Abstract
Accurate detection of breast malignancy from histologically normal cells (“field effect”) has significant clinical implications in a broad base of breast cancer management, such as high-risk lesion management, personalized risk assessment, breast tumor recurrence, and tumor margin management. More accurate and clinically applicable tools to detect markers characteristic of breast cancer “field effect” that are able to guide the clinical management are urgently needed. We have recently developed a novel optical microscope, spatial-domain low-coherence quantitative phase microscopy, which extracts the nanoscale structural characteristics of cell nuclei (i.e., nuclear nano-morphology markers), using standard histology slides. In this proof-of-concept study, we present the use of these highly sensitive nuclear nano-morphology markers to identify breast malignancy from histologically normal cells. We investigated the nano-morphology markers from 154 patients with a broad spectrum of breast pathology entities, including normal breast tissue, non-proliferative benign lesions, proliferative lesions (without and with atypia), “malignant-adjacent” normal tissue, and invasive carcinoma. Our results show that the nuclear nano-morphology markers of “malignant-adjacent” normal tissue can detect the presence of invasive breast carcinoma with high accuracy and do not reflect normal aging. Further, we found that a progressive change in nuclear nano-morphology markers that parallel breast cancer risk, suggesting its potential use for risk stratification. These novel nano-morphology markers that detect breast cancerous changes from nanoscale structural characteristics of histologically normal cells could potentially benefit the diagnosis, risk assessment, prognosis, prevention, and treatment of breast cancer.
Keywords: Field effect, Nanoscale structure, Phase microscopy, Nuclear nano-morphology markers
Introduction
The “field effect” or “field cancerization”, a term that denotes the presence of cancer-like signatures in histologically normal tissues surrounding the primary tumor, is a well-documented process in breast cancer and many other tumor types [1–4]. It has been well recognized that the identification of signatures characteristic of “field effect” in breast cancer with a high accuracy plays a significant role in a wide range of breast cancer management [3, 5]. For example, the presence of cancer-like signatures in histologically normal tissue could suggest the presence of malignancy missed by tissue biopsy, thus enhancing the detection of malignancy. It could also serve as a predictor of early stage carcinogenesis to guide personalized risk assessment to identify truly high-risk patients for close surveillance or aggressive adjuvant treatment. Further, the cancer-like signatures indicative of “field effect” in normal tissue can also provide important guidance to predict the local recurrence following lumpectomy, as the risk of local recurrence remains despite histologically negative margins.
Substantial evidence supports the biological plausibility of “field effect” in breast carcinogenesis, such as loss of heterozygosity [6], microsatellite instability [7, 8], gene mutations [9], epigenetic alterations [10–12], and gene expression aberrations [3, 13, 14], which have been identified in histologically normal tissue adjacent to the tumor and in cancer cells themselves. Although all of these markers have shown a statistically significant correlation between biomarkers and breast neoplasia (i.e., field effect), their performance characteristics are suboptimal for clinical practice. Further, the implementation of these molecular markers to routine clinical use has been challenging, in part due to the special requirement for sample preparation, high cost and time-consuming nature. More accurate, quick, simple and clinically applicable tools to identify cancer-like signatures from normal tissue characteristic of breast cancer “field effect” are urgently needed.
The inability of conventional micron-scale morphologic features to detect the “field effect” is due, in large part, to the limited resolution (~500 nm) of conventional light microscopy. But profound nanoscale architectural alterations have been reported in histologically normal epithelial cells undergoing field carcinogenesis [15–17]. The nanoscale structural properties have shown great promise to detect the field cancerization in many organs including colon, lung and pancreas [17–20]. However, the identification of nano-structural properties using routine tissue histology specimens remains a challenge and the nanoscale structural markers characteristic of mammary field carcinogenesis are as yet unreported.
We recently developed spatial-domain low-coherence quantitative phase microscopy (SL-QPM) that can assess subtle, nanoscale structural changes (i.e., nano-morphology markers) within the cell nuclei on standard histology slides. SL-QPM takes advantage of the ultra-high sensitivity of light interference effect to achieve nanoscale sensitivity not attainable with a conventional microscope [21, 22]. In this article, we investigate the SL-QPM-derived nano-morphology markers from cell nuclei as potential signatures to detect the breast cancer “field effect”. We investigate the histology slides from 154 patients and analyze the nano-morphology markers from normal breast tissue, histologically normal tissue adjacent to breast tumors, and invasive breast carcinoma. To further evaluate the potential of these nuclear nano-morphology markers for risk stratification, we compare them with a broad spectrum of breast pathology entities with progressively increased risk for breast cancer.
Materials and methods
Spatial-domain low-coherence quantitative phase microscopy
Our previous publications have detailed reports about SL-QPM instrument and data analysis methods [21–25]. In brief, a broadband white light from Xe-arc lamp was collimated by a 4f imaging system and focused onto the sample by a low-numerical aperture objective. The reflectance- mode image was collected by a scanning imaging spectrograph (Acton Research, MA) and a charge-coupled device camera (Andor Technology, CT) that recorded a three-dimensional spatial-spectral intensity cube I(x, y, k) (k is the 8 free-space wavenumber), which arises from the interference between the scattered wave propagating inside the sample and the reference waves. We then mathematically transformed the data I(x, y, k) to obtain a two-dimensional optical path length difference (OPD) map from the cell nucleus.
