Abstract
β-glucosidase A (bglA) in Clostridium thermocellum 27405 was increased by expression from shuttle vector pIBglA in attempts to increase cellulase activity and ethanol titer by lowering the end product inhibition of cellulase. Through a modified electrotransformation protocol C. thermocellum transformant (+MCbglA) harbouring pIBglA was produced. The β-glucosidase activity of +MCbglA was 2.3- and 1.6-fold greater than wild-type (WT) during late log and stationary phases of growth. Similarly, total cellulase activity of +MCbglA was shown to be 1.7-, 2.3- and 1.6-fold greater than WT during, log, late log and stationary phases of growth. However, there was no significant correlation found between increased cellulase activity and increased ethanol titers for +MCbglA compared with the WT. C. thermocellum has industrial potential for consolidated bioprocessing (CBP) to make a more cost effective production of biofuels; however, the hydrolysis rate of the strain is still hindered by end product inhibition. We successfully increased total cellulase activity by increased expression of bglA and thereby increased the productivity of C. thermocellum during the hydrolysis stage in CBP. Our work also lends insights into the complex metabolism of C. thermocellum for future improvement of this strain.
Keywords: Clostridium thermocellum, bioethanol, cellulase activity, end-product inhibition, β-glucosidase
Introduction
The production and commercialization of biofuels has gained a great deal of attention and support yet there remain some major bottlenecks with its current status; namely, there is a lack of biocatalysts that can work efficiently at high temperatures and under extreme pH conditions. Moreover, few microorganisms produce all the required enzymes for efficient hydrolysis of hemicellulose and cellulose. Additionally, hydrolysis and fermentation of lignocellulosic biomass-derived sugars requires separate steps (i.e., fungal enzymes and Saccharomyces cerevisiae, respectively), which in turn is less cost-effective.1 However, Clostridium thermocellum is a great potential candidate for biofuel production due to its ability to combine cellulase production and saccharification of biomass with fermentation in a process referred to as consolidated bioprocessing (CBP).2 In addition, C. thermocellum is a Gram positive, anaerobic, thermophilic, ethanologenic and cellulosome-producing bacteria. This means that during the production of biofuels from cellulosic and hemicellulosic biomass, C. thermocellum could decompose the biomass using highly versatile cellulosomes in addition to free cellulases and hemicellulases, and ferment 6-carbon sugars to ethanol without the addition of oxygen. Also, the thermophilic nature, (growth optimum 60°C), would allow easier extraction of the ethanol which requires higher temperatures to volatilize and precipitate.3 Currently, C. thermocellum has yet to be widely exploited in the production of biofuels due to several limiting factors. For example, C. thermocellum can hydrolyze both cellulose and hemicellulose; however, it can only ferment 6-carbon sugars and thus 5-carbon sugars are not being utilized. Additionally, end products of fermentation such as lactic and acetic acids, as well as ethanol can be toxic to C. thermocellum. Also, stresses such as toxicity and oxygen exposure can cause sporulation because it is a spore-forming bacterium.4,5 Further, cellulase activity in Clostridium cellulolyticum is shown to be inhibited by end products such as cellobiose.6 Inhibition of cellulase activity ultimately represents one of the greatest limitations to using C. thermocellum for biofuels. Without maximum hydrolysis of cellulosic biomass we cannot begin to consider optimum ethanol production and we cannot begin to change the economic viability of biofuels. Researchers have suggested and shown that by adding exogenous β-glucosidase isolated from Aspergillus niger one can increase the total cellulase activities of the cellulosome from C. thermocellum by 10-fold, in vitro.7 However, the purification and then addition of exogenous β-glucosidase would not be cost-effective for large-scale biofuel production. The genetic modification of the thermophilic anaerobe C. thermocellum, has been limited due to the strict restriction endonuclease system, which is described as a Dam+ phenotype.8,9 Also, many Gram positive bacteria with their thick cell walls have been reported as difficult to electrotransform.10,11 In this study, we have increased the copy number of β-glucosidase A gene (bglA) in C. thermocellum 27405 by electrotransforming it with a newly constructed shuttle vector (pIBglA) to ultimately increase cellulase activity and evaluate the overall effects on valuable end product formation such as ethanol.
Results
Construction of plasmid pIBglA
Plasmid pIBglA was constructed and transformed into Dam+ E. coli JM109. Due to the enzymatic limitations observed during experiments using SacI, which according to the distributer notes has several inhibitors causing low cutting efficiency, transformation of ligation products resulted in 1 positive bglA-containing transformant from 68 ampicillin positive transformants. Plasmid extraction of pIBglA followed by 1% agarose gel electrophoresis confirmed an approximate size of 7 kb. Sequencing revealed the complete cloned sequence of bglA of approximately 1,800 bp.
