In this review, Roca et al. provide a comprehensive overview of splice site selection. They review the research on 5′ splice site selection, how base-pairing strength determines splicing outcomes, and the factors and regulatory elements involved in this process.
Keywords: splicing, U1 snRNA, pre-mRNA, 5′ splice sites, exons
Abstract
Splice site selection is fundamental to pre-mRNA splicing and the expansion of genomic coding potential. 5′ Splice sites (5′ss) are the critical elements at the 5′ end of introns and are extremely diverse, as thousands of different sequences act as bona fide 5′ss in the human transcriptome. Most 5′ss are recognized by base-pairing with the 5′ end of the U1 small nuclear RNA (snRNA). Here we review the history of research on 5′ss selection, highlighting the difficulties of establishing how base-pairing strength determines splicing outcomes. We also discuss recent work demonstrating that U1 snRNA:5′ss helices can accommodate noncanonical registers such as bulged duplexes. In addition, we describe the mechanisms by which other snRNAs, regulatory proteins, splicing enhancers, and the relative positions of alternative 5′ss contribute to selection. Moreover, we discuss mechanisms by which the recognition of numerous candidate 5′ss might lead to selection of a single 5′ss and propose that protein complexes propagate along the exon, thereby changing its physical behavior so as to affect 5′ss selection.
Questions about the mechanisms by which 5′ splice sites (5′ss) are selected are deeply rooted in the history of research on pre-mRNA splicing. Identification of the sequences associated with 5′ss triggered the first key insights into splicing mechanisms, efforts that are reflected now in the widespread use of genomic methods to quantify the contributions of other sequences and their cognate factors. The first factors shown to modulate alternative splicing affected 5′ss selection, and the difficulties of working out the molecular mechanisms involved provided a foretaste of the complexities awaiting investigations into other regulatory proteins. Despite the many insights resulting from such studies over the years, it is clear that our conceptual frameworks are not yet adequate. New ideas and models are needed for studies on splice site selection. One purpose of this review is to emphasize that developing new ideas may involve first the challenge of uprooting commonsense but unsubstantiated preconceptions hidden in established models.
The 5′ss is involved in both steps of splicing. In the first step, the 2′-hydroxyl group of the branchpoint adenosine attacks the phosphodiester bond at the 5′ss and displaces the 5′ exon; in the second step, the 3′-hydroxyl group of the 5′ exon attacks the phosphodiester bond at the 3′ splice site (3′ss) and displaces the lariat intron. Splicing was discovered just before new, gel-based DNA-sequencing methods transformed molecular biology. Thus, although the original discoveries were made without the benefit of sequence information (Berget et al. 1977; Chow et al. 1977), sequences emerged rapidly thereafter and revealed clear similarities among 5′ss. Moreover, the “consensus” sequence (comprising, at each position, the nucleotide most commonly found there) was complementary to the sequence at the 5′ end of U1 small nuclear RNA (snRNA), which immediately suggested a mechanism for recognition of the 5′ss (Fig. 1A; Lerner et al. 1980; Rogers and Wall 1980). An additional short stretch of complementarity between U1 snRNA and the 3′ss region does not mediate an interaction (Seraphin and Kandels-Lewis 1993). There were two interesting and important ways in which early preconceptions shaped the subsequent development of the field: The base-pairing between all 5′ss and U1 snRNA was assumed to be in a constant register, and the “consensus” sequence was assumed to be an optimal 5′ss (Rogers and Wall 1980).
