Abstract
Optogenetic strategies to control genetically distinct populations of neurons with light have been rapidly evolving and widely adopted by the neuroscience community as one of the most important tool sets to study neural circuit function. Although optogenetics have already reshaped neuroscience by allowing for more precise control of circuit function compared with traditional techniques, current limitations of these approaches should be considered. Here, we discuss several strategies that combine optogenetic and contemporary pharmacological techniques to further increase the specificity of neural circuit manipulation. We also discuss recent advances that allow for the selective modulation of cellular function and gene expression with light. In addition, we outline a novel application of optogenetic circuit analysis for causally addressing the role of pathway-specific neural activity in mediating alterations in postsynaptic transcriptional processing in genetically defined neurons. By determining how optogenetic activation of specific neural circuits causally contributes to alterations in gene expression in a high-throughput fashion, novel biologic targets for future pharmacological intervention may be uncovered. Lastly, extending this experimental pipeline to selectively target pharmacotherapies to genetically defined neuronal populations or circuits will not only provide more selective control of neural circuits, but also may lead to the development of neural circuit specific pharmacological therapeutics.
I. Introduction
The field of optogenetics, termed for its use of light to modulate function within genetically defined populations of neurons, has evolved at an unprecedented pace since the initial introduction and expression of microbial opsin genes in mammalian neurons to permit the precise control of neural activity on a physiologically relevant, millisecond timescale (Boyden et al., 2005). Optogenetic strategies were rapidly adopted to investigate the underlying neural circuit mechanisms of many diverse processes, including somatosensation, sleep, fear, reward, and synaptic plasticity (Adamantidis et al., 2007; Huber et al., 2008; Zhang et al., 2008; Johansen et al., 2010; Tsai et al., 2009; Tye et al., 2011). Optogenetics permits greater specificity and precision for the systematic dissection of neural circuit mechanisms over established neuroscience techniques (i.e., electrical stimulation, lesion/ablation, and classic pharmacological manipulations) while circumventing many of the limitations of conventional methods. Although the enthusiasm for optogenetic tools is well deserved, these methods also have some inherent limitations. We propose that the specificity of optogenetic manipulations is enhanced when used in combination with pharmacological methodologies. Furthermore, we outline an innovative future application, which may produce clinically relevant information about neural circuit dysfunction, by experimentally perturbing neural activity and assaying the resulting transcriptional changes in postsynaptic neurons. When integrated with next-generation RNA sequencing technologies for high-throughput transcriptome analysis, determining how neural circuit activity induces neuroadaptive changes in the transcriptional landscape of genetically defined postsynaptic neurons is within reach. In this way, the neural circuit and cell-type specificity of optogenetic analysis can be leveraged in support of pharmacogenetic research efforts and accelerate the discovery of novel biologic targets, to which future small molecule pharmacotherapies that act in a cell-type and neural circuit specific manner can be applied.
New research approaches to develop therapeutics that more effectively treat neuropsychiatric and neurologic diseases are needed. These debilitating disorders exact a huge cost both in terms of suffering and daily life disruption to those afflicted individuals and as an economic burden to society in the form of lost productivity and high treatment cost for these chronic conditions (Kessler et al., 2005; Pillay and Stein, 2007). Despite the need for novel treatments, many pharmaceutical companies have retreated from psychiatric disease drug development because of the perceived risk of investing in complex diseases with incompletely understood pathologic mechanisms (Karayiorgou et al., 2012). At the same time, the dominant pharmaceuticals prescribed for anxiety-related, mood, and psychotic disorders fail to produce measurable improvements for a large percentage of individuals and carry a high potential risk of adverse effects. In addition, although we have a general mechanistic understanding of the effect of pharmacotherapy on different neurotransmitter systems in the brain, a significant gap remains in our knowledge of the precise neural and circuit-specific alterations that manifest in neuropsychiatric disorders and how pharmacological treatments specifically alter neural circuit function. The aim of this review is to highlight research that exploited the strengths of optogenetic strategies to investigate neural circuit function relevant to neuropsychiatric disease and to outline a novel experimental approach that integrates optogenetics and RNA sequencing technologies to provide new perspectives on the mechanistic underpinnings of neurologic disorders that may translate into novel treatments.
II. Optogenetics Overview
Neuroscience has traditionally relied on lesion/ablation, electrical stimulation, and pharmacological activation and inactivation to decipher relationships between neural function and behavior. Although essential for defining basic neuroanatomical and functional relationships between brain and behavior, these techniques have fundamental limitations that preclude their use to discern cell-type or pathway-specific function in a behavioral response. Optogenetic manipulations circumvent many of the weaknesses of traditional methods by enabling precise cell-type and circuit-specific investigation of neural function. A comprehensive overview of the entire field of optogenetics is beyond the scope of this review; therefore, we direct the reader to recent reviews that thoroughly document the history and development of the field and provide the requisite conceptual foundation to design and conduct research implementing these methods (Zhang et al., 2010; Bernstein and Boyden, 2011; Fenno et al., 2011; Yizhar et al., 2011a; Tye and Deisseroth, 2012). Here, we selectively highlight the principal strengths of optogenetic manipulations to uncover circuit level mechanisms relevant to neuropsychiatric disorders.
A. Strengths of the Optogenetic Approach
The novel research approach that is the central to this review aims to achieve the ultimate goal of identifying candidate genes or proteins to target with novel cell-type and circuit-specific pharmacotherapies for neuropsychiatric disorders. Several critical advances of the optogenetic approach, which we briefly review below, have enabled neural circuit function relevant to neuropsychiatric disorders to be investigated with greater specificity and precision (Fig. 1).
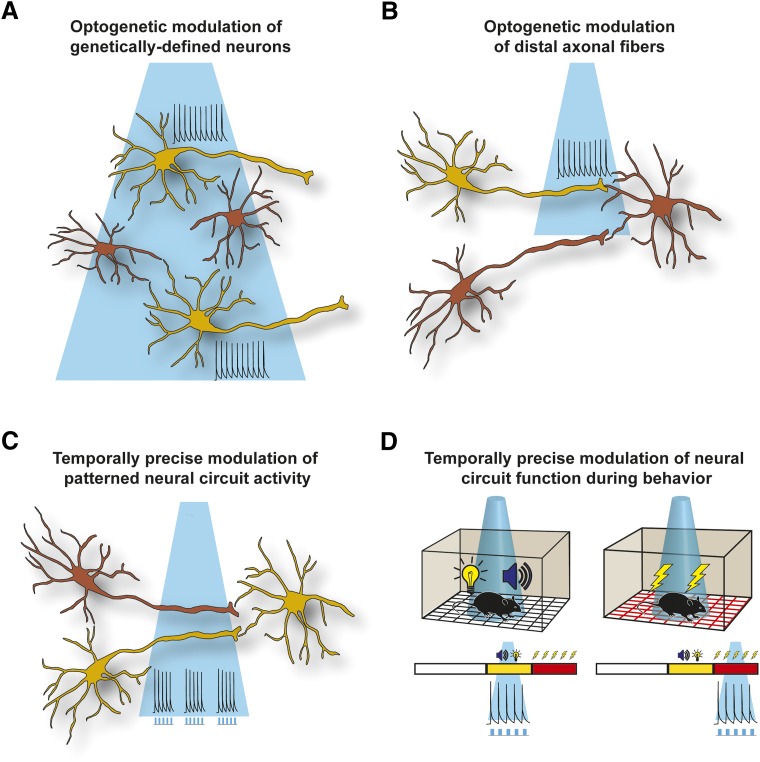
Fig. 1.
Strengths of the optogenetic approach for investigating neural circuit function. (A) Optogenetic tools permit activity in genetically defined cell types within complex circuits to be modulated by light without affecting nearby cell populations of a different genetic lineage. (B) Delivery of light to axon fibers at the projection targets of a genetically defined neuronal population permits the investigation of synaptic function in an input-specific fashion. (C) The spatiotemporal precision of light allows activity within specific neuronal populations to be driven with physiologically relevant millisecond precision to test the causal role of specific patterns of electrical activity for encoding information or driving a behavioral response. (D) The temporal specificity of optogenetic manipulations also allows for the time-locked activation or inhibition of specific neural pathways in vivo during precise moments of a behavioral sequence or during presentation of specific environmental cues, such as tone-light–conditioned stimuli or delivery of aversive shocks. The colored bar depicts the stages of a hypothetical fear-conditioning experiment, where white is the variable inter-trial interval, yellow is the tone-light–conditioned stimulus presentation, and red represents the shock delivery phase of the experiment.
