Summary
Biofilm formation by Vibrio fischeri is a complex process involving multiple regulators, including the sensor kinase (SK) RscS and the response regulator (RR) SypG, which control the symbiosis polysaccharide (syp) locus. To identify other regulators of biofilm formation in V. fischeri, we screened a transposon library for mutants defective in wrinkled colony formation. We identified LuxQ as a positive regulator of syp-dependent biofilm formation. LuxQ is a member of the Lux phosphorelay and is predicted to control bioluminescence in concert with the SK AinR, the phosphotransferase LuxU, and the RR LuxO. Of these, LuxU was the only other regulator that exerted a substantial impact on biofilm formation. We propose a model in which the Lux pathway branches at LuxU to control both bioluminescence and biofilm formation. Furthermore, our evidence suggests that LuxU functions to regulate syp transcription, likely by controlling SypG activity. Finally, we found that, in contrast to its predicted function, the SK AinR has little impact on bioluminescence under our conditions. Thus, this study reveals a novel connection between the Lux and Syp pathways in V. fischeri, and furthers our understanding of how the Lux pathway regulates bioluminescence in this organism.
Keywords: Vibrio fischeri, biofilm, quorum sensing, phosphorelay, sensor kinase, phosphotransferase
Introduction
Bacteria readily adapt to changing environmental conditions by sensing and integrating different cues present in their surroundings to produce a response appropriate for survival or growth. One mechanism by which bacteria coordinate such responses is through the use of two-component systems, with the sensor kinase (SK) component involved in detecting and relaying an environmental cue, and the response regulator (RR) component promoting the response, often a change in gene expression (Stock et al., 2000, West & Stock, 2001). A classic example of signal integration via two-component regulators is the Lux pathway in the marine bioluminescent bacterium Vibrio harveyi (Ng & Bassler, 2009). This organism integrates distinct signals (small molecules termed autoinducers or AIs) using specific SKs that funnel their activities into a common phosphorelay pathway to control the production of cellular bioluminescence.
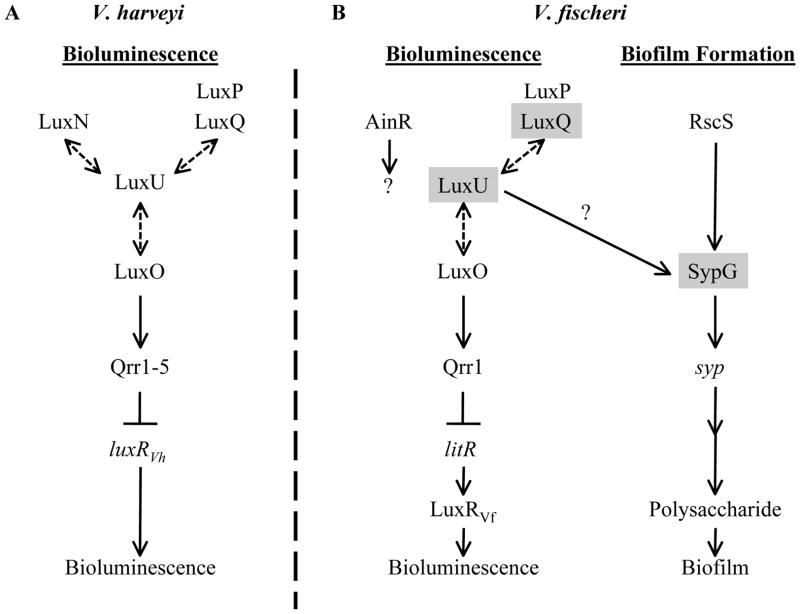
In V. harveyi, the Lux pathway (Fig. 1A) is composed of the SKs LuxQ (in association with the periplasmic protein LuxP), LuxN, and CqsS (not depicted), the histidine phosphotransferase LuxU, and the RR LuxO (reviewed in (Ng & Bassler, 2009)). Under low cell densities (low AI concentrations), the SKs exhibit net kinase activity and serve as phosphoryl-donors to LuxU, which serves as a phosphoryl-donor to the RR LuxO. Phosphorylated LuxO (LuxO~P) then promotes the transcription of five sRNAs (qrr1-5), which bind to and destabilize (in conjunction with Hfq) the transcript for the master transcriptional regulator LuxRVH. This regulator promotes the transcription of the lux operon, which encodes the proteins necessary for light production. Without LuxRVH, light is not produced. However, as the cell density increases, the AI concentrations also increase, causing a shift in the equilibrium of SK activity (i.e., from net kinase to net phosphatase activity). This switch to net phosphatase activity promotes the removal of phosphoryl groups from LuxO through LuxU. Without LuxO~P, LuxRVH is produced and promotes the expression of the lux operon, ultimately leading to light production.
Fig. 1. Models for the regulation by the Lux and Syp pathways.
(A) In V. harveyi, bioluminescence is regulated by the Lux phosphorelay, composed of the sensor kinases (SKs) LuxQ (which interacts with the periplasmic protein LuxP), LuxN, and CqsS (not depicted), the phosphotransferase LuxU, and the response regulator (RR) LuxO. Phosphoryl-transfer (dashed, double-sided arrows) occurs between the SKs, LuxU, and LuxO. Under low cell density conditions, LuxO is phosphorylated by the kinase activity of the SKs and activates transcription of the qrr sRNAs, which bind to the transcript of luxRVH and prevent its translation. Without LuxRVH, the lux operon (not depicted) is not expressed and light (bioluminescence) is not produced. At high cell densities, LuxO is dephosphorylated by the phosphatase activity of the SKs, leading to subsequent production of LuxRVH and bioluminescence. Autoinducers (AIs) or the AI synthases (LuxM, LuxS, or CqsA) are not depicted for simplicity.
(B) In V. fischeri, the Lux phosphorelay functions in largely the same manner to regulate bioluminescence, with homologs of LuxP, LuxQ, LuxU, and LuxO, but not CqsS or LuxN; V. fischeri encodes another putative SK, AinR. Additionally, V. fischeri uses the LuxRVH homolog, LitR, to activate transcription of LuxRVF (not similar to LuxRVH), which promotes transcription of the lux operon (when bound to the AI produced by LuxI) (not depicted), leading to subsequent light production (bioluminescence). Regulators shaded in gray indicate those found in this study to be involved in biofilm formation. RscS is an SK known to control biofilm formation. Phosphorylation of the RR SypG is predicted to activate transcription of the syp locus, which encodes proteins thought to regulate, produce, and transport a polysaccharide necessary for biofilm formation. The specific activity of LuxU in activating biofilm formation is unknown, but it appears from the current study to work at or above the level of syp transcription, likely at the level of SypG activation (indicated by a question mark). This figure is adapted from (Visick, 2005).
Similar pathways exist in a variety of other Vibrio species, including V. parahemolyticus, V. cholerae, V. anguillarum, V. vulnificus, and V. fischeri (Milton, 2006, Zhang et al., 2012). Like V. harveyi, the V. fischeri Lux pathway also controls bioluminescence (reviewed in (Stabb et al., 2008, Miyashiro & Ruby, 2012)). The V. fischeri Lux components appear similar to those in V harveyi (Fig. 1). For example, V. fischeri encodes (or is predicted to encode) homologs of LuxP, LuxQ, LuxU, and LuxO, which are proposed to function in a manner similar to their counterparts in V. harveyi (Miyamoto et al., 2000, Visick, 2005, Stabb et al., 2008, Miyashiro et al., 2010); however, differences between the Lux pathways in V. fischeri and V. harveyi exist. For example, V. fischeri does not encode a homolog of CqsS or LuxN, but encodes another sensor kinase, AinR, predicted to function at the same level as LuxQ (Gilson et al., 1995, Stabb et al., 2008, Miyashiro & Ruby, 2012). Furthermore, LuxO~P controls the transcription of a single qrr sRNA, which likely binds to and destabilizes the transcript of the transcriptional regulator LitR (a LuxRVH homolog) (Miyamoto et al., 2003, Miyashiro et al., 2010). Finally, LitR controls the transcription of an additional downstream regulator, LuxRVF (not similar to LuxRVH) (Fidopiastis et al., 2002), which activates the lux operon when bound by its AI (Stevens et al., 1994, Sitnikov et al., 1995). Thus, V. fischeri integrates AI cues to not only regulate the phosphorylation state of the RR LuxO, but also to regulate the transcription of the lux operon, adding another level of control to the production of cellular bioluminescence in this organism.
In addition to controlling bioluminescence, the Lux pathway in V. fischeri impacts other processes such as acetate metabolism (Studer et al., 2008) and motility (Lupp & Ruby, 2005, Hussa et al., 2007, Cao et al., 2012). Lux regulates a variety of processes in other Vibrios as well. For example, V. cholerae, V. anguillarum, V. vulnificus, and V. parahaemolyticus utilize the Lux pathway to control biofilm formation via regulators downstream of LuxO (Croxatto et al., 2002, Hammer & Bassler, 2003, Enos-Berlage et al., 2005, Lee et al., 2007, Zhang et al., 2012). For V. fischeri, no clear connection between the Lux pathway and biofilm formation has been observed, with the exception that a litR mutant exhibits a change in colony morphology (from translucent to opaque) consistent with a possible alteration in biofilm formation (Fidopiastis et al., 2002). Thus, it seems likely that V. fischeri could utilize the Lux pathway to regulate biofilm formation, but the exact role, if any, has yet to be determined.
