Abstract
Strawberry (Fragaria×ananassa) is one of the most important berry crops in the world. Root rot of strawberry caused by Rhizoctonia spp. is a serious threat to commercial strawberry production worldwide. However, there is no information on the genetic diversity and phylogenetic status of Rhizoctonia spp. associated with root rot of strawberry in Australia. To address this, a total of 96 Rhizoctonia spp. isolates recovered from diseased strawberry plants in Western Australia were characterized for their nuclear condition, virulence, genetic diversity and phylogenetic status. All the isolates were found to be binucleate Rhizoctonia (BNR). Sixty-five of the 96 BNR isolates were pathogenic on strawberry, but with wide variation in virulence, with 25 isolates having high virulence. Sequence analysis of the internal transcribed spacers of the ribosomal DNA separated the 65 pathogenic BNR isolates into six distinct clades. The sequence analysis also separated reference BNR isolates from strawberry or other crops across the world into clades that correspond to their respective anastomosis group (AG). Some of the pathogenic BNR isolates from this study were embedded in the clades for AG-A, AG-K and AG-I, while other isolates formed clades that were sister to the clades specific for AG-G, AG-B, AG-I and AG-C. There was no significant association between genetic diversity and virulence of these BNR isolates. This study demonstrates that pathogenic BNR isolates associated with root rot of strawberry in Western Australia have wide genetic diversity, and highlights new genetic groups not previously found to be associated with root rot of strawberry in the world (e.g., AG-B) or in Australia (e.g., AG-G). The wide variation in virulence and genetic diversity identified in this study will be of high value for strawberry breeding programs in selecting, developing and deploying new cultivars with resistance to these multi-genetic groups of BNR.
Introduction
Strawberry (Fragaria×ananassa) is one of the most economically important berry crops in the world, with production reaching about 4.4 million tons in 2010 [1], [2]. Root rot of strawberry caused by Rhizoctonia spp. is a serious threat to commercial strawberry production worldwide and is associated with severe economic losses, such as have been reported in Japan [3], [4], the USA [5]–[8], Australia [9], [10], South Africa [11], Israel [12] and Italy [13].
Rhizoctonia spp. are soil-borne fungal pathogens with a worldwide distribution [14], [15]. They can cause severe damage to a wide range of economically important agricultural and horticultural crops [16]–[18]. Disease caused by Rhizoctonia spp. is difficult to manage due to the soil-borne nature and wide host range of Rhizoctonia spp. [19], [20]. Management of Rhizoctonia root rot on strawberry remains reliant on soil fumigation [13], [21]–[23]. However, the phasing-out of some broad-spectrum pre-planting fumigants due to environmental and health concerns has fostered keen interest in developing alternative non-chemical ways to manage root rot and other soil-borne diseases more effectively and sustainably [23]–[26]. It is now accepted that identifying and deploying resistant cultivars is the most cost effective and environmentally sustainable strategy to control soil-borne disease on strawberry [24]–[27]. However, if novel management strategies, including the breeding of Rhizoctonia-resistant cultivars, are to be developed, it is essential to understand the genetic diversity within Rhizoctonia and how this diversity pertains to the virulence of isolates.
Rhizoctonia spp. have been classified into uninucleate, binucleate and multinucleate Rhizoctonia based on the cell nuclear condition [28], [29]. Binucleate Rhizoctonia (BNR) have been divided into 21 anastomosis groups (AG-A to AG-U), and multinucleate Rhizoctonia (MNR) have been divided into 13 anastomosis groups (AG-1 to AG-13) based on hyphal anastomosis reactions [15], [17], [18], [28]–[30]. Both BNR and MNR have been reported as pathogen groups causing root rot on strawberry. BNR AG-A and AG-G are the two most common groups associated with root rot on strawberry in the world [5], [6], [11]–[13]. BNR AG-I are also found to be associated with root rot on strawberry in the USA and South Africa, while BNR AG-F and AG-K are also found to be associated with root rot on strawberry in Israel [5], [6], [11], [12]. MNR AG-4, AG-5 and AG-6 are also associated with root rot of strawberry in Israel, the USA and South Africa, respectively [5], [11], [12].
The sequence of the internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA) in particular has been found to be very useful for evaluating genetic diversity and characterizing AG groups of Rhizoctonia spp. [12], [13], [15], [16], [29], [31], [32]. The ITS sequences lie between the 18S and 5.8S rRNA genes (ITS1) and between the 5.8S and 28S rRNA genes (ITS2). The ITS sequences are present in high copy numbers in all eukaryotic genomes, have a high rate of evolution, and are flanked by highly conserved nucleotide sequences [33]. The grouping of Rhizoctonia spp. using molecular analysis based on ITS sequences supports the grouping of Rhizoctonia spp. based on classical hyphal anastomosis reactions. Moreover, sequence analysis is rapid, provides higher resolution of relationships and is less technically challenging than grouping of Rhizoctonia spp. using classical hyphal anastomosis reactions [12], [20], [29].
Rhizoctonia spp. were frequently recovered from diseased plants during outbreaks of root rot on strawberry in Western Australia [10]. However, there is no information on the genetic diversity and phylogenetic status of the Rhizoctonia spp. that infect strawberry in Australia. This paper reports studies conducted to (i) characterize the nuclear condition of Rhizoctonia spp. isolates recovered from diseased strawberry in Western Australia; (ii) determine their pathogenicity and relative virulence; (iii) define their genetic diversity based on ITS1-5.8S rRNA-ITS2 sequence analysis; (iv) determine if there is an association between genetic diversity and virulence; and (v) determine the phylogenetic relationships of these isolates with those from strawberry and other crops across the world.
Materials and Methods
Fungal isolates
A total of 96 Rhizoctonia spp. isolates were recovered from discolored root or crown tissues of diseased strawberry plants collected from nine representative commercial strawberry fields in major strawberry production areas in Western Australia. All necessary permits were obtained for the described field studies. The fields were privately owned. The owner of each field permitted us to collect the plant samples. The field studies did not involve endangered or protected species. These isolates were purified by selecting a single hyphal tip (less than 1.5 mm) of each isolate growing on water agar and transferring to fresh water agar to obtain pure cultures, and preserved as lyophilized cultures on colonized millet seeds in glass ampoules. Whenever required, isolates were sub-cultured onto fresh potato dextrose agar (PDA) for subsequent studies.
Nuclear condition
The numbers of nuclei per hyphal cell in each of the 96 Rhizoctonia spp. isolates from strawberry plants were determined according to procedures of Yang et al. [34]. Briefly, a 1 mm2 of colonized PDA agar containing mycelia of each isolate from 2 to 3 day-old culture was placed on the middle of a sterilized glass slide coated with a thin layer of PDA. Slides were then maintained on sterile filter papers moistened with sterilized deionized (DI) water in Petri dishes at 25°C for 2 days in the dark. Nuclei were stained with 0.48% (w/v) Hoechst Dye 33258 (Sigma-Aldrich, Australia) in 0.1 M KH2PO4 buffer (pH 7.8) and in 0.025 M H3BO3 buffer (pH 10.5). The number of nuclei per cell of each isolate was determined under a fluorescence microscope and photographed (Axioplan 2 microscope, AxioCam Digital photograph system, Carl Zeiss, NSW Australia). The nuclei of 20 cells per isolate were counted to confirm the nuclear status of each isolate.
Pathogenicity test
Pathogenicity of the 96 Rhizoctonia isolates was determined on strawberry seedlings, including eight isolates which had been previously determined [35]. Millet seed-based inoculum of each isolate was prepared using a modified procedure of Fang et al. [35]. Briefly, 200 g millet seed (Panicum miliaceum) was soaked in 200 mL DI water in a 1 L flask for 12 h, excess water drained and subsequently autoclaved at 121°C for 20 min on three consecutive days. Six 3 mm-diameter disks from margins of one-week-old colonies of each isolate growing on PDA plates were added to each flask containing sterilized millet seeds. Flasks were shaken every 2 days to ensure uniform colonization and incubated at 22°C for 2 weeks.
A tissue culture system was developed to aseptically produce strawberry seedlings (cv. Camarosa) for this study based on methods described in Fang et al. [25]. Strawberry seedlings were removed from culture tubes after 4 weeks and washed with sterilized DI water. The seedlings were transplanted to plastic pots containing potting mix which were watered daily to free draining with DI water. The potting mix was finely crushed pine bark∶coco peat∶sand at 2.5∶1.0∶1.5 (w/w) and was pasteurized using aerated steam on three consecutive days at 65°C for 90 min prior to use. All pots were maintained in controlled environment rooms (22°C, 16 h photoperiod, 60% relative humidity) and covered for one week with transparent plastic bags to maintain high humidity. After 3 weeks, seedlings were removed from pots and washed with sterilized DI water before using for the pathogenicity tests.
Seedlings were transplanted into plastic pots (9 cm×9 cm) containing pasteurized potting mix infested with millet seed-based inoculum of each isolate at a rate of 0.5% (w/w). Control seedlings for comparison were transplanted into pots containing uninfested potting mix. Uninoculated sterilized millet seed was deliberately not used as a control comparison as it is known to have a ‘baiting-out’ effect on other potential pathogens in the soil, especially Pythium spp. when introduced uncolonized into soil [36]. There were four seedlings per pot and two replicate pots for each treatment arranged in a randomized block design. All pots were maintained in the same controlled environmental conditions (27°C, 16 h photoperiod, 60% relative humidity). Pots were watered daily to free draining with DI water. Plants were harvested 3 weeks later and the severity of root rot was assessed on a 0 to 5 disease severity scale, where: 0 = root well developed, no discoloration/rot; 1 = <25% root discolored/rotted; 2 = ≥25%, <50% root discolored/rotted; 3 = ≥50%, <75% root discolored/rotted; 4 = ≥75% root discolored/rotted; 5 = all root rotted. Following disease assessment, to fulfill Koch's postulates, pathogen re-isolations were made from pieces of freshly harvested discolored root pieces onto PDA. Re-isolated cultures were examined microscopically to confirm morphological similarities with inoculated isolates and demonstrate that infection was from the inoculated isolates.
