Abstract
Nannochloropsis species have emerged as leading phototrophic microorganisms for the production of biofuels. Several isolates produce large quantities of triacylglycerols, grow rapidly, and can be cultivated at industrial scales. Recently, the mitochondrial, plastid and nuclear genomes of Nannochloropsis gaditana were sequenced. Genomic interrogation revealed several key features that likely facilitate the oleaginous phenotype observed in Nannochloropsis, including an over-representation of genes involved in lipid biosynthesis. Here we present additional analyses on gene orientation, vitamin B12 requiring enzymes, the acetyl-CoA metabolic node, and codon usage in N. gaditana. Nuclear genome transformation methods are established with exogenous DNA integration occurring via either random incorporation or by homologous recombination, making Nannochloropsis amenable to both forward and reverse genetic engineering. Completion of a draft genomic sequence, establishment of transformation techniques, and robust outdoor growth properties have positioned Nannochloropsis as a new model alga with significant potential for further development into an integrated photons-to-fuel production platform.
Keywords: Nannochloropsis, algae, biofuels, genomics, lipids
Microalgae are among the most promising renewable feedstocks for the production of fuels and chemicals. They produce a variety of bioenergy carriers, including lipids and carbohydrates; however, biological productivities need to be improved before algae are leveraged in economically viable biofuel production processes.1-3 A promising strategy for increasing productivities is to “domesticate” algal strains via genetic engineering to improve lipid yields and to produce biomolecules tailored for biofuel applications.4 Currently, only a few algal systems with publicly available genomes are genetically tractable, including the green algae Chlamydomonas reinhardtii and Ostreococcus tauri,5 as well as the diatoms Phaeodactylum tricornutum6 and Thalassiosira pseudonana.7 These organisms were selected as model systems because of their phylogenetic or ecological significance and/or their ease of culturing in the laboratory. Unfortunately, these organisms do not natively exhibit the high lipid or biomass productivities required from a biofuel feedstock and would likely require extensive genetic modification to increase their bioenergy carrier yields to the necessary levels.
As an alternative to these model algal systems, we took the approach of studying an algal strain that natively produces large quantities of lipids and is cultivated at commercial scale outdoors. After evaluating a variety of organisms, Nannochloropsis gaditana CCMP526 emerged as a leading candidate that produces large quantities of lipid and sustains high biomass accumulation rates. Species of Nannochloropsis are successfully cultivated at industrial-scale by several commercial interests including Aurora Algae, Solix Biofuels and Seambiotic. To begin to develop Nannochloropsis as a genetically tractable, industrially relevant alga, we sequenced the N. gaditana genome and transcriptome, and developed a robust transformation protocol using endogenous promoters. These results were recently published in Radakovits et al.8 Importantly, homologous recombination was reported for Nannochloropsis sp W2JB3,9 further establishing Nannochloropsis as among the most promising oleaginous algae for metabolic engineering, and the recent genetic advances will enable Nannochloropsis to be developed into a model system for optimizing algal lipid accumulation.
Nannochloropsis for Biofuel Production
There are six recognized species in the Nannochloropsis genus that are phylogenetically divided into two clades; one consisting of N. gaditana and N. salina, and the second of N. granulata, N. limnetica, N. oceanica, and N. oculata. Nannochloropsis species in commonly accessed culture collections have been isolated from around the world and in every ocean with the notable exception of sites south of the equator (Table 1). Nannochloropsis is generally regarded as a marine species, but one species, N. limnetica, is a fresh water isolate. Additionally, we have adapted N. gaditana CCMP526 to grow in 10% of the salinity in seawater by gradually adapting cultures to lower salt levels, which illustrates Nannochloropsis’ ability to thrive in a variety of culture conditions.