V. fischeri is known to promote biofilm formation through the symbiotic polysaccharide (syp) locus (Fig. 1B). The syp locus is set of 18 genes thought to be involved in the regulation, production, and transport of a polysaccharide involved in biofilm formation (Yip et al., 2005, Yip et al., 2006, Shibata & Visick, 2012). This locus is transcriptionally controlled by the RR SypG, encoded within the syp locus, and σ54 (Yip et al., 2005). Another regulator of the syp locus is the SK RscS, which functions upstream of SypG to promote syp transcription (Yip et al., 2006, Hussa et al., 2008). Under standard laboratory conditions, V. fischeri does not form robust biofilms (i.e., wrinkled colony formation and pellicle production). However, robust biofilms that are dependent on the syp locus can be induced by overexpression of either rscS (Yip et al., 2006) or sypG; for SypG to induce biofilm formation, the biofilm inhibitor protein SypE must be absent or inactivated (Hussa et al., 2008, Morris et al., 2011). This enhanced biofilm production correlates with a competitive advantage for colonization by V. fischeri of its symbiotic host, the squid Euprymna scolopes (Yip et al., 2006, Morris et al., 2011).
In the current study, we sought to identify other components involved in biofilm formation by V. fischeri and found that the Lux pathway plays a role in this phenotype. Regulation of biofilm formation by the Lux pathway in V. fischeri is novel, as LuxU plays an important role but LuxO involvement is minimal. Our data indicate that the Lux pathway in V. fischeri branches at LuxU to regulate both bioluminescence and syp-dependent biofilm formation. Additionally, we have evidence to suggest that the SK AinR plays a minimal role in regulating light production and no role in regulating biofilm formation, under our conditions. Thus, this work provides new insight into the mechanism by which the Lux pathway functions in V fischeri, and also helps further our understanding of the complex regulatory network involved in controlling biofilm formation in this organism.
Results
Transposon mutagenesis reveals a regulatory connection between syp and lux
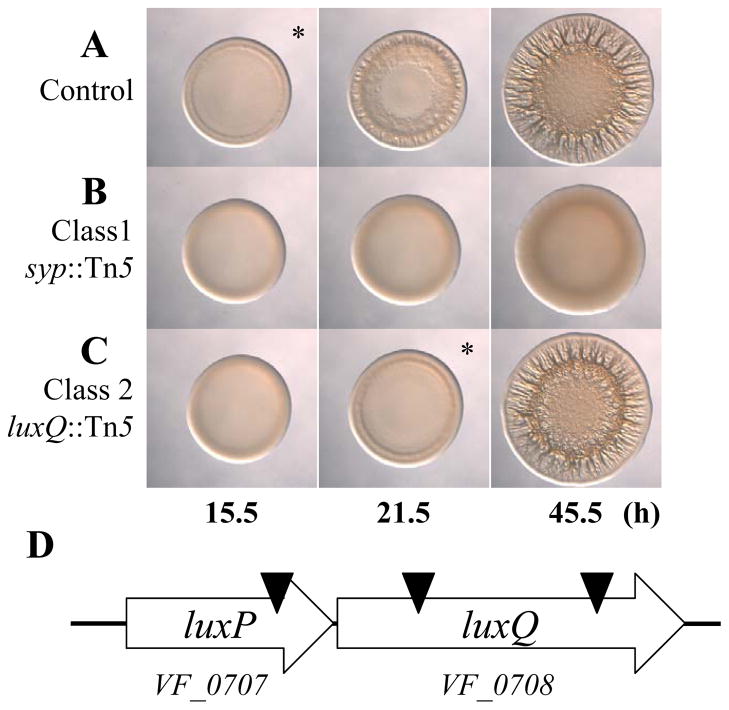
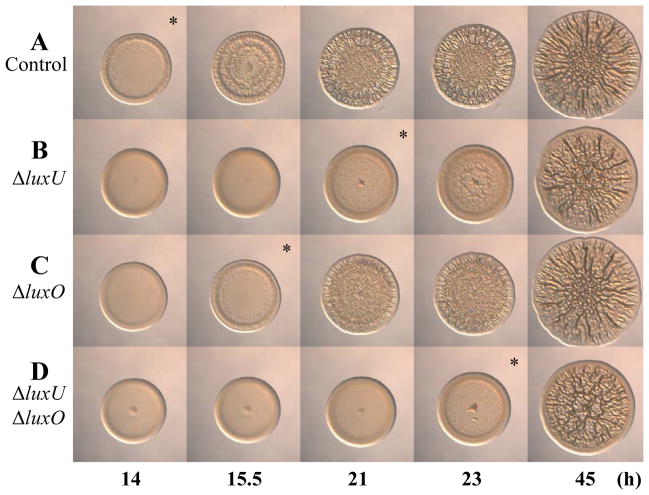
To better understand the requirements for biofilm formation in V. fischeri, we performed a random transposon mutagenesis of KV3299, a strain that lacks the syp biofilm inhibitor protein SypE. We then induced biofilm formation by introducing the sypG overexpression plasmid pEAH73; under these biofilm-inducing conditions, V. fischeri forms wrinkled colonies instead of smooth colonies (Hussa et al., 2008). We screened approximately 5000 mutants for those exhibiting a defect in wrinkled colony formation and found 27 that appeared to form smooth colonies. To verify the phenotypes of these mutants, we cured them of their sypG overexpression plasmid and then re-introduced it. All of the mutants remained defective in wrinkled colony formation and fell into two classes. Class 1 mutants (24 total) exhibited smooth colony morphology (compare Fig. 2A and B), while class 2 mutants (3 total) exhibited a substantial delay (approximately 6 h) in the start of wrinkled colony formation, but appeared similar to that of the parent strain at later times (compare Fig. 2A and C; asterisks indicate the time at which wrinkled colony formation is apparent, typically identified by ridge formation around the outer edge of the spot). To further evaluate these mutants, we performed Southern blot analysis on each mutant to determine whether the transposon had inserted within the syp locus, a location predicted to disrupt wrinkling. We found that the class 1 (smooth) mutants mapped within this locus, while the class 2 (delayed) mutants were unaltered in their syp regions (data not shown). These results confirm the assumption that wrinkled colony formation induced by sypG overexpression depends on the syp locus. Because our goal was to identify novel (non-syp) factors involved in biofilm formation, we pursued characterization of the class 2 mutants. Upon cloning and sequencing the DNA flanking the site of the Tn insertion in each class 2 mutant, we found that one insertion mapped near the end of VF_0707, while the other two were within VF_0708 (Fig. 2D). These genes are predicted to encode LuxP and LuxQ, respectively, two proteins proposed to be involved in controlling bioluminescence in V. fischeri (Fig. 1B).
Fig. 2.
Transposon mutagenesis reveals other regulators of biofilm formation in V. fischeri. (A–C). Time-course assays of wrinkled colony formation induced by sypG overexpression using plasmid pEAH73. Cultures were spotted onto LBS medium containing Tet and incubated at 28°C. Wrinkled colony formation was monitored up to 45.5 h post-spotting for the following strains: ΔsypE control (pEAH73/KV3299) (A), a representative class 1 mutant (pEAH73/KV5872; syp::Tn5 ΔsypE) (B), and a representative class 2 mutant (pEAH73/KV4431; luxQ::Tn5 ΔsypE) (C). An * indicates the time at which wrinkled colony formation was apparent, typically identified by the presence of ridges around the outer edge of the spot. Data are representative of at least three independent experiments. (D). A graphical depiction of the predicted luxPQ genes (block arrows) and Tn insertion sites (black triangles). There are 2 bp between the predicted translational stop site of luxP and the predicted translational start site of luxQ.
Loss of LuxQ affects bioluminescence and biofilm formation
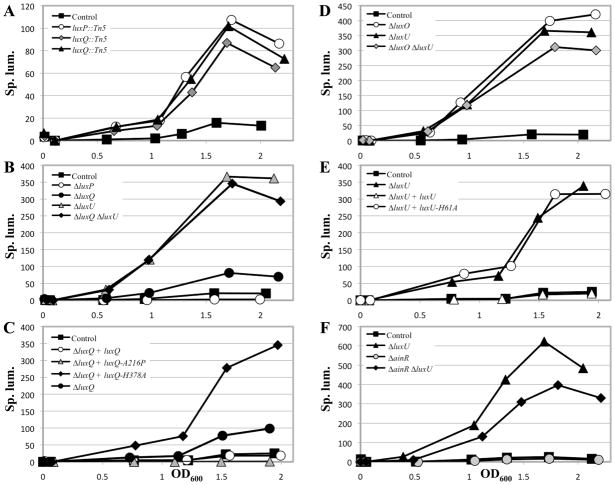
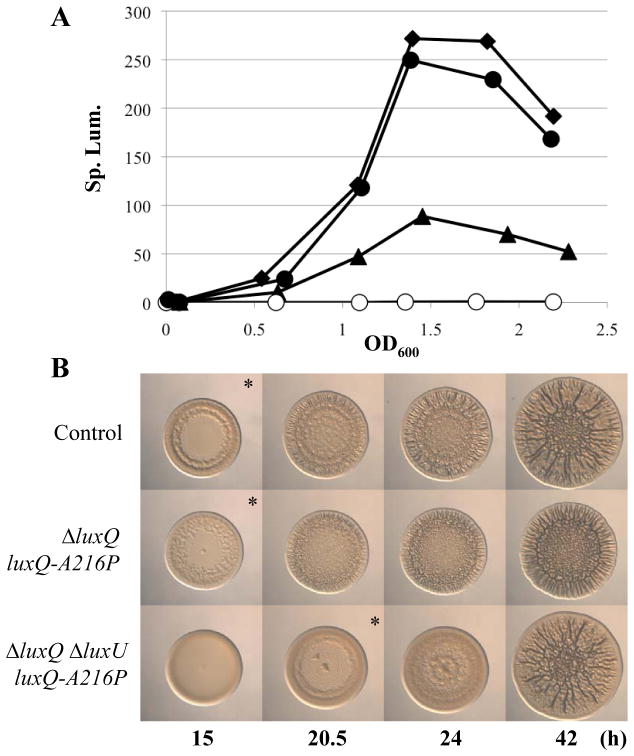
In V. fischeri, LuxP and LuxQ are predicted to regulate bioluminescence due to their sequence similarity to the well-characterized proteins of V. harveyi (58% and 44% identical, 72% and 67% similar, respectively) and to the functional conservation of other members of the lux regulatory pathway between V. harveyi and V. fischeri (reviewed in (Stabb et al., 2008)); however, the functions (bioluminescence or otherwise) of these two proteins in V. fischeri have not yet been assessed through mutagenesis studies. Thus, to understand the functions of these putative regulators, we asked whether these genes were involved in controlling bioluminescence, in addition to probing their role in controlling biofilm formation. Since the Tn insertions were in a ΔsypE background, it was first necessary to ask whether loss of SypE impacted luminescence; we found that it did not substantially impact luminescence (Fig. S1A), regardless of whether we used OD600 to estimate cell number (Fig. S1B) or determined the number of colony forming units (CFU) (Fig. S1C) to calculate the specific luminescence. Next, we assessed the impact of the Tn mutations on luminescence. The model (Fig. 1B), generated from work in V. harveyi (Bassler et al., 1994, Neiditch et al., 2005), predicts that a luxP mutant should fail to transmit the AI signal to LuxQ, causing LuxQ to remain a kinase; as a result, the levels of LuxO~P should be higher and luminescence should be lower. The model also predicts that the luxQ mutant should exhibit a decrease in LuxO~P levels, leading to increased luminescence. We found that all three mutants exhibited an increase in luminescence relative to their parent (ΔsypE) (Fig. 3A).