This experiment was repeated once. Isolates were considered to be pathogenic if they caused visible root rot symptoms on seedlings in both repeat experiments. Percent disease index (%DI) was calculated for each pathogenic isolate in each experiment by the following formula:
and where a, b, c, d, e and f represent the number of plants with disease scores of 0, 1, 2, 3, 4 and 5, respectively. Data on %DI from the two repeat experiments of each pathogenic isolate were combined and mean %DI was calculated. For all pathogenic isolates, the mean %DI was used to categorize their relatively virulence, where isolates were categorized as high virulence if they showed a mean %DI ≥67, moderate virulence if they showed a mean %DI ≥33 and <67, and low virulence if they showed a mean %DI <33. Only the BNR isolates that were pathogenic on strawberry were subsequently utilized in the molecular studies.
DNA extraction
Genomic DNA of each of the 65 pathogenic BNR isolates was extracted using the method described by Cenis [37]. Briefly, a 1.5 mL tube was filled with 500 µL autoclaved potato dextrose broth. The culture was started by sub-culturing some mycelia of each isolate into the broth and then kept at 25°C for 72 h. The mycelial mat was removed and washed with TE buffer and pelleted by centrifugation for 5 min at 13,000 rpm, and decanting the supernatant. After adding in 300 µL extraction buffer (200 mM Tris-HCl pH 8.5, 250 mM NaCl, 25 mM EDTA and 0.5% (w/v) SDS), the mycelia was crushed with a conical grinder. 150 µL 3 M sodium acetate (pH 5.2) was added, and tubes were placed at −20°C for 10 min. The supernatant was transferred to a fresh tube after centrifugation for 5 min at 13,000 rpm, and an equal volume of isopropanol was added. After at least 5 min at abient temperature, the precipitated DNA was collected by centrifugation for 10 min at 13,000 rpm. The pellet was vacuum dried for several minutes after washing with 70% (w/v) ethanol and re-suspended in 50 µL of TE buffer. The concentration and quality of the extracted DNA was determined using spectrophotometer (NanoDrop 1000 Spectrophotometer, Thermo Scientific).
PCR amplification and DNA sequencing
PCR amplification of the ITS region of the rDNA, including ITS1, 5.8S rDNA and ITS2 (ITS1-5.8S rDNA-ITS2), for each BNR isolate was performed using primers ITS1 (5′ TCC GTA GGT GAA CCT GCG G 3′) and ITS4 (5′ TCC TCC GCT TAT TGA TAT GC 3′) [38]. Amplification was conducted in 50 µL reaction mixtures containing 2 µL template DNA (about 100 ng), 25 µL 2× master mix (GoTaq Green Master Mix, Promega Corp.), and 1 µL of each primer (10 µM). Amplification was carried out in a thermal cycler (Bio-Rad Laboratories Pty Ltd) with the following program: an initial denaturation at 95°C for 2 min; 35 cycles consisting of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s; a final extension at 72°C for 3 min. A 5 µL aliquot of each PCR product was separated by electrophoresis on 1.5% (w/v) agarose gels stained with ethidium bromide and visualized using a UV transilluminator. The remaining 45 µL of each PCR product were purified (QIAquick PCR Purification Kit, Qiagen Inc., Valencia, CA) according to the manufacture's instructions and directly sequenced in both directions using the same primers used for the PCR amplification (Macrogen Inc., Korea). For all PCR reactions, sterilized ultra-pure water was used as a control in place of fungal genomic DNA to test for contamination of the reagents. PCR amplifications were performed three times. The size of each amplification product was determined by electrophoresis on agarose gels followed by staining with ethidium bromide to ensure the reproducibility of PCR amplification.
Sequence alignment, analysis and AG determination
The nucleotide sequences generated by sequencing the ITS1-5.8S rDNA-ITS2 region of each isolate in both directions were edited and assembled using BioEdit 7.1 [39] with manual adjustment. Sequences from all these isolates were compared with those in the GenBank nucleotide database provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the BLAST algorithm to determine sequence identity and find the closest match based on maximal percent identity. The ITS1-5.8S rDNA-ITS2 sequences of the 65 BNR isolates generated from this study were deposited in GenBank (Table 1). Additional reference sequences of 55 representative isolates from known AG groups of BNR from strawberry or other crops across the world were retrieved from GenBank database (Table 2). All but two of these reference sequences of the BNR isolates used in this study are associated with publication in a peer-reviewed scientific journal (Table 2). The ITS1-5.8S rDNA-ITS2 sequence of Athelia rolfsii FSR-052 (GenBank Accession No. AY684917) was used as the outgroup [12], [20], [29], [31].
Table 1. Virulence of the 65 pathogenic binucleate Rhizoctonia isolates obtained from diseased strawberry plants in Western Australia that were used in this study.
| Isolate | GenBank accession number | Isolated from | Mean %DI±SDa | Virulenceb |
| WUF-ST-Rhw2 | JQ859847 | Root | 55.9±5.9 | Moderate |
| WUF-ST-Rhw21 | JQ859848 | Root | 45.8±3.2 | Moderate |
| WUF-ST-Rhwf1 | JQ859849 | Root | 27.4±3.7 | Low |
| WUF-ST-Rhwf9 | JQ859850 | Crown | 18.9±5.4 | Low |
| WUF-ST-Rhwf10 | JQ859851 | Root | 20.0±4.3 | Low |
| WUF-ST-Rhwf12 | JQ859852 | Root | 10.9±2.6 | Low |
| WUF-ST-Rhwf13 | JQ859853 | Root | 100 | High |
| WUF-ST-Rhwf14 | JQ859854 | Crown | 84.7±4.9 | High |
| WUF-ST-Rhwf15 | JQ859855 | Crown | 100 | High |
| WUF-ST-Rhwf16 | JQ859856 | Crown | 100 | High |
| WUF-ST-Rhwf17 | JQ859857 | Root | 100 | High |
| WUF-ST-Rhwf18 | JQ859858 | Root | 78.3±4.6 | High |
| WUF-ST-Rhwf19 | JQ859859 | Root | 87.8±6.7 | High |
| WUF-ST-Rhwf20 | JQ859860 | Root | 100 | High |
| WUF-ST-RhT1-1 | JQ859861 | Root | 75.5±4.9 | High |
| WUF-ST-RhT1-2 | JQ859862 | Root | 80.8±6.3 | High |
| WUF-ST-Rhb5 | JQ859863 | Crown | 65.1±4.1 | Moderate |
| WUF-ST-Rhwf4 | JQ859864 | Root | 100 | High |
| WUF-ST-Rhwf5 | JQ859865 | Root | 89.3±5.2 | High |
| WUF-ST-Rhwf24 | JQ859866 | Root | 70.7±3.7 | High |
| WUF-ST-Rhwf25 | JQ859867 | Root | 100 | High |
| WUF-ST-Rhwf26 | JQ859868 | Root | 100 | High |
| WUF-ST-Rhwf27 | JQ859869 | Root | 77.1±8.6 | High |
| WUF-ST-RhT4-8 | JQ859870 | Root | 28.3±2.5 | Low |
| WUF-ST-RhT4-9 | JQ859871 | Root | 11.8±4.0 | Low |
| WUF-ST-RhT4-10 | JQ859872 | Root | 25.7±5.2 | Low |
| WUF-ST-RhT4-12 | JQ859873 | Root | 13.6±2.1 | Low |
| WUF-ST-RhT4-13 | JQ859874 | Root | 31.3±3.2 | Low |
| WUF-ST-RhT4-14 | JQ859875 | Root | 21.6±5.0 | Low |
| WUF-ST-Rhw1 | JQ859876 | Root | 61.0±5.7 | Moderate |
| WUF-ST-Rhw15 | JQ859877 | Root | 100 | High |
| WUF-ST-Rhw16 | JQ859878 | Root | 95.8±6.0 | High |
| WUF-ST-Rhwf2 | JQ859879 | Root | 45.2±5.4 | Moderate |
| WUF-ST-Rhwf3 | JQ859880 | Root | 36.1±1.6 | Moderate |
| WUF-ST-Rhwf6 | JQ859881 | Root | 59.4±3.7 | Moderate |
| WUF-ST-Rhwf7 | JQ859882 | Root | 53.3±2.5 | Moderate |
| WUF-ST-Rhwf8 | JQ859883 | Root | 37.9±3.0 | Moderate |
| WUF-ST-RhT4-2 | JQ859884 | Crown | 27.7±3.3 | Low |
| WUF-ST-RhT4-3 | JQ859885 | Crown | 28.3±4.7 | Low |
| WUF-ST-RhT4-6 | JQ859886 | Root | 58.2±5.9 | Moderate |
| WUF-ST-RhT3-10 | JQ859887 | Root | 51.5±4.9 | Moderate |
| WUF-ST-RhT3-12 | JQ859888 | Root | 34.3±3.8 | Moderate |
| WUF-ST-RhT3-13 | JQ859889 | Crown | 50.2±2.5 | Moderate |
| WUF-ST-RhT3-14 | JQ859890 | Root | 36.9±4.1 | Moderate |
| WUF-ST-RhT3-15 | JQ859891 | Root | 58.5±5.0 | Moderate |
| WUF-ST-RhT3-16 | JQ859892 | Root | 40.1±1.6 | Moderate |
| WUF-ST-RhT3-17 | JQ859893 | Root | 44.6±6.4 | Moderate |
| WUF-ST-RhT4-1 | JQ859894 | Crown | 64.4±3.7 | Moderate |
| WUF-ST-Rhw17 | JQ859895 | Root | 92.8±4.0 | High |
| WUF-ST-Rhw18 | JQ859896 | Root | 100 | High |
| WUF-ST-Rhw19 | JQ859897 | Root | 100 | High |
| WUF-ST-RhT2-6 | JQ859898 | Root | 88.7±5.2 | High |
| WUF-ST-RhT2-7 | JQ859899 | Root | 76.4±5.1 | High |
| WUF-ST-RhT2-8 | JQ859900 | Root | 72.3±4.7 | High |
| WUF-ST-RhT2-9 | JQ859901 | Root | 70.9±1.6 | High |
| WUF-ST-Rhw4 | JQ859902 | Root | 58.7±5.2 | Moderate |
| WUF-ST-Rhw5 | JQ859903 | Root | 44.8±6.0 | Moderate |
| WUF-ST-Rhw6 | JQ859904 | Root | 64.7±3.8 | Moderate |
| WUF-ST-Rhw7 | JQ859905 | Root | 43.4±4.7 | Moderate |
| WUF-ST-Rhw14 | JQ859906 | Root | 53.7±3.8 | Moderate |
| WUF-ST-RhT2-1 | JQ859907 | Root | 31.3±2.5 | Low |
| WUF-ST-RhT2-2 | JQ859908 | Root | 27.5±4.9 | Low |
| WUF-ST-RhT2-5 | JQ859909 | Root | 25.6±3.4 | Low |
| WUF-ST-RhT4-4 | JQ859910 | Root | 22.3±4.7 | Low |
| WUF-ST-RhT4-7 | JQ859911 | Root | 18.7±1.8 | Low |
Percent disease index (%DI).
