Abstract
As principal degrading enzymes of the extracellular matrix, metalloproteinases contribute to various pathologies and represent a family of promising drug targets and biomarker candidates. However, multiple proteases and endogenous inhibitors interact to govern metalloproteinase activity, often leading to highly context-dependent protease function that unfortunately has impeded associated clinical utility. We present a method for rapidly assessing the activity of multiple specific proteases in small volumes (<20μl) of complex biological fluids such as clinical samples which are only available in very limited amounts. We have developed a droplet-based microfluidic platform that injects the sample into thousands of picoliter-scale droplets from a bar-coded droplet library containing mixtures of unique moderately selective FRET-based protease substrates and specific inhibitors and monitors hundreds of the reactions thus initiated simultaneously by tracking these droplets. Specific protease activities in the sample are then inferred from the reaction rates using a deconvolution technique, Proteolytic Activity Matrix Analysis (PrAMA). Using a nine-member droplet library with three inhibitors and four FRET substrates, we apply the method to the peritoneal fluid of subjects with and without the invasive disease of endometriosis. Results show clear and physiologically relevant differences with disease; in particular, decreased MMP-2 and ADAM-9 activities.
Extracellular proteases participate in myriad physiological and disease processes, most prominently by degrading extracellular matrix components. In particular, matrix metalloproteinases (MMPs) and A Disintegrin and Metalloproteinases (ADAMs) have been investigated as potential drug targets and diagnostic biomarkers. Metalloproteinase activities are regulated through a tight network of multiple proteolytic enzymes and inhibitors (especially Tissue Inhibitors of Metalloproteinases, TIMPs), frequently resulting in highly context-dependent behavior that has hampered their usefulness in the clinic. Existing approaches such as zymography1, activity-based enzyme-linked immunosorbent assays (ELISAs)2, peptide microarrays3, and activity-based probes4 have been limited by trade-offs including throughput, simultaneous measurement of multiple activities (multiplexing), cost, and direct kinetic measurement. Alternatively, FRET-based polypeptides have been used in recently developed techniques3,5 including Proteolytic Activity Matrix Analysis (PrAMA) to simultaneously ascertain multiple specific protease activities5. The PrAMA technique interprets reaction rates from panels of moderately selective fluorogenic substrates combined with specific protease inhibitors to infer a profile of protease activities from relatively unprocessed physiological samples. Unfortunately, this approach involves performing separate parallel biochemical reactions and consequently carries large liquid-handling and material requirements, presenting a challenge in clinical applications with limited sample quantities.
In this work we report the development and use of an integrated droplet-based microfluidics platform for initiating and simultaneously observing hundreds of protease enzyme activity reactions for hours (up to around eight hundred individual droplets using nine different reaction conditions) using limited quantities (<20μl) of biological/clinical samples and then deconvolving the observed reaction rates using PrAMA. Compartmentalization of chemical reagents in picoliter-scale aqueous droplets allows for a potential 106-fold reduction in reagent consumption compared to standard methods, and facilitates the rapid monitoring of thousands of droplets, each of which may contain unique experimental conditions6. Droplet-based technology has recently been applied to a variety of biological applications7 and pico-injectors have recently been developed to efficiently perform multistep experiments for large-scale multiplexing8. Integration of these capabilities with PrAMA confers particular synergy: the droplet microfluidics create large scale parallel measurements of multiple protease activity reactions, while PrAMA efficiently interprets the high-dimensional kinetic data to infer multiple specific proteolytic activities.
We applied this method to study the invasive disease of endometriosis, which is generally defined by the presence of endometrial-like tissue residing outside the uterus and strongly associates with pain and infertility. Metalloproteinases have been implicated as important enzymes in endometriosis9, but their activities in the context of dysregulated endogenous inhibitors remain less clear9, 10. Using the droplet based multiplexed activity assay, we were able to analyze minimal amounts of clinically-obtained peritoneal fluid from patients with and without endometriosis, and found distinct patterns of protease activity between disease and control samples. In particular, we discovered that MMP-2 and ADAM-9 enzymatic activity decreased with disease and concluded that MMP and inhibitor (TIMP) protein concentrations alone failed to accurately describe the altered proteolytic turnover of specific enzymes. The multiplexing capability achieved through the microfluidic assay not only improved discrimination between control and disease samples, but also supported inference of multiple, specific protease activities that otherwise would have been ambiguous or sample-limited using traditional approaches.
