SUMMARY
Pinpointing a specific cell from within a relatively uniform cell population to determine its chemical content presents a challenging bioanalytical task. Immunocytochemistry is the classical method used to localize specific molecules and hence, selected cells. Mass spectrometry also probes endogenous molecules such as neuropeptides within a cell. Here, these two approaches are hyphenated to allow microchemical analysis of immunocytochemical-selected peptidergic neurons. This two-step strategy utilizes antibody-based localization of cells containing selected biomarkers to isolate the cell(s) of interest, followed by peptidomic analysis via mass spectrometry. Applicable to a broad range of analyte and cell types, the strategy was used to successfully profile neuropeptides from individual immunostained insect neurons stored for up to two weeks as well as from tissues preserved for 42 weeks.
Keywords: peptidomics, paraformaldehyde-fixed immunolabeled (PFIL) samples, single-cell analysis, MALDI-TOF mass spectrometry, cell culture, neuropeptides, insect
INTRODUCTION
Individual cells play crucial roles in the embryogenesis, regeneration, and organization of neuronal networks. Understanding the role of single-cells in the etiology and development of disease can be essential for the development of pharmacological treatments for a variety of illnesses, including cancer. Single-cell microchemical analysis is becoming increasingly important in modern biomedical genomic, metabolomic and proteomic research (e.g., Garden et al. 1996; Rubakhin et al. 2003; Neupert and Predel 2005; Predel et al. 2007; Morano et al. 2008; Rubakhin and Sweedler, 2008; Wang and Bodovitz, 2010; Rubakhin et al. 2011), and has recently been selected as the focus of several new NIH Common Fund initiatives [http://commonfund.nih.gov/singlecell/]. The success of single-cell characterization studies oftentimes depends on the availability of ‘omics’ technologies that are capable of analyzing micrometer-sized structures and compounds at the attomole level, as well as the ability to target a specific cell among thousands to millions of similar cells.
Single-cell ‘omics’ has made exceptional progress in recent years (Wang and Bodovitz, 2010; Rubakhin et al., 2011). Multiple unlabeled and labeled analytes can be detected in both a qualitative and quantitative manner (Morano et al., 2008; Rubahkin and Sweedler, 2008), with compounds located in single-cells often identified by direct tandem mass spectrometry (MS/MS) and fragment analysis (Neupert et al., 2005a). Nonetheless, despite the significant progress in single-cell mass spectrometry (SCMS), locating target cells among morphologically similar cells remains a challenge. A key factor for successful cell sampling is the availability and accessibility of sufficient amounts of analytes for desorption and ionization processes. There are several effective methods of target cell identification and visualization typically applied in SCMS that allow identification and visualization of specific cells by means of dye delivery or endogenous expression of fluorescent proteins. Options include retrograde cell tracing with dyes (Neupert et al., 2005a), dye injections into electrophysiologically identified cells (Romanova et al. 2004), dye injections to projection areas of neuronal ganglia (Predel et al., 2007), and molecular biology-assisted expression of intracellular molecular markers (Neupert et al. 2007). However, these approaches do have some limitations in their ability to detect cells that express smaller molecules (e.g., metabolites and peptides) as biomarkers.
Affinity probes such as antibodies and aptamers are widely used to determine the localization of a large variety of intracellular compounds ranging from small metabolites to large protein complexes (Zeng et al. 2010). Unfortunately, the majority of these probes are large in size and have to enter and leave the cell of interest to achieve specific labeling. This leads to a need to permeabilize the cellular membranes — performed by crosslinking using different chemical agents — while keeping the molecules of interest in vivo at their original location. One of most common technologies employed to visualize target cells is immunocytochemistry (ICC), which often uses formalin or paraformaldehyde (PFA) fixation to preserve the spatial localization of analytes. However, crosslinking makes many compounds inaccessible for MS-based peptidomics and proteomics. An additional effect of formaldehyde fixation is the formation of Schiff-bases, resulting in the appearance of multiple signals with a mass difference of +12 Da which complicates mass spectra interpretation as well as reduces the signal intensity of the major ion signal (Lemaire et al., 2007; Wisztorski et al. 2010). Although recent progress has been demonstrated in MS analyte detection in formalin-fixed tissues (Lemaire et al., 2007; Wisztorski et al., 2010; Rahimi et al. 2006; Stauber et al., 2010; Groseclose et al., 2008; Ronci et al. 2008), to our knowledge, no universal method has yet been demonstrated for microchemical analysis of fixed and stored single-cells.
In this study, we introduce a strategy for single-cell analysis utilizing ICC-guided cell recognition and visualization, followed by SCMS-enabled single-cell peptidomics. This strategy was developed for paraformaldehyde-fixed immunolabeled (PFIL) neuronal tissues as well as PFIL peptidergic cells of the pars intercerebralis of an insect brain, but can be applied to many cell types after appropriate method optimization. To demonstrate the concept, neurons were cultured on indium tin oxide (ITO) glass slides, immunostained, and then examined with matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS). Two critical steps were optimized for obtaining reproducible ion signals from PFIL tissues and cells. The first is heat treatment (95–100 °C), used to unmask otherwise inaccessible cross-linked peptides. This procedure worked well for PFA-fixed immunoreactive (ir) tissues but not always for single-cells. The heating step is imperative for profiling peptides in samples older than 42 weeks. The second step includes direct application of a reactive mixture of the MALDI matrix α-cyano-4-hydroxycinnamic acid (CHCA) and 2,4-dinitrophenylhydrazine (2,4-DNPH) onto the biological sample. The resulting mass spectra presented signal intensities of putative neuropeptides that were nearly identical to the intensities of the same signals in preparations of freshly isolated, untreated cells and tissues. Furthermore, almost no Schiff-bases were observed in the single-cell MS data. Our strategy also allows identification of analytes in single-cells using MS/MS. As a result of this work, these protocols enable ICC-assisted localization of rare cells, their sampling followed by their characterization including the identification of multiple endogenous peptides via single-cell MS.
Results
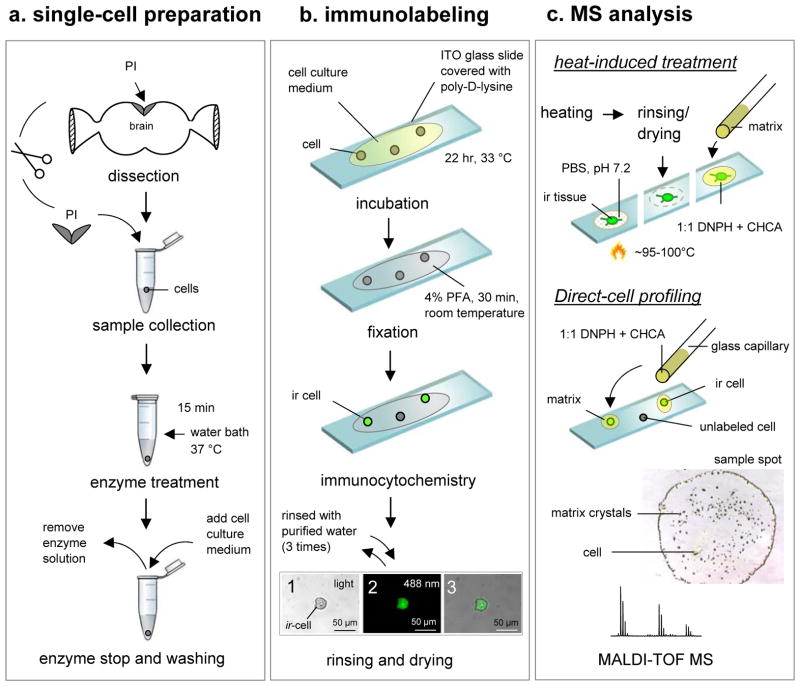
Although highly specific antibodies are often selected for ICC, the combination of ICC and MS allows nonspecific antibodies to be used, such as those that recognize specific functional domains or unusual posttranslational modifications that may be present in multiple antigens. Using the approaches outlined here, cells containing these moieties can now be detected and characterized using SCMS. Due to the small sample sizes and complexity of the intracellular content, sample preparation is a critical step for successful SCMS assays. The complete procedure used to analyze PFIL single neurons is summarized schematically in Figure 1.