Isolates were categorized as high virulence if they showed a mean %DI ≥67; moderate virulence if they showed a mean %DI ≥33 and <67; low virulence if they showed a mean %DI <33.
Table 2. Details of reference sequences that were retrieved from GenBank database to evaluate the phylogenetic relationships of the 65 pathogenic binucleate Rhizoctonia isolates in this study.
| Isolate | GenBank accession number | Anastomosis Group | Host plant | Geographic origin | Reference |
| Am1 | DQ102403 | AG-A | Strawberry | USA | [12] |
| Am2 | DQ102414 | AG-A | Strawberry | USA | [12] |
| YWK-83 | FJ440196 | AG-A | Corn | China | Unpublished |
| SIR-2 | AF354091 | AG-A | Sweet potato | Japan | [50] |
| RU56-8 | DQ102417 | AG-A | Soil | USA | [12] |
| C-662 | AF354092 | AG-A | Soil | Japan | [12] |
| Str3 | DQ102421 | AG-A | Strawberry | Israel | [12] |
| Str8 | DQ102422 | AG-A | Strawberry | Israel | [12] |
| Str22 | DQ102423 | AG-A | Strawberry | Israel | [12] |
| Str23 | DQ102424 | AG-A | Strawberry | Israel | [12] |
| Str34 | DQ102425 | AG-A | Strawberry | Israel | [12] |
| R2 | AY927315 | AG-A | Strawberry | Italy | [13] |
| R38 | AY927337 | AG-A | Strawberry | Italy | [13] |
| R55 | AY927347 | AG-A | Strawberry | Italy | [13] |
| FCR2604GB | HM623626 | AG-K | Unknown | China | Unpublished |
| Str24 | DQ102429 | AG-K | Strawberry | Israel | [12] |
| F523 | FJ492158 | AG-K | Sugar beet | Idaho, USA | [18] |
| AC-1 | AB122145 | AG-K | Onion | Hokkaido, Japan | [51] |
| 56D17 | AB286932 | AG-K | Sugar beet | Hokkaido, Japan | [31] |
| SH-10 | AB196652 | AG-K | Soil | Hokkaido, Japan | [31] |
| FA59209 | AJ242900 | AG-K | Unknown | Spain | [31] |
| Gm1 | DQ102395 | AG-G | Strawberry | USA | [12] |
| Gm2 | DQ102397 | AG-G | Strawberry | USA | [12] |
| Str16 | DQ102401 | AG-G | Strawberry | Israel | [12] |
| Str31 | DQ102399 | AG-G | Strawberry | Israel | [12] |
| Str35 | DQ102400 | AG-G | Strawberry | Israel | [12] |
| R1 | AY738627 | AG-G | Strawberry | Italy | [13] |
| R9 | AY927317 | AG-G | Strawberry | Italy | [13] |
| R18 | AY927325 | AG-G | Strawberry | Italy | [13] |
| R22 | AY927327 | AG-G | Strawberry | Italy | [13] |
| R25 | AY927329 | AG-G | Strawberry | Italy | [13] |
| R56 | AY927348 | AG-G | Strawberry | Italy | [13] |
| AH-9 | AB196646 | AG-G | Peanut | Chiba, Japan | [31] |
| Su-1 | AB196647 | AG-G | Peanut | Tokyo, Japan | [30] |
| RU18-1 | DQ102430 | AG-Bo | Soil | USA | [12] |
| RU89-1 | DQ102431 | AG-Bo | Soil | USA | [12] |
| C-302 | AB219143 | AG-Bo | Soil | Fukuoka, Japan | [31] |
| C-484 | AB196641 | AG-Ba | Rice | Miyagi, Japan | [31] |
| Scl-2 | AB286930 | AG-Ba | Rice | Japan | [31] |
| C-314 | AB286931 | AG-Ba | Soil | Fukuoka, Japan | [31] |
| C-350 | AB122144 | AG-Bb | Rice | Fukuoka, Japan | [31] |
| C1 | AJ000192 | AG-Bb | Rice | Malaysia | [52] |
| C6 | AJ000194 | AG-Bb | Rice | Japan | [52] |
| Im1 | DQ102443 | AG-I | Strawberry | USA | [12] |
| Im2 | DQ102444 | AG-I | Strawberry | USA | [12] |
| Ibs | DQ102442 | AG-I | Soil | Israel | [12] |
| 55D21 | AB290023 | AG-I | Sugar beet | Hokkaido, Japan | [31] |
| AV-2 | AJ419932 | AG-I | Mugwort | Japan | [53] |
| 55D25 | AB290021 | AG-C | Sugar beet | Japan | [31] |
| Str47 | DQ102441 | AG-F | Strawberry | Israel | [12] |
| Str51 | DQ102437 | AG-F | Strawberry | Israel | [12] |
| Str56 | DQ102438 | AG-F | Strawberry | Israel | [12] |
| Str110 | DQ102432 | AG-F | Strawberry | Israel | [12] |
| Str111 | DQ102433 | AG-F | Strawberry | Israel | [12] |
| FSR-052 | AY684917 | Athelia rolfsii | Lily | Taiwan | [12], [31] |
Alignment of ITS1-5.8S rDNA-ITS2 sequences of the 65 BNR isolates from this study was performed using the multiple alignment program in Clustal W [40]. The alignment was checked by visual inspection and manual adjustment where appropriate. Phylogenetic trees were constructed based on the multiple alignments (Molecular Evolutionary Genetics Analysis software, Version 5, MEGA 5) [41]. Maximum Likelihood (ML) [42], Maximum Parsimony (MP) [43] and Neighbor-Joining (NJ) [44] were used to construct the phylogenetic trees. The ML analysis was conducted using the Tamura-Nei model [42]. The MP analysis was obtained using the Close-Neighbor-Interchange algorithm [43]. NJ analysis was conducted based on the distance matrix produced by the p-distance model [43]. The robustness of each phylogenetic tree was determined by bootstrapping on the basis of 1000 random samples taken from the multiple sequence alignments. All positions containing gaps and missing data were eliminated. Only nodes with bootstrap values ≥70% were considered to be significant. These processes were repeated after incorporating the sequences of the isolates from this study with reference sequences of isolates from known BNR AG groups from strawberry or other crops across the world. The percent sequence identities of the 65 BNR isolates from this study were determined by analyzing the ITS1-5.8S rDNA-ITS2 sequence of each of these isolates by direct pairwise comparisons. The percent sequence identities of the 65 BNR isolates from this study with reference isolates were also determined by direct pairwise comparisons.
Two isolates were randomly selected from within each of the six different genetic groups revealed by sequence analysis of the 65 BNR isolates from this study to determine if they belonged to different AGs by conducting AG tests using the clean-slide technique [45]. Briefly, a 5 mm-diameter disk from the edge of a 3 day-old colony of a BNR isolate was placed on a sterilized glass slide coated with a thin layer of PDA and a mycelial disk from a similarly grown BNR isolate was placed on the slide at a distance of 2 cm from the first disk. Slides were then maintained on sterile filter papers moistened with sterilized DI water in Petri dishes at 25°C for up to 2 days in the dark. The overlapping hyphae were stained with 0.5% (w/v) safranin O (Sigma-Aldrich, Australia) and 3% (w/v) KOH. Hyphal branches of the stained area were examined microscopically to observe the anastomosis reaction. Further, pairs of the two isolates from between the different genetic groups were also similarly tested.