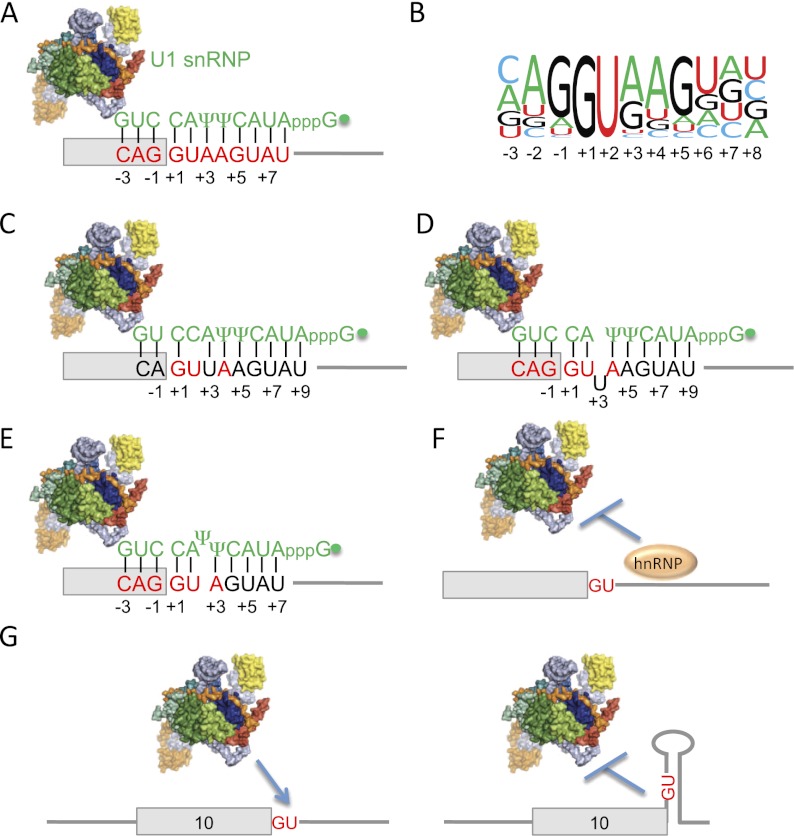
Figure 1.
Recognition of 5′ss by base-pairing to the 5′ end of U1 snRNA. (A) Mammalian consensus 5′ss sequence. Consensus 5′ss nucleotides are shown in red, and odd positions are numbered. (Box) Exon; (horizontal line) intron; (vertical lines) base pairs; (filled dot) trimethylguanosine cap. The U1 snRNP is represented by a space-filling model based on a crystal structure (Pomeranz Krummel et al. 2009). (B) Updated 5′ss motif from a recent collection of 201,541 human 5′ss (Roca et al. 2012). The height of each nucleotide corresponds to its conservation at the corresponding position. (C) Shifted base-pairing register in which the 5′ end of U1 base pairs to the 5′ss by shifting one position downstream. (D) Bulge +2/+3 register. This 5′ss base-pairs more efficiently to U1 by bulging out a U at either position +2 or +3 (only one is shown for clarity). (E) Bulge Ψ register. This 5′ss base-pairs more efficiently by bulging out one of the two Ψs at the 5′ end of U1. (F) Competition between U1 and hnRNPs at 5′ss. An hnRNP with a binding site overlapping with the 5′ss can reduce U1 binding at that 5′ss. (G) Regulation of 5′ss selection by RNA secondary structure in MAPT exon 10. See the text for details.
The original sequence compilations suggested that a limited variety of sequences might be recognized as 5′ss, the sequence within the consensus region being sufficient to both define sites and ensure their usage. However, such a simple view became implausible with the ever-expanding lists of actual mammalian 5′ss sequences: A human 5′ss compilation now contains >9000 sequence variants in the −3 to +6 region of the 5′ss (Roca et al. 2012). Moreover, three observations showed the inadequacy of such a model. First, pre-mRNAs were found to contain sequences that matched the 5′ss consensus as well as or better than the actual 5′ss but were not used (now termed pseudo-5′ss), demonstrating that either sequence could not be the only determinant of use or the consensus might not be optimal. Second, some sequences in β-globin that resemble 5′ss were used when a natural 5′ss was inactivated (termed cryptic 5′ss) (Treisman et al. 1983; Wieringa et al. 1983), demonstrating that the use or avoidance of 5′ss could depend on other sites and that it was not an intrinsic property of any given sequence. Finally, an adenovirus gene (E1A) was shown to use both of two alternative 5′ss, and the ratio of use depended on the sequences of the sites—meaning that 5′ss were in competition (Montell et al. 1982). These findings had important consequences.
The first consequence of the discovery of competitive alternative splicing for studies on 5′ss selection was that it allowed a genetic test in mammals of the role of U1 snRNA. By transfecting cells with a “suppressor” U1 gene containing appropriate mutations at positions hypothesized to be complementary to one of two alternative 5′ss, it was possible to shift the relative use of the two 5′ss in adenovirus E1A transcripts (Zhuang and Weiner 1986). While it was not possible to complement bases at all positions in the 5′ss, the results confirmed the hypothesis that the 5′ end of U1 snRNA recognizes 5′ss by base-pairing and showed that the extent of base-pairing affects competition. Earlier tests of the role of the 5′ end of U1 snRNA by RNase H cleavage had shown that it was required for splicing (Kramer et al. 1984), but the absence of U1 small ribonucleoprotein particles (snRNPs) from some spliceosome preparations had raised doubts (Konarska and Sharp 1986). The role of U1 snRNA in yeast was not clear at first because the 5′ end of U1 snRNA is perfectly conserved even though the consensus 5′ss has a mismatch to U1 snRNA (Siliciano et al. 1987). The suppression of mutations in 5′ss by mutant U1 snRNA genes confirmed that U1 snRNA is essential in yeast as well (Seraphin et al. 1988; Siliciano and Guthrie 1988).