Figure 1. Strategy for ICC-guided peptidomics of individual cells.
The strategy has three experimental stages: (a) single-cell preparation (isolation and culturing), (b) immunolabeling, and (c) individual cell and tissue MS-based characterization, including peptide profiling and identification. Abundant peptides that were observed in the mass spectra are subsequently fragmented by tandem mass spectrometry to reveal the amino acid sequence. The three images in panel (b) show (1) a single RFamide ir PI cell with a cell size of 30 μm under ambient light condition, (2) the same cell as in (a) visualized with a fluorescence microscopy, and (3) merged image of (1) and (2). Scale bar = 50 μm.
MS Detection of Neuropeptides in PFIL tissues
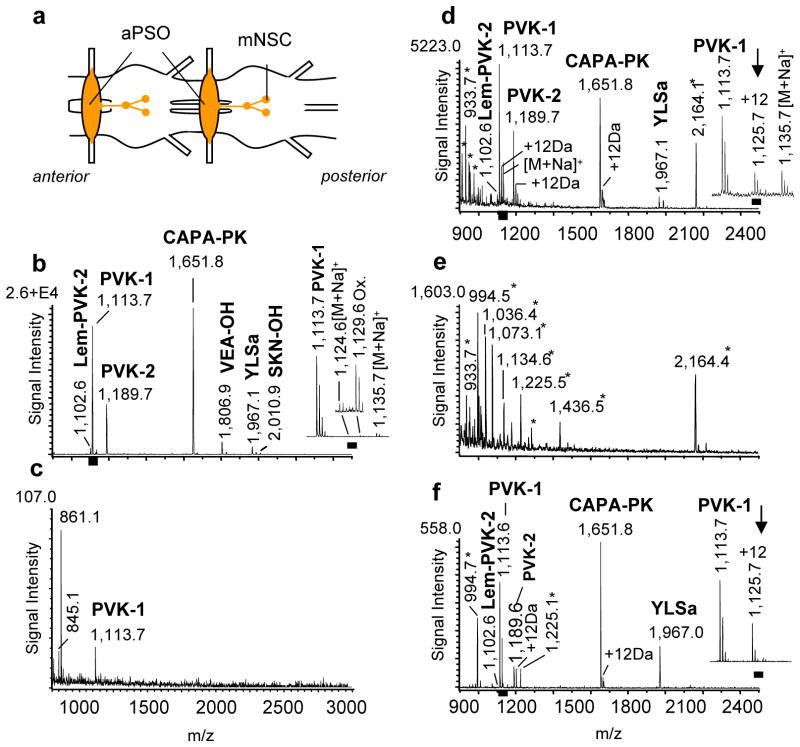
We have chosen to work with the remarkable neuronal insect, the American cockroach Periplaneta americana. The peptides in this model have been well-characterized and its cell morphology thoroughly investigated; it contains many neurons with cell body sizes that are appropriate for manual manipulations. Specifically, the abdominal perisympathetic organs (aPSOs) were used in developing a strategy to accomplish the mass spectrometric characterization of neuropeptides in immunostained biological samples. aPSOs are metameric neurohemal hormone release sites that are synthesized in median neurosecretory cells (with about 20–25 cells), which are located in all unfused abdominal ganglia (Figure 2a). Direct MS analysis of freshly isolated aPSOs confirmed the presence of multiply known peptides (n>100; Figure 2b). In total, seven capa-gene-related ion signals were detected. Three of these were identified as CAPA-periviscerokinins (CAPA-PVKs) (Pea-PVK-1 [M+H]+ 1,113.6; NCBI, GenBank Accession number P41837; Pea-PVK-2 [M+H]+ 1,189.6; NCBI, GenBank Accession number P81555; Lem-PVK-2 [M+H]+: 1,102.6 NCBI, GenBank Accession number P83926), one as CAPA-pyrokinin (CAPA-PK ([M+H]+ 1,651.8), and three structurally unrelated peptides, designated as Pea-YLSamide ([M+H]+ 1,967.0), Pea-VEA-OH ([M+H]+ 1,806.9) and Pea-SKN-OH ([M+H]+ 2,010.9) (Predel et al, 1999b).
Figure 2. MS analysis of peptides from immunolabeled tissues.
Comparative MS peptidomic analyses of freshly isolated and paraformaldehyde-fixed immunolabeled (PFIL) tissues demonstrated that endogenous peptides can be successfully detected in both types of samples. PFIL samples needed to be treated with high temperature (for 42 weeks old samples) before matrix application or a 2,4-DNPH/CHCA mixture (for samples up to 22 weeks) in order to detect peptides. (a) Simplified schematic of a region of the abdominal ventral nerve cord (aVNC) of P. americana. Peptides are produced in median neurosecretory cells (mNSC) in each abdominal ganglion and transported for storage and release to the abdominal perisympathetic organs (aPSO). Scale bar: 500 μm (b) Mass spectrum of freshly prepared aPSO (MALDI matrix solution contains saturated CHCA solution diluted in a 2:1 ratio with 50 % MeOH/water) (n > 100); Scale bar: 100 μm; (c) Mass spectrum of PFIL aPSO using only CHCA as the MALDI matrix; (n = 20); (d) Representative mass spectrum of a 1 to 22 weeks old PFIL aPSO stored in purified water at 4 °C and treated with the 2,4-DNPH/CHCA MALDI matrix solution (2,4-DNPH dissolved in 70% acetonitrile/water, 0.5% TFA mixed with saturated CHCA dissolved in 60% methanol/water, 0.01% TFA in a 1:1 ratio); (n = 15). (e) Mass spectrum of PFIL-treated aPSO stored for 42 weeks in purified water at 4 °C and treated with the same 2,4-DNPH/CHCA matrix used in (d). No known peptide ion signals were detected in any of the preparations (n = 3). (f) Mass spectrum of a 42 weeks old PFIL aPSO from that VNC which provided also the PFIL aPSO for MS analysis shown in (e), however, that sample was heat-treated (2–5 min at 95–100 °C in PBS, ph 7.2) before 2,4-DNPH/CHCA matrix mixture application. Labeled signals correspond to peptides from the putative capa-gene of P. americana. CAPA-PVKs (Pea-PVK-1 [M+H]+ 1,113.62, Pea-PVK-2 [M+H]+ 1,189.63, Lem-PVK-2 [M+H]+ 1,102.60), CAPA-pyrokinin (CAPA-PK, [M+H]+ 1,651.76) and Pea-YLSamide ([M+H]+ 1,967.00; n = 5. Ion signals marked with an asterisk are unspecific and could be only detected from ir tissues (see also supplemental Figure 1).
For the ICC experiments, the abdominal ventral nerve cord with attached aPSOs was fixed over 12–14 h in 4 % PFA to preserve and stabilize the fine structural details of the tissues and the neuronal morphology, as well as to prepare tissues for antibody labeling. A monomer of PFA, formaldehyde, reacts with the amino group of the N-terminal amino acid residue and side chains of arginine, cysteine, histidine, and lysine residues to form methylol and methylene bridges, as well as Schiff-bases(Metz et al. 2004). Therefore, not surprisingly, the MS results yielded either weak (n=20; Figure 2c) or no peptide signals in the PFA-treated samples (n=10).
PFA-fixation is only one aspect of a typical ICC protocol that can complicate MS detection of analytes in PFIL cells and tissues. The addition of antibodies, formation of antibody-antigen complexes, and prolonged washes also reduce the effectiveness of MS. For a successful direct MALDI-TOF MS analysis of PFA-treated biological material, the crosslinks formed during fixation need to be removed. We take advantage of the finding by Lemaire at el. (Lemaire et al. 2007) that a mixture of 2,4-DNPH and CHCA used as the MALDI matrix removes formaldehyde-formed crosslinks. Here, PFA fixed tissues were immunostained with primary antiserum (anti-Pea-PVK-2) and a secondary antibody conjugated with fluorescence Cy2 or Cy3 markers, and then exposed to the 2,4-DNPH/CHCA mixture. This resulted in the detection of endogenous peptides and revealed striking similarities in the peptide profiles from both the ICC-treated and freshly prepared samples, with the most intense signals being observed from expected peptides (Figure 2b, d). Interestingly, the mass spectra of the PFIL samples (stored from 1 up to 22 weeks) had signals from the capa-gene-encoded C-terminal amidated peptides only (n = 15; Figure 2d). The two non-amidated peptides, namely VEA-OH and SKN-OH, were not detectable as either unmodified or modified molecular ions. Schiff-bases were observed for a majority of the detected peptides (Δm = +12 Da). The formation of adduct ions lowers the amount of unmodified peptides and therefore decreases the total signal intensities of the unmodified molecular ions. Signals specific for tissues processed with the PFIL treatment also appeared in the mass spectrum (Figure 2e, e.g., [M+H]+ 2164.1).