The basic outline of the transformation is as follows: after removing the bias term, we took the pixel-wise Fourier transform of I(x, y, k) along the k dimension, giving us IF(x, y, z′), where z′ is the optical path length. We then extract the OPD for a fixed optical path length of interest, zp, using the equation OPD(x, y) = ∠I(x, y, z′)|z′=zp/(2k), where ∠I(x, y, z′) is the phase term and the free-space wavenumber k corresponds to λ0 = 550 nm. As a result, the OPD map OPD(x, y) for the cell nucleus can be obtained.
Standard operating protocol
All research was performed with the approval of the institutional review board at University of Pittsburgh. We used standard histology specimens from breast tissue biopsies. An experienced pathologist with expertise in breast pathology (RB) evaluated each slide and marked cells of interest from the target pathology.
We have established a standard operating procedure for processing histology specimens and acquiring, and analyzing the SL-QPM data. We extensively evaluated the contribution of various confounding factors, such as (1) variation in the staining level, (2) variation of tissue section thickness, (3) number of cells to be analyzed, (4) age of the sample, and (5) inter-user variability and found that the variation can be minimized to be within the system sensitivity (see Supplementary Methods for details).
First, all the tissue biopsies were processed according to the following standard clinical protocol: formalin-fixation, paraffin-embedding, sectioning at 4 micron thickness, paraffin-removal, hematoxylin and eosin (H&E)-staining, and coverslipping. We then evaluated factors that may alter the nano-morphology markers, including effect of stain variation (H&E), tissue section thickness, and number of cell nuclei required for SL-QPM analysis. We developed a correction model to account for the stain-induced variations in SL-QPM analysis, described in detail in our earlier publication [24] and in the Supplementary Methods. We confirmed that the variation among different microtomes and tissue section thickness has minimal effect on the SL-QPM analysis, within the system sensitivity (shown in Supplementary Methods). We also determined the minimum number of cell nuclei required for SL-QPM analysis: upon calculating the average value of each marker for different numbers (from 10 to 120) of cell nuclei, we found that a minimal of 40–60 cell nuclei were required to obtain a reliable statistical average for each SL-QPM marker. We also examined the histology slides ranging from 2004 to 2010 having identical diagnosis classification and found that the age of the slide did not significantly affect the SL-QPM-derived markers (P = 0.5, data not shown). Further, the SL-QPM data acquisition and analysis performed by two users produced the same SL-QPM marker values for the same set of specimens (P = 0.8).
Statistical analysis
We extracted the nano-morphology markers from the quantitative analysis of the OPD map from each cell nucleus, using two simple statistical parameters: average nuclear OPD over the two-dimensional OPD map OPDc (x, y) of the entire cell 〈OPD〉, which correlates with nuclear density, and intra-nuclear standard deviation of OPD σOPD, which describes the structural heterogeneity within the cell nucleus. To obtain the characteristic value for an individual patient, we obtained the mean value of 〈OPD〉 and σOPD by taking the average value of 40–60 cell nuclei from each patient, denoted as “(〈OPD〉)p” and “(σOPD)p”, respectively.
The statistical comparison between two patient groups was obtained using Wilcoxon’s rank-sum test at 95 % confidence interval, and two-sided P values were used for all analyses. A P value of 0.05 or less is considered as statistical significance. We developed a multivariate logistic regression prediction model using two variables (〈OPD〉)p and (σOPD)p. The cross-validated receiver operating characteristic (ROC) curves were calculated using the logistic regression model and leave-one-out cross-validation [26]. Specifically, we utilized logistic regression to calculate the probability of cancer by combining the two optical markers (〈OPD〉)p and (σOPD)p. Logistic regression is a statistical approach that constructs the probability of a positive diagnosis (i.e., cancer) using the equation: , where I is the constructed index, p is the probability of a positive diagnosis (i.e., cancer), and β0, β1, and β2 are the coefficients determined by the logistic regression model fit provided by SAS version 9.0 (SAS Institute). To avoid optimistically biased estimates, we used leave-one-out cross-validation on a logistic regression model. In each logistic regression model fit, we left one observation (i.e., one patient) out and utilize all other observations to obtain the regression coefficients. These coefficients and the two optical markers (with the left-out observation) were then used to calculate the probability of cancer, according to the above equation. This process was repeated for all of the observations to produce a set of predicted probabilities. Then at various probability threshold settings (from 0 to 1), we calculated the fraction of true positives out of the positives (TPR = true positive rate) and the fraction of false positives out of the negatives (FPR = false positive rate). The ROC curve was created by plotting TPR vs. FPR. The discriminant power of the model was assessed by computing the area under the ROC curve.
Results
Categorization of human specimens
We analyzed cells on the glass slides of histology specimens from a total of 154 patients classified in 6 categories according to criteria in the literature [27, 28] (shown in Table 1), ranked according to the progressively increasing risk for breast cancer [28]: (1) normal patients from reduction mammoplasty; (2) patients with non-proliferative benign breast lesions; (3) patients with proliferative breast lesions without atypia; (4) patients with proliferative breast lesions with atypia; (5) patients with invasive carcinoma, in which histologically normal-appearing cells located within 1 cm from malignant tumor, referred to as “malignant-adjacent” normal were analyzed; and (6) patients with invasive carcinoma in which malignant cells were analyzed.
Table 1.