Table 1. Strains of Nannochloropsis and status of genomic sequencing efforts.
| Species | Strain | Year Isolated | Location isolated | Genomic sequencinga |
|---|---|---|---|---|
|
Nannochloropsis gaditana |
CCMP526 |
1985 |
Lagune de Oualidia, Morocco |
completed |
|
Nannochloropsis gaditana |
CCMP527 |
1952 |
Great South Bay, Long Island, New York, USA |
in progress |
|
Nannochloropsis gaditana |
CCMP532 |
1956 |
Milford, Connecticut, USA |
|
|
Nannochloropsis gaditana |
CCMP536 |
1965 |
Sayville, New York, USA |
|
|
Nannochloropsis gaditana |
CCMP1775 |
|
Cadiz Bay, Cadiz, Spain |
|
|
Nannochloropsis gaditana |
CCMP1894 |
1995 |
Comacchio Lagoons, Ferrara, Italy |
|
|
Nannochloropsis granulata |
CCMP529 |
1958 |
Continental Shelf, North Atlantic, off of USA East coast |
in progress |
|
Nannochloropsis granulata |
CCMP534 |
1986 |
Bigelow Laboratory dock, West Boothbay Harbor, Maine, USA |
|
|
Nannochloropsis granulata |
CCMP535 |
1965 |
Sayville, New York, USA |
|
|
Nannochloropsis granulata |
CCMP1662 |
1993 |
Skagerrak, North Atlantic |
|
|
Nannochloropsis limnetica |
CCMP505 |
1971 |
Morehead City, North Carolina, USA |
in progress |
|
Nannochloropsis limnetica |
CCMP2260 |
1996 |
Arrowwood Lake, North Dakota, USA |
|
|
Nannochloropsis limnetica |
CCMP2267 |
1996 |
Arrowwood Lake, North Dakota, USA |
|
|
Nannochloropsis limnetica |
CCMP2271 |
1996 |
Jim Lake, North Dakota, USA |
|
|
Nannochloropsis limnetica |
CCMP2272 |
1996 |
Arrowwood Lake, North Dakota, USA |
|
|
Nannochloropsis oceanica |
CCMP531 |
|
Qingdao, China |
in progress |
|
Nannochloropsis oceanica |
CCMP1779 |
1979 |
Kuwait Institute for Scientific Research, Kuwait |
in progress |
|
Nannochloropsis oceanica |
LAMB0001 |
|
|
completed |
|
Nannochloropsis oceanica |
OZ-1 |
|
|
in progress |
|
Nannochloropsis oculata |
CCMP525 |
|
|
in progress |
|
Nannochloropsis oculata |
CCMP2195 |
1968 |
Tunis, Tunisia |
|
|
Nannochloropsis salina |
CCMP369 |
1986 |
Narragansett Bay, Rhode Island, USA |
|
|
Nannochloropsis salina |
CCMP537 |
1986 |
Narragansett Bay, Rhode Island, USA |
in progress |
|
Nannochloropsis salina |
CCMP538 |
1964 |
Pamlico Sound, North Carolina, USA |
|
|
Nannochloropsis salina |
CCMP1776 |
1965 |
Skate Point, Isle of Cumbrae, Scotland, UK |
in progress |
|
Nannochloropsis salina |
CCMP1777 |
1965 |
Skate Point, Isle of Cumbrae, Scotland, UK |
|
|
Nannochloropsis salina |
CCMP1778 |
1966 |
Skate Point, Isle of Cumbrae, Scotland, UK |
|
| Nannochloropsis sp |
CCMP821 |
1969 |
Narragansett Bay, Rhode Island, USA |
|
| Nannochloropsis sp |
CCMP1780 |
|
|
|
| Nannochloropsis sp |
CCMP1997 |
1994 |
Sargasso Sea |
|
| Nannochloropsis sp |
CCMP2001 |
1998 |
Great South Bay, Long Island, New York, USA |
|
| Nannochloropsis sp | CCMP2904 | 2006 | Microcystis cove, Klamath Lake, Oregon, USA | |
a Genome status from NCBI.
Nannochloropsis gaditana natively exhibits high photoautotrophic biomass and lipid productivities. Over a period of three months we achieved an average biomass production rate of 0.65 g l-1 d-1, or ~20 g m-2 d-1 on an extrapolated aerial basis, in 1 l Roux flasks sparged with air/2% CO2 with a portion of the culture being harvested weekly. Lipid production in these conditions averaged 0.31 g l-1 d-1, and made up almost 50% of the algal biomass.8 The primary storage lipid in Nannochloropsis is triacylglycerol (TAG). The fatty acid composition of N. gaditana is relatively simple, the most abundant chain lengths being 14:0, 16:0, 16:1, 18:1, 18:2 and 20:5(n-3) (EPA), with 16:0 and 16:1 typically dominating. Many algae, such as Chlamydomonas, produce little TAG during logarithmic growth and require nutrient starvation to initiate significant lipid body formation.10,11Nannochloropsis gaditana has constitutive TAG droplets even in nutrient replete media during linear growth, and when deprived of nitrogen a single large oil droplet often encompasses the majority of the cellular volume.