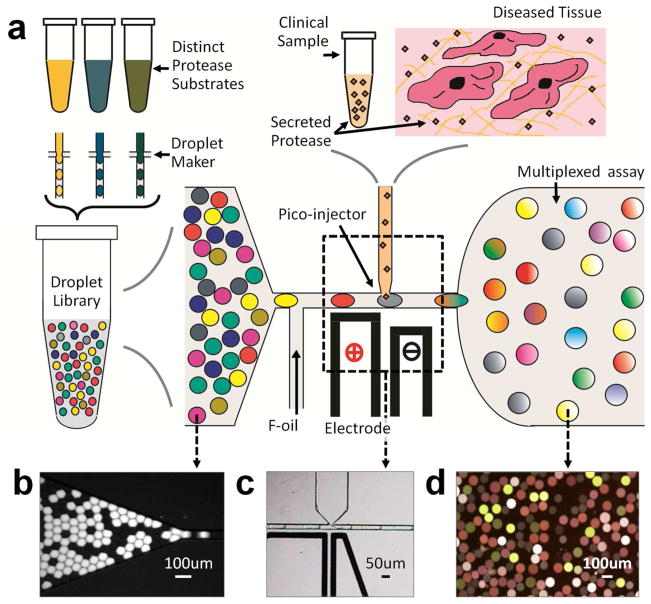
The complete method developed is schematized in Fig. 1A. The details of microfluidic device design, fabrication and operation are described in Supplementary-1. We first prepared protease substrate libraries consisting of 50μm diameter, monodisperse water-in-oil emulsions using droplet generator chips. We formulated droplets to encapsulate unique biochemical assays comprising aqueous solutions of particular protease substrates and, in some cases, protease inhibitors. The PrAMA methodology describes strategies for optimally selecting panels of substrates and inhibitors for accurately inferring specific protease activities. In brief, multiple unique FRET-substrates with distinct enzyme selectively profiles can be utilized in parallel to permit computational inference of specific enzyme activities. This inference can be additionally strengthened by incorporating the comparison of reaction rates in the presence or absence of specific inhibitors. In this application, we identify specific droplet compositions by optically “barcoding” them using specific concentrations of one or more indicator dyes (Alexa-405 and Alexa-546)6. The barcoded droplets are stabilized using an oil-phase surfactant, and then are mixed in a single tube where they remain stable for more than one week. Once the droplet library has been generated, it is flowed into to the device (Fig. 1B) where individual, single-file droplets (containing protease substrates) are mixed 1:1 with a fixed volume of biological sample (containing proteases) using a pico-injector (Fig. 1C)8. After mixing with the sample, droplets are flowed to an integrated incubation chamber (Fig. 1D) where they are monitored via time-lapse fluorescence microscopy for over three hours (see Supplementary Movie MV1 and the corresponding Supplementary Movie MV2 showing only the FRET kinetics). Hundreds of droplet reactions can be simultaneously monitored for hours using automated droplet tracking software, enabling both multiplexed capacity and accurate inference of reaction rates across multiple replicate droplets.
Figure 1.
The microfluidic device. (a) Various protease activity substrates, and in some cases inhibitors, are encapsulated into the droplets distinguished by optical dye labeling. (b) The droplet library flows into a pico-injector device (c) with high volume fraction order, where they merge with the biological sample. (d) Using automated image processing, the protease activity reactions in each droplet are tracked over time in an observation chamber.