The second consequence of competitive alternative splicing was that it suggested that the relationships between 5′ss sequences, their strength, and their U1 base-pairing potential could be explored systematically. Previous work had shown that introducing mutations into a single 5′ss might inactivate it or lead to the use of cryptic 5′ss, but it was difficult to quantify the strength of each sequence (Aebi et al. 1986). However, if there were an alternative 5′ss that was used to some extent, then it could provide a reference site against which test sites could be calibrated. The first system used a rabbit β-globin gene in which test sequences of 16 nucleotides (nt) were inserted to create potential alternative sites 25 nt upstream of the normal 5′ss. When the test sequence was a duplicate of the normal site's sequence, both 5′ss were used after transient transfection of HeLa cells with the construct. The first experiments showed that the consensus sequence (CAG/GUAAGU) was the most potent and that it silenced the normal 5′ss (Eperon et al. 1986). Much more extensive details on the 5′ss motif and its interaction with U1 as well as the influence of proteins in this recognition event are described below in “5′ss Preferences and U1 Base-Pairing Potential,” “5′ss Recognition Is Not Always Dependent on U1 snRNA Base-Pairing,” and “Extrinsic Factors Affecting 5′ss Choices.” However, even these initial results raised an interesting question: How can a perfectly good natural 5′ss, the reference site, be silenced? What, in fact, is the mechanism by which recognition by U1 snRNPs is turned into selection? These questions are addressed below in “How Does 5′ss Recognition Turn into Selection.”
5′ss preferences and U1 base-pairing potential
As outlined above, the role of base-pairing between the 5′ss and the 5′ end of U1 snRNA was firmly established in the mid-1980s (Zhuang and Weiner 1986). Around the same time, 5′ss competition experiments provided the first functional tests to estimate 5′ss strength by comparing the splicing efficiency of test 5′ss sequences relative to a reference 5′ss. In these experiments, multiple 5′ss could be ranked based on their splicing efficiency, and their ranks correlated reasonably well with base-pairing potential to U1 snRNA, as estimated by thermodynamic parameters. Such experiments provided a firm basis for models involving 5′ss recognition by U1 snRNA.
The dsRNA helix that forms upon base-pairing between a 5′ss and the 5′ end of U1 snRNA has a maximum length of 11 base pairs (bp) (Fig. 1A), as the 12th nucleotide of U1 is already engaged in an internal base pair in stem I. Not all base pairs at different 5′ss positions are equally important, and their contribution to splicing roughly correlates with their conservation (Fig. 1B). The most conserved 5′ss positions lie at the first two intronic nucleotides (+1 and +2), which determine the 5′ss subtype. The GU subtype, with Watson-Crick complementarity with A7 and C8 in U1, accounts for ∼99% of 5′ss. The minor subtypes have a mismatch to U1 at either +1 or +2 and include the GC (0.9%) and the very rare AU 5′ss recognized by the major spliceosome (only 15 cases in humans) (Sheth et al. 2006). The next most conserved 5′ss positions (>75% in humans) are −1G (the last exonic nucleotide) and +5G, which form strong G-C base pairs with U1, with three hydrogen bonds (Fig. 1B). Consensus nucleotides −2A, +3A, +4A, and +6U are also conserved and have a lesser yet important contribution to 5′ss strength because their base pairs to U1 contribute only two hydrogen bonds. Gs at positions −2, +3, and +4 can also establish weaker wobble base pairs (G-U or G-Ψ) (see below), which are very frequent at position +3. The consensus −3C forms a C-G base pair with U1, but the conservation of this nucleotide and its contribution to splicing are less important, probably because this base pair is weakened by the adjacent U1 stem I. In invertebrates, the 5′ss motif is very similar yet with reduced conservation of exonic nucleotides (Sheth et al. 2006). In budding yeast, exonic nucleotides are not conserved at all, the intronic positions +1 to +6 are nearly invariant, and the +4A is replaced by +4U, which constitutes a mismatch to U1.