Because of the exemplary lipid productivity in N. gaditana, we examined lipid metabolism genes, including those involved in fatty acid biosynthesis, TAG assembly, and lipid degradation. The number of genes involved in lipid metabolism in N. gaditana is expanded compared with the model algae, C. reinhardtii, P. tricornutum and Ectocarpus siliculosus (Fig. 1). To find additional gene expansions relative to C. reinhardtii and P. tricornutum, we used Fisher’s exact test to compare the prevalence of Gene Ontology (GO) terms found in each respective genome. Several GO-terms are over-represented in N. gaditana, which may be of importance for the lipid and biomass phenotypes observed. These terms include acyl-carrier protein biosynthetic processes, auxin biosynthesis, pyruvate metabolism, carbon utilization and acetyl-CoA catabolic processes. Interestingly, GO-terms involved in the regulation of growth rate, transcription factor activity, and vitamin binding are under-represented.8
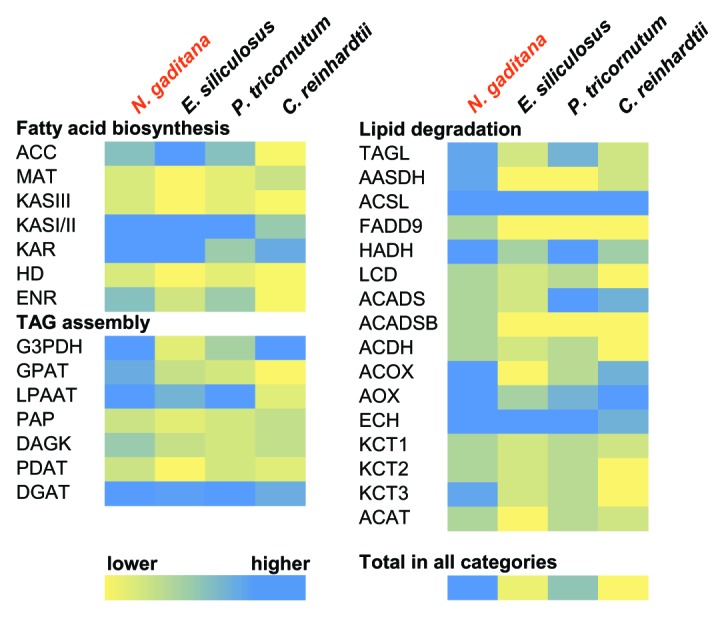
Figure 1. Selected lipid metabolism genes as a fraction of total genes. The number of gene homologs involved in lipid metabolism in each genome was normalized to the total number of genes in each organism. Total gene models in each organism: N. gaditana, 9,052; E. siliculosus, 16,256; P. tricornutum, 10,402; C. reinhardtii, 15,143.
Many species of algae require an exogenous source of vitamin B12 for use as a cofactor in vitamin B12-dependent enzymes. This micronutrient is often acquired through symbiosis with bacteria.12 Although Nannochloropsis does not require vitamin B12 supplemented media for growth, it does contain several vitamin B12 dependent enzymes. It is estimated that about half of all algae can only produce methionine with a vitamin B12-dependent methionine synthase.12 Nannochloropsis contains both a vitamin B12-dependent and a vitamin B12-independent methionine synthase. Due to higher catalytic efficiency, the vitamin B12-dependent enzyme is likely preferred when vitamin B12 is available;13 however, methionine synthesis can occur in the absence of vitamin B12 via the vitamin B12-independent methionine synthase. Nannochloropsis also contains a vitamin B12-dependent class II ribonucleotide reductase (RNR) which catalyzes the de novo synthesis of deoxyribonucleoside triphosphates (dNTPs). This enzyme is rarely found in eukaryotes, but was described in Euglena gracilis and Dictyostelium discoideum,14 which suggests that the class II enzymes were present in a common, B12-dependent, eukaryotic ancestor. Eukaryotes, including Nannochloropsis, typically have class I RNRs, which are primarily active in the presence of oxygen, but the addition of a class II RNR which is oxygen-independent may facilitate deoxyribonucleotide biosynthesis during anaerobiosis in aquatic ecosystems. This suggests, along with the presence of an FeFe-hydrogenase, that Nannochloropsis experiences anaerobic conditions in its native environment and has retained the metabolic capacity to withstand anoxic challenges. A third vitamin B12-dependent enzyme, methylmalonyl coenzyme A mutase, is found in Nannochloropsis and catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA which requires a vitamin B12 derivative, adenosylcobalamin, to function. While Nannochloropsis does not require exogenous vitamin B12, the addition of this micronutrient can augment its metabolic capabilities. Further studies are required to determine if addition of this vitamin can be leveraged to stimulate growth or improve biofuel production.