Fig. 3.
Luminescence of lux mutants in culture. Cultures were grown in SWTO and incubated at 24°C with vigorous shaking. Luminescence and OD600 were measured over time until maximum luminescence was achieved (between OD600 1.5 and 2). All data are plotted as specific luminescence (Sp. lum.; relative luminescence divided by OD600) versus OD600 and are representative of at least 3 independent experiments.
A. ΔsypE control (black squares; KV3299), luxP::Tn5 ΔsypE (white circles; KV4430), luxQ::Tn5 ΔsypE (grey diamonds; KV4431), luxQ::Tn5 ΔsypE (black triangles; KV4432)
B. ΔsypE control (black squares; KV3299), ΔluxP ΔsypE (white circles; KV5347), ΔluxQ ΔsypE (black circles; KV5394), ΔluxU ΔsypE (grey triangles; KV4830), ΔluxQ ΔluxU ΔsypE (black diamonds; KV6008)
C. ΔsypE attTn7::erm control (black squares; KV4390), ΔluxQ ΔsypE attTn7::erm (black circles; KV5973), ΔluxQ ΔsypE attTn7::luxQ-FLAG (white circles; KV5902), ΔluxQ ΔsypE attTn7::luxQ-A216P-FLAG (grey triangles; KV5904), ΔluxQ ΔsypE attTn7::luxQ-H378A-FLAG (black diamonds; KV5903)
D. ΔsypE control (black squares; KV3299), ΔluxO ΔsypE (white circles; KV5468), ΔluxU ΔsypE (black triangles; KV4830), ΔluxO ΔluxU ΔsypE (grey diamonds; KV5472)
E. ΔsypE attTn7::erm control (black squares; KV4390), ΔluxU ΔsypE attTn7::erm (black triangles; KV5974), ΔluxU ΔsypE attTn7::luxU-FLAG (white triangles; KV5905), ΔluxU ΔsypE attTn7::luxU-H61A-FLAG (white circles; KV5906)
F. ΔsypE control (black squares; KV3299), ΔainR ΔsypE (grey circles; KV6169), ΔluxU ΔsypE (black triangles; KV4830), ΔainR ΔluxU ΔsypE (black diamonds; KV6259)
The data for KV3299 in panel B are the same as that shown in panel D. The data for KV4390 in panel C are the same as that shown in panel E.
Since the luxP Tn mutant did not exhibit the predicted luminescence phenotype, we hypothesized that the Tn insertion, which was located at the end of luxP, was polar on luxQ. To test this prediction, we constructed in-frame deletions of both luxP (ΔluxP) and luxQ (ΔluxQ) in both the ΔsypE and wild-type backgrounds. Neither mutation impacted growth of V. fischeri (data not shown). Regardless of the background, loss of LuxP decreased bioluminescence, while loss of LuxQ increased bioluminescence as predicted (Fig. 3B and data not shown). The luminescence of the ΔluxQ mutant could be restored to the level of the luxQ+ control by expression of an epitope-tagged version of luxQ (luxQ-FLAG) in single copy from the chromosome (Fig. 3C). Together, these data indicate that: 1) LuxP and LuxQ are involved in controlling bioluminescence, as predicted, 2) the Tn insertion within luxP was polar on luxQ, and 3) luxP and luxQ likely comprise an operon (Fig. 2D).
Since the Tn insertion in luxP was polar on luxQ, we predicted that the 6 h delay in biofilm formation initially observed from the Tn mutants was likely due to loss or disruption of luxQ. To test this prediction, we examined wrinkled colony formation by the ΔluxQ (ΔsypE) mutant that overexpressed sypG. Like the Tn mutants, loss of LuxQ resulted in a similar delay in biofilm formation relative to the control (compare Fig. 4A & B). This delay in biofilm formation could be complemented by expression of luxQ-FLAG in single copy from the chromosome of the ΔluxQ mutant (Fig. 4A–C). Thus, LuxQ appears to control both bioluminescence and biofilm formation in V. fischeri. In contrast, the luxP mutation exerted relatively little effect on biofilm formation (Fig. S2); thus, we focused our subsequent studies on LuxQ and other Lux regulators.
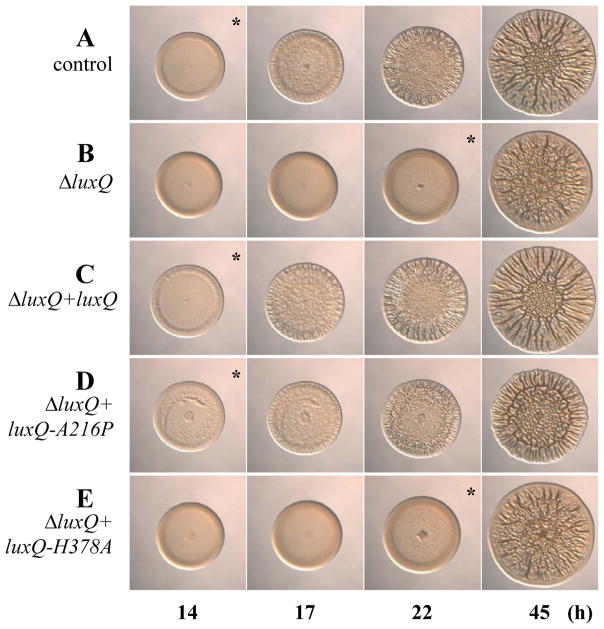
Fig. 4.
The effect of luxQ point mutations on biofilm formation. Time-course assays of wrinkled colony formation induced by sypG overexpression using plasmid pEAH73. Cultures were spotted onto LBS medium containing Tet and incubated at 28°C. Wrinkled colony formation was monitored up to 45 h post-spotting for the following strains: ΔsypE attTn7::erm control (pEAH73/KV4390) (A), ΔluxQ ΔsypE attTn7::erm (pEAH73/KV5973) (B), ΔluxQ ΔsypE attTn7::luxQ-FLAG (pEAH73/KV5902) (C), ΔluxQ ΔsypE attTn7::luxQ-A216P-FLAG (pEAH73/KV5904) (D), and ΔluxQ ΔsypE attTn7::luxQ-H378A-FLAG (pEAH73/KV5903) (E). An * indicates the time at which wrinkled colony formation was apparent, typically identified by the presence of ridges around the outer edge of the spot. Data are representative of at least three independent experiments.
LuxU exerts a more substantial impact on biofilm formation than LuxO
In V. harveyi, LuxQ functions through the phosphotransferase LuxU, to control the phosphorylation state of the RR LuxO (Fig. 1A) (Freeman & Bassler, 1999b, Freeman & Bassler, 1999a). Since LuxQ is involved in controlling biofilm formation in V. fischeri, we asked whether LuxU and LuxO were also involved. Thus, we generated deletions of both luxU (ΔluxU) and luxO (ΔluxO) in the ΔsypE background. However, it was necessary to first confirm that our mutants exhibited the predicted pattern of luminescence [i.e., increased bioluminescence; for luxO mutants, this has been previously reported (Lupp et al., 2003, Hussa et al., 2007)] (Fig. 1B). As expected, both mutants exhibited an increase in bioluminescence relative to their parent (Fig. 3D and Fig. S1). Neither mutant exhibited a growth defect (data not shown). Finally, a ΔluxU ΔluxO (ΔsypE) mutant exhibited a luminescence phenotype similar to that of the individual mutants (Fig. 3D). Overall, these data confirm that luxU functions to control bioluminescence in V. fischeri, as predicted.
Next, we introduced the sypG plasmid into the ΔluxU and ΔluxO mutants and assessed wrinkled colony formation. Similar to the loss of LuxQ, loss of LuxU resulted in a delay (about 7 h) in wrinkled colony formation (compare Fig. 5A & B). However, loss of LuxO resulted in only a slight, but reproducible delay (1.5 h) in wrinkled colony formation (compare Fig. 5A and C); we also observed the same slight delay for a luxO::kan mutant (data not shown), confirming the results of the ΔluxO mutant. These data suggest that, under our conditions, LuxU plays a more critical role than LuxO in controlling biofilm formation. These results also suggest that LuxU may function independently of LuxO to control biofilm formation. To investigate this possibility further, we evaluated biofilm formation by the ΔluxO ΔluxU (ΔsypE) mutant. We predicted that if LuxU functions through LuxO to regulate biofilm formation, then loss of both LuxU and LuxO would result in a phenotype similar to loss of LuxO alone (i.e., a 1.5 h delay). This was not the case: loss of both regulators resulted in an 8 h delay in wrinkled colony formation (compare Fig. 5A and D). This delay supports the hypothesis that the two regulators function independently to impact biofilm formation. Furthermore, this result contrasts with the luminescence results, in which the phenotypes of the luxU, luxO, and luxU luxO mutants were similar (Fig. 3D), and which suggest that LuxU likely functions through LuxO to control bioluminescence. Together, these data suggest that the Lux pathway branches at LuxU to control both bioluminescence and biofilm formation (Fig. 1B). Since loss of LuxU resulted in a more severe biofilm phenotype than loss of LuxO, we chose to pursue the role of LuxU (and its inputs) in the current study.