The 5′ss positions +7 and +8 do not exhibit substantial conservation in humans (Fig. 1B), yet several lines of evidence indicate that these positions can base-pair to U1 and contribute to splicing (Lund and Kjems 2002; Hartmann et al. 2008). In budding yeast, a hyperstable 5′ss/U1 helix with 10 or 12 bp (including those at positions +7 and +8) impedes splicing by reducing the off-rate of U1 (Staley and Guthrie 1999), whose displacement by U6 is a necessary step during spliceosome assembly. However, base pairs at +7 and +8 were shown to enhance splicing kinetics in human cells and extracts (Freund et al. 2005), indicating that the contribution of such 5′ss positions is species-specific.
The first 2 nt at the 5′ end of U1 have methylated riboses, but this modification is not expected to change the stability of the base pairs at these positions. A more important modification in this context is the replacement of the uridines at positions 5 and 6 with pseudouridines (Ψs) (Reddy et al. 1981). A Ψ is a regioisomer of uridine with analogous groups for hydrogen bond donors and acceptors and an extra imino group that contacts the sugar-phosphate backbone and stabilizes base stacking (Davis 1995). Consistently, thermodynamic experiments revealed a slightly higher stability of 5′ss/U1 helices with Ψs compared with those with unmodified Us (Hall and McLaughlin 1991; Roca et al. 2012).
Since the late 1980s, many algorithms have been developed to estimate 5′ss strength. These 5′ss scoring methods rely on either large-scale collections of genomic 5′ss or estimations of the 5′ss/U1 base-pairing stability. The methods in the first category assume that the most common 5′ss nucleotides and/or sequences are most efficient for splicing. The earliest and simplest algorithm used alignments of many 5′ss to derive position-weight matrices (PWMs), which account for the frequency of each nucleotide at each position (Shapiro and Senapathy 1987; Senapathy et al. 1990). Later PWMs were further processed using information content theory (Rogan and Schneider 1995). These methods assume independence between 5′ss positions, yet there is now ample evidence for complex associations between 5′ss positions (Burge 1998; Carmel et al. 2004; Roca et al. 2008). Other algorithms—like first-order Markov models, decision trees, and maximum entropy models—take into consideration these associations (Yeo and Burge 2004). Machine-learning approaches based on neural networks use overall sequence patterns to infer 5′ss strength (Brunak et al. 1991; Krawczak et al. 2007). Methods considering the frequency of the whole 5′ss sequence (excluding positions +7 and +8) in the collection of natural human 5′ss have also proved useful to estimate 5′ss strength (Sahashi et al. 2007). The second class of methods is based on the assumption that U1 binding is the only force governing 5′ss selection. The most common method estimates the minimum free energy of each 5′ss/U1 helix using experimentally derived thermodynamic parameters known as nearest-neighbor “Turner” rules (Mathews et al. 1999), although another algorithm using hydrogen-bonding patterns also exists (Freund et al. 2003; Hartmann et al. 2008). Overall, the scores correlate well among different algorithms and are useful in estimating 5′ss strength, yet they all have their limitations in matching the 5′ss strengths derived experimentally. Partly explaining their limitations, most (but not all) of these methods ignore the contribution of positions +7 and +8.
Another limitation of most methods is that they assume that all 5′ss are recognized by U1 using the same “canonical” base-pairing register, defined by U1 C8 nucleotide base-pairing to 5′ss +1G, and without bulged nucleotides. Recently, however, mutational analyses and suppressor U1 experiments demonstrated that subsets of 5′ss are recognized by U1 using alternative base-pairing registers. First, certain presumptively weak 5′ss were shown to be efficiently used because U1 base-pairs to them in a register that is shifted by 1 nt, so that 5′ss +1G base-pairs to U1 C9 (Fig. 1C; Roca and Krainer 2009). Second, many other 5′ss are more stably bound by U1 when a nucleotide is bulged at either the 5′ss (various positions) or the 5′ end of U1 (only the Ψs), and these base-pairing schemes were collectively termed bulge registers (Fig. 1D,E; Roca et al. 2012). While the shifted register was only estimated to apply to a few 5′ss (59 in humans), the bulge registers appear to be much more frequent, potentially accounting for the recognition of 5% of all human 5′ss. These registers highlight the flexibility of the interaction between 5′ss and U1, allowing for many base-pairing arrangements to result in efficient splicing, and also provide a means for the efficient recognition of 5′ss that otherwise would be weakly bound by U1. Another implication of these registers is that the relevant 5′ss positions vary depending on the used register such that, for example, 5′ss +9 might be base-paired in shifted and some bulge registers. The redefinition of the length of the 5′ss motif as well as the consideration of these registers in new algorithms would certainly result in more accurate 5′ss scoring methods.