Statistical analysis
The Mantel test [46] was performed (R, Version 2.15.1, http://www.r-project.org/) with 1000 simulations to determine the association between genetic distance and virulence of the 65 BNR isolates found to be pathogenic on strawberry in this study. The matrix of genetic distance for the 65 BNR isolates was generated by analyzing the ITS1-5.8S rDNA-ITS2 sequence of these isolates using direct pairwise comparisons. The matrix of difference in virulence for the 65 BNR isolates was generated by analyzing the difference in disease index (%DI) of these isolates using direct pairwise comparisons.
Results
Nuclear condition and pathogenicity
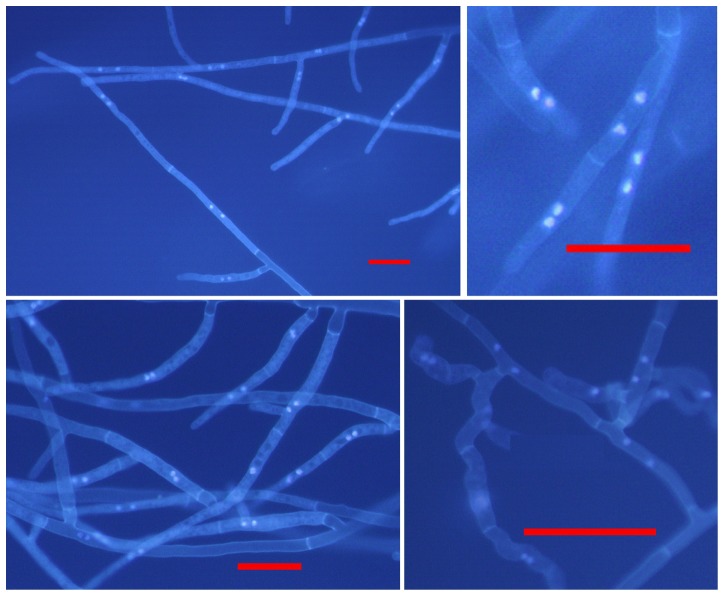
Nuclear staining of each Rhizoctonia isolate showed two nuclei per cell in the hyphae of all the 96 Rhizoctonia isolates tested (Fig. 1). This shows that BNR was the predominant Rhizoctonia recovered from diseased strawberry in Western Australia. Pathogenicity testing of the 96 BNR isolates showed that 65 isolates were pathogenic on strawberry. Plants in infested soil with these isolates showed root discoloration and/or rot symptoms in replicate experiments. In comparison, control plants in uninfested soil were healthy and did not have any root discoloration and/or rot symptoms. The remaining 31 isolates were non-pathogenic. Plants in infested soil with these isolates did not show any root discoloration and/or rot symptoms in replicate experiments and were similar to the control plants in uninfested soil. Of the 65 pathogenic isolates, 25 had high virulence on strawberry with a mean %DI of greater than 70%, 23 isolates had moderate virulence with a mean %DI of 34 to 65%, and 17 isolates were of low virulence with a mean %DI of 10 to 31% (Table 1).
Figure 1. Stained nuclei of representative Rhizoctonia isolates from strawberry in this study.
Fluorescence micrographs of Rhizoctonia isolates stained with 0.48% Hoechst Dye 33258 in 0.1 M KH2PO4 buffer (pH 7.8). Scale bars: 50 µm.
Genetic diversity of the 65 pathogenic binucleate Rhizoctonia isolates and AG determination
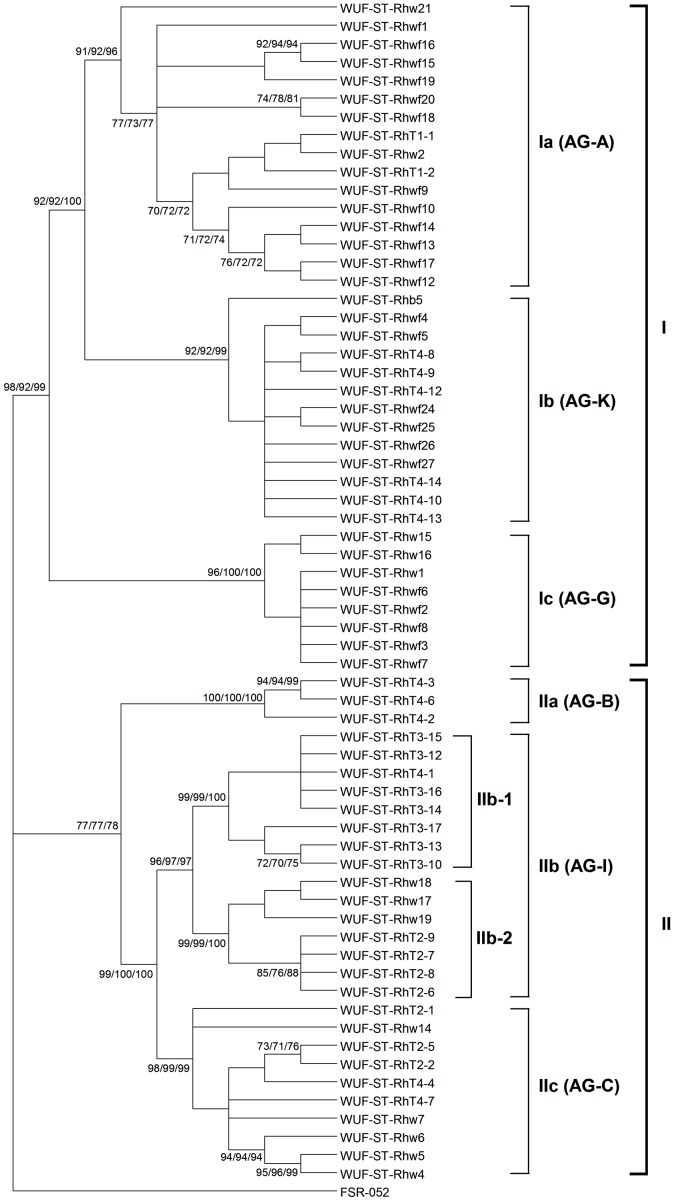
The ITS1-5.8S rDNA-ITS2 sequence from each of the 65 BNR isolates found to be pathogenic on strawberry (Table 1) was used to determine the genetic diversity of the isolates. Sequence analysis using ML, MP and NJ each found the same tree topology (Fig. 2). The 65 isolates clustered into two distinct clades, with clade I including 37 isolates and clade II including 28 isolates. The 37 isolates in clade I further clustered into three distinct subclades (Ia, Ib and Ic), with bootstrap support by ML, MP and NJ all greater than 90% for each subclade. Clade Ia included 16 isolates, clade Ib included 13 isolates and clade Ic included 8 isolates. The 28 isolates in clade II also clustered into three distinct subclades (IIa, IIb and IIc), with bootstrap support by ML, MP and NJ all greater than 95% for each subclade. Subclade IIa only included three isolates, while subclade IIb included 15 isolates and subclade IIc included 10 isolates. Moreover, subclade IIb was further sub-divided into two subclades (Ib-1 and IIb-2), with bootstrap support by ML, MP and NJ all greater than 98% for each.
Figure 2. Genetic relationships among the 65 pathogenic binucleate Rhizoctonia isolates from strawberry in this study based on the internal transcribed spacer sequences.
Trees were constructed using Maximum Likelihood (ML), Maximum Parsimony (MP) and Neighbor-Joining (NJ) analysis. All three methods generated the same tree topology. Tree branches were bootstrapped with 1,000 replications. Numbers at nodes indicate bootstrap values from ML/MP/NJ analysis, respectively. Only bootstrap values ≥70% are shown. The horizontal branch lengths do not represent genetic distance. The tree is rooted with Athelia rolfsii FSR-052 (GenBank Accession No. AY684917) as the outgroup. The AG grouping shown on the right of the figure was based on the closest match of the internal transcribed spacer sequences from the isolates in this study with sequences from isolates of known AG in GenBank using the BLAST algorithm.
Each ITS1-5.8S rDNA-ITS2 sequence of the 65 BNR isolates was compared with sequences in GenBank using the BLAST algorithm to identify the closest match based on maximal identity. None of the ITS1-5.8S rDNA-ITS2 sequences from the 65 BNR isolates were identical with a sequence in the GenBank. In each case, the closest match for every isolate within a clade was found to be in the same AG group (Fig. 2). The ITS1-5.8S rDNA-ITS2 sequences from isolates in clade I were most closely matched to sequences from isolates of AG-A, AG-K and AG-G, while the ITS1-5.8S rDNA-ITS2 sequences from isolates in clade II were most closely matched to sequences from isolates of AG-B, AG-I and AG-C.
The range of the ITS1-5.8S rDNA-ITS2 sequence identity within and between each subclade of the 65 BNR isolates was established by direct pairwise comparisons (Table 3). The sequence identity across all the isolates was 90.1 to 100%. Within the proposed subclades, all the AG-A closely related isolates within subclade Ia had the widest range of sequence identities of 94.4 to 100%, followed by AG-C closely related isolates within subclade IIc which had sequence identities of 96.3 to 100%, and AG-K closely related isolates within subclade Ib which had sequence identities of 98.4 to 100%. All the AG-G closely related isolates within subclade Ic, AG-B closely related isolates within subclade IIa and AG-I closely related isolates within subclade IIb had the sequence identity greater than 99%. Within clades I and II, all isolates had the same sequence identity of 93.0 to 100%. Between the proposed subclades, sequence identities ranged from were 90.1 to 99.3%.