Patients categorized from target tissue
| Categories | Tissue source (patient number, n) | Age (year, mean ± SD) | Most advanced pathological diagnosis (patient number, n) | |
|---|---|---|---|---|
| 1 | Reduction mammoplasty (24) | 38.3 ± 10.9 | Normal (24) | |
| 2 | Non-proliferative benign lesions (14) | 44.4 ± 15.9 | Fibrocystic changes (2) | |
| Fibroadenoma (9) | ||||
| Apocrine metaplasia (3) | ||||
| 3 | Proliferative lesions without atypia (10) | 49.5 ± 13.8 | Intraductal papilloma (3) | |
| Ductal epithelial hyperplasia (5) | ||||
| Sclerosing adenosis (2) | ||||
| 4 | Proliferative lesions with atypia (15) | 53.7 ± 11.9 | Atypical ductal hyperplasia (10) | |
| Atypical lobular hyperplasia (5) | ||||
| 5 | ‘‘Malignant-adjacent’’ normal (32) | 60.1 ± 14.5 | Malignant (32) | |
| Stage I (16) | ER-positive (26) | |||
| Stage II (10) | PR-positive (23) | |||
| Stage III (2) | HER2-positive (3) | |||
| No staging data (4) | ||||
| 6 | Invasive carcinoma (59) | 60.7 ± 12.2 | Malignant (59) | |
| Stage I (24) | ER-positive (50) | |||
| Stage II (15) | PR-positive (46) | |||
| Stage III (15) | HER2-positive (6) | |||
| Stage IV (1) | ||||
| No staging data (4) | ||||
OPD map from the cell nuclei
Figure 1 shows representative pseudo-color OPD maps from cell nuclei for each of the 6 categories. The color and spatial distribution in these OPD maps reveal a progressive change from Categories 1 to 6, which correlates with the status of the patient’s most advanced pathological diagnosis. The OPD maps from normal and benign categories (Categories 1–2) showed similar pattern and then modest difference from those of proliferative lesions (Categories 3–4). Most importantly, the OPD maps from “malignant-adjacent” normal cells (Category 5), although histologically normal-appearing, exhibited a great similarity to those of malignant cells (Category 6), indicative of cancer-like signatures in these “malignant-adjacent” normal cells.
Fig. 1.
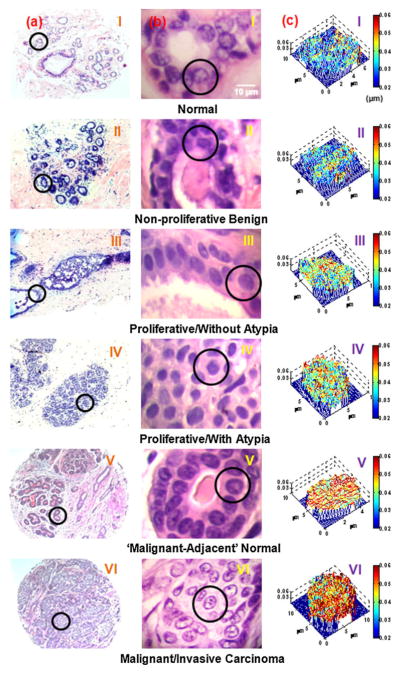
Representative conventional images of breast biopsies and structure-derived OPD map from a single cell nucleus. a Wide field histology images, b high-magnification histology images of breast tissue biopsies, and c corresponding OPD maps of the cell nuclei (marked in circles) from (I) normal cells from a healthy patient (Category 1); (II) cells labeled as fibrocystic changes from a non-proliferative benign patient (Category 2); (III) cells labeled as ductal epithelial hyperplasia from a patient with concurrent apocrine metaplasia and cystic changes (Category 3); (IV) cells labeled as atypical lobular hyperplasia (Category 4); (V) cells labeled by the expert breast pathologist as “normal” from a patient with invasive breast carcinoma (Category 5, “malignant-adjacent” normal); and (VI) cells labeled as “malignant” from a patient with invasive breast carcinoma (Category 6). Scale bar in the image indicates 10 μm. The color bar represents the OPD value from the cell nucleus
Nuclear nano-morphology markers of histologically normal cells detects the presence of malignancy
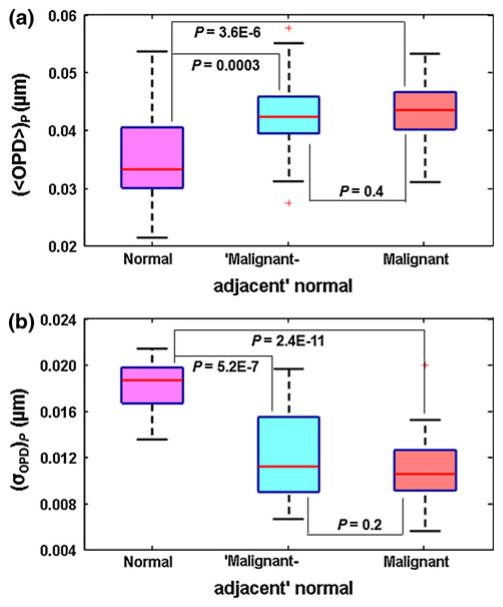
To investigate whether the signatures reflected in these OPD maps from “malignant-adjacent” normal cells can detect the “field effect” in breast cancer, we extracted two nano-morphology markers, average nuclear OPD 〈OPD〉 and standard deviation of OPD σOPD from the OPD map of each cell nucleus, and their mean value of 40–60 cell nuclei is used as the characteristic marker for each patient, denoted as “(〈OPD〉)p” and “(σOPD)p”, respectively. We compared the nano-morphology markers from patients with these “malignant-adjacent” normal cells to healthy patients (reduction mammoplasty, Category 1) and invasive cancer patients with malignant cells (Category 6). As shown by box-and-whisker plots in Fig. 2, although normal cells from healthy patients (Category 1) and normal cells from Category 5—“malignant-adjacent” normal were marked as “normal” by the expert breast pathologist, their nano-morphology markers ((〈OPD〉)p and (σOPD)p) exhibit distinct and highly statistically significant changes (P = 0.0003 for (〈OPD〉)p and P = 5.2E–7 for (σOPD)p). The difference between normal (Category 1) and malignant (Category 6) categories were even more pronounced (P = 3.6E–6 for (〈OPD〉)p and P = 2.4E–11 for (σOPD)p). The statistical analysis confirmed that the nano-morphology markers from “malignant-adjacent” normal cells are indeed distinct from normal tissue from healthy patients, with a great similarity to malignant cells. This result suggests that the nano-morphology markers detect the “field effect” in breast carcinogenesis. We also confirmed that this result was not due to the artifact of staining (See Supplementary Methods for details).