Verification of C. thermocellum electrotransformation with pIBglA
The transformation of C. thermocellum with pIBglA was completed and selection was based on a combined resistance to appropriate concentrations of ampicillin and lincomycin in semi-solid agar. Electrocompetent C. thercmocellum cells were transformed at a rate of 5.17 ± 3 transformants ml-1 of C. thermocellum media agar supplemented with appropriate concentrations of ampicillin and lincomycin. Thus, strain C. thermocellum+MCbglA (+MCbglA) was created. No spontaneous ampicillin-lincomycin resistant C. thermocellum cells were detected after 6 d incubation. PCR was performed with primers designed for the ampicillin gene using total DNA from the ampicillin-lincomycin resistant clones as a template. As shown in Figure 1, the presence of the ampicillin gene resulted in a ~530 bp product (lane 2), confirmed by pIBglA plasmid DNA as a positive control (lane 3) and negative control (WT total DNA) (lane 4).
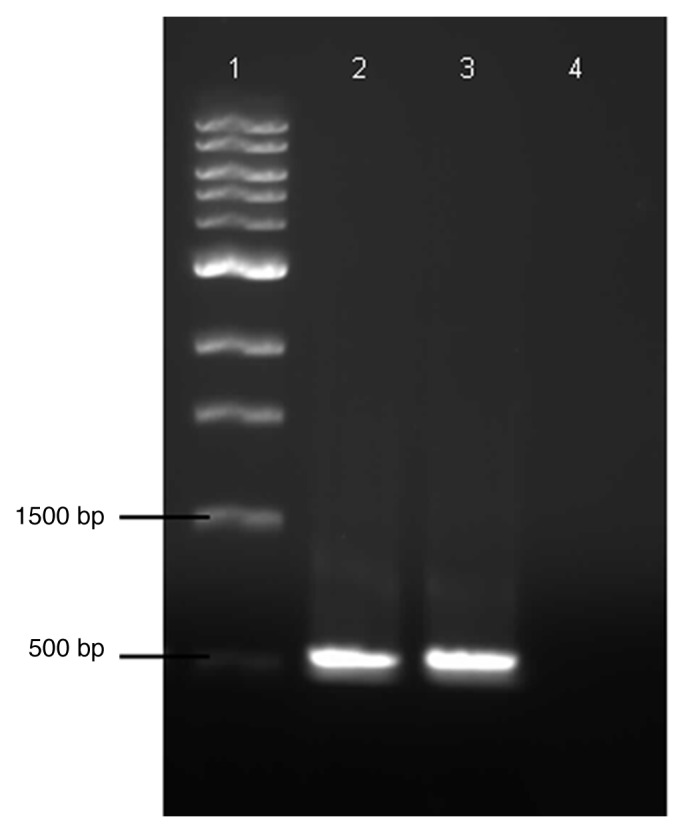
Figure 1. PCR amplification for conformation of ampicillin fragment (~500bp). Lanes: 1. 1kb ladder, 2. +MCbglA total DNA, 3. plasmid pIbglA DNA (positive control), 4. WT total DNA (negative control).
β-glucosidase activity of wild-type C. thermocellum and C. thermocellum+MCbglA
The β-glucosidase activity, was evaluated for WT- and +MCbglA- C. thermocellum and was found to be an average 1.9-fold greater in +MCbglA directly correlating with an increase in expression of β-glucosidase from plasmid pIBglA. As seen in Figure 2, the β-glucosidase activity increased during late log and stationary phases of growth for +MCbglA and were found to be 2.3- and 1.6-fold greater than WT with a statistical significance of p < 0.05 (Student’s t-test). However, biological and technical replicates revealed there was no significant difference in β-glucosidase activity during early log phases of growth for WT and +MCbglA.
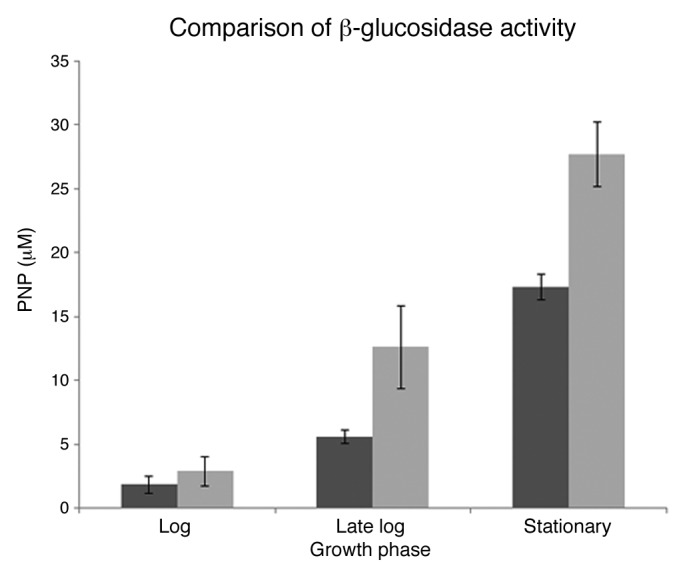
Figure 2. The β-glucosidase activity of WT (■) and MCbglA (■) during log, late log and stationary phases of growth and expressed as the amount of p-nitrophenol (µM) released after 30 min incubation with 4 µM PNPG.