Table 3. Ranges of percent sequence identity of the internal transcribed spacer sequences of isolates within and between clades (Fig. 2) of the 65 pathogenic binucleate Rhizoctonia isolates in this study.
| Ia | Ib | Ic | IIa | IIb | IIc | Outgroup | |||
| IIb-1 | IIb-2 | ||||||||
| Ia | 94.4–100 | ||||||||
| Ib | 93.0–99.1 | 98.4–100 | |||||||
| Ic | 93.3–97.2 | 96.5–97.2 | 99.8–100 | ||||||
| IIa | 90.5–95.6 | 94.0–96.0 | 93.7–94.5 | 99.3–100 | |||||
| IIb | IIb-1 | 90.8–94.7 | 93.7–95.1 | 94.4–94.7 | 95.1–96.0 | 99.8–100 | |||
| IIb-2 | 91.5–95.2 | 93.8–95.2 | 94.9–95.2 | 95.2–96.1 | 98.4–98.8 | 99.8–100 | |||
| IIc | 90.1–95.1 | 92.3–95.4 | 92.6–95.1 | 93.0–96.0 | 96.8–99.3 | 96.3–98.8 | 96.3–100 | ||
| Outgroup | 78.3–80.6 | 79.0–80.1 | 81.0–81.2 | 78.7–79.0 | 80.1–80.3 | 79.8–79.9 | 77.6–79.9 | 100 | |
AG tests were conducted by randomly selecting two isolates from within each of the six different genetic clades (viz. Ia, Ib, Ic, IIa, IIb and IIc) shown in Fig. 2. In each case, the two isolates selected from within the same genetic clade showed an anastomosis reaction, confirming their genetic similarity. In contrast, none of the pairs of the isolates selected from between the different genetic clades showed an anastomosis reaction, confirming their distinct genetic groupings.
Distribution of the 65 BNR isolates in each genetic clade based on virulence
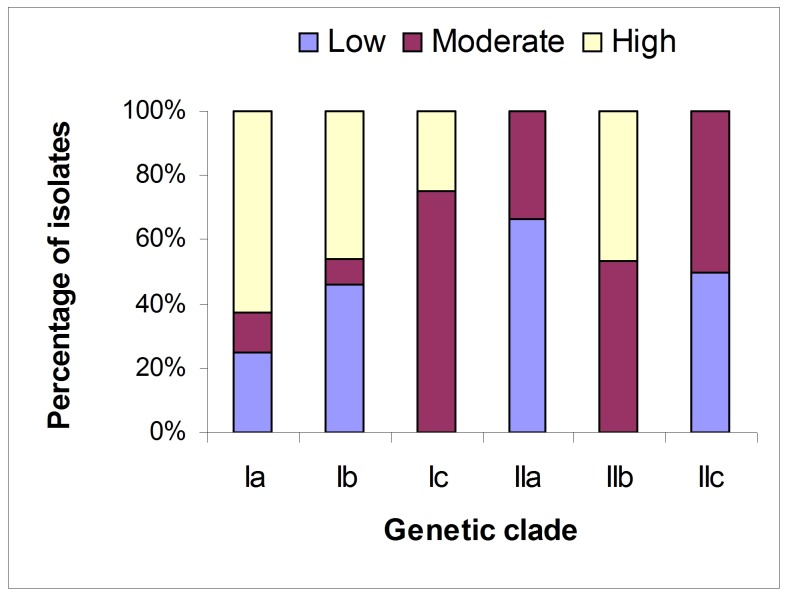
There was no significant association between genetic diversity and virulence of the 65 pathogenic BNR isolates (r = −0.0974, p = 0.6109, Mantel test). None of the observed genetic clades contained isolates from only one category of virulence (Fig. 3). Clades Ia and Ib contained representative isolates from all the three categories of virulence, while the other clades contained representative isolates from two categories of virulence. However, more than half of the isolates with high virulence were distributed in clade Ia, and more than half of the isolates with moderate virulence were distributed in clade Ic. Clades IIb and IIc had about a 50∶50 split between two virulence categories, with moderate/high and moderate/low virulence, respectively. Finally, no clades had only representative isolates from the high/low virulence categories.
Figure 3. Distribution of the 65 pathogenic binucleate Rhizoctonia isolates in each genetic clade based on virulence.
Low, moderate and high represent the virulence level of the isolates. Ia, Ib, Ic, IIa, IIb, and IIc represent genetic clades shown in Fig. 2.
Phylogenetic analysis of the 65 pathogenic binucleate Rhizoctonia isolates
The phylogenetic relationships of the 65 pathogenic BNR isolates from this study with the reference BNR isolates from across the world were determined. All the reference isolates included in the analysis were isolates for which the anastomosis group had been determined and the ITS1-5.8S rDNA-ITS2 sequence was available in GenBank. Preliminary analysis found that all reference BNR isolates belonging to AG-A, AG-K and AG-G grouped in clade I, while all reference BNR isolates belonging to AG-B, AG-I and AG-C grouped in clade II. Therefore, isolates in clade I and clade II were analyzed separately.
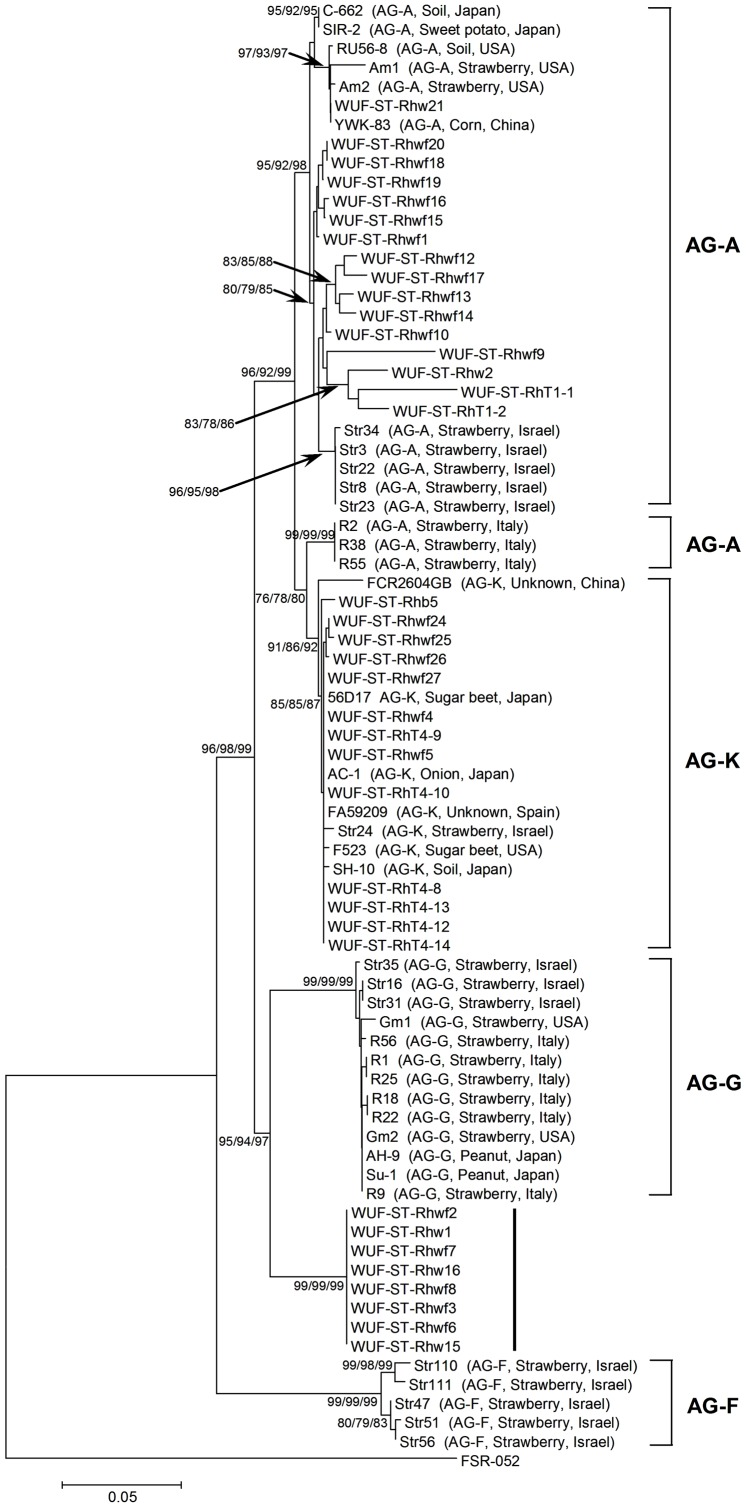
The phylogenetic relationships of the 37 BNR isolates in clade I (Fig. 2) and the reference BNR isolates belonging to AG-A, AG-K, AG-G and AG-F from across the world were determined. Phylogenetic analysis using ML, MP and NJ all found the same tree topology (Fig. 4). The 16 AG-A closely related isolates in subclade Ia clustered into a distinct clade containing nearly all of the reference isolates known to be AG-A. These included isolates from strawberry in the USA and Israel, corn in China, sweet potato in Japan and soil in the USA and Japan. This clade had bootstrap support of 95%, 92% and 98% by ML, MP and NJ, respectively, suggesting that these 16 isolates also belonged to AG-A. The 16 isolates had 94.8 to 99.6% sequence identity with the AG-A reference isolates from strawberry in Israel, and 93.7 to 100% sequence identity with the other reference isolates in this clade. Interestingly, three AG-A reference isolates from strawberry in Italy clustered into a clade that was sister to a clade containing AG-K reference isolates, with bootstrap support of 76%, 78% and 80% by ML, MP and NJ, respectively. The 13 AG-K closely related isolates in subclade Ib clustered into a distinct clade along with all reference isolates known to be AG-K from a variety of crops and geographic origins. This clade had bootstrap support of 91%, 86% and 92% by ML, MP and NJ, respectively, suggesting that these 13 isolates also belonged to AG-K. These 13 isolates had greater than 99.6% sequence identity with the AG-K reference isolates. The eight AG-G closely related isolates in subclade Ic clustered in a distinct clade along with the AG-G reference isolates from strawberry from a variety of geographic origins and peanut in Japan, with bootstrap support of 95%, 94% and 97% by ML, MP and NJ, respectively. However, these eight isolates formed a distinct rooted subclade with a bootstrap support of 99% by each ML, MP and NJ. This clear separation suggested that these eight isolates may represent a new subgroup of AG-G or a new AG group. The sequence identity between these eight isolates and the AG-G reference isolates were 96.0 to 96.7%. None of these BNR isolates in clade I from this study grouped with the AG-F reference isolates from strawberry in Israel.