Fig. 2.
Box-and-whisker plots showing the nuclear nano-morphology markers for healthy patients with normal cells (Category 1), invasive cancer patients with “malignant-adjacent” normal cells (Category 5), and invasive cancer patients with malignant cells (Category 6): a average nuclear OPD ((〈OPD〉)p), b intra-nuclear standard deviation ((σOPD)p). For each patient, we used the mean value of 〈OPD〉 and σOPD by averaging ~40–60 cell nuclei
Nuclear nano-morphology markers parallel breast cancer risk
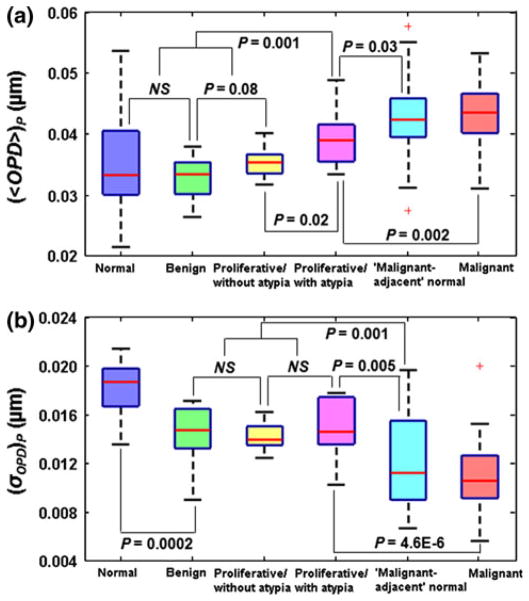
As the histologic appearance of benign lesions is strongly associated with the risk of breast cancer, we further evaluate these nuclear nano-morphology markers among the broad spectrum of breast pathology entities (Categories 1–6) with progressively increased cancer risk. Figure 3 shows the box-and-whisker plots of the nuclear nano-morphology markers of (〈OPD〉)p and (σOPD)p for all 6 categories. The statistical mean of both (〈OPD〉)p and (σOPD)p reveals a progressive change that parallels breast cancer risk. As shown in Fig. 3a, in Categories 1–3 (normal, non-proliferative benign, and proliferative lesions without atypia), the statistical mean values of (〈OPD〉)p show a progressive increase, but without statistical significance (ANOVA, P > 0.05). However, the value of (〈OPD〉)p from Category 4 (proliferative lesions with atypia) is significantly increased compared to those from Categories 1 to 3 (P = 0.001). This is in agreement with the clinical findings that no significantly increased risk (relative risk of 1–1.88, reported in Ref. [28]) are found in normal, non-proliferative benign and proliferative lesions without atypia, but the cancer risk significantly increases in proliferative lesions with atypia (relative risk of 4.24, reported in Ref. [28]). Further, the values of this marker from all of non-cancerous lesions (Categories 1–4) are significantly lower than those of “malignant-adjacent” normal category (Category 5) and malignant category (Category 6). In particular, for patients with proliferative lesions with atypia (Category 4), the value of (〈OPD〉)p is significantly lower than that of “malignant-adjacent” normal cells (Category 5) (P = 0.03), but significantly higher than those of proliferative lesions without atypia (Category 3) (P = 0.02). Given that proliferative lesions with atypia are often considered as high-risk lesions, this marker characteristic of breast cancer “field effect” shows the potential to detect patients who truly have malignant lesions in this high-risk group (proliferative lesions with atypia).
Fig. 3.
Box-and-whisker plots showing the changes in nuclear nano-morphology markers from all 6 categories with a broad range of breast pathology entities: a average nuclear OPD ((〈OPD〉)p) and b intra-nuclear standard deviation of OPD ((σOPD)p). For each patient, we used the mean value of 〈OPD〉 and σOPD by averaging ~40–60 cell nuclei. If P value is 0.1 or larger, “NS” is used
On the other hand, the intra-nuclear heterogeneity marker of (σOPD)p (Fig. 3b) distinguishes normal patients with reduction mammoplasty (Category 1) from patients with non-proliferative benign (Category 2) and proliferative lesions (Categories 3 and 4) with statistical significance (P = 0.0002), suggesting that (σOPD)p also is sensitive to abnormal changes in the breast tissue, even for low-risk benign conditions. The values of (σOPD)p do not show any statistical difference among non-proliferative benign and proliferative lesions (Categories 2–4), but they are significantly higher than those from “malignant-adjacent” normal (Category 5) (P = 0.001) and malignant categories (Category 6). Importantly, the (σOPD)p in proliferative lesions with atypia (Category 4) distinguishes well from “malignant-adjacent” normal cells (Category 5) with statistical significance (P = 0.005). This result indicates that these two nano-morphology markers provide complementary information that can be used as potential characteristic markers for the detection of breast cancer “field effect” and risk-stratification.
