100 mg/kg/day). At various days postinfection, trigeminal ganglia were harvested from each group of mice, and viral DNA loads were determined as a measure of the progression of the primary infection. As shown in Fig. 5A, OG-L002 reduced the levels of detectable viral genomes in the ganglia in a dose-dependent manner at both 3 and 5 days postinfection.
FIG 5 .
Treatment of mice with OG-L002 reduces HSV primary infection in vivo. (A and B) BALB/c mice were treated with vehicle, acyclovir (ACV) (100 mg/kg/day), or OG-L002 (indicated amounts) and infected with HSV-2 intranasally. At the indicated days postinfection, viral loads in the trigeminal ganglia were determined by qPCR. (A) Viral loads in trigeminal ganglia at days 3 and 5 postinfection at an LD90 (1.1 × 105 PFU/mouse) (n =3). (B) Viral loads in trigeminal ganglia at days 3, 5, and 10 postinfection at an LD50 (1.1 × 104 PFU/mouse). Mice (n = 5) were treated with OG-L002 at 20 mg/kg/day.
In a second experiment, mice were treated with vehicle, ACV (100 mg/kg/day), or OG-L002 (20 mg/kg/day), followed by HSV infection at a reduced LD50 level. Viral DNA loads in the trigeminal ganglia were determined as indicated above. As shown (Fig. 5B), OG-L002 clearly suppressed infection as determined by reduced viral genome accumulation at days 3, 5, and 10 postinfection. Interestingly, the impact of OG-L002 was more significant than the control antiherpesvirus pharmaceutical ACV at early times postinfection (Fig. 5, day 3) and comparable to ACV at later times (days 5 and 10). While the enhanced suppression by OG-L002 relative to ACV at day 3 was reproducible in these experiments, studies with significantly larger groups of animals would be required to statistically validate this interesting observation. Most importantly, the ability to suppress primary HSV infection even at an LD90 challenge further supports the critical role of LSD1 in determining the progression of lytic infection.
OG-L002 blocks HSV reactivation from latency in a mouse ganglion explant model.
In addition to the regulation of viral lytic replication, histone modification and chromatin remodeling play critical roles in regulating HSV latency and reactivation cycles. A number of studies have clearly correlated repressive marks associated with lytic viral genes during latency and the transition to activating marks upon reactivation (3, 4, 18–24). Notably, it was previously demonstrated that MAOIs (TCP) blocked viral IE gene expression during reactivation in a mouse latently infected ganglion explant model system. The data indicate that LSD1 was a critical component of the viral latency-reactivation cycle as well as lytic infection (11).
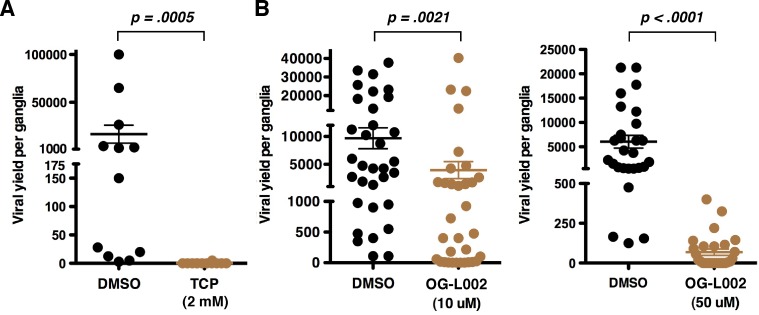
To determine the efficacy of OG-L002 in repression of HSV reactivation, trigeminal ganglia from HSV latently infected mice were bisected, and the halves were explanted in the presence of DMSO and TCP (2 mM) or DMSO and OG-L002 (10 to 50 µM) for 48 h. Yields of reactivated virus (viral yields) were determined by titration. As shown in Fig. 6, OG-L002 repressed viral reactivation at both 10 µM (P = 0.0225) and 50 µM (~40-fold reduction; P < 0.0001) at a significantly reduced concentration from that required for inhibition by TCP. Thus, as previously demonstrated using TCP (11), inhibition of LSD1 with OG-L002 supports the role of this protein in HSV reactivation and further demonstrates the potency of this novel compound.
FIG 6 .