Application of the 2,4-DNPH/CHCA mixture to the PFIL-treated samples improved the detection and signal intensity of endogenous peptides, even in samples that were stored for up to 22 weeks. However, longer storage of the samples, such as 42 weeks, prevented the detection of peptides using the 2,4-DNPH/CHCA mixture (Figure 2e; n=5); the detected compounds were specific to the PFIL process itself and not recognized as being related to known peptides, even in collision-induced dissociation (CID) experiments. As a way to address this issue, we considered the known property of PFA to depolymerize into formaldehyde monomers when dry-heated or heated in a water solution. The 42 weeks-old PFIL samples were heated to 95–100 °C for 2–5 min in phosphate buffered saline (PBS), pH 7.2. During the heat treatment, fresh PBS was continuously applied to the sample. After the PBS incubation time the samples were allowed to dry at room temperature. Next, the tissues were carefully rinsed multiple times with water to remove salts, covered with the 2,4-DNPH/CHCA matrix, and analyzed using MALDI-TOF MS. During optimization experiments, we tested different conditions in terms of temperature and incubation time on 42 weeks old ir tissue (n = 10 samples). The expected peptides were detected after the final procedure (95–100 °C for 2–5 min PBS [pH 7.2]) are shown in Figure 2f. (n = 3). Moreover, the amidated products of the capa-gene were also detected. Some decrease in absolute signal intensity but not mass resolution was noted for 42 weeks old samples (Figure 2f) compared to the 1 to 22 weeks old PFIL samples (Figure 2d). Surprisingly, the heat-induced treatment almost eliminated the signals specific to PFIL treatment such as [M+H]+ 2164.4. Also unexpected, the relative intensity of the YLSamide signal increased in the heat-treated samples compared to the freshly prepared control samples.
We also considered alternatives to PBS (pH 7.2) by testing heat-induced treatments using citrate, TRIS or bicarbonate buffers, each of which produced similar results. In all cases, the main issue was the formation of large salt crystals around the samples, particularly in the single-cell preparations, which prevented successful matrix application. These salt crystals need to be removed because their presence leads to decreased monoprotonated peptide signal intensities due to increases in [M+Na]+ and [M+K]+ salt adduct ion intensities. a total of 10–15 rinsing passes with purified water were needed to remove the salt crystals, particularly when using these alternate buffers. The rinses often caused the unintended mechanical removal of the tissue of interest, and thus should be avoided. In almost all cases, the resulting mass spectra revealed various ion signals that did not match any ion signals from peptides or other substances known to be in the freshly prepared biological material examined. Additional, we used the PBS standard solution under three different pH value conditions (PBS with 0.1 M HCl [pH 6.0] and PBS with 0.1M NaOH [pH 8.0 and pH 10]) for our heat-induced experiments. In all cases, no peptide signals were observed. Beside of different PBS modified solutions, we also tested purified water (pH 7.4), purified water plus 0.1 M HCl (pH 6.7) and purified water plus 0.1 M HCl (pH 4.6). Resulting MS experiments showed no CAPA peptide signals.
In addition to testing different buffers, we also experimented with different temperature condition to identify peptides from 42 week old samples. Heat treatment at less than 90 °C was not sufficient to disrupt PFA fixation-induced crosslinks and/or the effects of antigen binding by antibodies. Temperatures higher than 100 °C, however, rendered biological samples unusable. In both cases, no ion signals comparable to the putative peptide ions of P. americana could be detected. Based on these experiments, the useful temperature range was determined to be between 90–100 °C, with the overall best results obtained when tissues PBS, pH 7.2, as described above.
Detection of neuropeptides in PFIL single-cells
Our success with the tissue assays paved the way for the development of a protocol for assaying individual PFIL cultured cells. Single peptidergic neurons from the main neurosecretory center located along the anterior midline of the P. americana brain, the pars intercerebralis (PI), were used as our biological model in developing the PFIL single-cell MS peptidomics strategy (Figure 1). The neuronal soma from this brain region are ~25–30 μm in diameter and have been well characterized using ICC (Meola et al. 1991; East et al. 1997). These samples were more challenging, not only because of their small size but also due to the limited amount of analytes available.
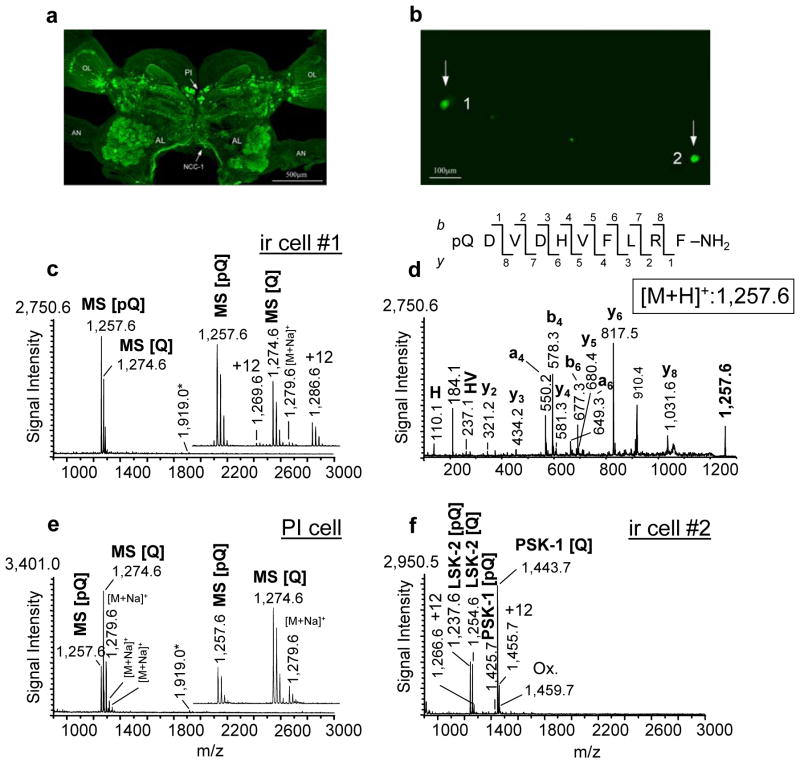
Among others, PI cells express RFamides encoded by different genes, namely myosuppressin, sulfakinin, short neuropeptide F, long neuropeptide F and extended fmrf. The RFamides are a large peptide family with many structural similarities, making it difficult to produce specific antibodies for each peptide. Here, we used an antibody that recognizes all peptides that possess the C-terminal RFamide (Figure 3a). Individual cells isolated from enzymatically treated cell clusters from the PI region were cultured on poly-D-lysine-coated ITO glass slides and immunostained using an antiserum against peptides with an RFamide C-terminus (Figure 3b). Efficient removal of the PFA crosslinks with negligible analyte loss is critical in PFIL single-cell sample preparation. Therefore, as with our 1 week up to 22 weeks-old PFIL tissues, application of the 2,4-DNPH/CHCA mixture directly onto the PFIL single-cells was tested. The optimization and testing of the protocol required more than 300 single-cell sections and the optimum protocols were then repeated on 80 individual PFIL-treated cells. A representative mass spectrum obtained from a PFIL single cultured neuron is shown in Figure 3c. The staining demonstrates that this ir cell expresses RFamide peptides. The mass spectrum shows two prominent ion signals [M+H]+ 1,257.6 and [M+H]+ 1,274.6 which representing the peptide myosuppressin (pQDVDHVFLRF-NH2) and its N-terminally unblocked form (QDVDHVFLRF-NH2). N-terminal modification of myosuppressin may occur due to the enzymatic (glutamyl cyclase) cyclization of glutamine to pyroglutamic acid (Garden et al. 1999) or due to a spontaneous chemical reaction (Fischer and Spiess, 1987). MS analysis of synthetic myosuppressin did not reveal the presence of the modified form; therefore, spontaneous cyclization of myosuppressin during sample preparation is unlikely (not shown; S. Neupert and R. Predel, unpublished). MS/MS fragmentation of [M+H]+ 1,257.6 Da detected from the sample preparation revealed its identity as myosuppressin (pQDVDHVFLRF-NH2) (Figure 3d). The mass spectra from the PFIL myosuppressin-expressing cells contained a low intensity ion signal at [M+H]+ 1,919.1 (Figure 3c). However, no detectable fragmentation of this substance was achieved in tandem MS mode, even employing collision-induced dissociation (CID). Intriguingly, this signal does not match known cockroach peptides by its m/z value, nor is it specific to the PFIL treatment; therefore, it may represent a novel peptide (Figure 3e).