Total cellulase activity of wild-type C. thermocellum and C. thermocellum+MCbglA
The total cellulase activity, the amount of glucose equivalents released (μM) from 1% Avicel, of WT- and +MCbglA- C. thermocellum was evaluated to determine if an increase in expression of bglA could increase the overall cellulase activity during batch fermentation trials. The results in Figure 3 show that total cellulase activity of the +MCbglA was observed to be 1.7-, 2.3- and 1.6-fold significantly greater than the activity of WT during log, late log and stationary phases of growth, respectively, p < 0.05 (Student’s t-test). Thus, total cellulase activity was an average 1.9-fold greater for +MCbglA compared with the WT.
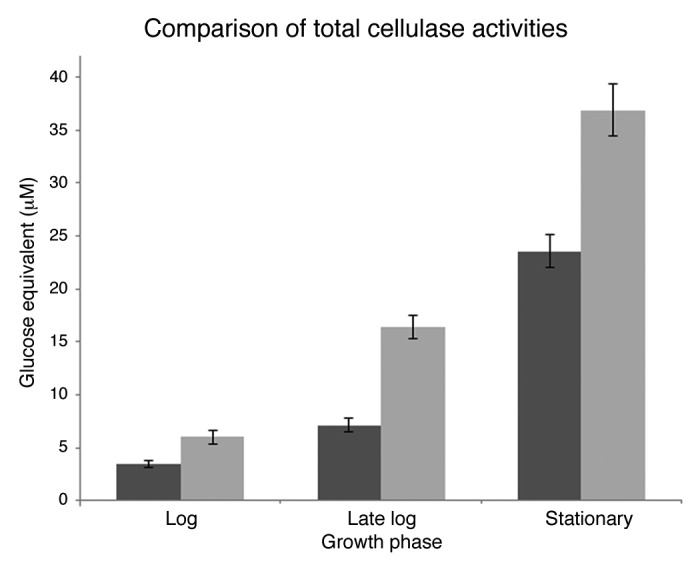
Figure 3. The total cellulase activity of WT (■) and MCbglA (■) during log, late log and stationary phases of growth and expressed in glucose equivalents (µM) released from 1% Avicel.
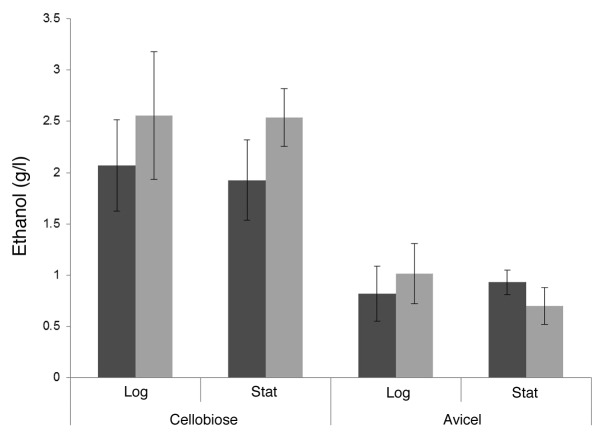
Analytical ethanol analysis for wild-type and +MCbglA C. thermocellum WT- and +MCbglA- C. thermocellum were grown in 1.5% cellobiose and 1% Avicel medium. Samples were taken when the cultures reached log and stationary phases of growth and analyzed for ethanol concentration (Fig. 4). When +MCbglA reached stationary phase, the average ethanol concentration in the media was 2.5 g/l. This is slightly higher than what was observed for the WT C. thermocellum (1.9 g/l), but this difference was deemed not to be statistically significant. Likewise, +MCbglA produced more ethanol during log phase growth, but it was not significantly different from the WT. Ethanol production by +MCbglA was also investigated in Avicel medium, and it yielded similar results to the cellobiose trials. No significant differences were observed between the bglA copy number mutant and the WT in both the log and stationary phases. Little difference was observed between ethanol concentrations at log and stationary phases.