Fig. 5.
Wrinkled colony formation by luxU and luxO mutants. Time-course assays of wrinkled colony formation induced by sypG overexpression using plasmid pEAH73. Cultures were spotted onto LBS medium containing Tet and incubated at 28 C. Wrinkled colony formation was monitored up to 45 h post-spotting for the following strains: ΔsypE control (pEAH73/KV3299) (A), ΔluxU ΔsypE (pEAH73/KV4830) (B), ΔluxO ΔsypE (pEAH73/KV5468) (C), and ΔluxO ΔluxU ΔsypE (pEAH73/KV5472) (D). An * indicates the time at which wrinkled colony formation was apparent, typically identified by the presence of ridges around the outer edge of the spot. Data are representative of at least three independent experiments.
LuxQ kinase activity promotes biofilm formation
Our current data suggest that LuxQ functions as a positive regulator of biofilm formation under our conditions. Because this SK is predicted to function as both a kinase and a phosphatase (Freeman & Bassler, 1999a, Neiditch et al., 2006), we asked whether the ability of LuxQ to positively regulate biofilm formation depended upon its kinase and/or phosphatase activity. Previous work from V. harveyi had demonstrated that certain point mutations cause the loss of one activity but not the other (i.e., kinase activity is lost, while phosphatase activity is retained and vice versa) (Neiditch et al., 2006). Thus, we generated point mutations in the V. fischeri luxQ gene that are predicted to cause either loss of phosphatase activity (luxQ-A216P; kin+/phos−) or loss of kinase activity (luxQ-H378A; kin−/phos+), while retaining the other activity, respectively. We then expressed these luxQ alleles in single copy from the chromosome of the ΔluxQ mutant. To confirm that these LuxQ derivatives were functional, we examined their ability to control light production. According to the model (Fig. 1B) and work from V. harveyi (Neiditch et al., 2006), a phosphatase mutant (LuxQ-A216P, kin+/phos−) should exhibit a decrease in bioluminescence (due to an increase in LuxO~P), while a kinase mutant (LuxQ-H378A, kin−/phos+) should exhibit an increase in bioluminescence (due to a decrease in LuxO~P). Indeed, each mutant exhibited the expected pattern of luminescence (Fig. 3C), indicating that the proteins produced were functional and behaved as predicted.
We next assessed the ability of these alleles to complement the ΔluxQ mutant with respect to the timing of wrinkled colony formation. We found that the phosphatase mutant, LuxQ-A216P (kin+/phos−), could complement the luxQ mutant, restoring the timing of wrinkled colony formation to approximately that of the control strain (luxQ+) and the wild-type-complemented ΔluxQ mutant (compare Fig. 4A, C, and D). In contrast, the kinase mutant, LuxQ-H378A (kin−/phos+), failed to complement the luxQ mutant; this strain exhibited wrinkled colony formation that was indistinguishable from the ΔluxQ parent (compare Fig. 4A, B, and E). These data suggest that the kinase activity of LuxQ, but not its phosphatase activity, is necessary to regulate biofilm formation.
The impact of LuxQ on biofilm formation depends on LuxU
Our data indicate that LuxQ (specifically its kinase activity) and LuxU are necessary to regulate biofilm formation. According to the model (Fig. 1B), LuxQ is predicted to function through LuxU. To test this hypothesis, we first asked whether LuxQ functioned through LuxU to regulate bioluminescence. If this were the case, we would expect that a luxQ luxU mutant would phenocopy a luxU mutant, and indeed it did (Fig. 3B). To further evaluate this regulatory connection, we expressed the luxQ-A216P (kin+/phos−) allele in the luxQ luxU mutant. Whereas, in the context of the luxQ (luxU+) background this allele decreased luminescence, it failed to do so when luxU was also disrupted: the levels of luminescence produced by the luxQ luxU mutant expressing luxQ-A216P (kin+/phos−) were indistinguishable from that of the luxU mutant (Fig. 6A). These data suggest that LuxQ functions through LuxU to regulate bioluminescence.
Fig. 6. Luminescence and wrinkled colony formation by luxQ and luxU mutants.
(A) Luminescence of lux mutants in culture. Cultures were grown in SWTO and incubated at 24°C with vigorous shaking. Luminescence and OD600 were measured over time until maximum luminescence was achieved (between OD600 1.5 and 2) for the following strains: ΔluxQ attTn7::erm control (black triangles; KV5973), ΔluxU ΔsypE attTn7::erm (black diamonds; KV5974), ΔluxQ ΔsypE attTn7::luxQ-A216P-FLAG (white circles; KV5904), and ΔluxQ ΔluxU ΔsypE attTn7::luxQ-A216P-FLAG (black circles; KV6054). All data are plotted as specific luminescence (Sp. lum.; relative luminescence divided by OD600) versus OD600 and are representative of at least 3 independent experiments.
(B) Time-course assays of wrinkled colony formation induced by sypG overexpression using plasmid pEAH73. Cultures were spotted onto LBS medium containing Tet and incubated at 28°C. Wrinkled colony formation was monitored up to 42 h post-spotting for the following strains: ΔsypE attTn7::erm control (pEAH73/KV4390), ΔluxQ ΔsypE attTn7::luxQ-A216P-FLAG (pEAH73/KV5904), and ΔluxQ ΔluxU sypE attTn7::luxQ-A216P-FLAG (pEAH73/KV6054). An * indicates the time at which wrinkled colony formation was apparent, typically identified by the presence of ridges around the outer edge of the spot. Data are representative of at least three independent experiments.
Next, we asked whether LuxQ functioned through LuxU to control biofilm formation. We first evaluated biofilm formation by the luxQ luxU (sypE) mutant. We found that the double mutant exhibited a delay in wrinkled colony formation similar to that seen with the individual luxQ and luxU mutants (Fig. S3), rather than an additive delay. Thus, these results suggest that LuxQ and LuxU function in the same pathway to regulate biofilm formation. To probe this relationship further, we utilized the luxQ-A216P allele, which permits complementation of the luxQ mutation (Fig. 4). We hypothesized that if LuxU were necessary for LuxQ to regulate biofilm formation, then disruption of luxU in the luxQ mutant expressing luxQ-A216P should delay biofilm formation relative to the luxU+ strain. Indeed, this was the case (Fig. 6B). These data suggest that the ability of LuxQ to positively regulate biofilm formation depends upon LuxU.
Biofilm formation depends on the conserved site of phosphorylation in LuxU
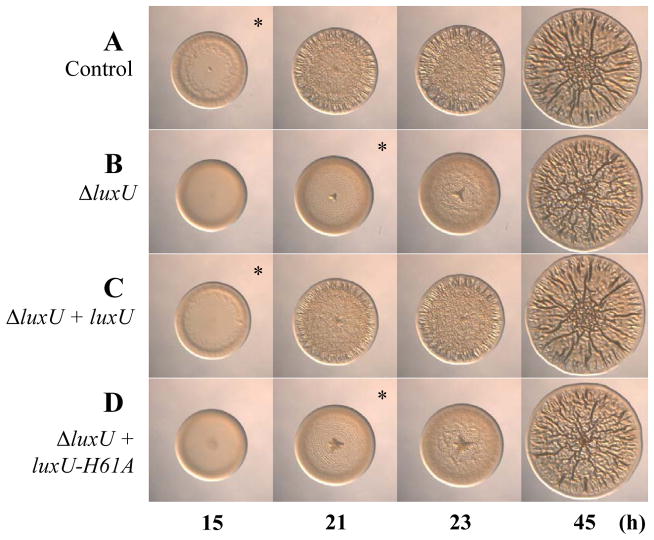
In V. harveyi, LuxU serves as a phosphotransferase, shuttling phosphoryl groups between the SKs and the RR LuxO (Fig. 1A). This role depends upon the conserved site of phosphorylation, His58 (Freeman & Bassler, 1999b). To determine whether the V. fischeri homolog functions in a similar manner, we first constructed an epitope-tagged version of luxU (luxU-FLAG). Expression of this allele in single copy from the chromosome of the ΔluxU mutant restored luminescence to that of the control (Fig. 3E), as well as the normal timing of wrinkled colony formation (compare Fig. 7A–C). Next, we substituted the predicted, conserved histidine for an alanine (H61A) in the luxU-FLAG construct and introduced this allele into the chromosome of the ΔluxU mutant. The ΔluxU mutant expressing the luxU-H61A allele failed to restore luminescence to the level of the parent (Fig. 3E) and exhibited the same 6 h delay in wrinkled colony formation as the uncomplemented ΔluxU mutant (compare Fig. 7B and D). To ensure that the lack of complementation was not due to a reduction or loss of the protein, we performed western blot analysis and found that protein was expressed from both alleles (Fig. S4). Together, these data suggest that the conserved site of phosphorylation in LuxU is necessary to regulate biofilm formation. Thus, it appears that key residues predicted to be involved in phosphotransfer are required for regulation of biofilm formation.
Fig. 7.
Wrinkled colony formation by complemented ΔluxU mutants. Time-course assays of wrinkled colony formation induced by sypG overexpression using plasmid pEAH73. Cultures were spotted onto LBS medium containing Tet and incubated at 28°C. Wrinkled colony formation was monitored up to 45 h post-spotting for the following strains: ΔsypE attTn7::erm control (pEAH73/KV4390) (A), ΔluxU ΔsypE attTn7::erm (pEAH73/KV5974) (B), ΔluxU ΔsypE attTn7::luxU-FLAG (pEAH73/KV5905) (C), and ΔluxU ΔsypE attTn7::luxU-H61A-FLAG (pEAH73/KV5906) (D). An * indicates the time at which wrinkled colony formation was apparent, typically identified by the presence of ridges around the outer edge of the spot. Data are representative of at least three independent experiments.