Figure 3. Single-cell mass spectrometry allows profiling and identification of analytes in individual PFIL cells.
(a) The distribution of anti-RFamide immunoreactivity in the whole brain of the cockroach P. americana as examined with confocal microscopy. Individual cells as well as different neuropil regions and axonal arborizations are visible (arrow PI, pars intercerebralis; arrow NCC-1; nervous corporis cardiaci-1). (b) Fluorescent image of two anti-RFamide-stained single-cells (ir cell#1 and ir cell#2; arrows) cultured onto an ITO glass slide(c) Mass spectrum of the cultured PFIL PI neuron (panel (b), cell #1). Note the concentration of the putative peptides in the mass range between 1,250 and 1,300 Da (see inset). An additional ion signal at [M+H]+ 1919.1 was observed in the mass spectra from myosuppressin-expressing neurons (arrow) represents hitherto an unknown product of the myosuppressin-gene remains an unanswered question because the myosuppressin-gene of P. americana is not known (see also Fig 3e). (d) CID mass spectrum of myosuppressin observed in the PI neuron at [M+H]+ 1,257.6. Prominent y-, b-, a-, an internal fragment ion and an immonium ion are labeled. (e) Mass spectrum acquired from a single freshly isolated myosuppressin-expressing PI cell (n = 15). No visible +12 Da ion adducts were observed (see inset). (f) Mass spectrum acquired from the cultured PFIL PI, (panel (b), cell #2) which expresses peptides encoded by a different RFamide neuropeptide family gene. Only unmodified and posttranslationally modified peptides encoded by the sulfakinin-(SK) gene are detected. LSK-2, Leucophaea maderae sulfakinin-2; PSK-1, Periplaneta americana sulfakinin-1; [Q], unblocked; [pQ], blocked form; ox, oxidation; OL, optical lobes; AL, antennal lobes; AN, antennal nerve.
The 2,4-DNPH/CHCA treatment of the PFIL single-cells often led to the formation of Schiff-bases (Δm = +12Da) (Figure 3c). However, the mass spectra acquired from some individual PFIL neurons had no visible +12 Da ion adducts. This lack of detectable Schiff-bases may be attributed to the shorter PFA-treatment time for individual cells (20–30 min) compared to the treatment of tissue preparations (12–14 hr), which always demonstrated these adducts. MS analysis of control single myosuppressin-expressing neurons from freshly isolated, untreated PIs revealed peptide profiles similar to those obtained from individual PFIL cells (Figure 3e; n = 15).
The utility of our strategy is illustrated by the SCMS-detected peptide profile of the RFamide ir cell #2 (Figure 3b). Despite similar immunostaining, cell #2 and cell #1 demonstrated distinct sets of peptides (Figure 3f). The neuropeptides found in cell #2 are products of the sulfakinin gene. Sulfakinins (SKs) are characterized by the active core sequence Y(SO3H)GHMRFamide, which is structurally and functionally similar to the peptides from the vertebrate gastrin-cholecystokinin peptide family. The mass spectra of the PFIL cells revealed that two SKs, Lem-SK2 and Pea-SK, were present as N-terminally unblocked and blocked amidated form (Figure 3f) (Veenstra, 1989; Predel et al. 1999a). The blocked form of Lem-SK2 ([M+H]+ 1,237.5 Da) and the unblocked form of Pea-SK ([M+H]+ 1,443.6 Da) produced the most intense signals. Besides pyroglutamate formation and amidation, the existence of a number of additional posttranslational modifications such as O-methylation of glutamic acid, and sulfation and oxidation were investigated for these peptides (Predel et al. 1999a). In the single-cell MS analysis of untreated as well as PFIL SK-expressing neurons, we did not confirm O-methylation of glutamic acid or the presence of tyrosine sulfation (MALDI MS experiments used positive mode) in the present study. A further form of modification in SKs is oxidation of methionine. This modification could be detected for Pea-SK as well as Lem-SK-2 in untreated SK-expressing cells and in ir cells, indicating its endogenous nature.
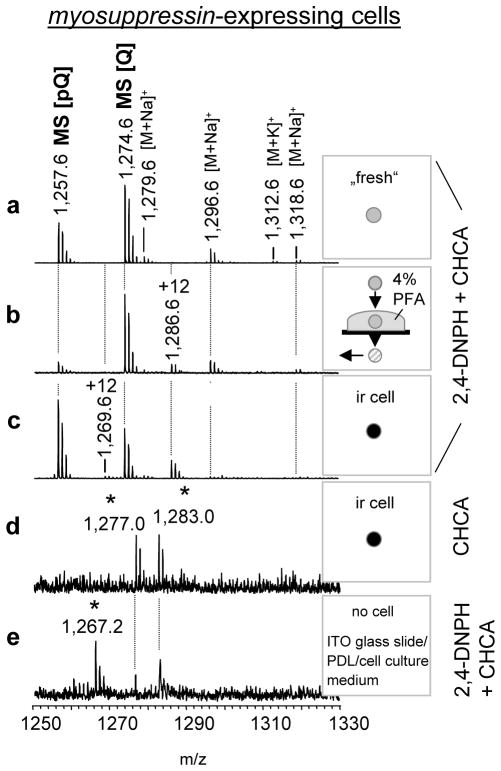
Several limitations to the approach have been noted. In a few ir cell samples, a number of ion signals were detected that did not match any known or predicted signals of endogenous peptides, their posttranslationally modified forms or alkali adducts e.g., m/z: 1,277.0; 1,283.0 and 1,267.2. Extensive comparisons of data from individual myosuppressin-expressing cells showed that these signals are related to the cell culture procedure (Figure 4a). For that, we tested different preparation conditions (Figure 4a–e) and obtained results with data from control experiments done onto a PDL treated ITO glass slide without cell samples but incubated with cell culture medium (Figure 4e). The signals could be reduced or completely eliminated by optimizing the parameters of sample preparation such as procedure of cell isolation, cell culturing, and fixation as well as by optimizing of the ratio of matrix molecule mixture and analyt molecules. However, optimization should be determined empirically for each cell preparation. For example, heat-induced treatment followed by 2,4-DNPH/CHCA application was found to be a less efficient approach for analyte MS detection in PFIL single-cells. Despite detection of a number of the peptides in single PFIL cells examined with 2,4-DNPH/CHCA without heat, the quality of the mass spectra need further improvement.
Figure 4. Comparative analysis of freshly prepared and PFIL treated myosuppressin-expressing cells.
The resulting mass spectra reveal the formation of several treatment-specific signals including formation of Schiff-bases. Mass spectra of (a) freshly isolated single PI cell; (b) single 4% PFA-treated cell; (c) cultured PFIL cell; (d) cultured RFamide-ir cell exposed to only CHCA saturated in a 1:1 methanol:water solution; (e) ITO glass surface coated with poly-D-lysine (PDL), exposed to cell culture medium for 18 h. Mass spectra of all rows except (d) were acquired from samples exposed to the 2,4-DNPH/CHCA mixture. Signals at [M+H]+ 1,267.2; 1,277.0; 1,283.0 are related to the cell culture procedure (asterisks). All samples were rinsed with purified water and air-dried before matrix application. +12 Da marks Schiff-bases (Δm = +12Da).