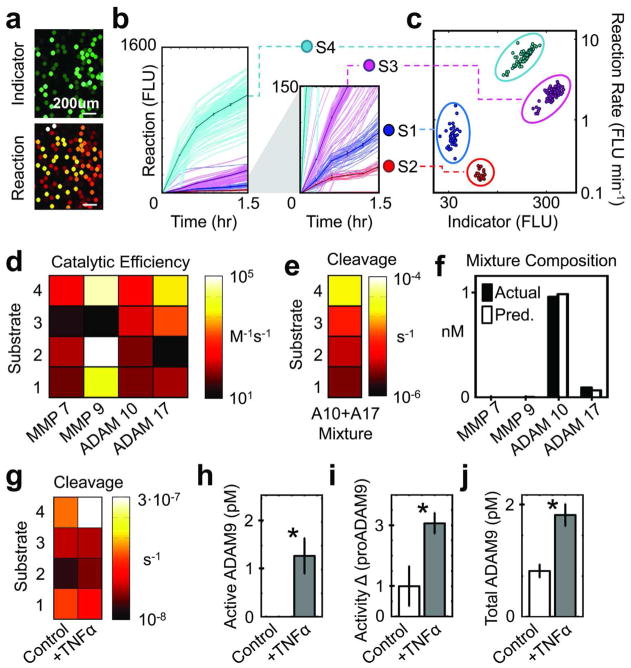
To establish microfluidic PrAMA accuracy, we first conducted reactions using recombinant enzymes. Purified enzyme solutions were injected into a four-component droplet library, consisting of four unique protease substrates (Supplementary-2) that were barcoded with Alexa-405. Sample-injected droplets were fluorescently imaged for 1.5 hours (Fig. 2A). Droplet tracking software (Supplementary-3) interpreted time-course images, and reaction rates were inferred from the increase in fluorescence resulting from substrate proteolysis (Fig. 2B)5. We gated the four droplet library components by their unique groupings of indicator fluorescence and reaction rate (Fig. 2C). Enzyme catalytic efficiencies inferred from these groupings (Fig. 2D) were compared to values obtained using a standard plate-reader assay (R2>95% between the two assay formats, Supplementary-4A). We inferred the composition of unknown enzyme mixtures based upon their observed patterns of substrate cleavage, and results indicated >95% accuracy (Fig. 2E–F, Supplementary-4B–C).
Figure 2.
Application to purified enzymes and cell-based assays. (a) Droplet image of indicator dye and reaction product fluorescence. (b) Proteolysis of substrates S1–S4 was monitored by fluorescence increase. Thick lines and error bars denote mean +/− S.E.M, after individual droplets (thin lines) were classified (c) by their indicator dye and reaction rate. (d) Catalytic efficiencies were determined for samples with known concentrations of purified recombinant enzyme. (e) Observed cleavage rates for a mixture of recombinant ADAM-10 and ADAM-17. (f) PrAMA inferred the enzyme mixture from (e), using parameters from (d). (g) Substrate cleavage measured from 12Z conditioned media. (h) ADAM-9 PrAMA results corresponding to (g). (i) The difference in substrate S4 cleavage measured in the presence or absence of proADAM-9. (j) ELISA result corresponding to g–i (*p<0.05).
After using purified enzymes to validate the device, we applied it to the in vitro study of an immortalized cell line (12Z) established from a peritoneal endometriotic biopsy11. To ascertain the proteolytic activity response of these cells to TNF-α (an implicated inflammatory cytokine11), we stimulated cells for 24hrs, collected and clarified supernatant, and analyzed the samples with the aforementioned four-component library (containing substrates S1–S4, Fig. 2G). Standard methods show here and in previous work that MMP-2, MMP-9, and ADAM-9 are all secreted by 12Zs11. We used microfluidic PrAMA to specifically analyze MMP-2, MMP-9, ADAM-9, and ADAM-10 (a known substrate of ADAM-912) activities. Of these enzymes, microfluidic PrAMA detected a significant increase only in ADAM-9 activity with TNF-α treatment (Fig. 2H, p<0.01, bootstrapping test5). To validate this result, we conducted activity assays in the presence or absence of the specific ADAM-9 inhibitor, proADAM-9 (Fig. 2I)12. The results, combined with ADAM-9 ELISA data (Fig. 2J), confirmed upregulation of active ADAM-9 secretion (p<0.05). By comparing PrAMA to ELISA results, we found roughly 50% of ADAM-9 to be active, compared to <10% of MMP-2. This discrepancy is consistent with high observed concentrations of the inhibitor TIMP-2, which does not inhibit ADAM-9 but does inhibit MMP-2 (Supplementary-5)13.