Nuclear areas
As the nuclear size is often used as one of the major criteria in conventional pathology, we evaluated whether the quantitative analysis of nuclear size can also detect the “field effect” of breast cancer or development of breast tumorigenesis. We calculated the average nuclear area as a parameter to measure the nuclear size and their mean value of 40–60 cell nuclei was used as the characteristic marker for each patient. As shown in Fig. 4, we found that the average nuclear size does not show any statistical significance between normal patients from reduction mammoplasty (Category 1) and patients with “malignant-adjacent” normal cells (Category 5). These “malignant-adjacent” normal cells also do not show any statistically significant difference, when compared to benign and proliferative lesions. Not surprisingly, the only significant change is seen between malignant cells (Category 6) and those cells from reduction mammoplasty, benign, proliferative lesions and “malignant-adjacent” normal cells (Categories 1–5) (P < 1E–9), in agreement with conventional pathology. This result confirms that breast cancer “field effect” is not detectable by quantitative evaluation of H&E slides by conventional light microscopy. The nuclear nano-morphology markers are the nanoscale structural information that is otherwise unattainable using conventional microscopy.
Fig. 4.
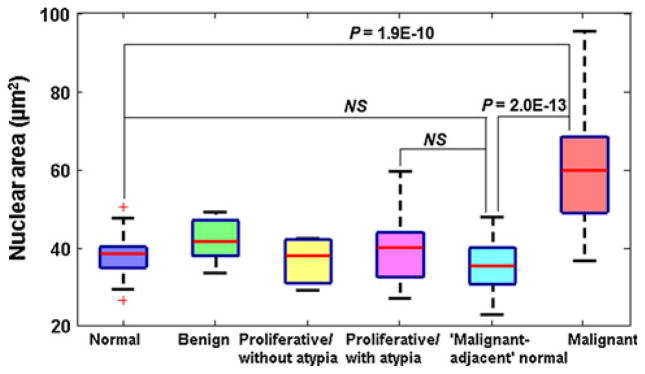
Box-and-whisker plots showing the average nuclear area from the 6 categories of breast pathology entities. The average nuclear area of normal cells and “malignant-adjacent” normal cells are not statistically significant (P = 0.2). If P value is 0.1 or larger, “NS” is used
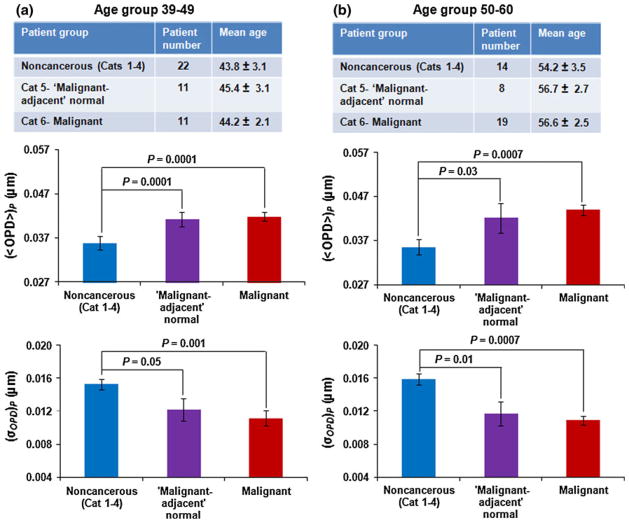
Nuclear nano-morphology markers do not reflect age-related changes
Because age can be an important confounding factor [29, 30], we wanted to ensure that our observed changes in nuclear nano-morphology markers reflected breast carcinogenesis rather than age difference. We compared (〈OPD〉)p and (σOPD)p for specimens divided into two age groups: 39–49 and 50–60 years (see tables in Fig. 5 for age break-down by Category). We combined data from Categories 1–4 into a non-cancerous group. As shown in Fig. 5, the nuclear nano-morphology markers of the non-cancerous group differ significantly from both “malignant-adjacent” normal (Category 5) and malignant categories (Category 6) (P < 0.05), with the similar progressive changes as shown in Figs. 2 and 3.
Fig. 5.
Statistical analysis of nuclear nano-morphology markers in age-matched patient groups: a 39–49-year-old patients and b 50–60-year-old patients. The non-cancerous group includes Categories 1–4. The error bar represents standard error
Nuclear nano-morphology markers are not sensitive to tumor subtype
We also looked for potential differences in nuclear nano-morphology markers based on the common breast tumor subtypes: estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). We used Spearman’s rank correlation method to analyze the correlation of (〈OPD〉)p and (σOPD)p from Categories 5 and 6 (“malignant-adjacent” normal and malignant) to the expression status of ER, PR, and HER2 (positive vs. negative). We found no correlation between these nuclear nano-morphology markers and ER, PR, and HER2 status (P > 0.1).