Figure 4. Phylogenetic relationships of the 37 pathogenic binucleate Rhizoctonia isolates in clade I ( Fig. 2 ) from this study with reference isolates from across the world based on the internal transcribed spacer sequences.
Trees were constructed using Maximum Likelihood (ML), Maximum Parsimony (MP) and Neighbor-Joining (NJ) analysis. All three methods generated the same tree topology. Tree branches were bootstrapped with 1,000 replications. Numbers at nodes indicate bootstrap values from ML/MP/NJ analysis, respectively. Only bootstrap values ≥70% are shown. Scale bar represents a genetic distance of 0.05 for horizontal branch lengths. The tree is rooted with isolate Athelia rolfsii FSR-052 (GenBank Accession No. AY684917) as the outgroup. The reference isolates are shown as isolate name followed by anastomosis group (AG), host and geographic origin of the isolate in parenthesis. The 37 isolates from this study are shown as isolate name only.
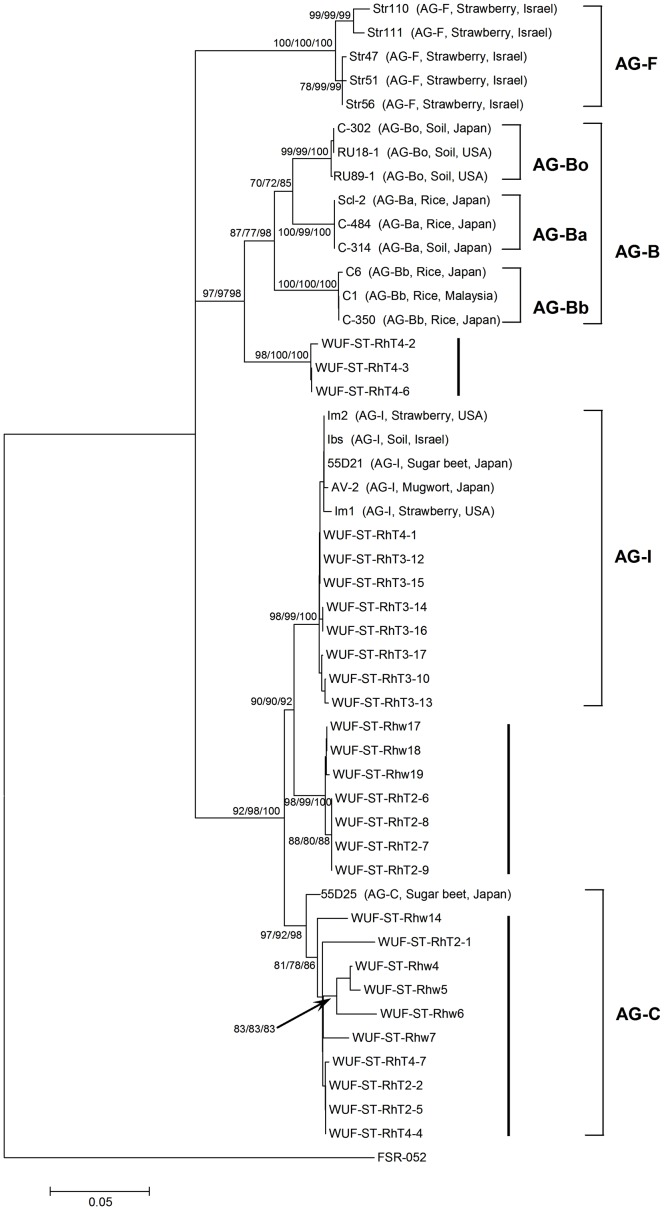
The phylogenetic relationships of the 28 BNR isolates in clade II (Fig. 2) and the reference BNR isolates belonging to AG-B, AG-I, AG-C and AG-F from across the world were determined. Phylogenetic analysis using ML, MP and NJ again all found the same tree topology (Fig. 5). The three AG-B closely related isolates in subclade IIa clustered into a distinct subclade that was sister to a distinct clade containing all the AG-B reference isolates, with the bootstrap support of 97% by both ML and MP, and 98% by NJ. These three isolates had 96.3 to 97.7% sequence identity with the AG-B reference isolates. The AG-B reference isolates clustered into three distinct subclades according to their known subgrouping (AG-Bo, AG-Ba and AG-Bb), with bootstrap support of 99 to 100% by ML, MP and NJ for each. This indicated that the three isolates from this study belonged to a new subgroup of AG-B or a new AG group. The 15 AG-I closely related isolates in subclade IIb clustered into two distinct subclades analogous to IIb-1 and IIb-2 with bootstrap support of 98 to 100% by ML, MP and NJ for each (Fig. 5). The eight isolates in clade IIb-1 clustered into a distinct clade along with all AG-I reference isolates from strawberry, sugar beet, mugwort or soil across a variety of geographic origins, and had greater than 99% sequence identity with the AG-I reference isolates, suggesting that these eight isolates belonged to AG-I. The subclade analogous to subclade IIb-2 contained another seven isolates from this study that may represented a new subgroup of AG-I. These seven isolates had 98.6 to 98.8% sequence identity with the AG-I reference isolates. The 10 AG-C closely related isolates from subclade IIc clustered into a distinct clade along with one AG-C reference isolate from sugar beet in Japan, with bootstrap support of 97%, 92% and 98% by ML, MP and NJ, respectively. This suggested these 10 isolates belong to AG-C. There was 97.3 to 100% sequence identity between these 10 isolates and the AG-C reference isolate. None of the BNR isolates in clade II from this study grouped with the AG-F reference isolates from strawberry in Israel.
Figure 5. Phylogenetic relationships of the 28 pathogenic binucleate Rhizoctonia isolates in clade II ( Fig. 2 ) from this study with reference isolates from across the world based on the internal transcribed spacer sequences.
Trees were constructed using Maximum Likelihood (ML), Maximum Parsimony (MP) and Neighbor-Joining (NJ) analysis. All three methods generated the same tree topology. Tree branches were bootstrapped with 1,000 replications. Numbers at nodes indicate bootstrap values from ML/MP/NJ analysis, respectively. Only bootstrap values ≥70% are shown. Scale bar represents a genetic distance of 0.05 for horizontal branch lengths. The tree is rooted with isolate Athelia rolfsii FSR-052 (GenBank Accession No. AY684917) as the outgroup. The reference isolates are shown as isolate name followed by anastomosis group (AG), host and geographic origin of the isolate in parenthesis. The 28 isolates from this study are shown as isolate name only.
Discussion
All Rhizoctonia spp. isolates recovered from diseased strawberry plants in this study were binucleate Rhizoctonia (BNR). BNR isolates have been reported as the main isolates recovered from diseased strawberry plants in the USA [5], [6], South Africa [11], Israel [12] and Italy [13]. BNR isolates associated with root rot of strawberry are generally referred to as R. fragariae [11], [12], but it is still not clear if BNR only contains one species or rather should be classified into several additional species. While MNR has also been reported recovered from diseased strawberry plants in the USA, South Africa and Israel, this was only at a very low frequency compared with that of BNR [6], [11], [12].
In this study, 65 of the 96 BNR isolates recovered from diseased strawberry were pathogenic on strawberry. It has been reported that BNR isolates recovered from diseased strawberry are also pathogenic on strawberry in other countries, such as in the USA [5], [6], South Africa [11], Israel [12] and Italy [13]. Therefore, this study supports the view that BNR constitutes the major pathogen associated with root rot of strawberry in the world. However, the pathogenic BNR isolates in this study showed a wide variation in virulence, with about 38% of the isolates having high virulence.
It is noteworthy that BNR isolates from this study with the closest match to isolates of the same AG were clustered into the same clade, and the same clade included all the reference isolates from the same AG when reference isolates were included in the analysis. AG determinations confirmed that the BNR isolates from this study that grouped within the same clade belonged to the same AG, while isolates in other clades belonged to different AGs. It has been demonstrated previously that grouping of Rhizoctonia isolates by molecular analysis based on the ITS sequences supports the AG grouping of Rhizoctonia isolates based on classical hyphal anastomosis reactions [12], [15], [16], [30]. This study confirms that molecular analysis based on the ITS sequences is appropriate for evaluating genetic diversity and characterizing potential AG groups of Rhizoctonia isolates.