Loss of AinR has little impact on bioluminescence and no impact on biofilm formation
It has been predicted that, like the V. harveyi Lux pathway, multiple SKs feed into LuxU to control bioluminescence in V. fischeri (Visick, 2005, Stabb et al., 2008, Miyashiro & Ruby, 2012). In particular, the SK AinR is proposed to function at the same level as LuxQ to control bioluminescence in V. fischeri (Fig. 1B) (Gilson et al., 1995, Stabb et al., 2008). To determine whether AinR is involved in controlling bioluminescence and biofilm formation, we generated a ΔainR (ΔsypE) mutant and first assessed its luminescence phenotype; no study of AinR has assessed its role in controlling bioluminescence in liquid culture. The model (Fig. 1B) predicts that, similar to loss of LuxQ, loss of AinR would result in an increase in luminescence. However, this was not the case: loss of AinR resulted in a consistent but very slight decrease in luminescence as compared to the control (Fig. 3F and Fig. S5). To determine whether AinR functioned through the known phosphorelay pathway (i.e., through LuxU), we generated a ΔainR ΔluxU (ΔsypE) mutant and assessed its luminescence phenotype. We expected that the double mutant would exhibit the luminescence phenotype of the luxU single mutant. Surprisingly, this mutant consistently resulted in an intermediate luminescence phenotype: the ΔainR ΔluxU mutant was brighter than the ΔainR mutant, but not as bright as the ΔluxU mutant (Fig. 3F). These data suggested that AinR may play only a minor role in controlling bioluminescence under these conditions.
As a putative SK, AinR is predicted to recognize and respond to the autoinducer (AI) N-octanoyl-homoserine lactone (C8-HSL) (Gilson et al., 1995). Thus, the diminished luminescence phenotypes of the ΔainR and ΔainR ΔluxU mutants could result from a failure of this mutant to respond to C8-HSL. Alternatively, deletion of ainR could impact expression of the upstream gene ainS, which encodes the C8-HSL synthase protein. An impact on AinS synthesis could lead to decreased amounts of C8-HSL and decreased light production, potentially via direct control of the lux operon, as previously demonstrated (Kuo et al., 1996, Egland & Greenberg, 2000). To distinguish between these possibilities, we added exogenous C8-HSL to the ainR mutants and controls. We predicted that, if the former hypothesis were true, the ainR mutants would retain diminished luminescence relative to their controls. This appeared not to be the case, however, as addition of C8-HSL to the ΔainR and ΔainR ΔluxU mutants increased their luminescence levels to those of the control strain and the ΔluxU mutant, respectively (Fig. S5B). These data suggest that the decrease in luminescence by both the ΔainR and ΔainR ΔluxU mutants is likely due to decreased levels of C8-HSL, whose activity in promoting luminescence is largely or fully independent of the function of AinR, at least under our conditions. Thus, the role of AinR in controlling luminescence remains unclear.
Although AinR did not function as predicted in controlling bioluminescence, we wondered whether loss of AinR would impact biofilm formation. This was not the case: the ainR mutant exhibited no defect in biofilm formation, while the ΔainR ΔluxU mutant exhibited the defect of the luxU mutant (Fig. S6) and could be complemented when the wild-type allele of luxU-FLAG was expressed in single copy from the chromosome (data not shown). Thus, AinR has no impact on biofilm formation, and its role in controlling bioluminescence remains unclear. Further work will be necessary to determine what role AinR plays, if any, in controlling luminescence in V. fischeri.
LuxU, but not RscS, is necessary to regulate syp-dependent biofilm formation under SypG-inducing conditions
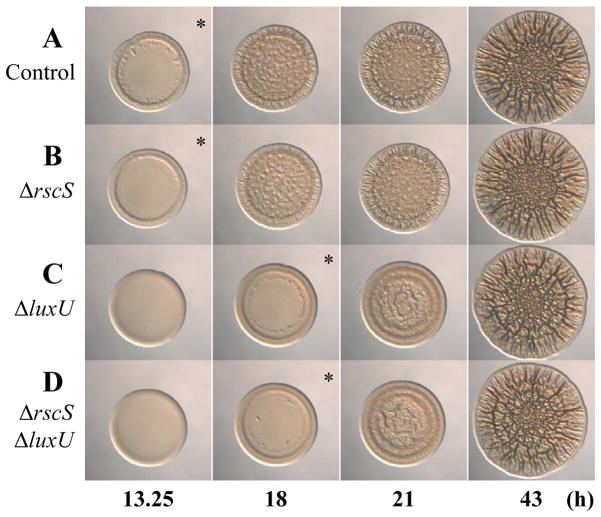
Since the only known role of LuxU is to serve as a phosphoryl-donor (Freeman & Bassler, 1999b, Shikuma et al., 2009) and LuxU impacts syp-dependent biofilm formation, we hypothesized that it could function upstream of SypG, a RR known to be required for transcription of the syp locus (Yip et al., 2005). Since previous studies had demonstrated that the SK RscS functions upstream of the RR SypG to control syp-dependent biofilm formation (Fig. 1B) (Yip et al., 2006, Hussa et al., 2008), we questioned the relative importance of these two potential inputs, RscS and LuxU, on SypG-induced biofilm formation. We thus generated sypE mutants with deletions in luxU, rscS, or both and evaluated SypG-induced biofilm formation. Surprisingly, only loss of LuxU exerted an impact: whereas the luxU mutant exhibited a delay in wrinkled colony formation (compare Fig. 8A and C), the rscS mutant showed no significant defect in biofilm formation under these conditions (compare Fig. 8A and B). Even when the rscS and luxU mutations were combined, this mutant exhibited the same delay as the luxU mutant alone could be complemented when the wild-type allele of luxU-FLAG was expressed in single copy from the chromosome (compare Fig. 8C and D and data not shown). Overall, these data indicate that LuxU plays a more important role than RscS in controlling biofilm formation when sypG is overexpressed.
Fig. 8.
Wrinkled colony formation by luxU and rscS mutants. Time-course assays of wrinkled colony formation induced by sypG overexpression using plasmid pEAH73. Cultures were spotted onto LBS medium containing Tet and incubated at 28°C. Wrinkled colony formation was monitored up to 43 h post-spotting for the following strains: ΔsypE control (pEAH73/KV3299) (A), ΔrscS ΔsypE (pEAH73/KV6268) (B), ΔluxU ΔsypE (pEAH73/KV4830) (C), and ΔrscS ΔluxU ΔsypE (pEAH73/KV6269) (D). An * indicates the time at which initiation of wrinkled colony formation was apparent, typically identified by the presence of ridges around the outer edge of the spot. Data are representative of at least three independent experiments.
LuxU functions at or above SypG to impact syp transcription
Since LuxU is necessary to promote syp-dependent biofilm formation, we sought to determine whether LuxU functioned upstream of SypG to control its activation (phosphorylation). If so, then we would expect that a phosphorylation-independent allele of SypG would be “blind” to the presence or absence of LuxU. We thus overexpressed a version of SypG in which the conserved site of phosphorylation, D53, was substituted for a glutamate (D53E). This substitution in RRs has previously been shown to promote the active state of the RR (Sanders et al., 1989, Sanders et al., 1992, Freeman & Bassler, 1999a). Indeed, this substitution in SypG caused an increase in SypG activity, as measured by syp transcription (Hussa et al., 2008). Consistent with this increased activity, when overexpressed in the ΔsypE mutant, sypG-D53E induced wrinkling sooner than when the wild-type allele of sypG was overexpressed (9–10 h vs. 13–15 h, respectively). When sypG-D53E was overexpressed in the ΔluxUΔsypE mutant, the timing of wrinkled colony formation was not delayed, but rather was similar to that of the luxU+ control (Fig. 9A). These data are consistent with a model in which LuxU functions at or above the level of SypG.
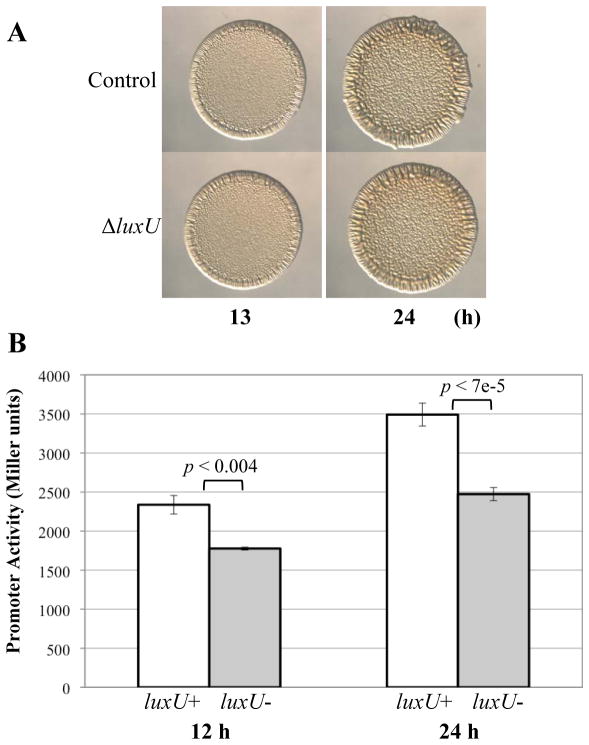
Fig. 9. The role of LuxU in syp activation.
(A) Time-course assays of wrinkled colony formation induced by sypG-D53E overexpression using plasmid pKV276. Cultures were spotted onto LBS medium containing Tet and incubated at 28°C. Wrinkled colony formation was monitored up to 24 h post-spotting for the following strains: ΔsypE control (pKV276/KV3299) and ΔluxU ΔsypE (pKV276/KV4830). Data are representative of at least three independent experiments.