Performance characteristics
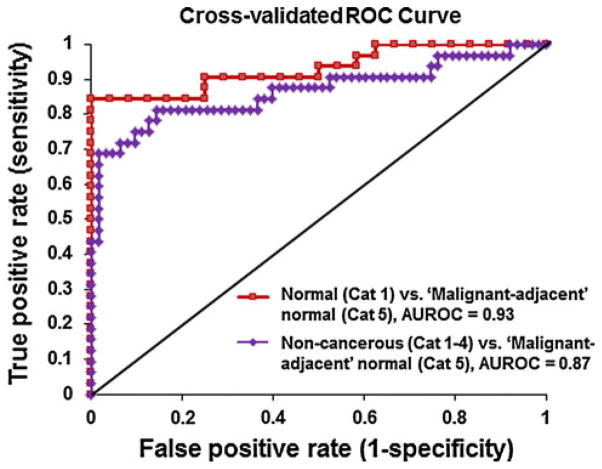
Our results showed statistically significant correlation between the nuclear nano-morphology markers and breast cancer “field effect”. To evaluate the performance characteristics of using nuclear nano-morphology markers to detect breast cancer “field effect” for potential clinical use, we calculated ROC curves. To avoid optimistically biased estimates, we used leave-one-out cross-validation on a logistic regression model using variables (〈OPD〉)p and (σOPD)p. The discriminant power of the model was assessed by calculating the area under the ROC curve (AUROC). As shown in Fig. 6, the nano-morphology markers from “malignant-adjacent” normal cells (Category 5) are well distinguished from those from normal patients (Category 1) with a high level of discriminatory accuracy of 0.93, as measured by AUROC. Further, these nano-morphology markers from the “malignant-adjacent” normal cells (Category 5) can also be distinguished from a broad spectrum of non-cancerous lesions, including those from non-proliferative benign, proliferative lesions without and with atypia (Categories 1–4), with a discriminatory accuracy of 0.87. These results support the ability of these nano-morphology markers to detect the “field effect” of breast cancer with a high level of accuracy, which may have a potential clinical use in early detection of malignancy and risk stratification.
Fig. 6.
Performance characteristics of nuclear nano-morphology markers described by cross-validated ROC curves to distinguish “malignant-adjacent” normal (Category 5) from normal (Category 1) and non-cancerous lesions (Categories 1–4). The discriminatory accuracy was assessed by the area under the ROC curve (AUROC)
Discussion
Exploitation of the mammary field cancerization to improve cancer diagnosis, risk stratification, prevention and treatment is a well-recognized clinical goal [3–5]. As we previously discussed, the current tools lack sufficient performance characteristics and the applicability for routine clinical use. Thus, the finding of an accurate and clinically applicable marker for breast cancer “field effect” would be of major clinical importance.
The application of the newly developed microscopy system, SL-QPM, to analyze standard histology specimens, has a great potential due to its simplicity and its superior sensitivity to nanoscale structural changes from cell nuclei. In this proof-of-concept study, we show, for the first time, that the SL-QPM-derived nano-morphology markers from cell nuclei can detect the presence of breast malignancy in histologically normal-appearing cells adjacent to the tumor (“field effect”), with a discriminatory accuracy of 0.93. Such superior performance to detect breast cancer “field effect” shows a great potential for routine clinical use. Furthermore, the nuclear nano-morphology markers sequentially progress in parallel with breast cancer risk, from normal breast epithelial tissue, to non-proliferative benign lesions, to proliferative lesions without atypia, to proliferative lesions with atypia, to “malignant-adjacent” normal tissue, and finally to malignant tumor. Thus, the progressive changes we observed in nuclear nano-morphology markers reflect early events during the neoplastic transformation of breast epithelial cells and support their relevance to breast carcinogenesis [27, 28, 31], suggesting their potential use for breast cancer risk stratification.
The genetic and epigenetic alterations of field carcinogenesis can lead to significant nanoscale structural consequences [15]. As numerous molecular changes associated with breast carcinogenesis have been reported in the “malignant-adjacent” histologically normal tissue, they could alter cellular processes during neoplastic transformation such as cell growth, repair, apoptosis, and cell cycle regulation and result in the observed changes in our nuclear nano-morphology markers. We noted that the significantly higher average nuclear OPD (〈OPD〉) has been associated with the subtle nanoscale increase in the nuclear density [32]. On the other hand, the alterations in the intra-nuclear standard deviation of OPD (σOPD) quantify the structural heterogeneity within the cell nucleus and may also be one of the earliest changes during breast carcinogenesis. Similar nano-structural changes have also been reported to detect the early stage carcinogenesis as a consequence of genetic alterations [16] and to be evidence of “field effect” in colon, lung, and pancreatic cancers [17, 18, 25].
Since SL-QPM can be directly applied to standard histology specimens and detect the breast cancer “field effect” with a high accuracy, it has a potential to be rapidly integrated into clinical practice. For example, as the absence of malignant cells often preclude the diagnosis of cancer, the histologically normal cells analyzed using SL-QPM that show cancer-like “field effect” signatures may suggest either an adjacent malignancy missed by core needle biopsy or that these “normal” cells have a higher cancer risk. However, we would like to emphasize that we do not intend to use this technique as a quantitative version of conventional pathology to distinguish the different pathological entities of breast lesions. Instead, we aim to utilize the ultra-high sensitivity of these SL-QPM-derived nano-morphology markers to detect breast field cancerization for clinical use to guide various aspects of breast cancer management.