Figure 4. The ethanol titers (g/l) of WT (■) and MCbglA (■) produced from batch fermentation trials on cellobiose and avicel containing mediums.
Discussion
The opportunity to use C. thermocellum for CBP in the biorefining industry has exceptional potential if we can overcome some of the challenges facing its development. The majority of difficulty working with thermophilic anaerobic bacterial systems arises from the slow progress in genetic manipulation of these systems. Plasmid pIKM1 bearing a kanamycin cassette and with Gram negative and positive origins of replication was constructed by Mai et al. in 1997 and transformed into Thermoanaerobacterium sp strain JW/SL-YS485 a close thermophilic anaerobic strain to C. thermocellum.17 Then, it was not until 2004, that Tyurin et al. developed an efficient protocol to transform C. thermocellum 27405 among two other C. thermocellum strains DSM 1313 and 4150 with plasmid pIKM1 using a uniquely designed electroporation system.14 More recently in 2010, Lin et al. developed a minimally invasive ultrasound-based sonoporation method for simple and rapid transformation of thermophilic Gram positive anaerobes.18 In doing so, they transformed Thermoanaerobacterium sp strain X514 with pIKM2 harboring a C. thermocellum β-1-4-glucanase gene, endoglucanase activity was observed in both electroporated and sonoporated X514 samples. Now for one of the first times reported, we expressed a functional gene (β-glucosidase A) from a Dam methylated plasmid pIBglA (constructed from pIKM1) in C. thermocellum 27405 using a modified electroporation protocol. In this research article, we strived to develop a C. thermocellum strain which can have greater total cellulase activity by reducing substrate specific end product inhibition. Thus, we proposed increasing the copy number of β-glucosidase. The BglA gene was chosen because it is fully sequenced and also because it’s native to C. thermocellum. Thus, this gene is suitable for expression in its thermophilic host. Due to the current lack of confident recombinant systems for C. thermocellum and for a concern in disruption of vital genes we chose to use plasmid pIKM1 shuttle vector with low copy number to increase expression of BglA without overburdening the cell. Growth in ampicillin remained constant; however, the loss of ampicillin resistance when continuously subculture without antibiotics was observed at approximately the 5th generation. One of the greatest limitations to using any whole microorganism for hydrolysis of cellulose and hemicellulose is the end product inhibition. Cellobiose is an inhibitor of cellulase activity in C. thermocellum and a previous report showed that the exogenous addition of β-glucosidase purified from Aspergillus niger could increase cellulase activity by 10-fold in C. thermocellum.7 Moreover, in Trichoderma reesei transformants the heterologous expression of a β-glucosidase gene from Penicillium decumbens resulted in an average 30% increase in filter paper activity (representing total cellulase activity).19 Therefore one may hypothesize that if there is an increase in copy number of β-glucosidase in C. thermocellum, there would also be an increase in β-glucosidase activity and this would ultimately increase total cellulase activity. In this study, the increase in copy number of bglA significantly increased both β-glucosidase activity as well as total cellulase activity during late log and stationary phases of growth by an average of 2.0-fold and 1.9-fold, respectively, for +MCbglA C. thermocellum over WT. Thus it appears the increase in β-glucosidase activity of C. thermocellum is also proportional to the observed increase in total cellulase activity. However, it was hypothesized here that the increase in total cellulase activity would also result in a likewise increase in ethanol production during batch fermentation trials, due to an increase in glucose catabolism. Nonetheless, there was no significant difference in ethanol production between WT- and +MCbglA- C. thermocellum suggesting that total cellulase activity is not directly proportional to fermentative metabolism of glucose under batch fermentation conditions. We suggest that this could be due to a metabolic overburden in +MCbglA from the presence of the shuttle vector pIBglA. This could potentially decrease the resistance of +MCbglA to toxic end products such as ethanol, lactic and acetic acids. The toxicity of end products such as ethanol in the fermentation of glucose for ethanogenic microorganisms such as C. thermocellum has gained a lot of attention in research for biotechnological implications. Ethanol is known to inhibit glycolytic enzymes and cause damage to cell membranes, thus inhibiting cell growth.20,21 From a commercial perspective titers of ethanol greater than 40 g/l are desired, for cost effective recovery.22 Thus, researchers have found the need to develop greater ethanol tolerant C. thermocellum strains such as by adaptation to levels as high as 50–55 g/l.22,23 However, to date, there remains a discrepancy whether increased ethanol tolerance can attribute to greater ethanol titers. The research presented here, also explored whether increased cellulase production could lend to an increase in ethanol. However, due to the lower levels of ethanol reported in this study, we suggest +MCbglA strain be produced in the future using recombinant DNA techniques to eliminate the stress of maintaining a plasmid.