1. Cell-Type Specificity
Genetic targeting strategies to introduce light-gated opsins into neuronal populations to selectively manipulate them represent a principal advance of optogenetic approaches. Neural tissue comprises a heterogeneous mixture of phenotypically diverse cell types that vary with respect to their morphologic, physiologic, synaptic, and molecular properties. Because of this complexity, parsing the specific contribution of any distinct neuronal population embedded within a network of heterogeneous cell types was formerly an insurmountable challenge. Before the introduction of optogenetic techniques, the tools available to record and manipulate neural activity nonspecifically altered activity within a broad volume of tissue, including fibers of passage, and were unable to discriminate between functionally distinct cell types. The optogenetic targeting of fast-spiking inhibitory (FS) interneurons in the mammalian neocortex exemplifies the strength of modulating activity in a functionally important and genetically distinct neuronal subtype. The cytoarchitectonically diverse neocortex contains many neuronal subtypes, including several phenotypically distinct classes of inhibitory interneurons and excitatory pyramidal neurons that each serve a specialized functional role in maintaining overall circuit activity (Freund, 2003; Markram et al., 2004; Isaacson and Scanziani, 2011). Of importance, cortical inhibitory interneuron dysfunction has been heavily implicated as a candidate neural mechanism underlying the manifestation of symptoms in psychiatric disorders, such as schizophrenia, autism, and various intellectual disorders (Rubenstein and Merzenich, 2003; Benes, 2010; Uhlhaas and Singer, 2010; Yizhar et al., 2011b; Marin, 2012). More specifically, dysfunctional circuit mechanisms within the FS interneurons that selectively express the calcium-binding protein, parvalbumin, are hypothesized to underlie a range of symptoms of these neuropsychiatric diseases. Optogenetic manipulations have allowed the functional role of FS interneurons in normal cortical circuit processing to be probed more specifically (Cardin et al., 2009; Sohal et al., 2009). Targeted delivery of light to optically activate the parvalbumin-positive FS neurons enhanced cortical network oscillations in the gamma band, which causally affected sensory processing and signal transmission within cortex (Cardin et al., 2009; Sohal et al., 2009). These studies provided the first evidence that a specific cortical cell type implicated in the pathogenesis of neuropsychiatric disorders could be selectively modulated to test causal relationships between brain function and behavior.
2. Temporal Specificity
The ability to deliver precise activation or inhibition events to defined neural circuit elements at a millisecond timescale, which is consistent with the temporal dynamics of endogenous neural activity, represents the second key strength of optogenetic manipulations. Although neural circuit function is temporally modulated over a range of time intervals, the moment-to-moment processing of sensory cues and adaptive behavioral responses are mediated by rapid electrical and synaptic signals that occur on a millisecond timescale. Therefore, tools to measure and manipulate electrical activity and synaptic neurotransmission with physiologically relevant precision are essential to understand the neural basis of normal and maladaptive behavior. Traditional electrophysiological tools operate with sufficient temporal resolution but nonselectively activate large volumes of tissue, including fibers of passage and heterogeneous cell types, making the unambiguous interpretation of electrical recordings a challenge.
The precise temporal control of neural activity enabled by optogenetic tools has been capitalized on with two types of experiment that each has distinct advantages for establishing causality between activity in specific neural circuits and behavioral consequences. First, optogenetics allow for neurons to be activated at consistent firing rates across a range of frequencies. This is important because, historically, electrical stimulation studies have shown that neuronal subtypes are capable of firing over a broad range of frequencies and that specific neuronal populations encode information by changing their firing rate. For example, dopaminergic neurons of the ventral tegmental area (VTA) have two characteristic firing patterns: a low-frequency tonic activity (∼3–8 Hz) and a higher frequency phasic burst firing pattern (∼10–20 Hz) (Grace and Bunney, 1984a,b). The phasic bursting mode was hypothesized to encode cue saliency and reward prediction errors based on traditional electrophysiological correlations (for review see Schultz, 1997; Wanat et al., 2009), but the ability to mimic this firing pattern in vivo selectively and exclusively in dopaminergic neurons of the VTA required an optogenetic approach. Selective stimulation of VTA dopamine neurons in a burst-like fashion was sufficient to produce reward-seeking behavior, whereas tonic stimulation patterns did not (Tsai et al., 2009). In addition, phasic optogenetic stimulation of dopamine neurons reinforces behavioral responding (Witten et al., 2011). Collectively, these studies highlight that controlling patterned neural activity in genetically defined neurons is a key advantage of implementing these tools to study neural circuits and behavior.
Temporally precise optogenetic manipulations also permit patterned activation of neural circuit elements time-locked to discrete environmental events or at key moments during behavior. Our group has used this approach to study how activation or inhibition of key neural circuits within the mesolimbic system control conditioned rewarding or aversive behavioral phenotypes (Stuber et al., 2011; van Zessen et al., 2012; Stamatakis and Stuber, 2012). To accomplish this, we have applied optogenetic stimulation or inhibition time-locked to salient events, such as presentation of reward predictive cues. Restricting optogenetic manipulations to a brief time window surrounding an event tests whether neural circuit activity at a key time point is sufficient or necessary to produce a particular behavioral outcome. This strategy also allows for testing of whether optogenetic manipulations of the same duration outside a given time point can produce similar or different results. Because many neuropsychiatric disorders result, at least in part, from an underlying circuit dysfunction that leads to overactive emotional responses to salient environmental cues, the temporal precision of time-locked optogenetic activations should enhance our understanding of the underlying neural mechanisms of these diseases (Bishop, 2007; Hartley and Phelps, 2010; Koob and Volkow, 2010; Price and Drevets, 2010).
3. Pathway-Specific Synaptic Function
Optogenetic tools have also enabled the investigation of afferent-specific synaptic neurotransmission and the input-specific modulation of projection target activity during behavior or paired with slice electrophysiology (Petreanu et al., 2009; Stuber et al., 2011; Tye et al., 2011; Pascoli et al., 2012). Pathway-specific synaptic neurotransmission at isolated efferent projection targets is necessary to gain a complete functional understanding of any specific brain region, which may differentially regulate downstream activity in a projection target-specific manner. In brief, virally transduced neurons within a given brain region achieve high opsin membrane insertion and efficient trafficking that results in high expression levels throughout the soma and axon terminals (Gradinaru et al., 2010). As a consequence, light delivery to fiber terminals within projection targets will activate input-specific synaptic transmission. This level of specificity is necessary to determine the downstream physiologic or behavioral effect of activity deriving from a common circuit node, such as the basolateral amygdala (BLA), which can vary in a target-specific manner and likely depends on differential effects of specific inputs that contribute to postsynaptic responses. Two recent studies that used projection-specific targeting to study amygdala function highlight how activity in one brain region may mediate distinct behavioral responses depending on the strength and pattern of activity that it communicates to specific downstream targets (Stuber et al., 2011; Tye et al., 2011). The amygdala is a critical structure for processing negative emotions, such a fear (Fanselow and Gale, 2003; Pare et al., 2004; Ehrlich et al., 2009; LeDoux, 2012), but also plays an integral role in the encoding and behavioral responses to salient environmental cues of both negative and positive valence (Paton et al., 2006; Ambroggi et al., 2008; Ishikawa et al., 2008; Tye et al., 2008; Shabel and Janak, 2009; Gardner, 2011). Pathway-specific optogenetic modulation of the glutamatergic projection from the BLA to the nucleus accumbens (NAc), including the activation and inactivation of BLA terminals in the NAc, confirmed that this circuitry is both necessary and sufficient for cue-driven motivated behavior (Stuber et al., 2011). In a separate study, a sophisticated project-targeting approach was used to isolate the circuit between the BLA and the central amygdala. Optical modulation of this pathway bidirectionally influenced anxiety behavior in mice, whereas activation of BLA cell bodies had no effect (Tye et al., 2011). Taken together, these studies demonstrate the strength of optogenetic methods to reveal precise neuronal projections that are relevant to neuropsychiatric disease.
III. Enhancing Circuit Specificity by Combining Optogenetic and Pharmacological Approaches
The optogenetic approaches reviewed above have already had a transformative effect on neuroscience research. However, it is often necessary to integrate multiple tools and/or approaches to achieve a more comprehensive understanding of neural circuit function. Optogenetic tools, despite overcoming many limitations of conventional methods, also have weaknesses that are beginning to be addressed with innovative strategies. Below, we discuss two principal strategies used to enhance the utility of optogenetics. First, combining pharmacological manipulations with optogenetic analysis in vivo and with slice physiology has greatly extended the strength of either individual approach. Second, novel approaches are extending the power of optogenetics beyond alterations in membrane physiology and control of spiking activity to selectively probe cell function and biologic processes at the level of signal transduction pathways, protein interactions, and gene regulation.