(B) SypG-induced syp transcription from PsypA-lacZ reporter strains. Cultures of sypG overexpressing strains ΔsypE attTn7::PsypA-lacZ (white bars; pEAH73/KV4926) and ΔluxU ΔsypE attTn7::PsypA-lacZ (grey bars; pEAH73/KV5516) were inoculated in LBS containing Tet and grown at 28°C with shaking. Samples were collected at 12 and 24 h and assessed for β-galactosidase activity (in Miller units) as a measure of promoter activity. All experiments were performed in triplicate. Data are a combination of two independent experiments with error bars representing the standard error. The P-value refers to the variation between the two samples as indicated by the brackets.
It remains formally possible that the accelerated wrinkling effects of the sypG-D53E allele, combined with the delayed wrinkling caused by the loss of LuxU, results in a strain with a net timing of biofilm formation similar to the wild-type strain. Thus, to further probe the level at which LuxU exerts its impact on biofilm formation, we asked whether loss of LuxU affected transcription of the SypG-controlled sypA gene using a lacZ reporter fusion. We assayed β-galactosidase activity from the reporter expressed from the chromosomes of the ΔluxU and the luxU+ strains that overexpressed the wild-type allele of sypG. Loss of LuxU resulted in a decrease in syp transcription at two time points tested (12 and 24 h) (Fig. 9B). Thus, these data suggest that LuxU functions at or above the level of syp transcription, potentially due to an impact on SypG activation (Fig. 1B).
Discussion
In this study, we identified a novel connection between the Lux pathway and biofilm formation in V. fischeri. Specifically, we found that disruption of either the gene encoding the SK LuxQ or the gene encoding the phosphotransferase LuxU caused a delay in SypG-induced biofilm formation. Surprisingly, this effect was independent of LuxO, which exerted only a minor impact on biofilm formation. However, LuxU does seem to function through LuxO to regulate bioluminescence. Thus, the Lux pathway appears to branch at LuxU to regulate bioluminescence through LuxO and biofilm formation via a SypG-dependent pathway.
From the data presented in this work, we propose a model in which LuxQ functions through LuxU to regulate syp-dependent biofilm formation through activation of the RR SypG (Fig. 1B). Support for the idea that LuxQ and LuxU serve as phosphoryl-donors to a downstream regulator of biofilm formation is as follows: 1) the kinase activity of LuxQ is necessary to promote biofilm formation, 2) the predicted, conserved site of phosphorylation in LuxU is necessary to regulate biofilm formation, 3) the only known role of LuxU in the literature is as a histidine phosphotransferase (HPt) (Freeman & Bassler, 1999b, Shikuma et al., 2009), 4) LuxQ depends on LuxU to regulate biofilm formation, and 5) the downstream RR of the Lux pathway, LuxO, is not required for the effect of LuxU on biofilm formation. Together, these data suggest that phosphotransfer is necessary for LuxQ and LuxU to regulate biofilm formation via a regulator distinct from LuxO. In support of the idea that LuxU serves as an input to regulate the activity of the RR SypG, we found that: 1) a “constitutively active” allele of SypG overcomes the requirement for LuxU, and 2) LuxU functions at or above the level of syp transcription. Overall, these data suggest that LuxU functions at or above the level of SypG, potentially at the level of SypG phosphorylation. This possibility is further supported by the fact that SypG and LuxO have similar domain structures (both are σ54-dependent RRs) and exhibit 50% identity to each other. However, proof of such a possibility awaits additional biochemical experimentation; to date, attempts to examine the phosphorylation state of SypG have been unsuccessful. Thus, while our data support the hypothesis that LuxU could serve as a phosphoryl-donor to SypG, the regulation is clearly complex and may include currently unknown regulators.
It has been previously proposed, in two other Vibrio species, that LuxU can function independently of LuxO to control the activity of downstream targets of the Lux pathway. The first example is from the fish pathogen Vibrio anguillarum. In this organism, VanU functions through VanO to regulate the expression of the LuxRVH homolog, VanT, by activating expression of qrr1-4 (Croxatto et al., 2004). VanU also appears to act through a VanO-independent mechanism to inhibit the expression of qrr1-4 (Croxatto et al., 2004, Weber et al., 2011). Weber et al. (2011) thus hypothesize that VanU functions through another RR to repress expression of qrr1-4. Similarly, in Vibrio alginolyticus, Liu et al (2011) propose that LuxU functions, at least in part, independently of LuxO to control expression of a downstream regulator, LuxT, likely through another RR. However, in neither case has a downstream RR been identified.
There are at least a couple of examples in the literature in which a single domain HPt protein interacts with more than one target RR. Caulobacter crescentus ChpT is one such example. ChpT phosphorylates the RRs CtrA and CpdR with equal affinity in vitro (Biondi et al., 2006); these phosphorylation events are critical during cell cycle progression. Phosphorylation of CtrA activates this protein, permitting it to bind DNA and control, among other things, DNA replication (Domian et al., 1997, Quon et al., 1998, Jacobs et al., 2003). In contrast, it is the unphosphorylated form of CpdR that is active; in this state, CpdR indirectly promotes degradation of (unphosphorylated) CtrA (Iniesta et al., 2006), permitting the cell to replicate its DNA. Thus, the same phosphorelay controls two separate RRs to exert opposite effects on protein activity.
Another well-studied example of an HPt protein interacting with two RRs occurs in the yeast Saccharomyces cerevisiae. In this organism, the HPt protein YPD1 serves as a phosphoryl-donor to the RRs SSK1 and SKN7 under hypo-osmotic conditions (Li et al., 1998). However, YPD1 interacts differently with each RR. For example YPD1 stabilizes the phosphorylated state of the RR SSK1 via protein-protein interactions, but does not form stable complexes with the RR SNK1 (Janiak-Spens et al., 2000). Phosphorylation of SSK1 inactivates this regulator until the cell experiences hyperosmotic conditions, in which case SSK1 is rapidly dephosphorylated and activates a downstream pathway involved in controlling osmotic stress genes (Posas et al., 1996, Posas & Saito, 1998). In contrast, phosphorylation of SNK1 promotes activation of a downstream pathway involved in controlling genes for the cell wall and cell cycle (Morgan et al., 1995, Li et al., 1998, Bouquin et al., 1999). These activities of YPD1 allow for the coordinated regulation of multiple pathways in S. cerevisiae. It is possible that LuxU similarly provides a mechanism for coordination of two distinct pathways in V. fischeri.
One question that remains is why only the kinase activity of LuxQ, but not its phosphatase activity, is important for biofilm formation. Furthermore, making LuxQ a “constitutive” kinase through three predicted routes (LuxQ-A216P mutation, deletion of luxP, or deletion of luxS) did not (reproducibly, in the case of LuxQ-A216P) promote accelerated biofilm formation by V. fischeri (Fig. 4D, Fig. S2, and Ray and Visick, unpublished data). Potentially, similar to the yeast system described above, LuxU could interact differently with LuxO and SypG, serving as a phosphoryl-donor to both, but only removing the phosphoryl groups from LuxO. Additional work is necessary to better understand the role of the Lux pathway in influencing biofilm formation.
Our work provides insight into the control of biofilm formation by V. fischeri, but also challenges the current model of how the Lux pathway regulates bioluminescence in V. fischeri. While LuxP, LuxQ, LuxU, and LuxO appeared to function to regulate bioluminescence as predicted (or as previously shown for LuxO (Miyamoto et al., 2000)) (Figs. 1B & 3B and D), the SK AinR did not. Loss of AinR led to a slight decrease in luminescence compared to the control strain (Fig. 3F and S5), while loss of both AinR and LuxU resulted in an intermediate level of luminescence compared to the ainR and luxU mutants (Fig. 3F). However, the decreased luminescence of the ainR and ainR luxU mutants could be overcome by the addition of exogenous C8-HSL, which is normally produced by the AI synthase AinS. From these data, we conclude that deletion of ainR impacts expression of the gene encoding ainS (located directly upstream of ainR), and that loss of AinR itself has little impact on bioluminescence. Furthermore, it seems possible that AinR may not function through the known phosphorelay, or at least not through LuxU, to control bioluminescence. These data are not inconsistent with those reported by Lyell et al., who showed that, on solid media, loss of AinS resulted in an increased luminescence phenotype that did not depend on AinR function (Lyell et al., 2010). Thus, additional work is necessary to understand what role AinR may have in controlling this process in V. fischeri.
If AinR doesn’t function through the known phosphorelay to regulate bioluminescence, does LuxQ serve as the only input? Our data suggest that this is not the case: loss of LuxU resulted in a greater increase in luminescence than loss of LuxQ (Fig. 3B). In both V. cholerae and V. harveyi, three SKs feed into LuxU (LuxQ, CqsS, and VpsS, and LuxQ, LuxN, and CqsS, respectively) (Ng & Bassler, 2009). In V. fischeri, no gene for CqsS exists, but one for VpsS is present (Shikuma et al., 2009). Thus, it is possible that VpsS may also feed into LuxU and serve as another input to regulate light production and possibly biofilm formation. Identifying a missing SK(s) would be an interesting future direction.
This is the first study to examine the role of RscS in biofilm formation under SypG-inducing conditions (overexpression of sypG in a ΔsypE background). Previous studies have already demonstrated that RscS functions upstream of SypG to induce syp transcription in a manner that depends on sypG (Hussa et al., 2008) and that RscS is critical in symbiotic biofilm (aggregate) formation and colonization (Visick & Skoufos, 2001, Yip et al., 2006); these previous studies have only explored how loss or overexpression of rscS impacts biofilm formation or how its loss impacts colonization. Our findings here indicate that LuxU is more important than RscS for biofilm formation under the conditions we used here. In contrast to these results, our preliminary data for the impact of a luxU mutation on the ability of V. fischeri to colonize squid revealed, at most, a mild defect due to loss of LuxU (Ray and Visick, unpublished data). These data suggest, perhaps not surprisingly, that our biofilm-inducing (sypG overexpression) conditions do not fully reflect the dynamics in nature (during colonization). It is of interest to note, however, that not all symbiosis-competent strains of V. fischeri encode a functional RscS protein (Mandel et al., 2009, Gyllborg et al., 2012). Therefore, it is not unreasonable to imagine that another pathway such as Lux could contribute to syp induction and biofilm formation during colonization. This work thus provides an important framework for deepening our understanding of the complex regulatory control over processes critical to colonization by V. fischeri.