Anaerobic catabolism is a challenging yet refined process in C. thermocellum where the modest availability of ATP needs to support growth, cellulase production and the uptake; a complex process which still requires exploration.24,25 It has been shown that the uptake of oligosaccharides with the combined intracellular phosphorolytic cleavage of β-1,4-glucosidic bonds is bioenergetically favorable when C. thermocellum is specifically grown on cellulose.25 Despite the lack of regeneration of ADP co-factors produced through the hydrolytic activity of BglA, a greater overall cellulase activity could be observed in +MCbglA compared with the WT due to a reduction in the cellulase end product inhibitor cellobiose. Additionally, the lack of increased ethanol production shown here further lends insights into the complex metabolism of glucose in C. thermocellum.
Materials and Methods
Media, strains and cultivation conditions
The strain Clostridium thermocellum (ATCC 27405) was obtained from the American Type Culture Centre through Cedarlane in Canada. C. thermocellum cells were grown at 60°C in Clostridium thermocellum Medium (ATCC media 1191). Either cellobiose or Avicel were included as the carbon source for appropriate experiments at a concentration of 0.5% and 1%, respectively.12,13 The following antibiotics were used for all growth, experiments transformant to maintain selection: 100 μg/ml ampicillin and 20 μg/ml lincomycin (Sigma-Aldrich). All of the work in this study using C. thermocellum cells was done inside a Coy Anaerobic Chamber (Coy Laboratories) under 5% hydrogen, 95% nitrogen mixed atmosphere, except applying the electric potential to the cells, centrifugation of cells and genomic DNA or enzyme extraction. The other strain used in this study Escherichia coli JM1O9, was grown at 37°C in Luria Bertani broth or on Luria Burtani agar containing 100 μg/ml of ampicillin for selection of transformants when appropriate.
Cloning β-glucosidase A from C. thermocellum
The genomic DNA of C. thermocellum was extracted from 3 ml of 48 h broth cultures using the Fungi/Yeast Genomic DNA Isolation Kit (Norgen Biotek Corporation) according to the instructions for Gram positive bacteria provided by the supplier. Two primers, BglPFW and BglSRV (shown in Table 1), were used to amplify the complete sequence of bglA from C. thermocellum with promotor and temerinator. The primers were designed to contain the restriction cut sites for PstI and SacI for BglPFW and BglSRV, respectively, in addition to nucleotides complementary to bglA. The PCR reaction mixtures contained 10 ng of C. thermocellum genomic DNA, 10 pmol of both forward and reverse primers, 10× Taq buffer with 500 mmol KCl, 25 mmol/l MgCl2, 0.2 mmol deoxynucleoside triphosphate, and 5 U Pfu DNA polymerase per 50 μl reaction. The PCR program was as follows: primary denaturation 3 min at 95°C, followed by 35 amplification cycles consisting of denaturing at 95°C for 1 min, annealing for 1 min at 54°C, and extension at 72°C for 1 min, upon completion of 35 amplification cycles a final extension step was done at 72°C for 10 min. The resulting amplicon of approximately 1.3 kb was confirmed by sequencing on ABI 3730xl automatic sequencer (Eurofins MWG Operon). The complete bglA was confirmed by complementation and alignment of sequencing results using DNAMAN software. The bglA product was then digested with restriction enzymes PstI and SacI (Fermentas, Canada) by combining 5 μl of bglA PCR product with 0.6 μl of SacI and PstI and 2 μl of 10× Tango buffer, incubated for 3 h 37°C.
Table 1. Primers used in this study.