OG-L002 blocks HSV-1 reactivation from latency. (A and B) Halves of bisected trigeminal ganglia from HSV-1 latently infected mice were explanted in the presence of control DMSO or TCP (2 mM) (A) and DMSO or OG-L002 (10 µM or 50 µM) (B) for 48 h. Viral yields were determined by titration on Vero cells. Statistical analyses were done using Wilcoxon matched-pair signed-rank test (DMSO versus TCP, P = 0.0005, n = 12; DMSO versus OG-L002 [10 µM]), P = 0.0021, n = 33; DMSO versus OG-L002 [50 µM], P < 0.0001, n = 27).
OG-L002 results in reduced expression of hCMV IE genes and adenovirus E1A gene.
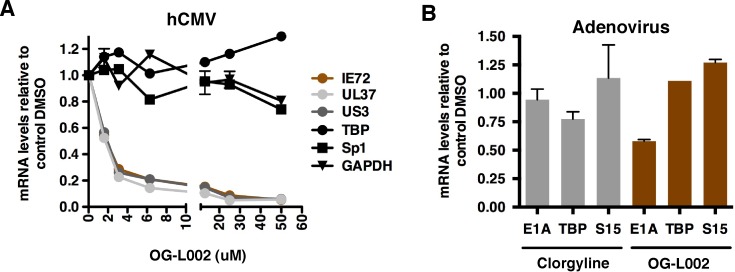
In addition to alphaherpesviruses, gene transcription of other DNA viruses (i.e., human cytomegalovirus [hCMV], adenovirus, and papillomavirus) is also subject to chromatin-mediated regulation (7, 25, 26). For hCMV, initial repression of the genomes is also evident via the assembly of nucleosomes bearing repressive marks (7), and viral IE gene expression is suppressed by the nonspecific LSD1 inhibitor TCP (27). These observations raise the possibility that chromatin modulation components such as LSD1 might be a required component for the initiation of lytic infection by multiple distinct viral pathogens. To test this hypothesis, cells were treated with control DMSO or the indicated concentrations of OG-L002, followed by infection with hCMV or adenovirus type 5. The levels of the viral mRNAs and cellular controls were determined by qRT-PCR and expressed as ratios to the levels for the DMSO control. The results clearly demonstrate that the expression of both hCMV (Fig. 7A) IE and adenovirus (Fig. 7B) early genes are substantially repressed by the LSD1 inhibitor. Thus, despite the distinct classes of these viruses, they share the requirement for some common chromatin modulation components, such as LSD1, to activate their early genes.
FIG 7 .
Inhibition of LSD1 by novel inhibitor OG-L002 results in reduced expression of hCMV IE genes and adenovirus E1A gene. (A) MRC-5 cells were treated with control DMSO or the indicated concentrations (micromolar) of OG-L002 for 3 h, followed by infection with hCMV (0.1 PFU/cell) for 5 h. The mRNA levels of the viral IE genes (IE72, UL37, and US3) and control genes (Sp1, GAPDH, and TBP) were determined by qRT-PCR and expressed as ratios to the levels in DMSO-treated cells. (B) HeLa cells were pretreated with control DMSO, OG-L002 (50 µM), or clorgyline (50 µM) for 4 h, followed by infection with adenovirus 5 (5.6 × 103 normalized infectious units [NIU]/cell) for 2 h. The mRNA levels of the viral E1A gene and cellular control genes (S15 and TBP) were determined by qRT-PCR and expressed as ratios to the levels of the DMSO control.
DISCUSSION
Epigenetic modifications, in conjunction with the cell type milieu of DNA binding regulators, control the transcriptional activity in healthy tissues and contribute to the temporal and spatial interpretation of the genome (28–30). Additionally, recent advances have demonstrated that this epigenetic overlay is often dysregulated in various cancers (31, 32), immunoinflammatory diseases (i.e., diabetes [33], lupus [34, 35], asthma [36]) and a variety of neurological disorders (37, 38). Importantly, epigenetic modifications, represented by DNA methylation, histone modifications, ATP-dependent nucleosomal remodeling, and the involvement of noncoding RNAs and organizational complexes (i.e., CTCF), are reversible, and changes in the epigenetic profile often precede disease pathology. Thus, components regulating the epigenetic landscape of the genome represent valuable indicators for disease progression and prognosis (39) and more importantly, new therapeutic targets.