ITS sequence analysis and AG determination indicated that there were at least six different genetic groups among the pathogenic BNR isolates associated with root rot of strawberry in Western Australia. These groups were inferred to be AG-A, AG-K, AG-G, AG-B, AG-I and AG-C. In addition, the potentials for several new AG groups/subgroups have been highlighted. The two most likely new groups/subgroups are those sister to AG-G and AG-B. The evidence that these may indeed be new groups/subgroups comes from the observation that the split of the new clade from AG-G is more deeply rooted than the split between AG-A and AG-K, with bootstrap support of 99% by each ML, MP and NJ; and that the split of the new clade from AG-B is more deeply rooted than the split between AG-I and AG-C, with bootstrap support of 98% by ML and 100% by both MP and NJ. Another possible new subgroup is that sister to AG-I. The split between this new clade and AG-I is slightly more deeply rooted than the split between AG-I and AG-C, with bootstrap support of 98%, 99% and 100% by ML, MP and NJ, respectively. Finally, the clade of Western Australian isolates that is sister to the clade that includes one AG-C reference isolate may also represent a new AG group/subgroup, although the argument is weaker in this case as there are very few sequences of known AG-C isolates in GenBank available for comparison. These results reveal that there is wide genetic diversity of the pathogenic BNR isolates associated with root rot of strawberry in Western Australia. For example, BNR associated with root rot of strawberry in the USA and South Africa only belong to three genetic groups (viz. AG-A, AG-G and AG-I) [5], [6], [11]. In Italy, only two genetic groups (viz. AG-A and AG-G) are associated with root rot of strawberry [13]. Four genetic groups (viz. AG-A, AG-G, AG-K and AG-F) are associated with root rot of strawberry in Israel [12], but this diversity is still lower than that observed in Western Australia. Therefore, BNR isolates associated with root rot of strawberry in Western Australia are of wide variation in genetic diversity.
This study also showed that there was wide variation in virulence of the pathogenic BNR isolates associated with root rot of strawberry in Western Australia. However, we only used pasteurized soil to test the virulence of the isolates. The virulence of pathogens such as multinucleate R. solani, and the activity of antagonistic soil microorganisms on suppressing disease are influenced by soil media [47], [48]. For this reason, it is possible that the observed virulence levels may have been somewhat elevated compared to those in non-pasteurized soil. Despite this consideration, we still did not find an association between genetic diversity and virulence of these BNR isolates. However, this was not surprising as the ITS sequencing in this study was done using generic primers that highlight genetic variation and that were not related to specific virulence or infection-related functions of these BNR isolates.
The lack of association between genetic diversity and virulence of these isolates makes it much more complex in terms of trying to define the distributions of different BNR groups and populations of varying virulence, increasing the challenge of effectively managing root rot of strawberry in Western Australia, such as in selecting, developing and deploying new cultivars with resistance to these different multi-genetic groups of BNR. The soil-borne nature of BNR [15], [20], and the ability of BNR to live also as a saprophyte [30], indicate that any selection associated with the infection of a plant host may not quickly reduce and/or eliminate virulent biotypes, especially those with low or moderate virulence. In addition to virulence on strawberry, biotypes with low or moderate virulence may have a selective advantage when growing as saprophytes. The saprophytic ability and virulence on other hosts may contribute to the observed population structure of the BNR isolates associated with root rot of strawberry in Western Australia.
Our study also highlighted a relatively wide range of ITS sequence identity among the 65 BNR isolates from this study, not only between the different groups of the BNR isolates, but also within the groups of AG-A and AG-C. There was 90.1 to 99.1% sequence identity among the different AG groups. There was 94.4 to 100% sequence identity among the BNR isolates within the group of AG-A, and 96.3 to 100% sequence identity among the BNR isolates within the group of AG-C. Some previous attempts have been made to determine the thresholds of ITS sequence identity for differentiating isolates belonging to different AG groups. However, robust thresholds could not be consistently obtained because there were overlaps between the ranges of ITS sequence identities for isolates within groups and the ranges between groups [12], [31]. The relatively wide range of sequence identity within a genetic group indicates the possible existence of subgroups [12], [31].
Of the six different genetic groups inferred from the BNR isolates associated with root rot of strawberry in Western Australia, we believe that this is the first record worldwide for isolates of AG-B being recovered from diseased strawberry plants and also being pathogenic on strawberry. Moreover, these three isolates showed two levels of virulence, with one isolate of moderate virulence and two others of low virulence. This disagrees with a previous report that isolates of AG-B (AG-Bo RU18-1 and RU89-1) from soil in the USA are non-pathogenic on strawberry [12]. For isolates of AG-C, we confirmed an earlier preliminary report of Fang et al. [35] who were the first to report AG-C as a pathogen of strawberry. These AG-C isolates showed two levels of virulence, with half having moderate virulence and half having low virulence, Isolates of AG-G recovered from diseased strawberry plants represent the first time this group has been shown as a pathogen on strawberry plants in Australia. Two of these AG-G isolates had high virulence, while the remainder had moderate virulence. AG-G has been reported as one common group associated with root rot of strawberry elsewhere in the world [5], [6], [11]–[13]. The majority of the AG-A isolates in Western Australia were of high virulence on strawberry; BNR AG-A is another common group associated with root rot of strawberry elsewhere in the world [5], [6], [11]–[13]. AG-I has been reported associated with root rot of strawberry in the USA and South Africa [5], [6], [11], and also was associated with root rot of strawberry in Western Australia. Previously, only a single isolate of AG-K had been found associated with root rot of strawberry in Israel [12], but in our study, AG-K was one of the main groups associated with root rot of strawberry in Western Australia. However, none of the BNR isolates associated with root rot of strawberry in Western Australia belong to AG-F, a group which only has previously been reported associated with root rot of strawberry in Israel [12].
While currently available ITS sequences from BNR isolates remain quite limited in public databases, increasing the number of sequences in the future should allow even more groups or subgroups of BNR isolates to be defined, and also will be useful for further studies on the genetic diversity of BNR isolates associated with root rot of strawberry worldwide. In particular, additional sequences from other DNA loci such as the beta-tubulin gene and the 28S large subunit of rDNA region, that have been used to determine the genetic diversity and phylogeny of multinucleate R. solani [49], could be useful in future not only to further confirm the genetic diversity of BNR isolates associated with root disease of strawberry. but perhaps more importantly in helping to further confirm and delineate the new AG groups or subgroups of BNR as highlighted in this study. Together, this will make it possible not only to improve the current knowledge about BNR as an agent of strawberry root rot worldwide, but also will help the development of novel management strategies on such a serious disease.
In conclusion, this study provides, for the first time, detailed information on the virulence, genetic diversity and phylogenetic status of BNR isolates associated with root rot of strawberry in Western Australia. ITS sequence analysis and AG determination indicated the existence of at least six genetically distinct groups. In addition, the potential for several new AG groups or subgroups of AG-B, AG-G, AG-I and AG-C have been highlighted. This study demonstrates that BNR isolates associated with root rot of strawberry in Western Australia are of wide genetic diversity, with the existence of some potential new genetic groups highlighted for the first time in the world. The wide variation in virulence and genetic diversity, and the lack of an association between genetic diversity and virulence of these BNR isolates associated with root rot of strawberry in Western Australia, together make this disease very challenging to manage. Despite this, the wide variation in virulence and genetic diversity identified in this study will be of high value for strawberry breeding programs in selecting, developing and deploying new cultivars with resistance to these multi-genetic groups of BNR.
Acknowledgments
We thank Dr. Xuanli Ma and Ms. Jessie Moniodis for their advice and discussion on sequence analysis. We also acknowledge the half salary support for MJB provided by the Department of Agriculture and Food Western Australia.