DISCUSSION
Single-cell microchemical analysis has made exceptional progress in recent years (Rubahkin et al. 2011). Expanding our knowledge of the single-cell peptidome, including the intracellular levels and spatial localization of biomolecules such as neuropeptides, is essential for achieving a detailed understanding of the mechanisms of neuronal processes. Ultimately, these insights promise to advance our ability to diagnose disease and contribute to the development of new pharmacological treatments for a variety of illnesses and disorders. Although SCMS has been proven capable of gathering such data, improved protocols for single-cell sample preparation are a key element to future advances in the technology. Targeted localization of specific cells among thousands to millions of similar cells is a crucial step for SCMS. We introduce a strategy for profiling biological molecules in individual cells based on direct single-cell MALDI MS; the protocols presented in this study are robust and flexible. Besides the inherent advantages of MS, incorporating the PFIL-treatment allows long-term sample storage (even for >42 weeks), provides repeatable results from cultured cells as well as the ability to work with the existing, extensive formalin- and PFA-preserved tissue banks, and is compatible with localized sampling and measurement at regional metabolomics measurement facilities. The ability to characterize tissues, cell cultures, and isolated rare cells is important for fundamental neuroscience, cancer and stem cell studies.
For small samples where the analytes are at low levels, efficient removal of the PFA crosslinks is critical. Considering that the methylene bonds and other formaldehyde modifications form strong crosslinks between molecules in biological samples stored over 22 weeks, we used heat-induced treatment with PBS (pH 7.2) at 95–100 °C before matrix application. Prior studies have demonstrated that heat successfully breaks down formalin-induced crosslinks, resulting in the successful detection of proteins from formalin-fixed, paraffin-embedded tissue by direct MS. However, those measurements were obtained in combination with endopeptidase enzymatic digestion (Hood et al., 2006; Nirmalan et al., 2008; Tian and Zhang, 2010). Here, we designed an analytical approach using heat-induced treatment without enzymatic digestions but rather, with direct 2,4-DNPH/CHCA matrix application (Lemaire et al., 2007). The 2,4-DNPH/CHCA mixture performs two roles in the MS analysis of PFIL samples. The first component, 2,4-DNPH, is commonly used in analytical chemistry as a qualitative test for carbonyl groups associated with aldehydes and ketones in solutions; thus, in MS analysis, it neutralizes aldehydes that have not reacted with the NH2 groups (Lemaire et al., 2007). CHCA is a MALDI matrix which provides the desired even crystallization pattern across the sample as well as enhances the degree and extent of the ionization of peptides in MALDI MS (Beaviset al., 1992).
How do we determine which peptides are present in individual cells? MS/MS is a powerful approach for analyte structure determination. Depending on cell size, a single-cell contains enough material to confirm the amino acid sequence of several analytes (Neupert and Predel, 2005; Neupert et al. 2007). However, it requires a higher amount of biological material to collect sufficient information for de novo sequencing of peptides or for structure determination of more than three compounds of interest per sample. In contrast, peptide fingerprinting (Li et al., 1994; Li et al. 2000) can be successfully applied in SCMS for identification of different peptides by matching exact measured molecular masses of peptides enzymatically formed from proteins, in silico predicted peptides, and synthesized related peptide standards. For example, in single-cells we detected seven prominent ion signals with m/z matching those of known peptides from one gene, the capa-gene: Pea-PVK-1, Pea-PVK-2, Lem-PVK-2, CAPA-PK, as well as three structurally unrelated peptides, designated as Pea-YLSamide, Pea-VEA-OH and Pea-SKN-OH (Predel et al. 1999b). This match is sufficient to assign identities to detected peptides according to commonly applied peptide fingerprinting approaches. Interestingly, even though a gene encoding Pea-YLSamide, Pea-VEA-OH and Pea-SKN-OH has yet to be characterized in P. americana, prior mass spectrometric analysis of single capa-expressing neurons (Predel et al. 2007), and immunocytochemical as well as electron microscopic studies (Eckert et al. 2002), have shown that these seven peptides are always co-localized, which suggests, but does not confirm, that they are encoded on the same peptide precursor.
Because of the limited amount of material, de novo sequencing of peptides detected from a single-cell is possible (Jiménez et al. 1998, Li et al. 1999) but not often attainable. Whether or not the additional ion signal at [M+H]+ 1919.1 we observed in the mass spectra from myosuppressin-expressing neurons represents hitherto an unknown product of the myosuppressin-gene remains an unanswered question because the myosuppressin-gene of P. americana is not known. No co-localizations with any of the other known neuropeptides of P. americana were detectable. Moreover, the screening of the myosupressin-precursors of distantly related species Diploptera punctata (Donly et al., 1996) and Blattella germanica (Vilaplana et al. 2004) did not aid in its identification.
SCMS is also a powerful approach for gaining insights into peptide posttranslational modifications. For example SK-expressing neurons revealed two major peptide modifications: Lem-SK2 and Pea-SK, which are present as N-terminally unblocked and blocked motifs (Veenstra, 1989; Predel et al. 1999a). The ratio of the signal intensity of the blocked motif was approximately 10% of the unblocked motif (Predel et al. 1999a), which indicates a more efficient cyclization of glutamine versus glutamate. Another observed modification in SKs is oxidation of methionine, which was detected for Pea-SK as well as Lem-SK-2 in untreated and ir treated SK-expressing cells. We could not distinguish between naturally occurring oxidized SKs and oxidation occurring during sample preparation.
Several parameters can be used to characterize and compare the herein described protocols for SCMS including signal intensity, signal resolution, and appearance or disappearance of ion signals. Similar to Lemaire et al. (2007) for formalin-fixed embedded tissues, we observed decreases in the intensities of signals but not in the mass resolutions. Several types of signals were also observed in PFIL tissues. One type is the expected +12 Da adduct for each [M+H]+ ion. This adduct is from the formalin fixation process and has been identified as a peptide-N=CH2 compound (Metz et al, 2004). The +12 ion signal intensity grows as a function of time during the PFA treatment and reaches ~80% of the parent [M+H]+ ion intensity. Interestingly, only the +12 Da adduct of CAPA-PK intensity is only 8% the intensity of the parent ion. We suggest that the distinctively small +12 Da CAPA-PK ion adduct is attributed to the preference in MS detection of unmodified Trp-containing [M+H]+ molecular ions.
Another set of unknown signals appear in the PFIL samples; prominent molecular ions with m/z 2,164.1; 1.277.0; 1.283.0 and 1,267.2 are among them. MS/MS fragmentation of these molecules did not reveal the structure of these compounds. Furthermore, we found no indication that these ions could be formaldehyde-related modifications of amino acid residues of known peptides of P. americana. We suggest that the observed compounds are not peptides related to the PFIL treatment.
SIGNIFICANCE
Cellular heterogeneity plays key roles in the normal and pathological functioning of multicellular organisms including organs and tissues. Achieving a comprehensive global assessment of the chemical heterogeneity among cells in a population of thousands to millions of morphologically similar cells is difficult unless specific, representative cells are localized for analysis. Recently, the utility of single-cell mass spectrometry (SCMS) has been demonstrated for metabolite and peptide profiling in a variety of neurons and endocrine cells. Here, we introduce a strategy for targeting and sampling single-cells for SCMS investigation. An SCMS-compatible preparation of immunostained target cells placed in cell culture is a key element of this strategy. Our results clearly demonstrate that paraformaldehyde-fixed immunolabeled (PFIL) neurons stored up to 2 weeks and tissues preserved for 42 weeks are suitable for direct MALDI MS peptidomics. However, sample preparation techniques may require optimization for cells and tissues of different sizes, types, and origins. Regardless, profiling of the single neuron peptidome and identification of individual peptides were accomplished in PFIL tissues and cells. The described protocols enable a broad range of studies with a variety of biological models and analytes where the immunocytochemical probes uncover specific individual cells and SCMS investigates their peptidomes and metabolomes. As an example, a robust, fast, and reproducible investigation of prohormone processing, differential gene expression resulted in expression of multiple similar peptide isoforms were determined in ICC identified individual cells using the protocols. The strategy also can be used for indirect validation of immunocytochemical results. Cells exhibiting unexpected immunoreactivity can be probed with SCMS and possible antigens determined. Our strategy and protocols are compatible with many other single-cell analyses approaches, including flow cytometry and laser capture microdissection, thus significantly broadening their potential use.