In recent years, significant advancements have been made in epigenetic drug discovery in oncology (40–42) and inflammatory immunology (43, 44). At this time, several inhibitors of histone deacetylation or DNA methylation are in clinical trials or have been approved for clinical use to treat hematological malignancies or myelodysplastic syndrome, respectively (39, 45, 46). More recently, histone methyltransferases and demethylases have emerged as new therapeutic targets, as these enzymes have been found to be overexpressed, altered, or mutated in some cancers or other diseases (47, 48).
LSD1, the first histone demethylase identified, is overexpressed in a number of malignancies, including some leukemia, breast, small-cell lung, colorectal, prostate, neuroblastoma, and bladder cancers (48–50). Furthermore, preclinical studies (51, 52) have demonstrated that the MAOI TCP is useful in the treatment of acute myeloid leukemia, likely via inhibition of the activity of this enzyme. Importantly, in addition to its role in these diseases, LSD1 is also a critical component of the transcriptional regulatory complexes that activate the initial stages of HSV and HIV gene expression (11, 53).
These results highlight the interesting possibility that LSD1 represents a novel target for the treatment of viral diseases. However, as noted, the LSD1 inhibitors presently utilized are members of the MAOI pharmaceuticals and have high Ki values for LSD1 and a broader spectrum of enzyme targets. Thus, for both oncology as well as for the treatment of infectious viral diseases, there remains a critical need to develop potent and specific LSD1 inhibitors.
In this report, a set of novel LSD1 inhibitors that were identified in in vitro inhibition assays were assessed for the ability to repress HSV IE gene transcription and lytic infection in vitro. One potent and specific inhibitor, OG-L002, was further characterized and demonstrated to (i) inhibit HSV IE gene expression in cell culture, (ii) increase repressive chromatin assembly on viral IE gene promoters, (iii) suppress viral lytic infection in vivo, and (iv) suppress HSV reactivation from latency in a mouse ganglion explant model system at significantly reduced compound levels relative to the MAOI TCP.
Importantly, LSD1 is critical at the initial stages of HSV infection, as the protein is required to limit the cell-mediated deposition of histone H3K9 repressive methylation of the invading viral genome. Corollary regulatory mechanisms in other viral pathogens (hCMV and adenovirus) led to the demonstration that OG-L002 was also effective at suppressing viral early gene expression from each of these distinct pathogens. Taken together, these results clearly define LSD1 as an important component of the lytic replication cycle of multiple DNA viruses in addition to the role in reactivation of HSV from latency.
Inhibition of initiation of infection or reactivation has distinct advantages in that it: (i) represents a stage of the viral infection that is distinct from the later viral DNA replication that is targeted by presently utilized inhibitors (i.e., ACV and derivatives); (ii) prevents the expression of viral gene products that may be toxic or oncogenic or result in inflammatory disease; (iii) may act synergistically with replication inhibitors for a more potent inhibition of lytic infection; and (iv) represents a cellular target that may circumvent the issues of viral escape and drug resistance. Most significantly, LSD1 represents a novel antiviral therapeutic target to suppress initiation of infection of a number of nuclear DNA viruses.
MATERIALS AND METHODS
Cells and viruses.
HeLa, MRC-5, and Vero cell lines were obtained from American Type Culture Collection (ATCC) and maintained according to standard procedures. Telomerase reverse transcriptase gene (TERT)-immortalized human foreskin fibroblast (HFF) cells were a gift of T. Shenk (Princeton University). Human simplex virus 1 (HSV-1) strains 17 and F were obtained from N. Fraser (University of Pennsylvania) and B. Roizman (University of Chicago), respectively. HSV-2 strain MS has been described elsewhere (54). Human cytomegalovirus (hCMV) Towne strain RC256 (VR-2356) and adenovirus type 5 (VR-1516) were obtained from ATCC, respectively.
Antibodies and primers.
Antibodies and the sequences of primers used in these studies are listed in Table S1 in the supplemental material.
MTT cytotoxicity assays.
HeLa or HFF cells were treated with the indicated concentrations of saponin (positive control) or compound OG-L002 for 12 h. Cytotoxicity was determined using conditions recommended by the manufacturer (BioAssay Systems) and expressed as ratios to the cytotoxicity of the DMSO control.
qRT-PCR.
cDNA was produced from equal amount of total RNA using RNAqueous-4PCR and RETROscript kits (Ambion) according to the manufacturer’s recommendations. cDNA was quantitated by qPCR using appropriate primers listed in Table S1 in the supplemental material and SYBR green master mix (Roche) on an ABI 7900HT system (Applied Biosystems; SDS 2.3 software). Primer sequences are listed in Table S1.