The 5′ss/U1 snRNA base-pairing appears to be the main determinant of 5′ss strength, yet this interaction can be affected by other factors binding at the same sequence. The U1C polypeptide, a specific protein of the U1 snRNP, has been shown to bind to 5′ss in the absence of U1 base-pairing (Du and Rosbash 2002). A recent crystal structure of the U1 snRNP (Pomeranz Krummel et al. 2009) revealed certain U1C amino acids contacting the minor groove of the base pairs established by A7 and C8 at the 5′ end of U1, possibly explaining the nearly universal conservation of 5′ss positions +1G and +2U. Certain heterogeneous nuclear RNPs (hnRNPs) like hnRNP A1 and hnRNP H, which are abundant proteins extrinsic to the U1 snRNP, have been proposed to bind some 5′ss sequences, thereby competing with U1 base-pairing (Fig. 1F; Buratti et al. 2004; De Conti et al. 2012).
5′ss can also be involved in internal base-pairing interactions with other pre-mRNA sequences, and such secondary structures would compete with U1. The first proof for such steric hindrance models was provided by 5′ss competition studies, which showed that a 5′ss in an internal stem had a selective disadvantage over a free 5′ss (Eperon et al. 1986, 1988), and others in which an entire exon was skipped when located in an internal RNA loop (Solnick 1985). After that, numerous studies provided evidence for the influence of secondary structures in 5′ss selection and splicing (Jin et al. 2011). A remarkable example is the MAPT (also known as tau) alternative exon 10, whose inclusion levels are determined by the efficiency of 5′ss recognition, which is compromised because of an internal base-pairing structure involving downstream intronic nucleotides (Fig. 1G; Donahue et al. 2006). Mutations weakening the structure increased exon 10 inclusion, thereby disrupting gene function and causing a neurodegenerative disease. The involvement of a pre-mRNA structure in a particular splicing event is typically modeled by RNA structure prediction tools (Jin et al. 2011). However, such predictions can be inaccurate because of the high number of different structures with similar stability, the interference of RNA-binding proteins—mainly from the hnRNP family—in structure formation (Solnick and Lee 1987), and the likely dependence of the structures on the rates of transcription (Eperon et al. 1988). Encouragingly, the traditional and labor-intensive methods for testing RNA structure, based on chemical or enzymatic probing, have been recently complemented by new technologies that allow high-throughput evaluation of structures (Underwood et al. 2010).
Overall, the pivotal role of 5′ss/U1 base-pairing in 5′ss recognition is well established. However, this short RNA duplex is far from a simple structure, as many variations and subtle determinants of 5′ss strength are being revealed. In “5′ss Recognition Is Not Always Dependent on U1 snRNA Base-Pairing,” “Extrinsic Factors Affecting 5′ss Choices,” and “How Does 5′ss Recognition Turn into Selection,” we discuss U1-independent mechanisms, the influence of proteins in 5′ss selection, and how all these processes are integrated to finally commit a 5′ss for splicing.
5′ss recognition is not always dependent on U1 snRNA base-pairing
Some studies have questioned the strict dependence of 5′ss use on U1 snRNA/snRNP in general or in particular cases. The most obvious examples of U1-independent 5′ss belong to the U12-type (or U12-dependent) introns (0.34% of all introns in humans) (Sheth et al. 2006). Splicing of U12-type introns is catalyzed by the minor spliceosome, comprising U11, U12, U4atac, and U6atac in lieu of U1, U2, U4, and U6 snRNPs, respectively, and sharing the U5 snRNP with the major or U2-type spliceosome (Will and Luhrmann 2005). The U12-type 5′ss conform to GU, AU, or noncanonical subtypes; have a distinct and highly conserved motif; and are recognized by base-pairing to U11 snRNA.