Metabolic Nodes in Fatty Acid Biosynthesis
Critical metabolic nodes for rationally engineering improvements in fatty acid biosynthesis are at the levels of acetyl-CoA and malonyl-CoA. Factors governing the synthesis and further metabolism of these metabolites are poorly understood in algae, and our research efforts with organisms such as Chlamydomonas clearly indicate that there are significant information deficits regarding the enzymes contributing to acetyl-CoA synthesis and how these enzymes are regulated in phototrophic microorganisms.15 In terrestrial plants, the acetyl-CoA used for fatty acid synthesis in the plastid is proposed to be derived primarily from the activity of a plastidic pyruvate dehydrogenase (PDH) complex, with additional potential contributions from acetyl-CoA synthetase and ATP-citrate lyase.16,17 Discrete plastidic and mitochondrial PDH complexes have been described in Arabidopsis thaliana,16-18 and genes encoding homologs for subunits corresponding to both of these distinct PDH complexes are present in N. gaditana. The plastidic PDH complex subunits are more similar to bacterial PDH complex subunits and lack several of the well-characterized posttranslational regulatory motifs described for mitochondrial PDH complexes. Linear photosynthetic electron transport from water oxidation is optimally configured to provide the necessary ATP/NADPH ratios for fixing CO2 to glyceraldehyde 3-phosphate (GAP) in the Calvin cycle. In principle, metabolism could be controlled to convert GAP, or even the primary product of the RuBisCO enzyme (3-phosphoglycerate) to acetyl-CoA using plastidic glycolytic enzymes and in the process attaining a net production of ATP (pyruvate kinase) and reducing equivalents (NAD(P)H) via plastidic PDH and GAP dehydrogenase for subsequent use in fatty acid biosynthesis. Acetyl-CoA can also be generated by ATP citrate lyase (ACL) which catalyzes the ATP dependent cleavage of citrate into acetyl-CoA and oxaloacetate. In Arabidopsis and Chlamydomonas ACL is a cytosolic enzyme made up of two discrete subunits that produces a cytosolic pool of acetyl-CoA.19 The N. gaditana genome encodes one monomeric ACL encoded by a single polypeptide that is similar to monomeric forms found in animals. As acetyl-CoA is at the nexus of respiratory ATP production (mitochondrion), the conversion of central metabolites to reducing equivalents, the entry point into citric acid cycle intermediates for biosynthetic precursors, and the key substrate for fatty acid biosynthesis in the plastid, control of acetyl-CoA biosynthesis in multiple cellular compartments and manipulation of its downstream metabolic fate is likely to enable improved biofuels phenotypes at the expense of protein biosynthesis and cell division. Establishing and characterizing the distinct metabolic enzymes/complexes that are able to generate acetyl-CoA, and an understanding of the competing pathways utilizing acetyl-CoA, is critical to this effort. By establishing that two discrete PDH complexes are present in Nannochloropsis, efforts can now be undertaken to begin characterizing and manipulating this important metabolic node.