Inhibitors.
The structures and IC50s of the relevant inhibitors are shown in Fig. 2A. Cells were pretreated with control DMSO, TCP (tranylcypromine) (catalog no. P8511; Sigma), or the indicated concentration(s) of inhibitors for the indicated times, followed by infection with HSV, hCMV, or adenovirus as described in the appropriate figure legends. Viral yields were determined by determining the titers of the homogenate/lysate of total infected cells on Vero cell monolayers.
ChIP assays.
Histone ChIP assays were done essentially as described previously (13, 55) with modifications. Briefly, HeLa cells pretreated with DMSO control, 2 mM TCP, 50 µM OG-L002 or 50 µM clorgyline (CLG) were infected with 0.1 PFU/cell of HSV for 3 h. The infected cells were cross-linked with 1% formaldehyde in phosphate-buffered saline (PBS). Then, chromatin from 5 × 106 cells was prepared, sheared by sonication, clarified, and incubated with 4 µg of the appropriate antibodies (Abs) (see Table S1 in the supplemental material) absorbed to 50 µl of Dynabeads (Invitrogen) at 4C for 4 h. The immunoprecipitated DNA was recovered and quantitated by qRT-PCR using appropriate primers listed in Table S1. The ChIP data shown are representative of at least two independent experiments.
Impact of compounds on HSV primary infection in vivo.
Groups of 4-week-old BALB/c female mice were pretreated with vehicle or OG-L002 (2 to 40 mg/kg of body weight/day; intraperitoneal) for 7 days followed by intranasal infection with HSV-2 (strain MS) at an LD50 or LD90 as described previously (54, 56, 57). Control animals were treated with acyclovir (ACV) (100 mg/kg/day; oral) beginning at 1 day postinfection according to standard protocols. Vehicle, OG-L002, or ACV treatments were continued daily for 8 to 10 days postinfection. Trigeminal ganglia were harvested from animals in each group at the indicated days postinfection. Total genomic DNA was isolated, and viral genomic DNA levels were determined, in triplicate, by qPCR using SYBR green mix (Roche) on an Eppendorf Realplex4 master cycler. The levels of viral DNA in each sample were normalized to the level of the cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene to control for variances in the isolated ganglion tissue.
Mouse ganglion explant-reactivation model.
BALB/c mice were infected with 2.5 × 105 PFU HSV-1 (strain F) per eye following corneal scarification. The trigeminal ganglia of latently infected mice were harvested 30 days after clearance of the primary infection. Rapidly explanted ganglia were divided in half, and each half was cultured in the presence of control DMSO or LSD1 inhibitor TCP or OG-L002 for 48 h. The resulting viral yields were determined by determining the titers of the homogenate of total trigeminal ganglia on Vero cell monolayers. All animal care and handling were done in accordance with the NIH Animal Care and Use Guidelines and as approved by the NIAID Animal Care and Use Committee. Statistical comparisons were made by using the Wilcoxon signed-rank test with a statistical significance of <0.05 (Prism V5.0a).
LSD1 demethylation assay.
Human recombinant LSD1 protein (BPS Bioscience Inc.) was incubated with dimethylated H3K4 peptide (Millipore) as the substrate in the presence of various concentrations of lead compound inhibitors (0 to 75 µM) or control tranylcypromine. The demethylase activity was measured by the release of H2O2 produced during the catalytic process, using the Amplex red peroxide/peroxidase-coupled assay kit. Each reaction was done in triplicate. The maximum LSD1 demethylase activity was obtained in the absence of inhibitor and corrected for background fluorescence. The Ki (IC50) of each inhibitor was calculated as half-maximal activity.
SUPPLEMENTAL MATERIAL
OG-L002 does not exhibit significant toxicity in HeLa and HFF cells. HeLa (A and B) or HFF (C and D) cells were incubated with the indicated concentrations (percentage or micromolar concentration) of positive toxicity control saponin or OG-L002 for 12 h. The cell viability-cytotoxicity was determined by MTT assays and expressed as ratios to that of the DMSO control. Download
Primers and antibodies used in these studies
ACKNOWLEDGMENTS
We thank M. Fyfe for OG-L002 design; T. Pierson, A. M. Turner, R. Alfonso, J. Arbuckle, A. Estiarte, and M. Maes for critical reading of the manuscript; members of the Molecular Genetics Section, Laboratory of Viral Disease for relevant discussions; and D. J. Collins and T. L. Rice (University of Alabama at Birmingham [UAB]) for expert technical assistance.