| DNA target | Forward 5′-3′ | Reverse 5′-3′ | Product (bp) |
|---|---|---|---|
| BglA |
BglPFW-ACAGAGCTCAGCTCCATTGTTGCTTAGC |
BglSRV-AATCTGCAGACTGGTAAGTGATTGCCGG |
1867 |
| pIKM1 |
KmFWD- CTGGGAAGAAGACACTCCA |
KmRV- TGGAGTGTCTTCTTCCCAG |
N/A |
| Ampicillin | AmpFW- CGTTCATCCATAGTTGC | AmpRV- GCACGAGTGGGTTACATCG | 534 |
Source and construction of plasmid pIBglA
Plasmid pIKm1 was a gift from Lee Lynd (Dartmouth College, USA). The pIKm1 DNA was isolated from E. coli using Ultra Clean 6 Minute Mini Plasmid Kit (Mo Bio Laboratories) following the directions provided by the supplier. No sequence information was available for plasmid pIKm1 therefore we designed the primers KmFW and KmRV, (Table 1). The primers were designed within the kanamycin cassette gene to sequence the flanking multiple cloning sites. Restriction maps were produced from resulting sequences using DNAMAN software and cross referenced to the sequence of bglA. Two restriction endonucleases were chosen and used for digestion of pIKm1: PstI and SacI (Fermentas), restriction digest was performed by combining 0.8 μg/μl of pIKm1 DNA with 6.3 U of SacI and 2 μl of 10× Tango Buffer (Fermentas), this mixture was then incubated at 37°C in a water bath for 1 h. After 1 h time, 6.3 U of PstI was added and then the mixture was incubated for an additional 2 h at 37°C. Digestion resulted in two bands, one approximately 1.5 kb and the other approximately 5 kb. The 5 kb representing the remainder of the vector minus most of the kanamycin cassette was gel extracted using the NucleoSpin Extraction II kit (Clonetech Laboratories,) following instructions provided by the distributor. Previously cloned, digested and cleaned bglA were ligated to the approximate 5 kb vector using T4 DNA ligase (Fermentas). The ligation reaction mixture contained 2 μl of ligation buffer, 5 U T4 DNA ligase, 15 μl of 80 ng μ/l bglA DNA and 2 μl of 100 ng μ/l of digested pIKm1 DNA, and allowed to incubate at ambient temperature for 3 h. After ligation, the resulting DNA was transformed to E. coli JM1O9 using 40 μl of prepared electrocompetent cells premixed with 1 μl of ligation reaction in a 0.4 cm cuvette at a capacitance of 25 μF using the Bio- Rad Gene Pulser apparatus (Bio-Rad Laboratories). Transformants were screened using colony PCR for the presence of two genes: bglA and ampicillin (amp) (Table 1).
Electrotransformation of C. thermocellum with pIBglA
Electrocompetent C. thermocellum cells were prepared precisely following the method described by Tyurin et al. in 2004, however no isoniacin was used.14 Final cell suspension was approximately 9x1010 cells/ml and 40 μl aliquots were divided into 0.4 cm gap electroporation cuvettes and chilled on ice for 5 min. One microlitre of ~2 μg of pIBglA DNA extract was premixed under anaerobic conditions with the chilled 40 μl cells. The cuvettes with cell + DNA on ice were removed from the anaerobic chamber 120 and immediately place under a constant stream of N2 where the Bio-Rad Gene Pulser apparatus was used to electroporate the cells at a capacitance of 25 μF. Still under N2, the cells were then immediately removed from the cuvette with a 1 ml syringe and injected through the rubber stopper of hungate tubes (Bellco Glass) in prewarmed (55°C) C. thermocellum media supplemented with 0.5% (w/v) cellobiose and 0.75% (w/v) agar and 100 μg/ml ampicillin and 20 μg/ml lincomycin.
β-glucosidase activity of wild-type C. thermocellum and C. thermocellum+MCbglA
The β-glucosidase activity of wild-type- (WT-) C. thermocellum and the transformant containing multiple copies of bglA (MCbglA-C. thermocellum) was assayed by measuring the increase of absorbance at 400nm via the release of p-nitrophenol from p-nitrophenyl-β-D-glucopyranoside (PNPG).15 Briefly, WT- and MCbglA- C. thermocellum were pre-cultured for 48 h in 6 ml of Clostridium broth with 1% (w/v) cellobiose as the sole carbon source (n = 3). Then 200 μl of each culture were subsequently inoculated in triplicate to 40 ml of the same broth. Growth was monitored over 48 h during batch fermentation and 3 ml of samples were collected for each strain at early exponential phase (0.25 O.D.600 nm), late exponential phase (0.5 O.D.600 nm) and stationary phase (0.7 O.D.600 nm) in triplicate for enzyme analysis. To extract total cellular β-glucosidase, cells were harvested at 17,000 g for 1 min; then washed twice with chilled 100 mM PBS (pH 7.0) before final resuspension in 1 ml of PBS. Microbeads were filled to 0.5 μl mark of the 1.5 ml microcentrifuge tubes then samples were vortexed at full speed for 5 min, then chilled in an ice bath for 5 min; this procedure was repeated 2 more times for a total of 15 min vortexing. Following, the samples were centrifuged at 17,000 g for 1 min. In a 96 well microtiter plate, 50 μl of 100 mM PBS (pH 7.0) containing 4 mM PNPG was loaded per well, plus the addition of 50 μl of enzyme extract for each strain and each growth phase in triplicate. The microtiter plate was then incubated at 55°C for 30 min. Thereafter, the reaction was stopped by adding 100 μl of chilled 1M Na2CO3, followed with 10 min incubation at 4°C. The absorbance was measured on a Bio-Rad Laboratories xMark spectrophotometer at 400 nm. The β-glucosidase activity was defined as the amount of p-nitrophenol (PNP) released per 30 min incubation.15
Total cellulase activity of wild-type C. thermocellum and C. thermocellum+MCbglA
Total cellulase activity was defined as the amount of glucose and cellobiose released from 1% Avicel and expressed in glucose equivalents (µM). Wild-type C. thermocellum (WT) and C. thermocellum+MCbglA (+MCbglA) were cultured for 48 h in 6 ml of C. thermocellum media supplemented with 1% Avicel (w/v) as the sole carbon source (n = 3), then 200 μl of each strain was subsequently transferred in triplicate to a fresh new 40 ml of the same media for batch fermentation. Samples were collected in 1.5 ml aliquots from each vial to determine total cellulase activity during exponential (0.25 O.D.600nm), late exponential (0.5 O.D.600nm) and stationary (0.7 O.D.600nm) phases of growth. A microplate assay using the di-nitrosalicylic acid (DNS) method to measure reducing sugars, modified from Xiao et al.16 was used. Briefly, 60 μl of supernatant from each strain during each growth phase was added to a well in triplicate, a 120 μl of DNS was also added to each well. The plate was then incubated at 95°C for 5 min. Finally, 36 μl was removed from each well and added to 160 μl of ddH2O, mixed, and then the absorbance was read on a Bio-Rad xMark spectrophotometer at 545 nm.