The pairing of optogenetics with patch-clamp electrophysiological techniques presents many possibilities for integration with pharmacological manipulations to enhance specificity for probing neural circuit mechanisms. At a basic level, optogenetic activation of genetically distinct cell types is already widely used to characterize input -specific synaptic neurotransmission at various postsynaptic cells within a microcircuit without perturbing the activity of other nearby nontarget cell types. Also, measuring the firing response of a specific cell type to various light delivery protocols (i.e., pulse trains of varying frequency, constant stimulation intervals) will yield important information for later targeting the same cell population during in vivo behavioral manipulations and can reveal important information about the electrical properties of that given cell type. Both transgenic mouse lines and cell type–specific promoters used in recombinant viruses allow for targeted opsin expression to a neuronal population defined by its neurotransmitter content (Yizhar et al., 2011a). However, functionally defining neuronal populations according to a single dimension, such as neurotransmitter content, or distinct genetic marker may be problematic. The neurotransmitters glutamate and GABA are ubiquitous throughout the nervous system, and often, their specific postsynaptic effects are modulated by the coincident release of a different chemical messenger (i.e., neurotransmitters, neuropeptides, or hormones) from converging inputs. The development of significantly red-shifted light-gated opsins, such as C1V1, permits the selective control of two input pathways to a postsynaptic target that allow for testing of the combinatorial effect of distinct inputs to a common projection source (Yizhar et al., 2011a). This technology should help to determine how pharmacological treatments, such as various neurotransmitter receptor agonists or antagonists, can differentially alter synaptic transmission in an input-specific fashion. In addition to the convergence of multiple chemical signals from distinct afferent pathways, single terminal fibers may corelease transmitters or peptides. Neurotransmitter corelease has been examined with optogenetics paired with slice physiology. Two recent studies highlight this phenomenon by showing that activation of one set of genetically defined presynaptic inputs resulted in two distinct neurotransmitter signaling events. Research has indicated an important role for DA and glutamate coincident signaling in a variety of motivated behaviors, including responding to motivationally significant cues (Di Ciano et al., 2001; Phillips et al., 2003; Stuber et al., 2008). The unsettled debate over possible glutamate release from DA neurons was resolved using optogenetics to selectively stimulate ChR2-positive DAergic terminals in the NAc shell, which resulted in excitatory postsynaptic currents that could be abolished by α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor antagonists (Stuber et al., 2010; Tecuapetla et al., 2010), confirming that at least some midbrain DA neurons are capable of coreleasing glutamate in the NAc. Pharmacological blockade of DA receptors did not significantly alter these glutamate-mediated currents, indicating that the detected glutamate release events were not attributable to polysynaptic signaling requiring DA receptor activation within local NAc microcircuitry.
Integrating pharmacological and optogenetic manipulations also allows for the dissection of complex local microcircuitry, such as what is seen in the neocortex. The use of subcellular ChR2-assisted circuit mapping (sCRACM) illustrates the integrative application of pharmacology and optogenetics to better understand cortical microcircuits (Petreanu et al., 2009; Mao et al., 2011). Cortical microcircuitry is exceedingly complex, such that the functional effect of a synaptic input to a single principal pyramidal neuron varies significantly depending on its precise spatial location on the dendrites or soma. In addition, because of the high degree of recurrent excitatory collaterals within cortical layers, it is often difficult to isolate monosynaptic responses arising from the activity of a specific input. sCRACM uses the pharmacological agents tetrodotoxin to block Na+ channels and 4-aminopyridine to block K+ channels, in conjunction with ChR2 terminal stimulation to isolate single monosynaptic glutamatergic inputs. This technique was applied to the rodent somatosensory cortex to map and define the functional effect of isolated inputs to pyramidal neurons in various layers of the cortex (Petreanu et al., 2009). Of interest, synapses were observed to spatially segregate according to the location of the structure providing the input, a fact that could have significant consequences for the induction of plasticity within cortical microcircuits. Thus, sCRACM combines the strengths of both pharmacological and optogenetic manipulations to more precisely map microcircuits, which may help to reveal dysfunctional connectivity patterns that have relevance for many neuropsychiatric and neurologic disorders.
The application of optogenetic manipulations to in vivo behavioral experiments has been a powerful approach for testing the necessity and sufficiency of activity in defined circuits for the production of a range of behavioral responses (Adamantidis et al., 2011; Letzkus et al., 2011; Stuber et al., 2011; Tye et al., 2011; Knobloch et al., 2012; Stamatakis and Stuber, 2012; van Zessen et al., 2012). For example, research has implicated both glutamatergic and dopaminergic signaling in the NAc as critical mediators of motivated behaviors (Di Ciano et al., 2001; Ambroggi et al., 2008; Stuber et al., 2008). In vivo optogenetic circuit analysis of the BLA to NAc glutamatergic pathway revealed that selective optogenetic activation of BLA fibers in the NAc was rewarding to mice and could reinforce nose-poke behaviors to achieve further stimulation (Stuber et al., 2011). To test whether coincident dopamine was mediating the effect of BLA glutamatergic inputs on motivated behavioral responding, local pharmacological administration of D1 and D2 receptor antagonists into the NAc prior to optical stimulation was performed and demonstrated that D1 receptor activation was necessary for the BLA glutamate to drive motivated behavior. This result underscores the benefit of applying an integrative approach to the analysis of neural circuit mechanisms and to enhance the strengths of any single approach to study neural function.
There are also important caveats and limitations that arise with in vivo optogenetic behavioral manipulations that can be addressed by pharmacological means. One caveat to the optogenetic projection targeting techniques arises because afferent fibers exiting a structure are frequently bundled together, and stimulating terminals in one region may also stimulate fibers of passage that may also be expressing opsins. For example, DA afferents from the VTA projecting to the PFC travel through the NAc (Beckstead et al., 1979; Herbert et al., 1997), and stimulation of DA terminals in the NAc will likely also stimulate PFC-projecting fibers. Another limitation to this method is the possibility of back-propagating action potentials. Optical stimulation of terminals in one region could trigger antidromic action potentials that activate the cell bodies, which may then activate axon collaterals projecting to other regions. This could in theory lead to behavioral effects unrelated to the specific input under investigation. This limitation can be partially dealt with by microinjecting tetrodotoxin or lidocaine in the cell bodies of the virally transduced population of neurons to eliminate neural activity that antidromically reaches the soma induced by terminal stimulation (Stuber et al., 2011). Currently, an intensive research effort is directed toward engineering or identifying new virus variants that permit more precise circuit mapping and control of activity in circuits and avoid these nonspecific effects. These alternative strategies typically exploit the ability of many viruses to spread trans-synaptically in either the anterograde or the retrograde direction, which will enable a single circuit between two structures to be selectively modulated without activating fibers that exit the same structure but target another brain area. Similar techniques involving a retrogradely traveling canine-adenovirus engineered to express Cre-recombinase have been used to restore dopamine function only in specific VTA dopaminergic pathways of dopamine-deficient mice and leave other dopamine projections unaffected (Hnasko et al., 2006). Newer approaches involving modified herpes simplex virus or pseudorabies are actively in development that enables the expression of opsins or reporters only in precise neural circuits (Neve et al., 2005; Hnasko et al., 2006; Robinson et al., 2007; Callaway, 2008; Osakada et al., 2011). Sophisticated advances such as these will inevitably lead to even more precise targeting of neural circuits for controlling neuronal activity or manipulation of gene and protein function.
IV. Optical Control of Intracellular Signaling Processes
Currently, optogenetic manipulations provide the best method for the spatiotemporal control of neuronal activity or presynaptic terminal neurotransmitter release in circuits, but the functional consequences of optical activation in downstream postsynaptic circuit elements remain largely undefined. Most opsins modulate neural activity by directly controlling ion conductance across the membrane, which leads to depolarization or hyperpolarization and a respective increase or silencing of spiking activity (Zhang et al., 2006; Fenno et al., 2011; Yizhar et al., 2011a). However, the pattern of action potential firing within discrete time intervals is only one aspect of the dynamic process of neural circuit function. Endogenous neural circuit activity also requires neurotransmitter release, postsynaptic receptor activation, the initiation of second messenger signaling cascades, protein-protein interactions, and ultimately, changes in patterns of gene expression. These intracellular processes that are associated with the endogenous circuit-level activity have not been amenable to study with optogenetic tools. Ongoing research aims to achieve an equivalent level of cell type and circuit specificity as optogenetic approaches for the selective manipulation of intracellular protein interactions and gene transcription processes (Kennedy et al., 2010; Wang et al., 2012). These technical advances may eventually aid in the identification of dysregulated postsynaptic intracellular mechanisms that contribute to maladaptive behaviors and the manifestation of disease states. As a whole, the combined application of these tools should broaden our understanding of the mechanistic underpinnings of neuropsychiatric disorders at multiple levels.
A novel class of synthetic genetically encoded optical tools, the optoXRs, which are chimeric proteins coupled to different intracellular G-protein–initiated signaling cascades, allow for G-protein receptor–initiated biochemical pathway activation in vivo (Airan et al., 2009; Zhang et al., 2010) (Fig. 2). In a study by Airan et al. (2009), two optoXR receptors with distinct postsynaptic signaling consequences were designed: one coupled to a β2-adrenergic initiated Gs cascade that lead to increased cyclic AMP levels and a second that coupled to an α1-adrenergic initiated Gq cascade that resulted in increased inositol trisphosphate and diacylglycerol levels. The behavioral consequences of precisely timed photoactivation of these signaling pathways within the NAc were tested in a place conditioning paradigm by pairing optical activation with one chamber on one day and quantified preference as mice freely explored the entire chamber the following day. Of interest, mice formed a significant preference for the stimulated chamber when the optoXR coupled to α1 was stimulated. Although early in development, these optical tools for modulating biochemical signaling pathways with cell- and pathway-specific precision are promising methods for interrogating neural circuit function beyond the level of spiking activity and to identify dysregulated intracellular processes that contribute to disease. Thus, we expect these and similar studies to provide critical information to guide the identification of molecular targets for novel small molecule pharmacotherapies.