Acetyl-CoA carboxylase (ACCase) is regarded as the committed and rate limiting step in fatty acid biosynthesis, converting acetyl-CoA to malonyl CoA. The N. gaditana genome encodes two monomeric ACCase homologs (> 2,000 AA each), as is commonly observed in other algal strains with secondary endosymbiotic ancestry.20 In Arabidopsis, two separate ACCase enzymes have been characterized. One is encoded by a single polypeptide and is targeted to the cytosolic face of the endoplasmic reticulum where it is proposed to be involved in fatty acid elongation reactions; whereas, the second ACCase complex (4 polypeptide subunits) is targeted to the plastid and is proposed to be the primary enzyme responsible for generating malonyl-CoA for plastidic fatty-acid biosynthesis.21,22 Attempts to overexpress ACCase for improved fatty-acid synthesis have met with limited success, implying that acetyl-CoA levels are limiting or that other regulatory features need to be considered.23 However, it must be stressed that some of these efforts focused on monomeric nonplastidic forms of ACCase,23 which may not function properly/optimally in the plastid, and that posttranslational regulatory features controlling enzyme activity in algae are not well established. As in the case of the PDH complexes, it is critical to develop a fundamental understanding of the physiological role, as well as the catalytic and regulatory properties of both of the Nannochloropsis ACCase enzymes, and then leverage this information in attempts to improve metabolic flux through this enzymatic node. The genome sequence has illuminated the requisite PDH and ACCase targets and directed studies are now underway to characterize these enzymatic systems, which influence fatty-acid synthesis and therefore ultimately TAG accumulation levels.
Genome Architecture and Gene Organization
Nannochloropsis gaditana has a haploid genome with an estimated size of 29 Mb.8 For genomic sequencing we used both 454 pyrosequencing and Illumina technologies. Sequence data were assembled into 2,087 scaffolds. Half of the genome is contained on 257 scaffolds (N50 statistic) with the 257th scaffold being 38 Kb in length (L50 statistic). The genomic sequence G+C content is 54.2%. All N. gaditana sequences were deposited in GenBank BioProject PRJNA73791 and have the accession numbers JH470562-JH472444.
In N. gaditana, we identified 9,052 gene models using two discovery methods: (1) gene models generated by Maker,24 which reconciled nine lines of EST, homology and ab initio evidence, and (2) ESTs with homologs in other algal lineages that were not included in the Maker gene model set. The majority of these gene models are supported by EST (92.3%) and homology (69.8%) evidence. Nannochloropsis genes on average have relatively few introns (1.62 per gene) when compared with model algae in the green lineage such as C. reinhardtii (7.33 per gene) or C variabilis NC64A (6.3 per gene). Many genes have no introns at all (more than 36% [3,102 genes]), which simplifies molecular biology and metabolic engineering tasks.
Genes in Nannochloropsis have unique orientations with respect to neighboring gene directionality and functionality. The majority of genes in N. gaditana are orientated such that genes diverge from a common locus and also converge with adjacent genes for termination, while having a smaller bias for genes that are sequentially transcribed in the same direction (Fig. 2A). The maximum number of consecutive genes observed with an alternating orientation is 15 (two instances), whereas the number of genes with a continuous orientation in a single direction is 9 (one instance). This preference is shared by other photosynthetic stramenopiles, but is not apparent in non-photosynthetic stramenopiles (Fig. 2B). This could constitute a mechanism used by Nannochloropsis to reduce genome size or to allow better control of gene transcription, as a single bi- directional promoter controls the transcription of two genes. The promoter for a violaxanthin/chlorophyll a-binding protein has been experimentally shown to be active in both directions in Nannochloropsis sp W2JB3.9 Many of these gene pairs have less than 1000 bp of intergenic sequence and bidirectional promoters can likely be exploited to drive the expression of two genes of interest, or to easily knockout two genes with one genomic insertion.
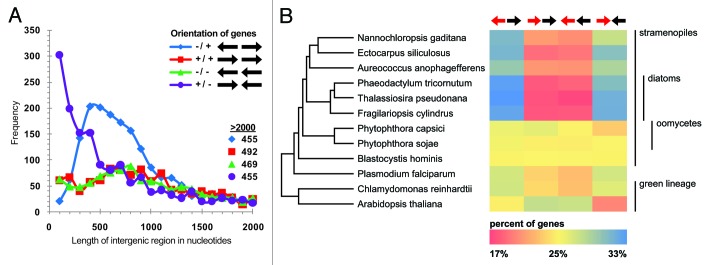
Figure 2. (A) Distance in nucleotides between Nannochloropsis gaditana gene models in relation to gene orientation. (B) A schematic phylogenetic tree of stramenopiles and other closely related eukaryotes, and a heat map indicating the percentage of genes with the specified relative orientation (diverging, converging, or same) to neighboring genes in the given organisms. Genes that are randomly oriented with respect to each other would be evenly distributed at 25% of genes in each orientation.