Experimental Procedures
Bacterial strains and media
V. fischeri strains utilized in this study are shown in Table 1. Strains used in this study were derived from strain ES114, a bacterial isolate from Euprymna scolopes (Boettcher & Ruby, 1990). For routine culturing, V. fischeri strains were grown in LBS medium (Graf et al., 1994). For luminescence studies, V. fischeri strains were grown in Sea Water Tryptone (SWT) (Yip et al., 2005) and SWTO (Bose et al., 2007). All derivatives of V. fischeri were generated via conjugation, as previously described (DeLoney et al., 2002). E. coli strains GT115 (InvivoGen, San Diego, CA), Tam1 λ pir (Active Motif, Carlsbad, CA), β3914 (Le Roux et al., 2007), π3813 (Le Roux et al., 2007), and CC118 (Herrero et al., 1990) were used for cloning and conjugation. All E. coli strains were grown in Luria-Bertani (LB) medium (Davis et al., 1980). Solid media were made using agar to a final concentration of 1.5%. Antibiotics were added to cultures when appropriate to the following final concentrations: ampicillin (Ap) at 100 μg ml−1 (E. coli), tetracycline (Tet) at 15 μg ml−1 (E. coli) or 5 μg ml−1 (V. fischeri), chloramphenicol (Cm) at 20 or 25 μg ml−1 (E. coli) or 5 μg ml−1 (V. fischeri), kanamycin (Kan) at 50 μg ml−1 (E. coli) or 100 μg ml−1 (V. fischeri), and erythromycin (Erm) at 5 μg ml−1 (V. fischeri). Along with any necessary antibiotics, diaminopimelate (DAP) was added to a final concentration of 0.3 mM for E. coli strain β3914 and thymidine was added to a final concentration of 0.3 mM for E. coli strain π3813.
Table 1.
V. fischeri strains used in this study.
| Strains | Relevant Genotype | Reference |
|---|---|---|
| ES114 | wild-type | (Boettcher & Ruby, 1990) |
| KV3299 | ΔsypE | (Hussa et al., 2008) |
| KV4390 | ΔsypE attTn7::erm | (Morris et al., 2011) |
| KV4430 | luxP::Tn5 ΔsypE | This study |
| KV4431 | luxQ::Tn5 ΔsypE | This study |
| KV4432 | luxQ::Tn5 ΔsypE | This study |
| KV4830 | ΔluxU ΔsypE | This study |
| KV4926 | ΔsypE attTn7::PsypA-lacZ (Ermr) | This study |
| KV5347 | ΔluxP ΔsypE | This study |
| KV5394 | ΔluxQ ΔsypE | This study |
| KV5468 | ΔluxO ΔsypE | This study |
| KV5472 | ΔluxO ΔluxU ΔsypE | This study |
| KV5516 | ΔluxU ΔsypE attTn7::PsypA-lacZ (Ermr) | This study |
| KV5872 | syp::Tn5 ΔsypE | This study |
| KV5902 | ΔluxQ ΔsypE attTn7::luxQ-FLAG | This study |
| KV5903 | ΔluxQ ΔsypE attTn7::luxQ-H378A-FLAG | This study |
| KV5904 | ΔluxQ ΔsypE attTn7::luxQ-A216P-FLAG | This study |
| KV5905 | ΔluxU ΔsypE attTn7::luxU-FLAG | This study |
| KV5906 | ΔluxU ΔsypE attTn7::luxU-H61A-FLAG | This study |
| KV5973 | ΔluxQ ΔsypE attTn7::erm | This study |
| KV5974 | ΔluxU ΔsypE attTn7::erm | This study |
| KV6008 | ΔluxQΔ luxU ΔsypE | This study |
| KV6054 | ΔluxQ ΔluxU ΔsypE attTn7::luxQ-A216P-FLAG | This study |
| KV6196 | ΔainR ΔsypE | This study |
| KV6259 | ΔainR ΔluxU ΔsypE | This study |
| KV6268 | ΔrscS ΔsypE | This study |
| KV6269 | ΔrscS ΔluxU ΔsypE | This study |
Transposon mutagenesis and identification of mutants with wrinkling defects
Transposon mutants were generated as described previously (Lyell et al., 2008). Briefly, plasmid pEVS170, containing the mini-Tn5 transposon, was introduced into V. fischeri strain KV3299 via conjugation. Ex-conjugates were then pooled and the sypG overexpression plasmid pEAH73 was introduced via conjugation. The resultant ex-conjugates were then screened for their ability to form wrinkled colonies. Any mutants found to be defective for wrinkled colony formation after 2 days were then cured of their sypG overexpression plasmid and the plasmid was re-introduced. Any mutant that still exhibited a defect was subject to further analysis as described below.
Southern blot analysis
Southern blot analysis was performed as described previously (Visick & Skoufos, 2001, Yip et al., 2005), except chromosomal DNA was digested with KpnI and probed for the syp locus or with PstI and probed for Tn sequences. All Tn mutants exhibited a pattern consistent with only one Tn insertion.
Molecular techniques
All plasmids were constructed using standard molecular biological techniques, with restriction and modification enzymes obtained from New England Biolabs (Beverly, MA) or Fermentas (Glen Burnie, MD). Plasmids utilized in this study are shown in Table 2. To identify the site of insertion of the 3 non-syp Tn mutants, we cloned the Tn, with flanking DNA, as previously described (Lyell et al., 2008). Unmarked deletions in V. fischeri were generated as previously described (Le Roux et al., 2007, Shibata & Visick, 2012). V. fischeri ES114 was used as the template in PCR amplifications to obtain the DNA containing or flanking the genes of interest using primers listed in Table S1. PCR products were cloned into the pJET1.2 cloning vector (Fermentas, Glen Burine, MD) or pCR1.2-TOPO (Life Technologies, Grand Island, NY), then subcloned into appropriate final vectors using standard molecular techniques. Site-directed mutagenesis was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). For complementation in single copy from the chromosome, luxQ and luxU alleles were cloned upstream of a PlacZ promoter in the mini-Tn7 delivery vector pEVS107. Insertion at the Tn7 site of the chromosome was performed via tetraparental mating, as previously described (McCann et al., 2003). All plasmids constructed in this study were sequenced at the Genomics Core Facility at the Center for Genetic Medicine at Northwestern University (Chicago, IL) or ACGT (Wheeling, IL) to ensure that the insertion contained the desired sequence or mutation.
Table 2.
Plasmids used in this study.
| Plasmids | ||
|---|---|---|
| pEAH73 | pKV69 carrying wild-type sypG; Cmr Tetr | (Hussa et al., 2008) |
| pEAH90 | pEVS107 the PsypA promoter region (generated with primers 714 and 782) upstream of promoterless lacZ | This study |
| pEVS104 | Conjugal helper plasmid (tra trb); Kanr | (Stabb & Ruby, 2002) |
| pEVS107 | Mini-Tn7 delivery plasmid; oriR6K, mob; Kanr, Ermr | (McCann et al., 2003) |
| pJET1.2 | Commercial cloning vector; Apr | Fermentas |
| pKV69 | Mobilizable vector; Cmr Tetr | (Visick & Skoufos, 2001) |
| pKV276 | pEAH73 with D53E mutation in sypG; Cmr Tetr | (Hussa et al., 2008) |
| pKV363 | Mobilizable suicide vector; Cmr | (Shibata & Visick, 2012) |
| pSW7848 | Mobilizable suicide vector; Cmr | Marie-Eve Val |
| pVAR17 | pSW7848 containing 2 kb sequence flanking sypE derived from pCLD19 | (Hussa et al., 2008) and this study |
| pVAR18 | pSW7848 containing 3.3 kb sequence flanking luxU using primers 995, 996, 1017, and 1018 | This study |
| pVAR29 | pKV363 containing 850 bp sequencing flanking luxQ using primers 1286, 1287, 1288, 1304 | This study |
| pVAR30 | pKV363 containing 1.1 kb sequence flanking luxP using primers 1282, 1283, 1284, and 1303 | This study |
| pVAR36 | pKV363 containing 1.1 kb sequence flanking luxO using primers 1319, 1320, 1344, and 1345 | This study |
| pVAR37 | pKV363 containing 1.1 kb sequence flanking luxO and luxU using primers 1319, 1321, 1344, and 1346 | This study |
| pVAR52 | pEVS107 with PlacZ containing 2.3 kb luxQ-FLAG allele using primers 1314 and 1437 | This study |
| pVAR53 | pEVS107 with PlacZ containing 2.3 kb luxQ-A216P-FLAG allele using primers 849 and 1425 | This study |
| pVAR54 | pEVS107 with PlacZ containing 2.3 kb luxQ-H378A-FLAG allele using primers 849 and 1426 | This study |
| pVAR55 | pEVS107 with PlacZ containing 400 bp luxU-FLAG allele using primers 1312 and 1422 | This study |
| pVAR56 | pEVS107 with PlacZ containing 400 bp luxU-H61A-FLAG allele using primers 849 and 1427 | This study |
| pVAR62 | pKV363 containing 1.1 kb sequence flanking ainR using primers 1323 and pr_NL35 (Lyell et al., 2010) | This study |
Luminescence assays
V. fischeri cultures were grown in SWT overnight at 24°C with shaking, then diluted to an optical density at 600 nm (OD600) of ~0.01 in 30 ml of SWTO and incubated at 24°C with vigorous shaking. Samples were taken every 30–60 minutes. At each time point, bioluminescence (using a Turner Designs TD-20/20 luminometer at the factory settings and a large, clear scintillation vial) and OD600 (using a cuvette) were measured for each sample. Maximum luminescence was observed at OD600 measurements between 1.5 and 2 for all strains. Specific luminescence was calculated as relative luminescence (the relative light units of 1 ml of culture integrated over a 6-second count) divided by the OD600.