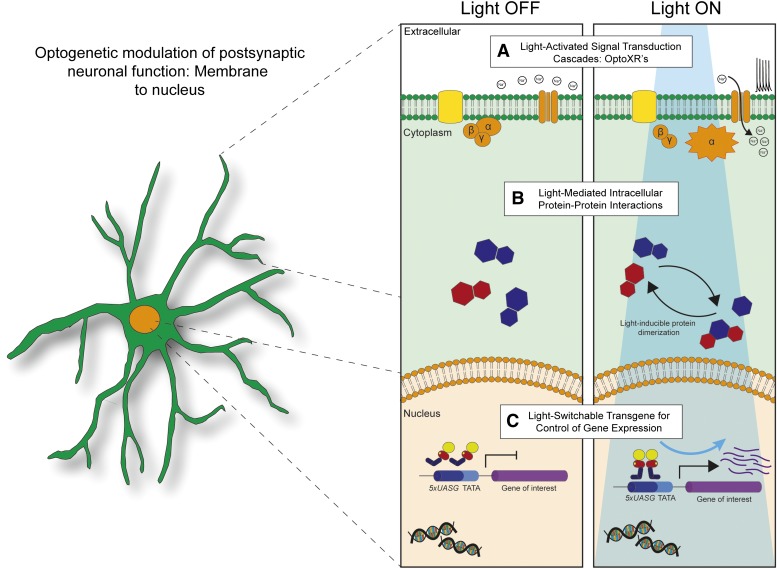
Fig. 2.
Optogenetic modulation of postsynaptic neuronal function at multiple levels of cellular activity. (A) Synthetic light-gated receptors, such as the OptoXR’s, are coupled to G-protein signaling cascades that alter postsynaptic function by activating intracellular second-messenger cascades that may lead to alterations in neuronal excitability. (B) Light-inducible protein dimerizers will permit the selective control of intracellular signaling cascades and other diverse cellular processes with subsecond temporal resolution and subcellular spatial resolution through light activation of protein-protein interactions. (C) Light-switchable transgenes will permit the selective control of target gene expression within genetically defined neuronal populations to test the causal role of gene activation on various behaviors. Shown in the figure is a schematic of the LightOn gene expression system, which involves light-induced homodimerization of GAVP, which then interact with upstream UASG promoter elements to initiate the transcription of a gene of interest.
Both neuropsychiatric disease states and optogenetic circuit manipulations to induce behavioral phenotypes undoubtedly involve altered intracellular pathways and require protein interactions, but the technology to selectively manipulate these protein-protein interactions and test their functional consequences has only just become available (Kennedy et al., 2010; Wang et al., 2012). Recent strategies have been devised that use genetically encoded dimerizers from plant photoreceptors and exploit the spatiotemporal precision of light to control protein-protein interactions. Initial demonstrations of these tools, such as those using the light oxygen voltage domain of Arabidopis thaliana (Wu et al., 2009) demonstrated proof of concept but suffered from slow light-activated kinetics or irreversibility of dimerization after light induction (Kennedy et al., 2010). Newer systems have been designed on the basis of dimerization modules of the basic helix-loop-helix protein Arabidopsis CIB1 and cryptochrome 2. This system can induce protein-protein dimerization after blue light stimulation with subsecond time and subcellular spatial resolution (Fig. 2). These properties were exploited to induce various biologic processes, including protein translocation and transcription in mammalian and yeast cells. The continued improvement of this technology will eventually permit intracellular signaling cascades within genetically defined cell types and pathways to be selectively modulated to test the contribution of alterations in specific protein function for the development of disease states.
Ultimately, for neuronal circuit activity to translate into long-term behavioral consequences, gene function must be altered in specific neuronal populations and pathways. Intracellular signaling cascades that are initiated by ligand-receptor interactions at the membrane and involve protein interactions, which can now be selectively manipulated with light, often result in changes in neuronal gene transcription. Activity-dependent changes in neuronal structure and function, which are the proposed neural mechanisms of learning and memory, are dependent on changes in gene regulation. Just as light-gated opsins have allowed for precise manipulation of electrical activity within defined circuits to test function, a tool for precise spatiotemporal control of gene expression within specific pathways would permit the systematic analysis of the effect of selective changes in gene function on behavior. Very recently, light-switchable transgene systems have been designed and implemented that exploit the spatiotemporal specificity of light to selectively trigger changes in gene function within specific cell types. One system, named LightOn, capitalized on the light dimerization property of Vivid, a light oxygen voltage domain containing protein, to form a synthetic light-switchable gene-promoter system. This transactivator was shown to bind promoters after blue light exposure and to rapidly initiate transcription of target transgenes in both mammalian cells and mice (Wang et al., 2012). LightOn and other systems that allow for precise spatiotemporal control of genes in a cell type–specific fashion with light are promising new tools for testing causal relationships between specific alterations in cellular processes and the manifestation of pathologic behavioral phenotypes (Fig. 2). However, it is important to highlight that these new optical approaches for controlling protein-protein interactions and gene transcription are still in the early stages of development. As with any new technique, a rigorous analysis of these new methods must be conducted to ensure that introduction of light-activated protein products or genes is not disruptive or toxic to normal cellular function. Nevertheless, we suggest that the potential benefits of selectively isolating and manipulating key components of signaling or gene transcription cascades that may reveal novel drug targets outweighs the initial challenge posed during refinement and optimization of a new technique.
V. The Future: Establishing Causal Relationships Between Neural Circuit Activity and Cell Type–Specific Changes in Gene Transcription
The dominant paradigm for understanding the genetic basis of psychiatric disorders has been to identify candidate gene polymorphisms through genome-wide analysis studies of clinical or at-risk populations that may be associated with the development of these diseases (Marian, 2012). Although there have been notable successes from this approach, the experimental process is laborious, and the candidate genes often explain very little of the variance in susceptibility and are correlative, not causal, in nature. We propose, as a complement to this top-down approach, an alternative strategy that takes a bottom-up approach that begins with optogenetic circuit activity perturbations followed by transcriptional sequencing to identify many candidate genes and potentially protein targets in a high-throughput, massively parallel fashion. Fundamentally, these two approaches underscore the more complex issue of whether the neural circuit alterations underlying neuropsychiatric and neurologic disorders arise from pathologic neural processing associated with aberrant experiences or whether genetic variants and predispositions play a greater causal role in their development. In reality, genetic factors, environmental experiences, and gene-environment interactions all synergistically contribute to the development of these complex disorders. However, the technological advances of optogenetics and other fields have provided a mechanism for selectively disrupting neural circuit activity at key circuit nodes to induce changes in gene expression.
The research strategy that we outline here dovetails well with advances and reduced costs of high-throughput sequencing. The critical gap that can be addressed with an integrative optogenetic-transcriptomics approach is how to translate the important research findings enabled by optogenetics regarding cell type– and pathway-specific neural function in mediating behaviors relevant to psychiatric disorders and expand on this information to produce data with real potential to inform the development of better treatments for these disorders.
An integrative optogenetic-transcriptomics approach may reveal novel candidate genes and, thus, proteins that are dysregulated after experimentally administered changes in neural circuit activity. The transcriptome, which is broadly defined as the entire RNA contents of a cell, is a dynamic feature of specific cell types that mechanistically serves to translate genotype information into an adaptive phenotype (Tang et al., 2011). Changes in cellular function that occur across development, in response to environmental stress, and with the development of disease states are all mediated by transcriptome alterations and changes in gene function. For this reason, investigating the transcriptome of targeted cell types within dysregulated neural circuits is likely to produce important insights into altered cellular function underlying disease. In addition, a logical extension of the pharmacogenomics effort that has been evolving in parallel is the effort in molecular genetics and nanotechnology to devise strategies for the delivery of more highly targeted, cell-specific small molecules that act precisely at the locus of dysfunction (Zhang et al., 2009; Paulo et al., 2011; Tian et al., 2012). It is within these current trends that we envision optogenetics being applied most innovatively for the identification of candidate genes and proteins within specific neuronal populations and circuits that subsequently can be targeted with cell-specific small molecule pharmacotherapies.
Our aim from the following research approach is not to provide a detailed experimental protocol but to present a flexible roadmap for guiding the implementation of similar studies (Fig. 3). However, we hope it will serve as a useful reference and to stimulate ideas for applying optogenetics toward the elucidation of novel cell- and circuit-specific mechanisms and for the identification of candidate genes to target with new selective pharmacotherapies. For clarity, we have subdivided the discussion into the following three sections that specifically address a different stage of the experimental process: 1) neural circuit selection and optogenetic investigation in animal models, 2) application of transcriptome sequence technologies to identify dysregulated gene and potentially protein targets associated with pathologic state, and finally, 3) the development of small molecule pharmacological agents to target candidate gene/protein function with cell and circuit specificity.