More than 20 metabolic pathway gene clusters were found by analyzing the spatial distribution of GO-terms. A nitrogen assimilation gene cluster was identified that includes a nitrate reductase, nitrite reductase, and a nitrate transporter. Additionally, a hydrogenase gene cluster with a unique configuration was found that contains an FeFe-hydrogenase (HYDA) and three genes needed for FeFe-hydrogenase maturation (HYDE, HYDF, and HYDG).25 This observed gene clustering can likely be used in some instances to infer the physiological function of neighboring proteins of unknown function.
Genomic Transformation and Homologous Recombination
For Nannochloropsis to emerge as an algal biofuel production platform, transformation of the nuclear and chloroplast genomes must be facile and routine. To date, the highest-efficiency transformations are achieved using electroporation with high electric field strengths and endogenous promoters driving the expression of a selection marker.8,9 Markers that confer resistance to zeocin,8 hygromycin B, and blasticidin S9 have been effectively used in Nannochloropsis. Higher transformation efficiencies are achieved when plasmids are linearized prior to transformation. Additionally, Nannochloropsis has the ability to take up multiple pieces of exogenous DNA. Using different selectable markers, cotransformation frequencies of about 50% for one unselected marker and about 30% for two unselected markers were seen in Nannochloropsis sp W2JB3.9
Without using homologous flanking regions, random integration into the genome can occur and multiple integrations are possible.8 When using homologous flanking regions, homologous recombination can proceed with high efficiency (up to 94%) in Nannochloropsis sp W2JB3.9 The ability to create random or targeted genomic insertions in Nannochloropsis can be leveraged in forward and reverse genetic approaches. These transformation techniques complement each other and will help facilitate functional genomics studies and allow for advanced metabolic engineering strategies.
Many eukaryotes, including the current model algal systems, preferentially use non-homologous end-joining (NHEJ) in double-strand break (DSB) repair and as a result exogenous DNA is ectopically inserted into the genome even if long stretches of homologous regions are present, resulting in very low homologous recombination (HR) efficiencies.4,26 Interestingly, the Nannochloropsis genome does not encode for Ku80, a protein that forms a heterodimer with Ku70 that binds to DSB ends and promotes NHEJ. It was demonstrated in several organisms that elimination of a Ku70 or Ku80 homolog greatly improves the frequency of homologous integration of exogenous DNA.27 The absence of Ku80 in Nannochloropsis may facilitate the observed high rates of homologous recombination. In an effort to increase homologous recombination efficiency, and further reduce ectopic insertions, the Ku70 homolog is a promising target for genetic disruption.
Molecular Tool Development and Synthetic Biology in Nannochloropsis
Chlamydomonas is the most developed model algal system and a variety of molecular tools and resources exist that expedite research. These include plasmid, cosmid, and bacterial artificial chromosome libraries, methods for tagging mutant alleles, map-based cloning, RNA interference, and reporter genes.28 Developing a similar foundation for Nannochloropsis is a priority. One resource that may be easier to establish in Nannochloropsis relative to Chlamydomonas is a whole genome knockout library. High through-put techniques have been employed to make targeted genome scale deletion collections in organisms that have high-rates of homologous recombination29 and a similar approach should be taken with Nannochloropsis. Auxotrophic strains can also be produced rapidly with homologous recombination, which will speed the development of endogenous selection markers.