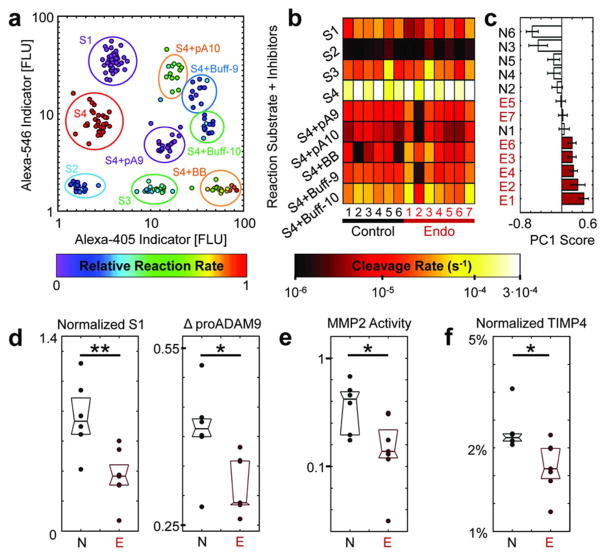
We then analyzed peritoneal fluid (PF) from patients with moderate/severe endometriosis (n=7) and compared them to PF from a control population without the disease (n=6) (Supplementary-6). Microfluidic PrAMA inference of specific protease activities revealed significant differences between disease and control samples. PF lines the pelvic cavity and comprises a heterogeneous mixture of leukocytes, cell debris, thousands of soluble proteins, and likely over 100 proteases and protease inhibitors that interact with endometriotic lesions14. We analyzed clarified PF samples using a nine-component substrate library consisting of the same four substrates used previously, but with the inclusion of droplets containing a broad spectrum metalloproteinase inhibitor (BB94), pro-domain inhibitors for ADAM-9 and ADAM-1012,15, and buffer controls for the pro-domain inhibitors. The nine types of droplets were distinguished by a ratio of two indicator dyes (Fig. 3A). Overall, the observed reaction rates (Fig. 3B) showed strongest activity with substrate S4, which can be efficiently cleaved by both MMPs and ADAMs (Supplementary-7A, C). Addition of the broad-spectrum inhibitor BB94 reduced the observed reaction rates by 90% on average, thereby confirming S4 cleavage to be principally the result of metalloproteinases. For most PF samples, droplets containing pro-domain inhibitors for ADAM-9 and ADAM-10 exhibited significantly lower reaction rates compared to their buffer controls. Across all samples, the ADAM-9 and ADAM-10 inhibitors reduced reaction rates by an average of 25% (σ=16%) and 80% (σ=3%), respectively (Supplementary-7D). For six of the thirteen PF samples, the summed proADAM-9/proADAM-10 inhibition accounted for roughly all observed S4 cleavage. We conducted PrAMA to infer specific protease activities from the cleavage measurements (Supplementary-3), and found ADAM-10 to be the most active protease in general. Of the MMPs, PrAMA results suggested MMP-2 activity to be the highest on average (Supplementary-7E–G).
Figure 3.
Clinical peritoneal fluid analysis. (a) Nine types of droplets were distinguished by two indicator dyes. (b) Peritoneal fluid samples were analyzed from six non-endometriotic patients (“Control”) and seven patients with severe disease (“Endo”). The nine protease activity reactions measured for each clinical sample were defined by substrate (S1–S4) and inhibitor (pA9=proADAM-9, pA10=proADAM-10, BB=BB94, Buff = buffer controls for pro-domain inhibitors). (c) PLSDA scores plot for classifying disease status using the measurements shown in (b), +/− standard error. (d) Box-plot of the two most significant variables identified by PLSDA, which both significantly decreased in endometriotic patients. (e) The most significant PrAMA result corresponding to data in (b) determined by PLSDA. (f) TIMP-4 concentrations decreased significantly with disease, especially when normalized to MMP-2 and TIMP-2 concentrations for each patient sample (**p<0.01,*p<0.05).
Significant correlation among the observed droplet reaction rates compelled multivariate statistical approaches for data interpretation. We used partial least squares-discriminant analysis (PLS-DA) to describe patient status as a statistical function of multiple input variables, which we initially defined as the substrate cleavage measurements. Droplet data was first normalized by dividing rates by their column averages (Fig. 3B). In an automated manner, PLS-DA iteratively selected variables that most accurately predicted disease status, which in this case were from three reactions: S1, S4 with proADAM-9, and S4 with the buffer control for proADAM-9 (Supplementary-8A). PLS-DA combined information from these three measurements to classify disease status with 95% accuracy (Fig. 3C, p=0.03, permutation test).