Although our data are compelling, this is a proof-of-concept study and a number of limitations in this study need to be acknowledged. First, our sample size is modest. It is encouraging that even with this modest sample size, our technique can still detect the breast cancer “field effect” with a high level of statistical significance and an excellent accuracy. To mitigate the possibility of overfitting, we only used two most significant nano-morphology markers in the prediction model and performed leave-one-out cross-validation to avoid biased estimates in their performance characteristics. Second, this proof-of-concept investigation focuses on evaluating the potential of SL-QPM-derived nuclear nano-morphology markers to detect breast cancer “field effect”, and its intended clinical use in the context of a specific clinical scenario has not been demonstrated. The clinical utility of this technique needs to be further validated with a larger patient population. Ultimately, a large multicenter study will need to be performed for a specific clinical implication to guide diagnosis, risk assessment, prognosis, prevention and treatment.
In conclusion, we report that nuclear nano-morphology markers, derived from a new optical microscopy system (SL-QPM), show great promise to detect breast cancer “field effect” with a high accuracy. These nuclear nano-morphology markers are based on the detection of nanoscale structural characteristics from the “field effect” of breast carcinogenesis, which otherwise cannot be appreciated using light microscopy or digital image analysis. As this simple optical microscopy system analyzes standard histology slides, it can be readily integrated with conventional pathologic techniques. The utilization of this highly sensitive, simple and clinically applicable tool to detect breast cancer “field effect” has potential clinical utilities in bringing a “personalized” approach for cancer diagnosis, prevention and treatment. We are continuing to evaluate the various clinical utilities of this technique and to examine other nano-structural properties that may further improve performance.
Supplementary Material
Acknowledgments
This study was supported by research grants from National Institute of Health (R21CA152935) and Wallace H. Coulter foundation. P. W. acknowledges the support from National Energy Technology Laboratory Research Participation Program sponsored by the U.S. Department of Energy and administered by the Oak Ridge Institute for Science and Education.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-012-2125-2) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Rajan K. Bista, Division of Gastroenterology, Hepatology and Nutrition, Department of Medicine, University of Pittsburgh, Pittsburgh, PA 15232, USA
Pin Wang, Division of Gastroenterology, Hepatology and Nutrition, Department of Medicine, University of Pittsburgh, Pittsburgh, PA 15232, USA.
Rohit Bhargava, Department of Pathology, Magee-Womens Hospital, University of Pittsburgh Medical Center, Pittsburgh, PA 15213, USA.
Shikhar Uttam, Division of Gastroenterology, Hepatology and Nutrition, Department of Medicine, University of Pittsburgh, Pittsburgh, PA 15232, USA.
Douglas J. Hartman, Department of Pathology, University of Pittsburgh School of Medicine, Pittsburgh, PA 15232, USA
Randall E. Brand, Division of Gastroenterology, Hepatology and Nutrition, Department of Medicine, University of Pittsburgh, Pittsburgh, PA 15232, USA
Yang Liu, Email: liuy@pitt.edu, Division of Gastroenterology, Hepatology and Nutrition, Department of Medicine, University of Pittsburgh, Pittsburgh, PA 15232, USA. Department of Bioengineering, University of Pittsburgh, Pittsburgh, PA 15219, USA. Departments of Medicine and Bioengineering, University of Pittsburgh, 5117 Centre Ave, HCCLB 2.32, Pittsburgh, PA 15232, USA.
References
- 1.Kopelovich L, Henson DE, Gazdar AF, Dunn B, Srivastava S, Kelloff GJ, Greenwald P. Surrogate anatomic/functional sites for evaluating cancer risk: an extension of the field effect. Clin Cancer Res. 1999;5(12):3899–3905. [PubMed] [Google Scholar]
- 2.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6(5):963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 3.Heaphy CM, Griffith JK, Bisoffi M. Mammary field cancerization: molecular evidence and clinical importance. Breast Cancer Res Treat. 2009;118(2):229–239. doi: 10.1007/s10549-009-0504-0. [DOI] [PubMed] [Google Scholar]
- 4.Chai H, Brown RE. Field effect in cancer-an update. Ann Clin Lab Sci. 2009;39(4):331–337. [PubMed] [Google Scholar]
- 5.Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274(5295):2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 7.Harada Y, Katagiri T, Ito I, Akiyama F, Sakamoto G, Kasumi F, Nakamura Y, Emi M. Genetic studies of 457 breast cancers. Clinicopathologic parameters compared with genetic alterations. Cancer. 1994;74(8):2281–2286. doi: 10.1002/1097-0142(19941015)74:8<2281::aid-cncr2820740812>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 8.Larson PS, de las Morenas A, Cupples LA, Huang K, Rosenberg CL. Genetically abnormal clones in histologically normal breast tissue. Am J Pathol. 1998;152(6):1591–1598. [PMC free article] [PubMed] [Google Scholar]