Analytical methods for ethanol
Broth samples for ethanol analysis were collected from batch cultures used to measure enzyme activities from trials of WT- and +MCbglA- C. thermocellum on both cellobiose and Avicel containing media previously mentioned, by aliquoting 1.5 ml of culture supernatant into 1.5 ml Eppendorf tubes. All samples were stored at -20°C until analysis. The samples were then analyzed using an Agilent 6850 Gas Chromatograph fitted with a Carbowax column (30 min × 0.32 mm, film thickness 0.25 μm) and flame ionization detector. In preparation for analysis, the samples were thawed at room temperature, centrifuged at 17,000 g for 5 min and 1 ml of each sample was transferred to a fresh 1.5 ml Eppendorf tube. Prior to injection, each sample was spiked with 100 μl n-butanol, which acted as an internal standard. Injection volume was 1μl and the inlet was run splitless. Nitrogen was used as a carrier gas with a flow rate of 1.5 ml/min and the run time was 5 min/sample. The injection port temperature was set at 250°C, the column temperature was isothermally set at 75°C, and the detector temperature was 300°C. Standards were prepared the day of analysis using anhydrous ethanol and they were also spiked with 100 μl n-butanol per 1 ml standard.
Conclusion
Our work clearly demonstrates that by increasing expression of native BglA there will be a nearly proportion increase in total cellulase activity of C. thermocellum leading to the production of a more industrially applicable C. thermocellum. The hydrolysis step is a rate limiting step in the production of biofuels; however, more work on +MCbglA strain is required to increase ethanol titer for CBP potential. We will continue to test the following hypothesis: adapted increased tolerance to toxic end products of +MCbglA will lead to increased ethanol production, in hopes to lend more insight into the application of C. thermocellum in biofuel production.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/21951
References
- 1.Wyman CE, Dale BE, Elander RT, Holtzapple M, Ladisch MR, Lee YY. Coordinated development of leading biomass pretreatment technologies. Bioresour Technol. 2005;96:1959–66. doi: 10.1016/j.biortech.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Maki M, Leung KT, Qin W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci. 2009;5:500–16. doi: 10.7150/ijbs.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demain AL, Newcomb M, Wu JHD. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev. 2005;69:124–54. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng TK, Ben-Bassat A, Zeikus JG. Ethanol Production by Thermophilic Bacteria: Fermentation of Cellulosic Substrates by Cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1981;41:1337–43. doi: 10.1128/aem.41.6.1337-1343.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mori Y. Characterization of a Symbiotic Coculture of Clostridium thermohydrosulfuricum YM3 and Clostridium thermocellum YM4. Appl Environ Microbiol. 1990;56:37–42. doi: 10.1128/aem.56.1.37-42.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petitdemange E, Tchunden T, Valles S, Pirson H, Raval G, Gay R. Effect of carbon sources on cellulase production by clostridium cellulolyticum. Biomass Bioenerg. 1996;3:393–402. [Google Scholar]
- 7.Lamed R, Kenig R, Morgenstern E, Calzada JF, De Micheo F, Bayer EA. Efficient cellulose solubilisation by a combined cellulosome-β-glucosidase system. Appl Biochem Biotechnol. 1990;27:173–83. doi: 10.1007/BF02921525. [DOI] [Google Scholar]
- 8.Klapatch TR, Demain AL, Lynd LR. Restriction endonuclease activity in Clostridium thermocellum and Clostridium thermosaccharolyticum. Appl Microbiol Biotechnol. 1996;45:127–31. doi: 10.1007/s002530050659. [DOI] [PubMed] [Google Scholar]