EXPERIMENTAL PROCEDURES
Animals
Periplaneta americana were maintained at a constant temperature of 23 °C. They were fed food pellets for rats and had free access to water. Adult males and females were used in all experiments. The animals which were used in this study were treated pursuant to the tenets of the Declaration of Helsinki.
Cell culture
Protocols for cell dissociation and culture from Petri and Stengl (1999) and Vömel and Wegener (2007) were used with modifications. For each experiment, the brains were dissected and transferred into a drop of modified Ca2+/Mg2+-free insect saline (Su and O’Dowd(2003): (in mM) 126 NaCl, 5.4 KCl, 0.17 NaH2PO4 and 0.22 KH2PO4, pH 7.4. For cell dissociation, isolated brains were pinned on a layer of Sylgard 184 (Dow Corning, Germantown, WI) with microneedles. The ganglionic sheath in the region of the protocerebrum was removed with ultra-fine forceps. Subsequently, the pars intercerebralis (PI) was selectively cut out using ultra-fine scissors. Using a glass capillary, the isolated PI was transferred into a 1.5 ml microtube (Eppendorf AG, Hamburg, Germany) filled with 500 μl of insect saline.
For enzymatic cell dissociation, we tested solutions of the following enzymes dissolved in insect saline supplemented with antibiotics: a) 4mg/ml dispase II (neutral protease, grade II) (Roche Diagnostics GmbH, Mannheim, Germany); b) a mixture of 1 mg/ml collagenase (Sigma-Aldrich, St. Louis, MO) and 4 mg/ml dispase with incubation times between 20–40 min; c) 10mg/ml protease IX (Sigma-Aldrich). The best results were obtained with 4 mg/ml dispase II. Therefore, tissues were treated in 500 μl 4 mg/ml of dispase II dissolved in 1 ml Ca2+/Mg2+-free insect saline for 10–15 min at 37 °C. The microtube was centrifuged at 370 rpm at 18 °C for 2 min, the enzyme solution removed, and 250 μl of ESF 921 Insect Cell Culture Medium, Protein-Free (Expression Systems, Woodland, CA) added. The culture medium was replaced 2–3 times and finally replaced with a medium supplemented with 50 μl of a mixture of 100 units/mL penicillin G, 100 units/mL streptomycin and 100 units/mL gentamicin. The tissue was allowed to settle for 20–30 min. The medium was replaced and the tissue triturated with a pipette to isolate single-cells. The cell suspension (120 μl) was deposited on each of two ITO microscope glass slides, pretreated with poly-D-lysine (Sigma-Aldrich, St. Louis, MO). We also tested poly-L-lysine alone as well as poly-L-lysine and poly-D-lysine in combination with concanavalin A (Calbiochem, La Jolla, CA) as a substrate for cell culture on the ITO slides, respectively. The best results (cell growth as well as MALDI-TOF MS analysis) were observed with poly-D-lysine alone. Insect cell culture medium (~100 μl) was added to each slide and the cells were cultured in a humidified incubator for 15–18 hr.
Surprisingly, we have found that successful culturing of the PI cells on ITO glass slides required a temperature of ~33 °C. This temperature is 30–35% higher than typically recommended (e.g. Petri and Stengl, 1994) for optimal insect neuron adhesion and growth on the glass in humidified incubators
Immunocytochemistry (ICC)
Cell culture
For ICC, cultured cells were fixed for 20–30 min at 4 °C with 4% PFA in PBS solution, pH 7.2, comprised of: (in mM) 84.1 ml of solution A: 57 Na2HPO4*2H20, and 15.9 ml of solution B: 101.5 NaH2P04*H2O, 145.5 NaCl, added to 900 ml purified water. As an alternative to 4% PFA, we tested a neutral buffered 10% formalin solution (Sigma-Aldrich, St. Louis, MO) as a fixative. The best results in immunostaining intensity were obtained with the 4% PFA. Subsequently, the preparations were carefully washed three times with PBS (10 min each) and preincubated with 5% normal goat serum (Jackson ImmunoResearch, West Grove, PA) dissolved in PBS for 30 min. The cells were carefully washed again three times (10 min each) in PBS and incubated overnight with anti-RF thyroglobulin-glutaraldehyde serum (1:500) diluted in PBS containing 10% goat normal serum (Jackson ImmunoResearch) at 4 °C. After incubation with primary antiserum, the cells were rinsed three times (10 min each) in PBS and subsequently incubated with either a Cy2- or a Cy3-tagged secondary antibody (Jackson ImmunoResearch) at 1:300 dilutions for 8–12 hr. Finally, the cells were rinsed again over a period of 2–48 hr in purified water.
Whole mount preparations
For whole mount preparation, brains as well ventral nerve cords were fixed overnight (12–14 h) at 4 °C with 4% PFA in PBS, pH 7.2. Subsequently, the samples were first washed in PBS containing 4% Triton X-100 (Sigma-Aldrich, St. Louis, MO) and then in PBS supplemented with 1% Triton X-100 at 4 °C for 24 hr using a laboratory shaker, respectively. The preparations were preincubated with 5% normal goat serum dissolved in PBS for 30 min and then incubated for 4 d at 4 °C in anti-RF thyroglobulin-glutaraldehyde serum (1:500) or anti-Pea-PVK-II serum (1:4000) diluted with PBS, 1% Triton X-100 and 10% normal goat serum. Both primary antibodies were kindly provided by Prof. R. Predel (Cologne, Germany). Following overnight washing in 0.1 mol/l TRIS-HCL, 3% NaCl, 1% Triton X-100, pH 7.6, the Cy2- or Cy3-tagged secondary antibodies were directly applied at a 1:3000 dilution for 4 d. The preparations were washed again overnight in 0.1 mol/l TRIS-HCL, 3%, NaCl, 1% Triton X-100, pH 7.6. Finally, the buffer was replaced with purified water. For visualization, tissues were dehydrated in ethanol, cleared in toluene (Sigma-Aldrich, St. Louis, MO) and mounted in Permount (Fisher Scientific, Fairlawn, New Jersey). The whole mount immunofluorescence labeling results were examined with a confocal laser scanning microscope (SP2 system, Leica, Bensheim, Germany), equipped with an Argon 2 laser (wavelengths 458, 477, 488, 514 nm). Scans of the Cy 2 fluorescence (Exmax = 490, Emmax = 508) were performed in scanning mode at a resolution of 1,024 × 1,024 pixels by using a 10× (HCPLFLUOTAR 10×/0.30 mm) and a 20× objective (HCPLAPO 20×/0.70 mm) (Leica, Bensheim, Germany). Serial optical sections were assembled into combined images.
Cultured cells, however, were investigated using a Zeiss (Carl Zeiss, Jena, Germany) digital camera mounted on an Axiovert 25 inverted microscope (Carl Zeiss, Jena, Germany) equipped with a filter set to visualize the Cy2 and Cy3 fluorescence labeling. Image acquisition and processing was performed using AxioVision software (Carl Zeiss, Jena, Germany). All images were exported to Adobe Photoshop 7.0 for further processing.
Sample preparation for MALDI-TOF mass spectrometry
Direct tissue profiling
The dissection procedure of the abdominal perisympathetic organs was followed as previously described in Predel (2001).
Direct single-cell analysis
Brains were dissected in insect saline and mounted with mirconeedles. After removal of the ganglionic sheath in front of the protocerebrum, the pars intercerebralis were nearly completely dissected using ultrafine scissors and subsequently transferred with a glass capillary into a drop of non-enzymatic cell dissociation solution (Sigma-Aldrich, St. Louis, MO). The cells were incubated for 20–30 min at room temperature. Thereafter, single-cells were separated from the cell cluster with a glass capillary and transferred onto a sample plate for MALDI-TOF MS (Neupert and Predel, 2010).