- 9.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7(12):937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 10.Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, Eng C, Yeatman TJ, Huang TH. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res. 2006;12(22):6626–6636. doi: 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- 11.Dworkin AM, Huang TH, Toland AE. Epigenetic alterations in the breast: implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol. 2009;19(3):165–171. doi: 10.1016/j.semcancer.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis CM, Cler LR, Bu DW, Zochbauer-Muller S, Milchgrub S, Naftalis EZ, Leitch AM, Minna JD, Euhus DM. Promoter hypermethylation in benign breast epithelium in relation to predicted breast cancer risk. Clin Cancer Res. 2005;11(1):166–172. [PubMed] [Google Scholar]
- 13.Heaphy CM, Bisoffi M, Fordyce CA, Haaland CM, Hines WC, Joste NE, Griffith JK. Telomere DNA content and allelic imbalance demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer. 2006;119(1):108–116. doi: 10.1002/ijc.21815. [DOI] [PubMed] [Google Scholar]
- 14.Kolquist KA, Ellisen LW, Counter CM, Meyerson M, Tan LK, Weinberg RA, Haber DA, Gerald WL. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat Genet. 1998;19(2):182–186. doi: 10.1038/554. [DOI] [PubMed] [Google Scholar]
- 15.Backman V, Roy HK. Light-scattering technologies for field carcinogenesis detection: a modality for endoscopic prescreening. Gastroenterology. 2011;140(1):35–41. doi: 10.1053/j.gastro.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian H, Pradhan P, Liu Y, Capoglu IR, Li X, Rogers JD, Heifetz A, Kunte D, Roy HK, Taflove A, Backman V. Optical methodology for detecting histologically unapparent nanoscale consequences of genetic alterations in biological cells. Proc Natl Acad Sci USA. 2008;105(51):20118–20123. doi: 10.1073/pnas.0804723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian H, Roy HK, Pradhan P, Goldberg MJ, Muldoon J, Brand RE, Sturgis C, Hensing T, Ray D, Bogojevic A, Mohammed J, Chang JS, Backman V. Nanoscale cellular changes in field carcinogenesis detected by partial wave spectroscopy. Cancer Res. 2009;69(13):5357–5363. doi: 10.1158/0008-5472.CAN-08-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy HK, Subramanian H, Damania D, Hensing TA, Rom WN, Pass HI, Ray D, Rogers JD, Bogojevic A, Shah M, Kuzniar T, Pradhan P, Backman V. Optical detection of buccal epithelial nanoarchitectural alterations in patients harboring lung cancer: implications for screening. Cancer Res. 2010;70(20):7748–7754. doi: 10.1158/0008-5472.CAN-10-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy HK, Hensing T, Backman V. Nanocytology for field carcinogenesis detection: novel paradigm for lung cancer risk stratification. Future Oncol. 2011;7(1):1–3. doi: 10.2217/fon.10.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bista RK, Brentnall TA, Bronner MP, Langmead CJ, Brand RE, Liu Y. Using optical markers of nondysplastic rectal epithelial cells to identify patients with ulcerative colitis-associated neoplasia. Inflamm Bowel Dis. 2011;17(12):2427–2435. doi: 10.1002/ ibd.21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, Bista R, Khalbuss WE, Qiu W, Staton K, Zhang L, Brentnall TA, Brand RE, Liu Y. Nanoscale nuclear architecture for cancer diagnosis beyond pathology via spatialdomain low-coherence quantitative phase microscopy. J Biomed Opt. 2010;15:066028. doi: 10.1117/1.3523618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, Bista RK, Qiu W, Khalbuss WE, Zhang L, Brand RE, Liu Y. An insight into statistical refractive index properties of cell internal structure via low-coherence statistical amplitude microscopy. Opt Express. 2010;18(21):21950–21958. doi: 10.1364/OE.18.021950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P, Bista R, Bhargava R, Brand RE, Liu Y. Spatialdomain low-coherence quantitative phase microscopy for cancer diagnosis. Opt Lett. 2010;35(17):2840–2842. doi: 10.1364/OL.35.002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uttam S, Bista RK, Hartman DJ, Brand RE, Liu Y. Correction of stain variations in nuclear refractive index of clinical histology specimens. J Biomed Opt. 2011;16(11):116013. doi: 10.1117/ 1.3650306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bista RK, Uttam S, Hartman DJ, Qiu W, Yu J, Zhang L, Brand RE, Liu Y. Investigation of nuclear nano-morphology marker as a biomarker for cancer risk assessment using a mouse model. J Biomed Opt. 2012;17(6):066014. doi: 10.1117/1.JBO.17.6.066014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonen M. Analyzing receiver operating characteristic curves with SAS. SAS Institute; Cary: 2007. [Google Scholar]
- 27.Dupont WD, Page DL. Risk factors for breast cancer in women with proliferative breast disease. N Engl J Med. 1985;312(3):146–151. doi: 10.1056/NEJM198501173120303. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann LC, Sellers TA, Frost MH, Lingle WL, Degnim AC, Ghosh K, Vierkant RA, Maloney SD, Pankratz VS, Hillman DW, Suman VJ, Johnson J, Blake C, Tlsty T, Vachon CM, Melton LJ, 3rd, Visscher DW. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353(3):229–237. doi: 10.1056/ NEJMoa044383. [DOI] [PubMed] [Google Scholar]
- 29.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7(4):536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 30.Issa JP. Epigenetic variation and human disease. J Nutr. 2002;132(8 Suppl):2388S–2392S. doi: 10.1093/jn/132.8.2388S. [DOI] [PubMed] [Google Scholar]
- 31.Santen RJ, Mansel R. Benign breast disorders. N Engl J Med. 2005;353(3):275–285. doi: 10.1056/NEJMra035692. [DOI] [PubMed] [Google Scholar]
- 32.Bista RK, Uttam S, Wang P, Staton K, Choi S, Bakkenist CJ, Hartman DJ, Brand RE, Liu Y. Quantification of nanoscale nuclear refractive index changes during the cell cycle. J Biomed Opt. 2011;16(7):070503. doi: 10.1117/1.3597723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.