‘Normalized S1’ rates statistically decreased with disease even when analyzed individually (Fig. 3D, p=0.042, Mann-Whitney test, Bonferroni correction). Based on the PLS-DA modeling, we also investigated the difference in reaction rates with and without the proADAM-9 inhibitor (ΔproADAM-9), and found a decrease with disease (Fig. 3D, p=0.035, Mann-Whitney test). Given pro-ADAM-9 specifically inhibits ADAM-9, reduced ΔproADAM-9 can be readily interpreted as a decrease in ADAM-9 activity with disease. However, interpretation of decreased S1 cleavage is less straight-forward because S1 can be cleaved by multiple proteases (particularly MMPs, including MMP-2, rather than ADAMs). To address this issue, we utilized PrAMA inference to reveal significant differences in specific protease activities, rather than ambiguous differences in substrate cleavage, between disease and control samples. We used PLS-DA to identify the most significant PrAMA descriptors of disease status, which yielded an equally accurate (Supplementary-8B–C, 93% accuracy, p=0.04, permutation test) four-component model that ranked MMP-2 activity as the most significant determinant (Supplementary-8B). Based on the PLS-DA result, we examined relative MMP-2 activity individually and found it to decrease with disease (Fig. 3E, p=0.02, Mann-Whitney test). We performed ELISAs on the PF samples for MMP-2 and two of its endogenous inhibitors, TIMP-2, and TIMP-4, to determine if changes in MMP-2 activity reflected trends in concentration (Supplementary-9). Results identified high levels of both MMP-2 and TIMP-2, but neither the absolute concentrations nor the ratio of MMP-2 to TIMP-2 significantly changed with disease. Rather, TIMP-4 concentrations significantly decreased with disease, particularly when divided by the average concentration of the three analytes for each patient sample (Fig. 3F, p=0.024, Mann-Whitney test, Bonferroni correction). The non-intuitive and concomitant decrease of both MMP-2 and ADAM-9 activities, in spite of reduced TIMP-4 inhibitor concentration, suggests that endometriosis perturbs multiple, overlapping protease-inhibitor interactions in the peritoneal environment. This complexity highlights the challenges associated with inferring enzyme activities from concentration alone, and emphasizes the need for multiplexed, direct activity measurements.
In summary this work creates a platform for assessing multiple specific protease activity assays with minimal liquid handling and sample-requirement by integrating several components, including a droplet generator6, a pico-injector8, and an analytical inference technique (PrAMA)5. Accomplishing the same multiplexed measurements in a 96-well microtiter plate would consume roughly 100-fold more biological sample and reagent (20μl sample for the multiplexed microfluidic assay, compared to a 96-well format requiring 80μl sample/well, in triplicate, with the nine reaction conditions) which would be a prohibitively high amount especially for clinical samples. It is further noteworthy that only a tiny fraction (~30nl) of the 20μl sample volume was actually utilized to generate droplets for multiplexed sensing, due to unoptimized world-to-chip interfacing. This leaves much room for efficiency improvement, either by drastically increasing the degree of multiplexing or further decreasing the sample volume needed. Integrated chip design that combines droplet generation with pico-injection, along with advanced droplet barcoding strategies, will be critical to advancing the platform multiplexing capabilities. Here we present proof-of-principle droplet libraries that utilize three barcode colors and nanoparticle dyes (Supplementary-10), and previous work has shown the potential of using microparticles, quantum dots, and hydrogels to optically distinguish potentially thousands of unique particles that have been shown as amenable to droplet encapsulation16. While such large droplet libraries may require additional optical setup, the future challenge for enhancing the multiplexing capabilities of our platform and its overall technological potential centers on the physical generation of the droplet-library itself17.
Our results underscore the value of multiplexing with microfluidic platforms for clinical sample analysis. When examined individually, substrate cleavage cannot generally be understood as relating to specific proteases. However, in the context of multiple reactions using inhibitors and distinct substrates, results emerged that clearly distinguished disease from control samples, while also enabling determination of specific enzyme activities. Furthermore, results from the multiplexed assay provided a novel perspective into MMP/TIMP regulation in endometriosis (Supplementary-11). Previous studies have reported conflicting observations regarding MMP-2 levels in endometriosis patients, and direct evidence of protease activity in the context of multiple interacting TIMPs has proved inconsistent9,10. Here we report that despite absence of detectable changes in MMP-2 concentration microfluidic PrAMA revealed a significant decrease in MMP-2 activity with disease, and these observations help explain the frequent clinical observation of isolated endometriotic cysts that do not invade the surrounding tissue10. The microfluidic platform developed here could be extended to various applications (Supplementary-12), and device modularity ultimately makes the platform highly customizable for a variety of applications.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. R. Sperling for help with surfactant synthesis, along with support from the NIH grants P50-GM68762, U54-CA112967, the DARPA Cipher Program, the NUS startup grant (R-397-000-137-133) and SMART Innovation grant (ING12043-BIO).