These studies were supported by the Laboratory of Viral Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health (T.M.K.) and contract HHSN272201000027I (D.Q.). The development of compounds was cofunded by CENIT-2008-1013 grant from the Spanish Ministry of Economy and Competitiveness and Oryzon Genomics SA.
The National Institutes of Health has the following patent application: Methods of preventing or treating viral infection or reactivation from latency in a host using inhibitors of the LSD1 protein. T. M. Kristie and Y. Liang, U.S. patent application 61/083,304; international patent application PCT/US2009/051557.
Footnotes
Citation Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. 2013. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 4(1):e00558-12. doi:10.1128/mBio.00558-12.
REFERENCES
- 1. Lieberman PM. 2006. Chromatin regulation of virus infection. Trends Microbiol. 14:132–140 [DOI] [PubMed] [Google Scholar]
- 2. Lieberman PM. 2008. Chromatin organization and virus gene expression. J. Cell. Physiol. 216:295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta 1799:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6:211–221 [DOI] [PubMed] [Google Scholar]
- 5. Paulus C, Nitzsche A, Nevels M. 2010. Chromatinisation of herpesvirus genomes. Rev. Med. Virol. 20:34–50 [DOI] [PubMed] [Google Scholar]
- 6. Günther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 6:e1000935 http://dx.doi.org/10.1371/journal.ppat.1000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sinclair J. 2010. Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim. Biophys. Acta 1799:286–295 [DOI] [PubMed] [Google Scholar]
- 8. Takacs M, Banati F, Koroknai A, Segesdi J, Salamon D, Wolf H, Niller HH, Minarovits J. 2010. Epigenetic regulation of latent Epstein-Barr virus promoters. Biochim. Biophys. Acta 1799:228–235 [DOI] [PubMed] [Google Scholar]
- 9. Tempera I, Lieberman PM. 2010. Chromatin organization of gammaherpesvirus latent genomes. Biochim. Biophys. Acta 1799:236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lacasse JJ, Schang LM. 2012. Herpes simplex virus 1 DNA is in unstable nucleosomes throughout the lytic infection cycle, and the instability of the nucleosomes is independent of DNA replication. J. Virol. 86:11287–11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 15:1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silva L, Cliffe A, Chang L, Knipe DM. 2008. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 4:e1000071 http://dx.doi.org/10.1371/journal.ppat.1000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Narayanan A, Ruyechan WT, Kristie TM. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. 2006. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 13:563–567 [DOI] [PubMed] [Google Scholar]
- 16. Schmidt DMZ, McCafferty DG. 2007. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 46:4408–4416 [DOI] [PubMed] [Google Scholar]
- 17. Suzuki T, Miyata N. 2011. Lysine demethylases inhibitors. J. Med. Chem. 54:8236–8250 [DOI] [PubMed] [Google Scholar]
- 18. Amelio AL, Giordani NV, Kubat NJ, O’Neil JE, Bloom DC. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 80:2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Creech CC, Neumann DM. 2010. Changes to euchromatin on LAT and ICP4 following reactivation are more prevalent in an efficiently reactivating strain of HSV-1. PLoS One 5:e15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kubat NJ, Amelio AL, Giordani NV, Bloom DC. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 78:12508–12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kubat NJ, Tran RK, McAnany P, Bloom DC. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78:1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83:8173–8181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. 2007. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J. Virol. 81:13248–13253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U. S. A. 102:16055–16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giberson AN, Davidson AR, Parks RJ. 2012. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res. 40:2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. You J. 2010. Papillomavirus interaction with cellular chromatin. Biochim. Biophys. Acta 1799:192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM. 2013. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci. Transl. Med. 5:167ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res. 21:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128:707–719 [DOI] [PubMed] [Google Scholar]