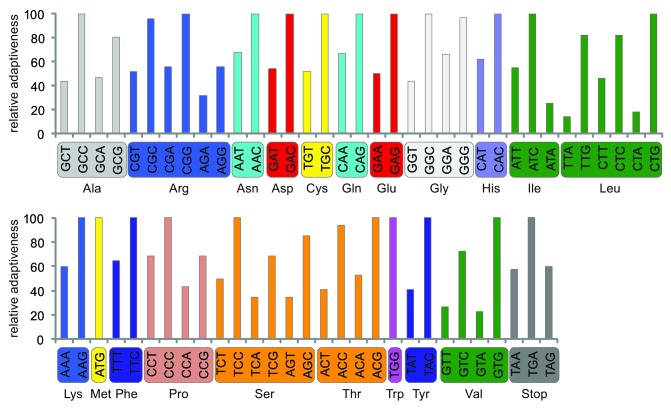
For synthetic gene design and optimization, we determined Nannochloropsis codon usage as gene expression levels are commonly correlated with codon usage in many organisms.30 The most frequently used codons were identified with a relative adaptiveness analysis and are indicated in Figure 3. Nannochloropsis coding regions have a slightly higher G+C content (58.0%) than the overall genomic DNA sequence (54.2%), and have a slight preference for A or T at the second base of the codon, with the average G+C content of the first, second, and third codon base being 61.0%, 45.9% and 67.0%, respectively.
Figure 3. Codon utilization in N. gaditana shown by relative adaptiveness, wij, which is defined as the ratio of the number of occurrences, xij, of a codon, j, for a given amino acid, i, compared with the number of occurrences of the codon used with the highest frequency for that amino acid, ximax, or wij = 100 * xij / ximax.30 Codon use was analyzed for 8,642 genes comprising more than 3 million codons.
Sexual recombination has not been reported to date in Nannochloropsis. The absence of a tractable reproductive system in which crosses can be generated is one remaining feature that would benefit the progression of Nannochloropsis into a model alga. In the N. gaditana genome, we found a set of genes that should be sufficient to facilitate meiosis. However, transcript evidence for the expression of these genes was never observed under any of the transcriptome conditions assessed which included plus and minus nitrate, heat and cold shock, linear and stationary phase culturing, supplemental CO2 and light/dark transistions.8 This suggests that (1) the correct environmental conditions to trigger meiosis were not evaluated, (2) multiple mating types may need to be present for meiosis to be initiated and/or (3) these genes are no longer transcribed because N. gaditana no longer undergoes meiosis. To determine whether Nannochloropsis can undergo a mating cycle, more environmental conditions need to be tested in addition to combining different strains that could possibly have an alternative mating type. Although difficult to establish, a tractable sexual recombination system in Nannochloropsis would complement the molecular tools already developed.
Conclusions
Algae can produce large quantities of lipid, have high photon to biomass conversion efficiencies, and can grow in a variety of water sources, but the lack of a genetically tractable, industrially relevant alga previously limited progress. The genomic DNA sequence of Nannochloropsis gaditana, in addition to efficient transformation protocols, will permit the rapid development of this organism into a biofuel production platform. Interrogation of the N. gaditana genome has revealed many features that contribute to our understanding of the oleaginous phenotype observed, but these findings are just a starting point for further investigations. The genomic sequence provides the basis for systems biology investigations and will serve as a platform for transferring knowledge attained from other algal systems to Nannochloropsis to improve biofuel phenotypes.31 An informed understanding of the metabolic pathways and their regulation in Nannochloropsis will allow for metabolic engineering strategies to reroute metabolites to biofuel precursors. The tools already developed for Nannochloropsis have positioned it for rapid strain improvements and advances are likely to emerge in the near future.
Acknowledgments
R.E.J. was supported by a Graduate Research Fellowship from the National Science Foundation. R.R. and this research were supported with funding provided by Conoco-Phillips through a grant to the Colorado Center for Biofuels and Biorefining (C2B2).