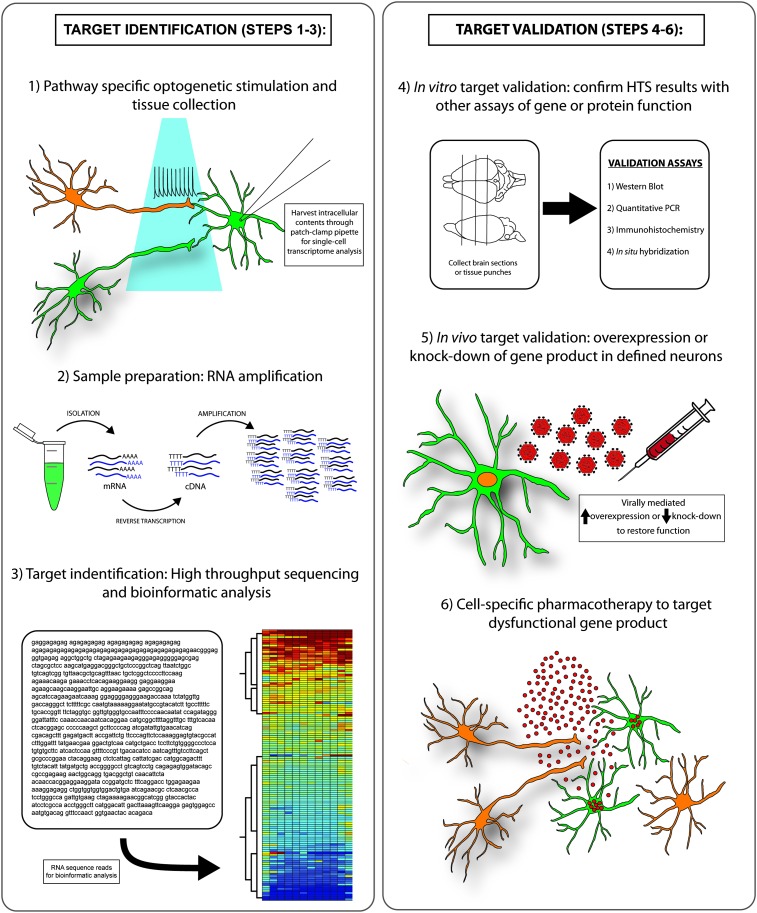
Fig. 3.
Optogenetic-transcriptomics approach to identify neural circuit-specific dysregulated genes or proteins that may be future therapeutic targets for the treatment of neuropsychiatric disorders. Target identification steps are described as follows: 1) optogenetic analysis of function within a specific neural pathway followed by the harvesting of postsynaptic cell contents to be subject to transcriptome analysis; 2) sample preparation involving the isolation of postsynaptic RNA, reverse transcription to cDNA library, and subsequent amplification; and 3) high-throughput RNA-seq analysis to identify and measure all transcripts within a specific cell to be followed by sophisticated bioinformatics analysis to identify which gene and protein levels have been altered by optogenetic circuit activation. Target validation steps as follows: 4) after RNA-seq analysis and target identification, results should be verified through brain tissue collection and protein or gene analysis with Western blot, quantitative polymerase chain reaction, or histologic methods; 5) target gene or protein function should be validated in vivo through viral-mediated overexpression or knockdown of target gene to reverse the behavioral effect of optogenetic circuit manipulation; and 6) development of small-molecule pharmacotherapy to target dysregulated gene or protein function within the affected cell population to restore normal cellular and behavioral function.
A. Circuit Selection and Optogenetic Interrogation
Although we present our approach as bottom-up to emphasize that the direction of the process contrasts that of the more conventional approach of first identifying single genetic polymorphisms, some top-down information about neural circuits that have been implicated in neuropsychiatric disorders is necessary to guide the selection of pathway to study. One conceptual hypothesis about the origin of many neuropsychiatric disorders that may guide circuit selection is that these diseases arise from altered function in limbic brain structures and circuits that process reward and aversion or in the cortical regions that provide top-down control of these processes (Kim et al., 2011). An extensive body of research has amassed within the past decade that has used functional magnetic resonance imaging on healthy and clinical populations and serves as an invaluable resource to guide research design and the selection of a clinically relevant neural circuit to investigate (Linden and Thome, 2011; Linden, 2012). Significant methodological and analytic advancements have enabled better assessment of global resting-state activity and measures of regional and functional connectivity and have produced vital circuit-level information in healthy and various clinical populations (Raichle and Snyder, 2007; Raichle, 2010; Pan et al., 2011). For simplicity, we limit the discussion to one prominent neural circuit connecting the amygdala and the prefrontal cortex. This pathway has been heavily implicated in a range of psychiatric disorders that span the spectrum, such as mood and anxiety related, addiction, autism, schizophrenia, and other cognitive disturbances (Bishop, 2007, 2008; Koob and Volkow, 2010; Price and Drevets, 2010; Shin and Liberzon, 2010). In addition, a concerted effort has been made to unify the vast knowledge acquired from animal research into the neural mechanisms of fear conditioning and extinction and the human psychology study of fear (Milad and Quirk, 2012), which has sharpened our understanding of the neural basis of anxiety- and fear-related disorders. In general, functional magnetic resonance imaging studies have identified a disrupted balance in activity between the prefrontal cortex and amygdala that is associated with anxiety. Specifically, this disruption is observed as a hyperactive amygdala and hypoactive prefrontal cortex that manifests behaviorally as an increased threat responsivity and diminished capacity for regulating emotional responses (Bishop, 2007, 2008). These findings fit well with the extensive body of research into the neural circuit mechanisms of associative fear conditioning and extinction processes, which have established the amygdala as the key neural structure for learning associations between cues and aversive outcomes and infralimbic regions of the medial prefrontal cortex for suppressing conditioned fear responses after extinction training (Quirk and Beer, 2006; Quirk and Mueller, 2008; Sotres-Bayon and Quirk, 2010). The choice of neural circuit to investigate will affect all subsequent stages of the experimental procedure and will likely determine the therapeutic potential of any candidate proteins identified for drug targeting.
The next stage of the approach entails a complete optogenetic analysis of the selected pathway involving slice physiology and in vivo circuit manipulations during various behavioral assays. Here, the goal is to successfully recapitulate some of the behavioral features of the focal psychiatric disorder using as many validated and established behavioral assays as is feasible. Next, the choice of cell type to target and the strategy, either the promoter specific or transgenic approach, for achieving cell type–restricted opsin expression must be made (see Yizhar et al., 2011a; Tye and Deisseroth, 2012, for comprehensive review of opsin targeting strategies and issues of experimental design). Because glutamatergic pyramidal projection neurons are known to strongly express the enzyme calcium-calmodulin kinase 2α and are the main output projection neurons in both the BLA and the medial PFC, viral gene expression tied to the calcium-calmodulin kinase 2α promoter would be a suitable approach for investigating glutamatergic inputs in either direction of this reciprocal circuit. Next, the pathway should be characterized in accordance with firing properties and synaptic function with use of optogenetic manipulations paired with brain slice electrophysiology to gain an understanding of the electrical properties of neurons in the pathway and to inform the choice of stimulation parameters for behavioral manipulations. Many validated and well-established paradigms for measuring behavior relevant to amygdalocortical function are available, including open-field assessment of locomotor and anxiety behavior, cued and contextual fear conditioning, conditioned place preference, elevated plus-maze, and social interactions. Of importance, all are easily interfaced with the necessary hardware and software for performing in vivo optogenetic manipulations (Zhang et al., 2010; Sparta et al., 2012). With use of a range of stimulation parameters and behavioral metrics, the detailed assessment of the symptoms of an anxious phenotype should be performed. This should be validated in a range of tests that may include optogenetic activation or inhibition time-locked to aversive events, cues that predict them, or long- or short-term stimulation to induce place aversion or other characteristic symptoms of an anxious phenotype.