Wrinkled colony assays
V. fischeri strains were cultured overnight at 28°C with shaking in LBS containing Tet, then sub-cultured 1:100 into fresh LBS containing Tet and grown under the same conditions for 3 to 4 h the next day. Sub-cultures were standardized to an OD600 of 0.2 and 10 μl aliquots were spotted onto LBS agar plates containing Tet and incubated at 28°C. Spotted cultures were then monitored from the time the start of wrinkled colony formation became apparent to the point at which wrinkled colony development ceased or the appropriate data set was collected. Each set of strains for a particular experiment was spotted onto the same plate to account for any minor plate-to-plate variations. Each assay was performed at least 2–3 times, and most were done much more than 3 times. To ensure that cultures spotted at an OD600 of 0.2 resulted in the same number of cells inoculated per spot, we evaluated the correlation between cell number and OD. Specifically, we determined the cell number of the pEAH73-containing strains ΔsypE, ΔsypEΔluxU, ΔsypEΔluxQ, and ΔsypEΔainR using cultures normalized to an OD600 of 0.2, and found no significant difference in the number of colony-forming units obtained from dilutions of the normalized cultures of these strains.
β-galactosidase assay
Cultures of the reporter strains KV4926 and KV5516 carrying the sypG overexpression plasmid pEAH73 were grown in LBS containing Tet at 28°C with shaking. Samples (50 μl) were collected at 12 and 24 hours and 50 μl of Pierce β-galactosidase Assay Reagent (Pierce Biotechnology, Rockford, IL) were added to each sample. Measurements were taken in a microtiter dish using an ELx800 Absorbance Microplate Reader (BioTek, Winooski, VT) with the appropriate settings. As a measure of syp transcription, β-galactosidase activity was determined as previously described (Miller, 1972). P-values were calculated using the student’s t-test.
Supplementary Material
Acknowledgments
We thank members of the Visick lab and Alan Wolfe for critical reading of the manuscript and helpful comments. We thank Beth Hussa for construction of pEAH90 and Kevin Quirke for construction of the precursors to pVAR29 and pVAR30. We thank Malcolm Winkler and Kyle Wayne for their assistance with phos-tag experiments designed to assess the phosphorylation state of SypG. We also thank our anonymous reviewers and Eric Stabb for the idea that loss of AinR could effect ainS expression. This work was funded by NIH Grant GM59690 awarded to K.L.V.
References
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Skerker JM, Arif M, Prasol MS, Perchuk BS, Laub MT. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2006;59:386–401. doi: 10.1111/j.1365-2958.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol. 2007;65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- Bouquin N, Johnson AL, Morgan BA, Johnston LH. Association of the cell cycle transcription factor Mbp1 with the Skn7 response regulator in budding yeast. Mol Biol Cell. 1999;10:3389–3400. doi: 10.1091/mbc.10.10.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Studer SV, Wassarman K, Zhang Y, Ruby EG, Miyashiro T. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. MBio. 2012;3:e00285–00211. doi: 10.1128/mBio.00285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A, V, Chalker J, Lauritz J, Jass J, Hardman A, Williams P, Camara M, Milton DL. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J Bacteriol. 2002;184:1617–1629. doi: 10.1128/JB.184.6.1617-1629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A, Pride J, Hardman A, Williams P, Camara M, Milton DL. A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum. Mol Microbiol. 2004;52:1677–1689. doi: 10.1111/j.1365-2958.2004.04083.x. [DOI] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. Advanced Bacterial Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1980. [Google Scholar]
- DeLoney CR, Bartley TM, Visick KL. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J Bacteriol. 2002;184:5121–5129. doi: 10.1128/JB.184.18.5121-5129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Egland KA, Greenberg EP. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J Bacteriol. 2000;182:805–811. doi: 10.1128/jb.182.3.805-811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol. 2005;55:1160–1182. doi: 10.1111/j.1365-2958.2004.04453.x. [DOI] [PubMed] [Google Scholar]
- Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol. 2002;45:131–143. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999a;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol. 1999b;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson L, Kuo A, Dunlap PV. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllborg MC, Sahl JW, Cronin DC, 3rd, Rasko DA, Mandel MJ. Draft genome sequence of Vibrio fischeri SR5, a strain isolated from the light organ of the Mediterranean squid Sepiola robusta. J Bacteriol. 2012;194:1639. doi: 10.1128/JB.06825-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Herrero M, de Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, Darnell CL, Visick KL. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, O’Shea TM, Darnell CL, Ruby EG, Visick KL. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J Bacteriol. 2007;189:5825–5838. doi: 10.1128/JB.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol Microbiol. 2003;47:1279–1290. doi: 10.1046/j.1365-2958.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, Sparling DP, West AH. Novel role for an HPt domain in stabilizing the phosphorylated state of a response regulator domain. J Bacteriol. 2000;182:6673–6678. doi: 10.1128/jb.182.23.6673-6678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A, Callahan SM, Dunlap PV. Modulation of luminescence operon expression by N-octanoyl-L-homoserine lactone in ainS mutants of Vibrio fischeri. J Bacteriol. 1996;178:971–976. doi: 10.1128/jb.178.4.971-976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux F, Binesse J, Saulnier D, Mazel D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol. 2007;73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Rhee JE, Park U, Ju HM, Lee BC, Kim TS, Jeong HS, Choi SH. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J Microbiol Biotechnol. 2007;17:325–334. [PubMed] [Google Scholar]
- Li S, Ault A, Malone CL, Raitt D, Dean S, Johnston LH, Deschenes RJ, Fassler JS. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998;17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Ruby EG. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J Bacteriol. 2005;187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Urbanowski M, Greenberg EP, Ruby EG. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol. 2003;50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- Lyell NL, Dunn AK, Bose JL, Stabb EV. Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J Bacteriol. 2010;192:5103–5114. doi: 10.1128/JB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyell NL, Dunn AK, Bose JL, Vescovi SL, Stabb EV. Effective mutagenesis of Vibrio fischeri by using hyperactive mini-Tn5 derivatives. Appl Environ Microbiol. 2008;74:7059–7063. doi: 10.1128/AEM.01330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol. 2003;69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Milton DL. Quorum sensing in vibrios: complexity for diversification. Int J Med Microbiol. 2006;296:61–71. doi: 10.1016/j.ijmm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Miyamoto CM, Dunlap PV, Ruby EG, Meighen EA. LuxO controls luxR expression in Vibrio harveyi: evidence for a common regulatory mechanism in Vibrio. Mol Microbiol. 2003;48:537–548. doi: 10.1046/j.1365-2958.2003.03453.x. [DOI] [PubMed] [Google Scholar]
- Miyamoto CM, Lin YH, Meighen EA. Control of bioluminescence in Vibrio fischeri by the LuxO signal response regulator. Mol Microbiol. 2000;36:594–607. doi: 10.1046/j.1365-2958.2000.01875.x. [DOI] [PubMed] [Google Scholar]
- Miyashiro T, Ruby EG. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol. 2012;84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol. 2010;77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Bouquin N, Merrill GF, Johnston LH. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 1995;14:5679–5689. doi: 10.1002/j.1460-2075.1995.tb00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Darnell CL, Visick KL. Inactivation of a novel response regulator is necessary for biofilm formation and host colonization by Vibrio fischeri. Mol Microbiol. 2011;82:114–130. doi: 10.1111/j.1365-2958.2011.07800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DA, Gillece-Castro BL, Burlingame AL, Koshland DE., Jr Phosphorylation site of NtrC, a protein phosphatase whose covalent intermediate activates transcription. J Bacteriol. 1992;174:5117–5122. doi: 10.1128/jb.174.15.5117-5122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL, Koshland DE., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- Shibata S, Visick KL. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J Bacteriol. 2012;194:185–194. doi: 10.1128/JB.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikuma NJ, Fong JC, Odell LS, Perchuk BS, Laub MT, Yildiz FH. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J Bacteriol. 2009;191:5147–5158. doi: 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnikov DM, Schineller JB, Baldwin TO. Transcriptional regulation of bioluminesence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- Stabb E, Schaefer A, Bose JL, Ruby EG. Quorum Signalling and Symbiosis in the Marine Luminous Bacterium Vibrio fischeri. In: Winans SC, Bassler BL, editors. Chemical Communication among Bacteria. Washington, DC: ASM Press; 2008. pp. 233–250. [Google Scholar]
- Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Stevens AM, Dolan KM, Greenberg EP. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc Natl Acad Sci U S A. 1994;91:12619–12623. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, V, Robinson L, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Studer SV, Mandel MJ, Ruby EG. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J Bacteriol. 2008;190:5915–5923. doi: 10.1128/JB.00148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL. Layers of signaling in a bacterium-host association. J Bacteriol. 2005;187:3603–3606. doi: 10.1128/JB.187.11.3603-3606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Skoufos LM. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol. 2001;183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Lindell K, El Qaidi S, Hjerde E, Willassen NP, Milton DL. The phosphotransferase VanU represses expression of four qrr genes antagonizing VanO-mediated quorum-sensing regulation in Vibrio anguillarum. Microbiology. 2011;157:3324–3339. doi: 10.1099/mic.0.051011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol. 2006;62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Grublesky BT, Hussa EA, Visick KL. A novel, conserved cluster of genes promotes symbiotic colonization and sigma(54)-dependent biofilm formation by Vibrio fischeri. Mol Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Qiu Y, Tan Y, Guo Z, Yang R, Zhou D. Transcriptional Regulation of opaR, qrr2-4 and aphA by the Master Quorum-Sensing Regulator OpaR in Vibrio parahaemolyticus. PLoS One. 2012;7:e34622. doi: 10.1371/journal.pone.0034622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.