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/21880
References
- Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Weyer K, Bush D, Darzins A, Willson B. Theoretical maximum algal oil production. BioEnergy Research. 2010;3:204–13. doi: 10.1007/s12155-009-9046-x. [DOI] [Google Scholar]
- 2.Work VH, D’Adamo S, Radakovits R, Jinkerson RE, Posewitz MC. Improving photosynthesis and metabolic networks for the competitive production of phototroph-derived biofuels. Curr Opin Biotechnol. 2012;23:290–7. doi: 10.1016/j.copbio.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 3.Pienkos PT, Darzins A. The promise and challenges of microalgal-derived biofuels. Biofuels. Bioproducts and Biorefining. 2009;3:431–40. doi: 10.1002/bbb.159. [DOI] [Google Scholar]
- 4.Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell. 2010;9:486–501. doi: 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corellou F, Schwartz C, Motta J-P, Djouani-Tahri B, Sanchez F, Bouget F-Y. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21:3436–49. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apt KE, Kroth-Pancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet. 1996;252:572–9. doi: 10.1007/BF02172403. [DOI] [PubMed] [Google Scholar]
- 7.Poulsen N, Chesley PM, Kröger N. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae) J Phycol. 2006;42:1059–65. doi: 10.1111/j.1529-8817.2006.00269.x. [DOI] [Google Scholar]
- 8.Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilian O, Benemann CSE, Niyogi KK, Vick B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc Natl Acad Sci U S A. 2011;108:21265–9. doi: 10.1073/pnas.1105861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell. 2009;8:1856–68. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, et al. Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell. 2010;9:1251–61. doi: 10.1128/EC.00075-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–3. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 13.González JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry. 1992;31:6045–56. doi: 10.1021/bi00141a013. [DOI] [PubMed] [Google Scholar]
- 14.Torrents E, Trevisiol C, Rotte C, Hellman U, Martin W, Reichard P. Euglena gracilis ribonucleotide reductase: the eukaryote class II enzyme and the possible antiquity of eukaryote B12 dependence. J Biol Chem. 2006;281:5604–11. doi: 10.1074/jbc.M512962200. [DOI] [PubMed] [Google Scholar]
- 15.Catalanotti C, Dubini A, Subramanian V, Yang W, Magneschi L, Mus F, et al. Altered fermentative metabolism in Chlamydomonas reinhardtii mutants lacking pyruvate formate lyase and both pyruvate formate lyase and alcohol dehydrogenase. Plant Cell. 2012;24:692–707. doi: 10.1105/tpc.111.093146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ. The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol. 2000;123:497–508. doi: 10.1104/pp.123.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver DJ, Nikolau BJ, Wurtele ES. Acetyl-CoA-Life at the metabolic nexus. Plant Sci. 2009;176:597–601. doi: 10.1016/j.plantsci.2009.02.005. [DOI] [Google Scholar]
- 18.Tovar-Méndez A, Miernyk JA, Randall DD. Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur J Biochem. 2003;270:1043–9. doi: 10.1046/j.1432-1033.2003.03469.x. [DOI] [PubMed] [Google Scholar]
- 19.Fatland BL, Nikolau BJ, Wurtele ES. Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell. 2005;17:182–203. doi: 10.1105/tpc.104.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huerlimann R, Heimann K. Comprehensive guide to acetyl-carboxylases in algae. Crit Rev Biotechnol. 2012;•••:1–17. doi: 10.3109/07388551.2012.668671. [DOI] [PubMed] [Google Scholar]
- 21.Rawsthorne S. Carbon flux and fatty acid synthesis in plants. Prog Lipid Res. 2002;41:182–96. doi: 10.1016/S0163-7827(01)00023-6. [DOI] [PubMed] [Google Scholar]
- 22.Ke J, Wen T-N, Nikolau BJ, Wurtele ES. Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme A carboxylase. Plant Physiol. 2000;122:1057–71. doi: 10.1104/pp.122.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roesler K, Shintani D, Savage L, Boddupalli S, Ohlrogge J. Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol. 1997;113:75–81. doi: 10.1104/pp.113.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–96. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posewitz MC, King PW, Smolinski SL, Zhang L, Seibert M, Ghirardi ML. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem. 2004;279:25711–20. doi: 10.1074/jbc.M403206200. [DOI] [PubMed] [Google Scholar]
- 26.Sodeinde OA, Kindle KL. Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1993;90:9199–203. doi: 10.1073/pnas.90.19.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci U S A. 2004;101:12248–53. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, et al. Novel metabolism in Chlamydomonas through the lens of genomics. Curr Opin Plant Biol. 2007;10:190–8. doi: 10.1016/j.pbi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–7. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp PM, Li W-H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–95. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jinkerson RE, Subramanian V, Posewitz MC. Improving biofuel production in phototrophic microorganisms with systems biology. Biofuels. 2011;2:125–44. doi: 10.4155/bfs.11.7. [DOI] [Google Scholar]