B. Candidate Gene Search Using High-Throughput Genome Sequencing Methods
In recent years, the methods for genomic sequencing and transcriptome analysis have improved exponentially in their power and specificity to measure gene expression with a high degree of cellular resolution (Tang et al., 2011). It is now possible to catalog all species of RNA within a single cell with is of powerful next generation RNA-sequencing technologies (RNA-seq), which will allow sophisticated and quantitative analysis of gene expression within specific cell types after different environmental exposures or disease states (Wang et al., 2009). The next stage of the approach exploits the strength of optogenetics to identify and functionally characterize neural circuits and combines it with the genetic specificity of next-generation sequencing technologies to identify disrupted gene expression patterns after experimental enhancement or reduction in pathway-specific neural activity. The inherent complexity of mental disorders, which undoubtedly depend on the interaction of many genes and biologic pathways with environmental circumstances to manifest as a phenotype, necessitates that a fully integrative approach be applied to deciphering these complicated disorders. Of importance, a key strength of this integrative approach comes from being nonbiased and not guided by expectations about potential candidate genes. Instead, the approach is driven by behavioral observations of pathologic phenotypes induced by circuit-specific optogenetic activation of precise neural pathways. These behavioral observations inform the subsequent sequencing stage, whereby we attempt establish causal relationships between evoked activity in a neural pathway and changes in gene transcription induced by the pathologic activation state. Ultimately, any candidate genes identified by sequence analysis need to be validated to determine whether restoring their expression to normal levels affects neural circuit activity and reverses the negative behavioral phenotype. Rigorous experimenter control is essential to ensure that the measured changes in gene transcription are the result of optogenetic activity manipulations and not preexisting differences that may confound the interpretation of the data.
After optogenetic pathway-specific stimulation, two primary options are available for tissue samples collection: large tissue punches encompassing an entire brain region or single cell collection of specific cell types or cells that are integrated within a specific microcircuit. Single cell resolution is preferable but may not be attainable in all circumstances. Quantification of global patterns and changes in gene expression in a target area after optogenetic stimulation is a viable first pass alternative for the identification of dysregulated genes. Three main techniques are available for achieving single cell resolution. First, fluorescent activated cell sorting is a well-established tool to separate cells by type according to level of fluorescent marker expression; however, this method is optimal for cells in culture but can be less effective for collecting neurons in brain tissue. Second, laser capture microdissection is a feasible option for collection of single cells for analysis of mRNA expression. The laser capture microdissection procedure does involve some chemical staining that can potentially affect the quality of the obtained mRNA, but it is usually sufficient for quantitative transcriptome analysis (McCullumsmith and Meador-Woodruff, 2011). The third option involves harvesting whole cell mRNA during patch clamp electrophysiology by extracting the contents of the patched cell and then applying sequencing technologies to amplified mRNA (Citri et al., 2012). This option is especially attractive, because the precise details of each cell’s anatomic position within the target region will be known, which is of great importance in the cortex. As mentioned, the mammalian cortex has a highly stereotyped laminar cytoarchitecture, with each layer being populated by specific cell types that have clearly defined functional roles and patterns of efferent and afferent connections. Therefore, the possibility exists that the cortical dysfunction underlying various neuropsychiatric disorders may involve changes in processing that are more associated with a specific layer or cell type in one layer. Therefore, it is important that single cell resolution be achieved for gene expression analysis to determine whether input-specific activity alters transcription in a layer-specific fashion.
The recommended method for whole-transcriptome analysis is the next-generation RNA-seq. RNA-seq has many advantages over older methods of transcriptome analysis, such as hybridization-based approaches and microarray technology. The major limitations of these traditional methods are that they depend on existing knowledge of the genome sequence, they have high background because of cross-hybridization, and the expression data are difficult to compare across studies (Wang et al., 2009). RNA-seq methods have the major advantage of requiring less starting material, which is ideal for single-cell analysis. In addition, this method is not limited by known genomic sequence data and, thus, provides a whole transcriptome analysis that detects both coding and noncoding regulatory RNAs, which could be involved in the pathologic mechanisms of various psychiatric disorders. Of most importance, with RNA-seq, the actual nucleotide sequence is determined, which allows for the detection of single nucleotide polymorphisms and alternative gene isoforms. An obvious caveat arising from these advantages is that the number of identified splice variants may be extensive, which could make subsequent bioinformatic analyses more challenging and may necessitate that complementary studies be performed using microarrays. Furthermore, the issue of defining a normal pattern of expression to serve as a baseline comparison for any observed transcriptional changes assessed by RNA-seq can become challenging. However, with our proposed approach, a control group that does not receive optical stimulation of the target pathway will always serve as a comparison condition. Despite these limitations, applying RNA-seq technology is likely to be the best strategy for identifying changes in gene function in a specific cell type of a neural circuit in which dysfunction is implicated in a neuropsychiatric disorder. After RNA-seq and the identification of candidate genes in which the function changes after optogenetic stimulation of the target neural circuit, it is advisable to validate the results with more traditional methods, such as quantitative polymerase chain reaction analysis, in situ hybridization to measure mRNA within brain sections, Western blots, or immunohistochemistry.
C. Cell Type and Circuit Selective Small Molecule Pharmacotherapies as Novel Treatments for Psychiatric Disorders
Targeted drug delivery to a specific site of action has the profound advantage of maximizing therapeutic efficiency and minimizing systemic toxicity and off-target effects, which is of great importance in the treatment of any disease. Disorders of brain function present a great challenge for treatment because of the extreme structural complexity and cellular heterogeneity of the nervous system. The final stage of the experimental approach aims to achieve the ultimate goal of developing novel pharmacotherapies that act with cell type and pathway specificity to restore normal cell function by targeting those genes that were found to be dysregulated during the RNA sequencing stage. The true potential for this stage of the experimental approach depends on the continued technological advancement is several fields that are intensely focused on developing novel methods for delivering small molecules to disrupt some aspect of cellular function in a cell-specific manner. Although the goal of small molecule delivery to specific cell populations to modulate intracellular function is being approached from a variety of angles, here, we discuss two strategies for targeted drug development: nanoparticles for intracellular drug delivery and selective enzyme substrate pairs, both of which hold great potential for the future treatment of neuropsychiatric disorders.
The possibility for nanotechnology to revolutionize pharmaceutical development is currently generating great enthusiasm among scientists and the general public. The initial impetus for the field of nanoparticles for delivery of small molecules emerged from prominent failures in gene therapy clinical trials and the realization that synthetic methods for intracellular delivery of DNA needed to be developed (Paulo et al., 2011). Nanotechnology has permitted the development of nanocarriers that can encapsulate drugs, proteins, nucleic acids, or vaccines for strategic delivery to various diseased tissue and cellular targets (Morachis et al., 2012). A variety of materials, with their respective advantages and drawbacks, have been used for nanocarrier design, including lipids, inorganic materials, and polymeric systems (Morachis et al., 2012). This rapidly evolving field currently has the capability to produce a variety of nanocarriers that can deliver small interfering RNAs, proteins, and small molecules to different intracellular regions, which has important implications for cancer therapeutics, stem cell differentiation, and therapeutic medicine (Paulo et al., 2011). To date, nanotechnology has not produced a means to target a precise genetically distinct cell type in the nervous system, but the methods for achieving specificity are rapidly evolving. For example, the surface of nanoparticles has been conjugated with specific ligands or antibodies to permit their binding to cell types expressing the complementary receptor, and subsequently, the drug contents can be endocytosed for action within the cell. One achievement of nanoparticle technology with great relevance for the treatment of neuropsychiatric disorders has been the use of antibody-conjugated nanoparticles to overcome the blood-brain barrier defense of the brain (Aktas et al., 2005). The blood-brain barrier has posed a persistent challenge for the pharmacological treatment of brain disorders, because this system of tight junctions between endothelial cells is extremely effective at restricting most proteins and small molecules from entering the brain (Morachis et al., 2012). The use of nanoparticles for intracellular delivery of small molecules is still in its infancy but is likely to transform the pharmaceutical industry and have an immense impact of the future treatment of neuropsychiatric disorders.
A second promising approach for delivery of cell-specific small molecule drugs exploits the interaction of enzyme-substrate pairs to achieve cell specificity. The underlying principle of this strategy is to mask a small molecule drug by the attachment of a cleavable blocking functional group that renders it unreactive to endogenous cellular enzymes but is easily removed by a specific exogenous enzyme (Zhang et al., 2009; Tian et al., 2012). Cell type specificity is achieved by the expression of this exogenous enzymatic protein within a target cellular population where cleavage of the pro-drug will occur and its therapeutic effect may be somewhat restricted. If optogenetic circuit analysis and RNA-seq identify a specific gene product that may perform an integral role in cellular signaling, therapeutics to inhibit or restore gene function or associated intracellular signaling modalities can then be designed. The technology for targeting genetically defined cells with small molecule drugs to restore normal functioning was demonstrated by a recent study from Tian et al. (2012) that identified a specific ester-esterase pair that could be used to deliver active small molecules to precisely defined cell types. The specific pair of porcine liver esterase (PLE) and a α-cyclopropyl ester was highly efficient at unmasking the small molecule and had negligible reactivity with endogenous esterases. Of most importance, this ester-esterase strategy was used to deliver the pharmacological agent monastrol in a cell-specific manner. Monastrol acts to inhibit a component of the mitotic motor and, thus, disrupts cell division, which produces a clear phenotype that can be used to assess the efficacy of the targeted drug delivery. HeLa cells that were either transfected with the exogenous esterase PLE (PLE+) or not transfected (PLE−) were incubated with the ester conjugated pharmacological agent and tested to determine the efficacy of the targeted drug delivery. Of importance, it was found that PLE+ cells displayed significantly more spindle disruptions, which indicated that the cell-specific targeting was successful. This elegant study offers a clear demonstration that the development of a generalizable cell-specific delivery strategy for small molecule pharmacotherapies is within reach. An obvious caveat to the application of this method to humans is the necessity for gene therapy or other mechanisms to introduce and express the exogenous esterase for unmasking the pharmacological agent. Although this will entail some risk, we feel it should not deter the effort to develop novel cell-specific pharmacotherapies or which the benefits are certain to outweigh any minimal risk that they carry. At present and likely in the future, the methods for achieving cell specificity inevitably entail some genetic manipulation; however, as technology and scientific understanding evolve, the prospect for bringing cell-specific pharmacotherapies into wide clinical use may become a reality and the safety of these currently highly experimental strategies will inevitably improve. The safety of gene therapy for clinical application is currently an area of active research. For example, recent studies with the adeno-associated virus serotype 9 (AAV9) have reported good distribution and transduction of the AAV9 vector in nonhuman primate central nervous system tissue. Of importance, for safety concerns, AAV9 delivery to the cerebral spinal fluid did not shield against AAV antibodies (Samaranch et al., 2012). Another effort that promises to improve safety and potential for cell-specific pharmacotherapies to achieve ready use in clinical settings is the Pleiades Promoter Project. This interdisciplinary research endeavor has used genome-wide bioinformatic databases to analyze genes with brain expression patterns of interest to inform the design of compact MiniPromoters that can drive gene expression in specific cell types and brain regions (Portales-Casamar et al., 2010). The stated goal of this research effort is to provide a publicly available resource to facilitate research on brain development and therapies and should prove to be an invaluable starting point for designing versatile and safer pharmacotherapies that act cell and circuit specifically in the central nervous system.