- 9.Ozkan M, Desai SG, Zhang Y, Stevenson DM, Beane J, White EA, et al. Characterization of 13 newly isolated strains of anaerobic, cellulolytic, thermophilic bacteria. J Ind Microbiol Biotechnol. 2001;27:275–80. doi: 10.1038/sj.jim.7000082. [DOI] [PubMed] [Google Scholar]
- 10.Dunny GM, Lee LN, LeBlanc DJ. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991;57:1194–201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löfblom J, Kronqvist N, Uhlén M, Ståhl S, Wernérus H. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J Appl Microbiol. 2007;102:736–47. doi: 10.1111/j.1365-2672.2006.03127.x. [DOI] [PubMed] [Google Scholar]
- 12.Brener D, Johnson BF. Relationship Between Substrate Concentration and Fermentation Product Ratios in Clostridium thermocellum Cultures. Appl Environ Microbiol. 1984;47:1126–9. doi: 10.1128/aem.47.5.1126-1129.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernardez TD, Lyford KA, Lynd LR. Kinetics of the extracellular cellulases of Clostridium thermocellum acting on pretreated mixed hardwood and Avicel. Appl Microbiol Biotechnol. 1994;41:620–5. doi: 10.1007/BF00178500. [DOI] [Google Scholar]
- 14.Tyurin MV, Desai SG, Lynd LR. Electrotransformation of Clostridium thermocellum. Appl Environ Microbiol. 2004;70:883–90. doi: 10.1128/AEM.70.2.883-890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hang YD, Woodams EE. Apple pomace: A potential substrate for production of β-glucosidase by Aspergillus foetidus. Food Sci Technol. 1994;27:587–9. [Google Scholar]
- 16.Xiao Z, Storms R, Tsang A. Microplate-based filter paper assay to measure total cellulase activity. Biotechnol Bioeng. 2004;88:832–7. doi: 10.1002/bit.20286. [DOI] [PubMed] [Google Scholar]
- 17.Mai V, Lorenz WW, Wiegel J. Transformation of Thermoanaerobacterium sp. strain JW/SL-YS485 with plasmid pIKM1conferring kanamycin resistance. FEMS Microbiol Lett. 1997;148:163–7. doi: 10.1111/j.1574-6968.1997.tb10283.x. [DOI] [Google Scholar]
- 18.Lin L, Song H, Ji Y, He Z, Pu Y, Zhou J, et al. Ultrasound-mediated DNA transformation in thermophilic gram-positive anaerobes. PLoS One. 2010;5:e12582. doi: 10.1371/journal.pone.0012582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L, Zhang J, Zou G, Wang C, Zhou Z. Improvement of cellulase activity in Trichoderma reesei by heterologous expression of a beta-glucosidase gene from Penicillium decumbens. Enzyme Microb Technol. 2011;49:366–71. doi: 10.1016/j.enzmictec.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Ingram LO. Ethanol tolerance in bacteria. Crit Rev Biotechnol. 1990;9:305–19. doi: 10.3109/07388558909036741. [DOI] [PubMed] [Google Scholar]
- 21.Jones RP. Biological principles for the effects of ethanol. Enzyme Microb Technol. 1989;11:130–51. doi: 10.1016/0141-0229(89)90073-2. [DOI] [Google Scholar]
- 22.Shao X, Raman B, Zhu M, Mielenz JR, Brown SD, Guss AM, et al. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl Microbiol Biotechnol. 2011;92:641–52. doi: 10.1007/s00253-011-3492-z. [DOI] [PubMed] [Google Scholar]
- 23.Williams TI, Combs JC, Lynn BC, Strobel HJ. Proteomic profile changes in membranes of ethanol-tolerant Clostridium thermocellum. Appl Microbiol Biotechnol. 2007;74:422–32. doi: 10.1007/s00253-006-0689-7. [DOI] [PubMed] [Google Scholar]
- 24.Strobel HJ, Caldwell FC, Dawson KA. Carbohydrate transport by the anaerobic thermophile Clostridium thermocellum LQRI. Appl Environ Microbiol. 1995;61:4012–5. doi: 10.1128/aem.61.11.4012-4015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YH, Lynd LR. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc Natl Acad Sci U S A. 2005;102:7321–5. doi: 10.1073/pnas.0408734102. [DOI] [PMC free article] [PubMed] [Google Scholar]