Immunolabeled tissue profiling
For MALDI-TOF analysis, ir aPSOs were rinsed over 24 hr or stored for 42 weeks in purified water at 4 °C, subsequently transferred to a stainless steel sample plate for MALDI-TOF MS, and air-dried at room temperature. Samples stored for a period of 42 weeks were heat-induced in PBS (pH 7.2) at 95–100°C for 2–5 min before matrix application (see below).
Ir single-cell analysis
For MS analysis, the purified water from the immunolabeled cells on the glass slides was completely removed. The samples were air-dried at room temperature. Only isolated ir cells samples that were not contaminated by other cells or axons were chosen for MS profiling.
Heat-induced treatment
42 weeks old ir PSOs were prepared in purified water under fluorescence microscope-assisted visual control, transferred with a glass capillary or an insect pin to an ITO glass slide or a stainless steel sample plate and air-dried. Depending on the sample size, approximately 2–10 μl (for tissue) or 10–50 nl (for single-cells) of PBS, pH 7.2, were loaded onto the samples and heated at 95–100 °C using a hot plate. During heat treatment, PBS was continuously added to single-cells (0.2–0.5 μl PBS drops) and tissue specimens (up to 1 μl PBS) over a period of 2–5 min. Optimal volumes of PBS need to be added to samples of different sizes have to be found empirically.
Subsequently, the samples were cooled and then rinsed with purified water up to 10 times, and air-dried again. Each washing is done by the covering of the samples with 0.5 to 1μl purified water. After a few seconds, the water was carefully removed using a glass capillary under visual control assisted with a stereo microscope. We also tested alternatives to the PBS treatment at 7.2 pH: PBS solutions with 0.1 M HCl (pH 6.0), with 0.1 M NaOH (pH 8.0 and ph 10); citrate buffer containing 9.9 mM Na3C6H5O7*2H2O, pH 6.0; bicarbonate buffer containing 100 mM Na2CO3, 500 mM NaCl, pH 8; 100mM TRIS-HCL buffer, pH 7.4; and purified water (pH 7.4) as well as under different pH conditions (pH 4.6; pH 6.7) using 0.1 M HCl respectively. The PBS treatment at pH 7.2 was the one most compatible with the MALDI-MS sample preparation and produced very good results.
MALDI matrix application
For mass spectrometric analysis of untreated (fresh) tissues and single-cells, a 2:1 mixture of the following solutions were loaded onto the dried samples: saturated CHCA (Sigma-Aldrich, St. Louis, MO) dissolved in 60% methanol/water, containing 0.01% TFA. Finally this matrix solution was again diluted in a ratio of two parts saturated matrix solution with one part 50% methanol/water. Using a glass capillary (Hilgenberg GmbH, Malsfeld, Germany) single-cells were covered with 10–20 nl and tissues with 300 nl of this final mixture. The samples were incubated for a few seconds, air-dried, and covered with purified water. After a few seconds, the water was removed again with a glass capillary. Rinsing with water is commonly used to reduce high inorganic salt content in biological samples.
For direct ir sample profiling, solutions of saturated CHCA dissolved in 60% methanol/water, 0.01% TFA, and 4mg/ml 2,4-Dinitrophenylhydrazine (2,4-DNPH; Sigma-Aldrich, St. Louis, MO) dissolved in 70% acetonitrile/water, 0.5% TFA, were prepared. Both solutions were centrifuged at 14,000 rmf at 4 °C for 15 min and mixed at 1:1 ratio. Approximately 0.3–1 μl (tissue) and 20–30 nl (single-cells) of the mixture were loaded onto the dried samples using a glass capillary (Hilgenberg GmbH, Malsfeld, Germany). Each preparation was air-dried over a period of 2–7 min at room temperature and then washed for a few seconds with purified water. Any remaining water was subsequently removed by cellulose paper.
MALDI – TOF mass spectrometry
The mass spectra were acquired in positive ion mode on an UltrafleXtreme TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). All MS acquisitions were made under manual control in reflectron mode. Laser fluency was adjusted to provide the optimal signal-to-noise ratio. The instrument was calibrated using a Bruker peptide standard kit. The data obtained in these experiments were processed with the FlexAnalysis 3.3 software package. MS/MS was performed with CID turned on. CID acceleration was set at 1kV. The number of laser shots used to obtain a spectrum varied from 2000–5000, depending on signal quality. The peptide ion signals identities were verified using MS/MS fragmentation of the molecules, determination of the molecular mass of the fragments, and alignment of predicted and experimentally obtained fragmentation patterns. The fragmentation pattern of peptides with masses corresponding to calculated masses of known Periplaneta peptides was compared with the theoretical fragments obtained from ProteinProspector (http://prospector.ucsf.edu).
Supplementary Material
HIGHLIGHTS.
Investigation of archived biological tissue at the single-cell level
Screening of neuropeptides from immunolabeled tissue preserved up to 42 weeks
Validation of antibody specificity using mass spectrometric approaches
Standardized protocols for comprehensive characterization of selected cells
Acknowledgments
We acknowledge the financial assistance of the German Academic Exchange Service (DAAD; [SN D/09/51168]). The project described was supported by Award No. 5RO1NS031609 from the National Institute of Neurological Disorders and Stroke (NINDS) and Award Number P30 DA018310 from the National Institute on Drug Abuse (NIDA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the awarding agencies. We thank Prof. M. Stengl (University of Kassel, Germany) and Xiying Wang (University of Illinois at Urbana-Champaign, USA) for help with cell culture and Prof. R. Predel (Biocenter, University of Cologne, Germany) and Dr. M. Eckert (Friedrich-Schiller University, Jena, Germany) for providing the anti-Pea-PVK-II and anti-RFamide sera.
Footnotes
AUTHOR CONTRIBUTIONS
S.N. conceived the project; S.N. performed experiments while getting appropriate advice from S.S.R. and J.V.S; S.N., S.S.R. and J.V.S. wrote the article.