VI. Conclusions
The overview and future research directions that we have presented here for pairing of optogenetic circuit analysis with current and next generation pharmacological approaches reveal how novel cellular pathophysiological mechanisms underlying psychiatric disorders can be uncovered. The current trend in biomedical research and the demands imposed by new technologies that require diverse technical expertise and produce enormous data sets inevitably leads to a climate in which integrative and collaborative approaches from diverse disciplines are required. Current research is still in the very early stages of developing and implementing multidisciplinary approaches for the treatment of complex brain disorders, but unparalleled potential is certainly apparent as revolutionary tools, such as those discussed here, contribute to the future.
Acknowledgments
The authors thank the members of the Stuber laboratory for helpful comments on this manuscript.
Abbreviations
- AAV9
adeno-associated virus serotype 9
- BLA
basolateral amygdala
- ChR2
channelrhodopsin-2
- DA
dopamine
- FS
fast-spiking inhibitory
- NAc
nucleus accumbens
- PFC
prefrontal cortex
- RNA-seq
RNA-sequencing
- sCRACM
subcellular ChR2-assisted circuit mapping
Authorship contributions
Wrote or contributed to the writing of the manuscript: Stuber, Mason.
Footnotes
This work was supported by the Brain and Behavior Research Foundation, The Foundation for Alcohol Research, The Whitehall Foundation, The Foundation of Hope, and the National Institutes of Health National Institute on Drug Abuse [Grants DA029325 and DA032750].
References
- Adamantidis AR, Tsai H-C, Boutrel B, Zhang F, Stuber GD, Budygin EA, Touriño C, Bonci A, Deisseroth K, de Lecea L. (2011) Optogenetic interrogation of dopaminergic modulation of the multiple phases of reward-seeking behavior. J Neurosci 31:10829–10835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. (2007) Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450:420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. (2009) Temporally precise in vivo control of intracellular signalling. Nature 458:1025–1029 [DOI] [PubMed] [Google Scholar]
- Aktaş Y, Yemisci M, Andrieux K, Gursoy RN, Alonso MJ, Fernandez-Megia E, Novoa-Carballal R, Quinoa E, Riguera R, Sargon MF, et al. (2005) Development and brain delivery of chitosan-PEG nanoparticles functionalized with the monoclonal antibody OX26. Bioconjug Chem 16:1503–1511 [DOI] [PubMed] [Google Scholar]
- Ambroggi F, Ishikawa A, Fields HL, Nicola SM. (2008) Basolateral amygdala neurons facilitate reward-seeking behavior by exciting nucleus accumbens neurons. Neuron 59:648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead RM, Domesick VB, Nauta WJ. (1979) Efferent connections of the substantia nigra and ventral tegmental area in the rat. Brain Res 175:191–217 [DOI] [PubMed] [Google Scholar]
- Benes FM. (2010) Amygdalocortical circuitry in schizophrenia: from circuits to molecules. Neuropsychopharmacology 35:239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JG, Boyden ES. (2011) Optogenetic tools for analyzing the neural circuits of behavior. Trends Cogn Sci 15:592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop SJ. (2007) Neurocognitive mechanisms of anxiety: an integrative account. Trends Cogn Sci 11:307–316 [DOI] [PubMed] [Google Scholar]
- Bishop SJ. (2008) Neural mechanisms underlying selective attention to threat. Ann N Y Acad Sci 1129:141–152 [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8:1263–1268 [DOI] [PubMed] [Google Scholar]
- Callaway EM. (2008) Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol 18:617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. (2009) Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature 459:663–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Pang ZP, Südhof TC, Wernig M, Malenka RC. (2012) Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protoc 7:118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ciano P, Cardinal RN, Cowell RA, Little SJ, Everitt BJ. (2001) Differential involvement of NMDA, AMPA/kainate, and dopamine receptors in the nucleus accumbens core in the acquisition and performance of pavlovian approach behavior. J Neurosci 21:9471–9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich I, Humeau Y, Grenier F, Ciocchi S, Herry C, Lüthi A. (2009) Amygdala inhibitory circuits and the control of fear memory. Neuron 62:757–771 [DOI] [PubMed] [Google Scholar]
- Fanselow MS, Gale GD. (2003) The amygdala, fear, and memory. Ann N Y Acad Sci 985:125–134 [DOI] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. (2011) The development and application of optogenetics. Annu Rev Neurosci 34:389–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund TF. (2003) Interneuron Diversity series: Rhythm and mood in perisomatic inhibition. Trends Neurosci 26:489–495 [DOI] [PubMed] [Google Scholar]
- Gardner EL. (2011) Addiction and brain reward and antireward pathways. Adv Psychosom Med 30:22–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace AA, Bunney BS. (1984a) The control of firing pattern in nigral dopamine neurons: burst firing. J Neurosci 4:2877–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace AA, Bunney BS. (1984b) The control of firing pattern in nigral dopamine neurons: single spike firing. J Neurosci 4:2866–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. (2010) Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141:154–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley CA, Phelps EA. (2010) Changing fear: the neurocircuitry of emotion regulation. Neuropsychopharmacology 35:136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert H, Klepper A, Ostwald J. (1997) Afferent and efferent connections of the ventrolateral tegmental area in the rat. Anat Embryol (Berl) 196:235–259 [DOI] [PubMed] [Google Scholar]
- Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PE, Kremer EJ, Palmiter RD. (2006) Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc Natl Acad Sci USA 103:8858–8863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Petreanu L, Ghitani N, Ranade S, Hromádka T, Mainen Z, Svoboda K. (2008) Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature 451:61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JS, Scanziani M. (2011) How inhibition shapes cortical activity. Neuron 72:231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Ambroggi F, Nicola SM, Fields HL. (2008) Contributions of the amygdala and medial prefrontal cortex to incentive cue responding. Neuroscience 155:573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen JP, Hamanaka H, Monfils MH, Behnia R, Deisseroth K, Blair HT, LeDoux JE. (2010) Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc Natl Acad Sci USA 107:12692–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Flint J, Gogos JA, Malenka RC, Genetic and Neural Complexity in Psychiatry 2011 Working Group (2012) The best of times, the worst of times for psychiatric disease. Nat Neurosci 15:811–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7:973–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. (2005) Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 62:593–602 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Loucks RA, Palmer AL, Brown AC, Solomon KM, Marchante AN, Whalen PJ. (2011) The structural and functional connectivity of the amygdala: from normal emotion to pathological anxiety. Behav Brain Res 223:403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch HS, Charlet A, Hoffmann LC, Eliava M, Khrulev S, Cetin AH, Osten P, Schwarz MK, Seeburg PH, Stoop R, Grinevich V, et al. (2012) Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron 73:553–566 [DOI] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. (2010) Neurocircuitry of addiction. Neuropsychopharmacology 35:217–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux J. (2012) Rethinking the emotional brain. Neuron 73:653–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzkus JJ, Wolff SBE, Meyer EMM, Tovote P, Courtin J, Herry C, Lüthi A. (2011) A disinhibitory microcircuit for associative fear learning in the auditory cortex. Nature 480:331–335 [DOI] [PubMed] [Google Scholar]
- Linden D, Thome J. (2011) Modern neuroimaging in psychiatry: towards the integration of functional and molecular information. World J Biol Psychiatry 12 (Suppl 1):6–10 [DOI] [PubMed] [Google Scholar]
- Linden DE. (2012) The challenges and promise of neuroimaging in psychiatry. Neuron 73:8–22 [DOI] [PubMed] [Google Scholar]
- Mao T, Kusefoglu D, Hooks BM, Huber D, Petreanu L, Svoboda K. (2011) Long-range neuronal circuits underlying the interaction between sensory and motor cortex. Neuron 72:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian AJ. (2012) Molecular genetic studies of complex phenotypes. Transl Res 159:64–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O. (2012) Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci 13:107–120 [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. (2004) Interneurons of the neocortical inhibitory system. Nat Rev Neurosci 5:793–807 [DOI] [PubMed] [Google Scholar]