Funding Statement
This work was supported by the Australia Research Council and the Department of Agriculture and Food Western Australia. XLF gratefully acknowledges the financial assistance of the China Scholarship Council and the University of Western Australia by a jointly awarded PhD Scholarship. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Faostat (2010) Available: http://faostat.fao.org/. [Google Scholar]
- 2. Bombarely A, Merchante C, Csukasi F, Cruz-Rus E, Caballero JL, et al. (2010) Generation and analysis of ESTs from strawberry (Fragaria xananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watanabe T (1977) Fungi associated with strawberry roots in Japan. T Mycol Soc Jpn 18: 251–256. [Google Scholar]
- 4. Matsumoto M, Yoshida T (2006) Characterization of isolates of binucleate Rhizoctonia spp. associated with strawberry black root rot complex using fatty acid methyl ester (FAME) profiles. J Gen Plant Pathol 72: 318–322. [Google Scholar]
- 5. Martin SB (1988) Identification, isolation frequency, and pathogenicity of anastomosis groups of binucleate Rhizoctonia spp. from strawberry roots. Phytopathology 78: 379–384. [Google Scholar]
- 6. Martin FN (2000) Rhizoctonia spp. recovered from strawberry roots in central coastal California. Phytopathology 90: 345–353. [DOI] [PubMed] [Google Scholar]
- 7. Wilhelm S, Paulus AO (1980) How soil fumigation benefits the California strawberry industry. Plant Dis 64: 264–270. [Google Scholar]
- 8. Yuen GY, Schroth MN, Weinhold AR, Hancock JG (1991) Effects of soil fumigant with methyl bromide and chloropicrin on root health and yield of strawberry. Plant Dis 75: 416–420. [Google Scholar]
- 9. Porter IJ, Brett RW, Wiseman BM (1999) Alternatives to methyl bromide: chemical fumigants or integrated pest management systems? Australas Plant Path 28: 65–71. [Google Scholar]
- 10. Fang XL, Phillips D, Li H, Sivasithamparam K, Barbetti MJ (2011) Severity of crown and root diseases of strawberry and associated fungal and oomycete pathogens in Western Australia. Australas Plant Path 40: 109–119. [Google Scholar]
- 11. Botha A, Denman S, Lamprecht SC, Mazzola M, Crous PW (2003) Characterisation and pathogenicity of Rhizoctonia isolates associated with black root rot of strawberries in the Western Cape Province, South Africa. Australas Plant Path 32: 195–201. [Google Scholar]
- 12. Sharon M, Freeman S, Kuninaga S, Sneh B (2007) Genetic diversity, anastomosis groups and virulence of Rhizoctonia spp. from strawberry. Eur J Plant Pathol 117: 247–265. [Google Scholar]
- 13. Manici LM, Bonora P (2007) Molecular genetic variability of Italian binucleate Rhizoctonia spp. isolates from strawberry. Eur J Plant Pathol 118: 31–42. [Google Scholar]
- 14.Cubeta MA, Vilgalys R (2000) Rhizoctonia. In: Lederberg J, editor. Encyclopedia of Microbiology. San Diego, CA : Academic Press. pp. 109–116 [Google Scholar]
- 15. Rinehart TA, Copes WE, Toda T, Cubeta MA (2007) Genetic characterization of binucleate Rhizoctonia species causing web blight on azalea in Mississippi and Alabama. Plant Dis 91: 616–623. [DOI] [PubMed] [Google Scholar]
- 16. Kuramae EE, Buzeto AL, Nakatani AK, Souza NL (2007) rDNA-based characterization of a new binucleate Rhizoctonia spp. causing root rot on kale in Brazil. Eur J Plant Pathol 119: 469–475. [Google Scholar]
- 17. Pannecoucque J, Van Beneden S, Hofte M (2008) Characterization and pathogenicity of Rhizoctonia isolates associated with cauliflower in Belgium. Plant Pathol 57: 737–746. [Google Scholar]
- 18. Strausbaugh CA, Eujayl IA, Panella LW, Hanson LE (2011) Virulence, distribution and diversity of Rhizoctonia solani from sugar beet in Idaho and Oregon. Can J Plant Pathol 33: 210–226. [Google Scholar]
- 19.Katan J (1996) Soil solarization for the control of diseases caused by Rhizoctonia spp. In: Sneh B, Jabaji-Hare S, Neate S, Dijst G, editors. Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control. Boston: Kluwer Academic Publisher. pp. 423–432 [Google Scholar]
- 20. Ohkura M, Abawi GS, Smart CD, Hodge KT (2009) Diversity and aggressiveness of Rhizoctonia solani and Rhizoctonia-like fungi on vegetables in New York. Plant Dis 93: 615–624. [DOI] [PubMed] [Google Scholar]
- 21. Himelrick D, Dozier W (1991) Soil fumigation and soil solarization in strawberry production. Advances in strawberry production 10. [Google Scholar]
- 22. Martin FN, Bull CT (2002) Biological approaches for control of root pathogens of strawberry. Phytopathology 92: 1356–1362. [DOI] [PubMed] [Google Scholar]
- 23. Subbarao KV, Kabir Z, Martin FN, Koike ST (2007) Management of soilborne diseases in strawberry using vegetable rotations. Plant Dis 91: 964–972. [DOI] [PubMed] [Google Scholar]
- 24. Fravel D, Olivain C, Alabouvette C (2003) Fusarium oxysporum and its biocontrol. New Phytol 157: 493–502. [DOI] [PubMed] [Google Scholar]
- 25. Fang X, Kuo J, You MP, Finnegan PM, Barbetti MJ (2012) Comparative root colonisation of strawberry cultivars Camarosa and Festival by Fusarium oxysporum f. sp. fragariae . Plant Soil 358: 75–89. [Google Scholar]
- 26. Fang XL, Phillips D, Verheyen G, Li H, Sivasithamparam K, et al. (2012) Yields and resistance of strawberry cultivars to crown and root diseases in the field and cultivar responses to pathogens under controlled environmental conditions. Phytopathol Mediterr 51: 69–84. [Google Scholar]
- 27. Particka CA, Hancock JF (2005) Field evaluation of strawberry genotypes for tolerance to black root rot on fumigated and nonfumigated soil. J Am Soc Hortic Sci 130: 688–693. [Google Scholar]
- 28.Sneh B, Burpee LL, Ogoshi A (1991) Identification of Rhizoctonia species. St. Paul, MN: APS Press. [Google Scholar]
- 29. Sharon M, Kuninaga S, Hyakumachi M, Sneh B (2006) The advancing identification and classification of Rhizoctonia spp. using molecular and biotechnological methods compared with the classical anastomosis grouping. Mycoscience 47: 299–316. [Google Scholar]
- 30. Hyakumachi M, Priyatmojo A, Kubota M, Fukui H (2005) New anastomosis groups, AG-T and AG-U, of binucleate Rhizoctonia spp. causing root and stem rot of cut-flower and miniature roses. Phytopathology 95: 784–792. [DOI] [PubMed] [Google Scholar]
- 31. Sharon M, Kuninaga S, Hyakumachi M, Naito S, Sneh B (2008) Classification of Rhizoctonia spp. using rDNA-ITS sequence analysis supports the genetic basis of the classical anastomosis grouping. Mycoscience 49: 93–114. [Google Scholar]
- 32. Lehtonen MJ, Ahvenniemi P, Wilson PS, German-Kinnari M, Valkonen JPT (2008) Biological diversity of Rhizoctonia solani (AG-3) in a northern potato-cultivation environment in Finland. Plant Pathol 57: 141–151. [Google Scholar]
- 33. Feng J, Hwang R, Chang KF, Hwang SF, Strelkov SE, et al. (2010) Genetic variation in Fusarium avenaceum causing root rot on field pea. Plant Pathol 59: 845–852. [Google Scholar]
- 34. Yang HA, Sivasithamparam K, O'Brien P (1991) An improved technique for fluorescence staining of fungal nuclei and septa. Australas Plant Path 20: 119–121. [Google Scholar]
- 35. Fang XL, Phillips D, Li H, Sivasithamparam K, Barbetti MJ (2011) Comparisons of virulence of pathogens associated with crown and root diseases of strawberry in Western Australia with special reference to the effect of temperature. Sci Hortic 131: 39–48. [Google Scholar]
- 36. Barbetti MJ, Sivasithamparam K (1987) Effects of soil pasteurization on root rot, seedling survival and plant dry weight of subterranean clover inoculated with six fungal root pathogens. Aust J Agric Res 38: 317–327. [Google Scholar]
- 37. Cenis JL (1992) Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res 20: 2380–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. New York: Academic Press. [Google Scholar]
- 39.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; 1999. pp. 95–98. [Google Scholar]
- 40. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial-DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526. [DOI] [PubMed] [Google Scholar]
- 43.Nei M, Kumar S (2000) Molecular Evolution and Phylogenetics. New York: Oxford University Press. [Google Scholar]
- 44. Saitou N, Nei M (1987) The Neighbor-Joining method - a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 45. Mathew FM, Lamppa RS, Chittem K, Chang YW, Botschner M, et al. (2012) Characterization and pathogenicity of Rhizoctonia solani isolates affecting Pisum sativum in North Dakota. Plant Dis 96: 666–672. [DOI] [PubMed] [Google Scholar]
- 46. Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220. [PubMed] [Google Scholar]
- 47. Krause MS, Madden LV, Hoitink HAJ (2001) Effect of potting mix microbial carrying capacity on biological control of Rhizoetonia damping-off of radish and Rhizoctonia crown and root rot of Poinsettia. Phytopathology 91: 1116–1123. [DOI] [PubMed] [Google Scholar]
- 48. Watanabe K, Matsui M, Honjo H, Becker JO, Fukui R (2011) Effects of soil pH on rhizoctonia damping-off of sugar beet and disease suppression induced by soil amendment with crop residues. Plant Soil 347: 255–268. [Google Scholar]
- 49. Gonzalez D, Cubeta MA, Vilgalys R (2006) Phylogenetic utility of indels within ribosomal DNA and beta-tubulin sequences from fungi in the Rhizoctonia solani species complex. Mol Phylogenet Evol 40: 459–470. [DOI] [PubMed] [Google Scholar]
- 50. Gonzalez D, Carling DE, Kuninaga S, Vilgalys R, Cubeta MA (2001) Ribosomal DNA systematics of Ceratobasidium and Thanatephorus with Rhizoctonia anamorphs. Mycologia 93: 1138–1150. [Google Scholar]
- 51. Toda T, Mghalu JM, Priyatomojo A, Hyakumachi M (2004) Comparison of sequences for the internal transcribed spacer region in Rhizoctonia solani AG 1-ID and other subgroups of AG 1. J Gen Plant Pathol 70: 270–272. [Google Scholar]
- 52. Johanson A, Turner HC, McKay GJ, Brown AE (1998) A PCR-based method to distinguish fungi of the rice sheath-blight complex, Rhizoctonia solani, R. oryzae and R. oryzae sativae1. Fems Microbiol Lett 162: 289–294. [DOI] [PubMed] [Google Scholar]
- 53. Gronberg H, Paulin L, Sen R (2003) ITS probe development for specific detection of Rhizoctonia spp. and Suillus bovinus based on Southern blot and liquid hybridization-fragment length polymorphism. Mycol Res 107: 428–438. [DOI] [PubMed] [Google Scholar]