References
- Beavis C, Chaudhary T, Chait BT. a-cyano-4-hydroxycinnamic acid as a matrix for matrix- assisted laser desorption mass-spectrometry. Organic Mass Spectrometry. 1992;27:156–158. [Google Scholar]
- Donly BC, Fuse M, Orchard I, Tobe SS, Bendena WG. Characterization of the gene for leucomyosuppressin and its expression in the brain of the cockroach Diploptera punctata. Insect Biochem Mol Biol. 1996;26:627–37. doi: 10.1016/s0965-1748(96)00015-x. [DOI] [PubMed] [Google Scholar]
- East PD, Hales DF, Cooper PD. Distribution of sulfakinin-like peptides in the central and sympathetic nervous system of the American cockroach, Periplaneta americana (L.) and the field cricket, Teleogryllus commodus (Walker) Tissue Cell. 1997;29:347–54. doi: 10.1016/s0040-8166(97)80010-9. [DOI] [PubMed] [Google Scholar]
- Eckert M, Herbert Z, Pollák E, Molnár L, Predel R. Identical cellular distribution of all abundant neuropeptides in the major abdominal neurohemal system of an insect (Periplaneta americana) J Comp Neurol. 2002;452:264–75. doi: 10.1002/cne.10382. [DOI] [PubMed] [Google Scholar]
- Fischer WH, Spiess J. Identification of a mammalian glutaminyl cyclase converting glutaminyl into pyroglutamyl peptides. Proc Natl Acad Sci U S A. 1987;84:3628–32. doi: 10.1073/pnas.84.11.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden RW, Moroz TP, Gleeson JM, Floyd PD, Li L, Rubakhin SS, Sweedler JV. Formation of N-pyroglutamyl peptides from N-Glu and N-Gln precursors in Aplysia neurons. J Neurochemistry. 1999;72:676–81. doi: 10.1046/j.1471-4159.1999.0720676.x. [DOI] [PubMed] [Google Scholar]
- Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. Excess salt removal with matrix rinsing: direct peptide profiling of neurons from marine invertebrates using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 1996;10:1126–30. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Groseclose MR, Massion PP, Chaurand P, Caprioli RM. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics. 2008;8:3715–24. doi: 10.1002/pmic.200800495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood BL, Conrads TP, Veenstra TD. Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics. 2006;6:4106–14. doi: 10.1002/pmic.200600016. [DOI] [PubMed] [Google Scholar]
- Jiménez CR, Li KW, Dreisewerd K, Spijker S, Kingston R, Bateman RH, Burlingame AL, Smit AB, van Minnen J, Geraerts WP. Direct mass spectrometric peptide profiling and sequencing of single neurons reveals differential peptide patterns in a small neuronal network. Biochemistry. 1998;37:2070–6. doi: 10.1021/bi971848b. [DOI] [PubMed] [Google Scholar]
- Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007;6:1295–305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- Li KW, Jiménez CR, Van Veelen PA, Geraerts WP. Processing and targeting of a molluscan egg-laying peptide prohormone as revealed by mass spectrometric peptide fingerprinting and peptide sequencing. Endocrinology. 1994;134:1812–9. doi: 10.1210/endo.134.4.8137747. [DOI] [PubMed] [Google Scholar]
- Li L, Garden RW, Romanova EV, Sweedler JV. In situ sequencing of peptides from biological tissues and single-cells using MALDI-PSD/CID analysis. Anal Chem. 1999;71:5451–8. doi: 10.1021/ac9907181. [DOI] [PubMed] [Google Scholar]
- Li L, Garden RW, Sweedler JV. Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 2000;18:151–60. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- Meola SM, Wright MS, Holman GM, Thompson JM. Immunocytochemical localization of leucomyosuppressin-like peptides in the CNS of the cockroach, Leucophaea maderae. Neurochem Res. 1991;16:543–9. doi: 10.1007/BF00974872. [DOI] [PubMed] [Google Scholar]
- Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJ, Jiskoot W. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–43. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- Morano C, Zhang X, Fricker LD. Multiple isotopic labels for quantitative mass spectrometry. Anal Chem. 2008;80:9298–309. doi: 10.1021/ac801654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert S, Predel R. Peptidomic analysis of single identified neurons. Methods Mol Biol. 2010;615:137–44. doi: 10.1007/978-1-60761-535-4_11. [DOI] [PubMed] [Google Scholar]
- Neupert S, Johard HA, Nassel DR, Predel R. Single-cell peptidomics of Drosophila melanogaster neurons identified by Gal4-driven fluorescence. Anal Chem. 2007;79:3690–4. doi: 10.1021/ac062411p. [DOI] [PubMed] [Google Scholar]
- Neupert S, Predel R, Russell WK, Davies R, Pietrantonio PV, Nachman RJ. Identification of tick periviscerokinin, the first neurohormone of Ixodidae: single-cell analysis by means of MALDI-TOF/TOF mass spectrometry. Biochem Biophys Res Commun. 2005a;338:1860–4. doi: 10.1016/j.bbrc.2005.10.165. [DOI] [PubMed] [Google Scholar]
- Neupert S, Predel R. Mass spectrometric analysis of single identified neurons of an insect. Biochem Biophys Res Commun. 2005b;327:640–5. doi: 10.1016/j.bbrc.2004.12.086. [DOI] [PubMed] [Google Scholar]
- Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Mining the archival formalin-fixed paraffin-embedded tissue proteome: opportunities and challenges. Mol Biosyst. 2008;4:712–20. doi: 10.1039/b800098k. [DOI] [PubMed] [Google Scholar]
- Petri B, Stengl M. Presumptive insect circadian pacemakers in vitro: immunocytochemical characterization of cultured pigment-dispersing hormone-immunoreactive neurons of Leucophaea maderae. Cell Tissue Res. 1999;296:635–43. doi: 10.1007/s004410051324. [DOI] [PubMed] [Google Scholar]
- Predel R. Peptidergic neurohemal system of an insect: mass spectrometric morphology. J Comp Neurol. 2001;436:363–75. [PubMed] [Google Scholar]
- Predel R, Brandt W, Kellner R, Rapus J, Nachman RJ, Gäde G. Post-translational modifications of the insect sulfakinins: sulfation, pyroglutamate-formation and O-methylation of glutamic acid. Eur J Biochem. 1999a;263:552–60. doi: 10.1046/j.1432-1327.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- Predel R, Eckert M, Holman GM. The unique neuropeptide pattern in abdominal perisympathetic organs of insects. N Y Acad Sci. 1999b;897:282–90. doi: 10.1111/j.1749-6632.1999.tb07899.x. [DOI] [PubMed] [Google Scholar]
- Predel R, Eckert M, Pollák E, Molnár L, Scheibner O, Neupert S. Peptidomics of identified neurons demonstrates a highly differentiated expression pattern of FXPRLamides in the neuroendocrine system of an insect. J Comp Neurol. 2007;500:498–512. doi: 10.1002/cne.21183. [DOI] [PubMed] [Google Scholar]
- Rahimi F, Shepherd CE, Halliday GM, Geczy CL, Raftery MJ. Antigen-Epitope Retrieval To Facilitate Proteomic Analysis of Formalin-Fixed Archival Brain Tissue. Anal Chem. 2006;78:7216–722. doi: 10.1021/ac060294s. [DOI] [PubMed] [Google Scholar]
- Ronci M, Bonanno E, Colantoni A, Pieroni L, Di Ilio C, Spagnoli LG, Federici G, Urbani A. Protein unlocking procedures of formalin-fixed paraffin-embedded tissues: application to MALDI-TOF imaging MS investigations. Proteomics. 2008;8:3702–14. doi: 10.1002/pmic.200701143. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Profiling metabolites and peptides in single-cells. Nat Methods. 2011;8:20–9. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin SS, Sweedler JV. Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal Chem. 2008;80:7128–36. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–80. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- Romanova EV, Fosser KA, Rubakhin SS, Nuzzo RG, Sweedler JV. Engineering the morphology and electrophysiological parameters of cultured neurons by microfluidic surface patterning. FASEB J. 2004;11:1267–9. doi: 10.1096/fj.03-1368fje. [DOI] [PubMed] [Google Scholar]
- Stauber J, Ayed ME, Wisztorski M, Salzet M, Fournier I. Specific MALDI-MSI: Tag-Mass. Methods Mol Biol. 2010;656:339–61. doi: 10.1007/978-1-60761-746-4_20. [DOI] [PubMed] [Google Scholar]
- Su H, O’Dowd DK. Fast synaptic currents in Drosophila mushroom body Kenyon cells are mediated by a-bungarotoxin-sensitive nicotinic acetylcholine receptors and picrotoxin-sensitive GABA receptors. J Neurosci. 2003;23:9246–9253. doi: 10.1523/JNEUROSCI.23-27-09246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Zhang H. Isolation of proteins by heat-induced extraction from formalin-fixed, paraffin-embedded tissue and preparation of tryptic peptides for mass spectrometric analysis. Curr Protoc Mol Biol. 2010;Chapter 10(Unit 10.26):1–7. doi: 10.1002/0471142727.mb1026s90. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Isolation and structure of two gastrin/CCK-like neuropeptides from the American cockroach homologous to the leucosulfakinins. Neuropeptides. 1989;14:145–9. doi: 10.1016/0143-4179(89)90038-3. [DOI] [PubMed] [Google Scholar]
- Vilaplana L, Castresana J, Belles X. The cDNA for leucomyosuppressin in Blattella germanica and molecular evolution of insect myosuppressins. Peptides. 2004;25:1883–9. doi: 10.1016/j.peptides.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Vömel M, Wegener C. Neurotransmitter-induced changes in the intracellular calcium concentration suggest a differential central modulation of CCAP neuron subsets in Drosophila. Dev Neurobiol. 2007;67:792–808. doi: 10.1002/dneu.20392. [DOI] [PubMed] [Google Scholar]
- Wang D, Bodovitz S. Single-cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010;28:281–90. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisztorski M, Franck J, Salzet M, Fournier I. MALDI direct analysis and imaging of frozen versus FFPE tissues: what strategy for which sample? Methods Mol Biol. 2010;656:303–22. doi: 10.1007/978-1-60761-746-4_18. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Zhang P, Zhao N, Sheehan AM, Tung CH, Chang CC, Zu Y. Using oligonucleotide aptamer probes for immunostaining of formalin-fixed and paraffin-embedded tissues. Mod Pathol. 2010;23:1553–8. doi: 10.1038/modpathol.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.