Footnotes
Detailed experimental procedures and expanded discussions are included in the online supporting information, available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kleiner DE, Stetler-Stevenson WG. Anal Biochem. 1994;218:325–29. doi: 10.1006/abio.1994.1186. [DOI] [PubMed] [Google Scholar]
- 2.Lauer-Fields JL, Nagase H, Fields GB. J Biomol Tech. 2004;15:305–16. [PMC free article] [PubMed] [Google Scholar]
- 3.Gosalia DN, Denney WS, Salisbury CM, Ellman JA, Diamond SL. Biotech Bioeng. 2006;94:1099–1110. doi: 10.1002/bit.20927. [DOI] [PubMed] [Google Scholar]
- 4.Sieber SA, Niessen S, Hoover HS, Cravatt BF. Nat Chem Biol. 2006;2:274–81. doi: 10.1038/nchembio781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, Lauffenburger DA. Integr Biol. 2011;3:422–438. doi: 10.1039/c0ib00083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Proc Natl Acad Sci U S A. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proc Natl Acad Sci U S A. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Zheng B, Roach LS, Ismagilov RF. J Am Chem Soc. 2003;125:11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]; (b) Miller OJ, et al. Proc Natl Acad Sci U S A. 2012;109:378–383. doi: 10.1073/pnas.1113324109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chen CH, Sarkar A, Song YA, Miller MA, Kim SJ, Griffith LG, Lauffenburger DA, Han J. J Am Chem Soc. 2011;133:10368–10371. doi: 10.1021/ja2036628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abate AR, Hung T, Mary P, Agresti JJ, Weitz DA. Proc Natl Acad Sci U S A. 2010;107:19163–19166. doi: 10.1073/pnas.1006888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szamatowicz J, Laudánski P, Tomaszewska I. Hum Reprod. 2002;17:284–288. doi: 10.1093/humrep/17.2.284. [DOI] [PubMed] [Google Scholar]
- 10.Gilabert-Estellés J, et al. Hum Reprod. 2003;18:1516–1522. doi: 10.1093/humrep/deg300. [DOI] [PubMed] [Google Scholar]
- 11.(a) Grund EM, Kagan D, Tran CA, Zeitvogel A, Starzinski-Powitz A, Nataraja S, Palmer SS. Mol Pharmacol. 2008;73:1394–1404. doi: 10.1124/mol.107.042176. [DOI] [PubMed] [Google Scholar]; (b) Banu SK, Lee JH, Starzinski-Powitz A, Arosh JA. Fertil Steril. 2008;90:972–987. doi: 10.1016/j.fertnstert.2007.07.1358. [DOI] [PubMed] [Google Scholar]
- 12.Moss ML, et al. J Biol Chem. 2011;286:40443–40451. doi: 10.1074/jbc.M111.280495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amour A, et al. FEBS Lett. 2002;524:154–158. doi: 10.1016/s0014-5793(02)03047-8. [DOI] [PubMed] [Google Scholar]
- 14.Amon LA, et al. PloS ONE. 2010;5:e11137. doi: 10.1371/journal.pone.0011137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss ML, et al. J Biol Chem. 2007;282:35712–35721. doi: 10.1074/jbc.M703231200. [DOI] [PubMed] [Google Scholar]
- 16.(a) Pregibon DC, Toner M, Doyle PS. Science. 2007;315:1393–96. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]; (b) Abate AR, Chen CH, Agresti JJ, Weitz DA. Lab Chip. 2009;9:2628–31. doi: 10.1039/b909386a. [DOI] [PubMed] [Google Scholar]
- 17.Guo MT, Rotem A, Heyman JA, Weitz DA. Lab Chip. 2012;12:2146–55. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.