- 30. Ong CT, Corces VG. 2011. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fouse SD, Costello JF. 2009. Epigenetics of neurological cancers. Future Oncol. 5:1615–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Villeneuve LM, Reddy MA, Natarajan R. 2011. Epigenetics: deciphering its role in diabetes and its chronic complications. Clin. Exp. Pharmacol. Physiol. 38:451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Javierre BM, Richardson B. 2011. A new epigenetic challenge: systemic lupus erythematosus. Adv. Exp. Med. Biol. 711:117–136 [DOI] [PubMed] [Google Scholar]
- 35. Pan Y, Sawalha AH. 2009. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl. Res. 153:4–10 [DOI] [PubMed] [Google Scholar]
- 36. Ito K, Charron CE, Adcock IM. 2007. Impact of protein acetylation in inflammatory lung diseases. Pharmacol. Ther. 116:249–265 [DOI] [PubMed] [Google Scholar]
- 37. Feng J, Fan G. 2009. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int. Rev. Neurobiol. 89:67–84 [DOI] [PubMed] [Google Scholar]
- 38. Urdinguio RG, Sanchez-Mut JV, Esteller M. 2009. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 8:1056–1072 [DOI] [PubMed] [Google Scholar]
- 39. Kelly TK, De Carvalho DD, Jones PA. 2010. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28:1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Best JD, Carey N. 2010. Epigenetic opportunities and challenges in cancer. Drug Discov. Today 15:65–70 [DOI] [PubMed] [Google Scholar]
- 41. Chi P, Allis CD, Wang GG. 2010. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10:457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ganesan A, Nolan L, Crabb SJ, Packham G. 2009. Epigenetic therapy: histone acetylation, DNA methylation and anti-cancer drug discovery. Curr. Cancer Drug Targets. 9:963–981 [DOI] [PubMed] [Google Scholar]
- 43. Jayaraman S. 2011. Epigenetics of autoimmune diabetes. Epigenomics 3:639–648 [DOI] [PubMed] [Google Scholar]
- 44. Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468:1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Copeland RA, Olhava EJ, Scott MP. 2010. Targeting epigenetic enzymes for drug discovery. Curr. Opin. Chem. Biol. 14:505–510 [DOI] [PubMed] [Google Scholar]
- 46. Verbrugge I, Johnstone RW, Bots M. 2011. Promises and challenges of anticancer drugs that target the epigenome. Epigenomics 3:547–565 [DOI] [PubMed] [Google Scholar]
- 47. Copeland RA, Solomon ME, Richon VM. 2009. Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 8:724–732 [DOI] [PubMed] [Google Scholar]
- 48. Rotili D, Mai A. 2011. Targeting histone demethylases: a new avenue for the fight against cancer. Genes Cancer 2:663–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R. 2011. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer 128:574–586 [DOI] [PubMed] [Google Scholar]
- 50. Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. 2012. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One 7:e35065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, Miller CJ, Ogilvie DJ, Somervaille TC. 2012. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21:473–487 [DOI] [PubMed] [Google Scholar]
- 52. Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, Casero RA, Jr, Marton L, Woster P, Minden MD, Dugas M, Wang JC, Dick JE, Muller-Tidow C, Petrie K, Zelent A. 2012. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 18:605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sakane N, Kwon HS, Pagans S, Kaehicke K, Mizusawa Y, Kamada M, Lassen KG, Chan J, Greene WC, Schnoelzer M, Ott M. 2011. Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog. 7:e1002184 http://dx.doi.org/10.1371/journal.ppat.1002184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quenelle DC, Lampert B, Collins DJ, Rice TL, Painter GR, Kem ER. 2010. Efficacy of CMX001 against herpes simplex virus infections in mice and correlations with drug distribution studies. J. Infect. Dis. 202:1492–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee TI, Johnstone SE, Young RA. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1:729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Prichard MN, Kern ER, Hartline CB, Lanier ER, Quenelle DC. 2011. CMX001 potentiates the efficacy of acyclovir in herpes simplex virus infections. Antimicrob. Agents Chemother. 55:4728–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Quenelle DC, Collins DJ, Rice TL, Rahman A, Glazer R. 2011. Efficacy of orally administered low dose N-methanocarbathymidine against lethal herpes simplex virus type-2 infections of mice. Antivir. Chem. Chemother. 22:131–137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OG-L002 does not exhibit significant toxicity in HeLa and HFF cells. HeLa (A and B) or HFF (C and D) cells were incubated with the indicated concentrations (percentage or micromolar concentration) of positive toxicity control saponin or OG-L002 for 12 h. The cell viability-cytotoxicity was determined by MTT assays and expressed as ratios to that of the DMSO control. Download
Primers and antibodies used in these studies