- McCullumsmith RE, Meador-Woodruff JH. (2011) Novel approaches to the study of postmortem brain in psychiatric illness: old limitations and new challenges. Biol Psychiatry 69:127–133 [DOI] [PubMed] [Google Scholar]
- Milad MR, Quirk GJ. (2012) Fear extinction as a model for translational neuroscience: ten years of progress. Annu Rev Psychol 63:129–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morachis JM, Mahmoud EA, Almutairi A. (2012) Physical and chemical strategies for therapeutic delivery by using polymeric nanoparticles. Pharmacol Rev 64:505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RL, Neve KA, Nestler EJ, Carlezon WA., Jr (2005) Use of herpes virus amplicon vectors to study brain disorders. Biotechniques 39:381–391 [DOI] [PubMed] [Google Scholar]
- Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM. (2011) New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron 71:617–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Epstein J, Silbersweig DA, Stern E. (2011) New and emerging imaging techniques for mapping brain circuitry. Brain Res Brain Res Rev 67:226–251 [DOI] [PubMed] [Google Scholar]
- Paré D, Quirk GJ, Ledoux JE. (2004) New vistas on amygdala networks in conditioned fear. J Neurophysiol 92:1–9 [DOI] [PubMed] [Google Scholar]
- Pascoli V, Turiault M, Lüscher C. (2012) Reversal of cocaine-evoked synaptic potentiation resets drug-induced adaptive behaviour. Nature 481:71–75 [DOI] [PubMed] [Google Scholar]
- Paton JJ, Belova MA, Morrison SE, Salzman CD. (2006) The primate amygdala represents the positive and negative value of visual stimuli during learning. Nature 439:865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo CSO, Pires das Neves R, Ferreira LS. (2011) Nanoparticles for intracellular-targeted drug delivery. Nanotechnology 22:494002. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, Svoboda K. (2009) The subcellular organization of neocortical excitatory connections. Nature 457:1142–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PE, Stuber GD, Heien ML, Wightman RM, Carelli RM. (2003) Subsecond dopamine release promotes cocaine seeking. Nature 422:614–618 [DOI] [PubMed] [Google Scholar]
- Pillay NS, Stein DJ. (2007) Emerging anxiolytics. Expert Opin Emerg Drugs 12:541–554 [DOI] [PubMed] [Google Scholar]
- Portales-Casamar E, Swanson DJ, Liu L, de Leeuw CN, Banks KG, Ho, Sui SJ, Fulton DL, Ali J, Amirabbasi M, Arenillas DJ, et al. (2010) A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc Natl Acad Sci USA 107:16589–16594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Drevets WC. (2010) Neurocircuitry of mood disorders. Neuropsychopharmacology 35:192–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk GJ, Beer JS. (2006) Prefrontal involvement in the regulation of emotion: convergence of rat and human studies. Curr Opin Neurobiol 16:723–727 [DOI] [PubMed] [Google Scholar]
- Quirk GJ, Mueller D. (2008) Neural mechanisms of extinction learning and retrieval. Neuropsychopharmacology 33:56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. (2010) Two views of brain function. Trends Cogn Sci 14:180–190 [DOI] [PubMed] [Google Scholar]
- Raichle ME and Snyder AZ (2007) A default mode of brain function: a brief history of an evolving idea. Neuroimage 37:1083–1090; discussion 1097–1089. [DOI] [PubMed]
- Robinson S, Rainwater AJ, Hnasko TS, Palmiter RD. (2007) Viral restoration of dopamine signaling to the dorsal striatum restores instrumental conditioning to dopamine-deficient mice. Psychopharmacology (Berl) 191:567–578 [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. (2003) Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav 2:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaranch L, Salegio EA, San Sebastian W, Kells AP, Foust KD, Bringas JR, Lamarre C, Forsayeth J, Kaspar BK, Bankiewicz KS. (2012) Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum Gene Ther 23:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W. (1997) Dopamine neurons and their role in reward mechanisms. Curr Opin Neurobiol 7:191–197 [DOI] [PubMed] [Google Scholar]
- Shabel SJ, Janak PH. (2009) Substantial similarity in amygdala neuronal activity during conditioned appetitive and aversive emotional arousal. Proc Natl Acad Sci USA 106:15031–15036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin LM, Liberzon I. (2010) The neurocircuitry of fear, stress, and anxiety disorders. Neuropsychopharmacology 35:169–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. (2009) Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459:698–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotres-Bayon F, Quirk GJ. (2010) Prefrontal control of fear: more than just extinction. Curr Opin Neurobiol 20:231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparta DR, Stamatakis AM, Phillips JL, Hovelsø N, van Zessen R, Stuber GD. (2012) Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits. Nat Protoc 7:12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AM, Stuber GD. (2012) Activation of lateral habenula inputs to the ventral midbrain promotes behavioral avoidance. Nat Neurosci 15:1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Hnasko TS, Britt JP, Edwards RH, Bonci A. (2010) Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J Neurosci 30:8229–8233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Klanker M, de Ridder B, Bowers MS, Joosten RN, Feenstra MG, Bonci A. (2008) Reward-predictive cues enhance excitatory synaptic strength onto midbrain dopamine neurons. Science 321:1690–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Sparta DR, Stamatakis AM, Stuber GD, Sparta DR, Stamatakis AM, van Leeuwen WA, Hardjoprajitno JE, Cho S, Tye KM, Kempadoo KA, Zhang F, Deisseroth K, et al. (2011) Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature 475:377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Lao K, Surani MA. (2011) Development and applications of single-cell transcriptome analysis. Nat Methods 8(Suppl):S6–S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecuapetla F, Patel JC, Xenias H, English D, Tadros I, Shah F, Berlin J, Deisseroth K, Rice ME, Tepper JM, et al. (2010) Glutamatergic signaling by mesolimbic dopamine neurons in the nucleus accumbens. J Neurosci 30:7105–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Yang Y, Wysocki LM, Arnold AC, Hu A, Ravichandran B, Sternson SM, Looger LL, Lavis LD. (2012) Selective esterase-ester pair for targeting small molecules with cellular specificity. Proc Natl Acad Sci USA 109:4756–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H-C, Zhang F, Adamantidis A, Stuber GD, Bonci A, de Lecea L, Deisseroth K. (2009) Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science 324:1080–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM, Deisseroth K. (2012) Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat Rev Neurosci 13:251–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM, Prakash R, Kim S-Y, Fenno LE, Grosenick L, Zarabi H, Thompson KR, Gradinaru V, Ramakrishnan C, Deisseroth K. (2011) Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature 471:358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM, Stuber GD, de Ridder B, Bonci A, Janak PH. (2008) Rapid strengthening of thalamo-amygdala synapses mediates cue-reward learning. Nature 453:1253–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. (2010) Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci 11:100–113 [DOI] [PubMed] [Google Scholar]
- van Zessen R, Phillips JL, Budygin EA, Stuber GD. (2012) Activation of VTA GABA neurons disrupts reward consumption. Neuron 73:1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanat MJ, Willuhn I, Clark JJ, Phillips PE. (2009) Phasic dopamine release in appetitive behaviors and drug addiction. Curr Drug Abuse Rev 2:195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen X, Yang Y. (2012) Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods 9:266–269 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten IB, Steinberg EE, Lee SY, Davidson TJ, Zalocusky KA, Brodsky M, Yizhar O, Cho SL, Gong S, Ramakrishnan C, et al. (2011) Recombinase-driver rat lines: tools, techniques, and optogenetic application to dopamine-mediated reinforcement. Neuron 72:721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. (2009) A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. (2011a) Optogenetics in neural systems. Neuron 71:9–34 [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O'Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. (2011b) Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477:171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, Deisseroth K. (2010) Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc 5:439–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang L-P, Boyden ES, Deisseroth K. (2006) Channelrhodopsin-2 and optical control of excitable cells. Nat Methods 3:785–792 [DOI] [PubMed] [Google Scholar]
- Zhang H, Ma Y and Sun X-L (2010) Recent developments in carbohydrate-decorated targeted drug/gene delivery. Med Res Rev 30:270–289. [DOI] [PubMed]
- Zhang Y-P, Holbro N, Oertner TG. (2008) Optical induction of plasticity at single synapses reveals input-specific accumulation of alphaCaMKII. Proc Natl Acad Sci USA 105:12039–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]