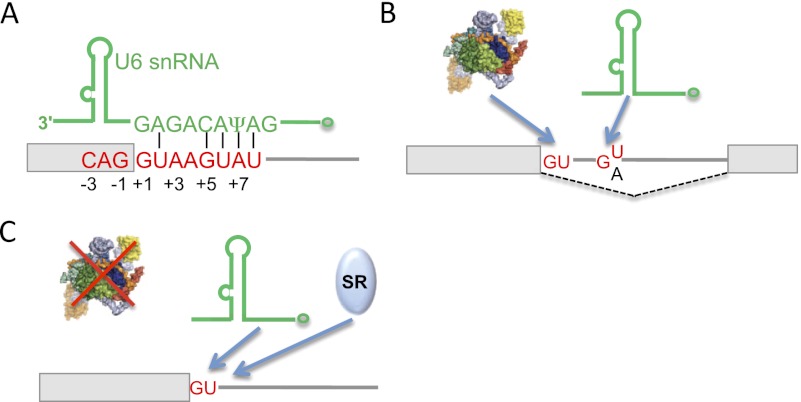
During the maturation of the spliceosome, U1 is replaced by U6 at the 5′ss (Wassarman and Steitz 1992; Kandels-Lewis and Seraphin 1993; Lesser and Guthrie 1993), with U6 establishing up to 5 bp with the consensus 5′ss (Fig. 2A). This change requires ATP and the DExD/H-box RNA helicase Prp28p (Staley and Guthrie 1999). The U6—but not U1—snRNP is an integral component of the active spliceosomal complexes and has been proposed to catalyze both transesterification reactions (Valadkhan 2010). U1 and U6 snRNAs can bind at nearby yet different sequences within the pre-mRNA, and the site of transesterification is ultimately determined by U6 and not U1. This phenomenon was shown first using artificial substrates (Cohen et al. 1994; Hwang and Cohen 1996) and later in the natural human FGFR1 pre-mRNA, in which U6 can support splicing via a noncanonical GA 5′ss in the presence of a nearby U1-binding site (Fig. 2B; Brackenridge et al. 2003). Despite these and other cases in which suppressor U6 snRNAs enhance use of a 5′ss (Kubota et al. 2011), in general, U6 is thought to play only a minor role in initial 5′ss selection.
Figure 2.
Exceptions to 5′ss recognition by U1. (A) Base-pairing between the mammalian consensus 5′ss and U6 snRNA (green, simplified structure). (Open dot) Monomethyl-phosphate cap. (B) For certain 5′ss, the U1- and U6-binding sites are separate, and U6 determines the final site of transesterification. (Dotted line) Splicing pattern. (C) In vitro depletion of U1 snRNA/snRNP can be compensated with excess SR proteins or enhanced complementarity to U6. See the text for details.
A few reports have questioned the necessity of either the U1 snRNP or the base-pairing of U1 snRNA to the 5′ss for certain U2-type introns. Strikingly, splicing that was abolished after either depletion of U1 snRNPs or addition of oligonucleotides complementary to the 5′ end of U1 snRNA could be restored by the addition of SR proteins (Fig. 2C; Crispino et al. 1994; Tarn and Steitz 1994). The efficiency of splicing after depletion was affected by the level of complementarity to U6 snRNA (Crispino and Sharp 1995) and by non-5′ss sequences in the intron (Crispino et al. 1996). The ability to circumvent a block on base-pairing might be attributed to U1C binding to 5′ss in the absence of base-pairing, as observed in yeast (Du and Rosbash 2002). This is supported by an in vitro SELEX experiment using extracts with either intact U1 snRNA or a cleaved 5′ end (by oligonucleotide-directed RNase H) that gave rise to nearly identical 5′ss winner motifs (Lund and Kjems 2002). Subsequent reports have provided more evidence for mechanisms entirely independent of U1 snRNA/snRNP. U1 depletion both in vitro and in Xenopus oocytes did not affect splicing of the human ATP5C1 intron 9, and spliceosome assembly assays showed that U1 was absent in the prespliceosomal E complex, which can form in the absence of ATP and normally includes U1 as the only snRNP (Fukumura et al. 2009). This U1-independent 5′ss recognition might also play a role on the alternative splicing of ATP5C1 exon 9. Another study showed that the 5′ss in human NF1 exon 29 is somehow less dependent on U1 base-pairing (Raponi et al. 2009). Further research is required to fully ascertain the complete U1 independence of such splicing events and reveal the prevalence of these mechanisms. Finally, many U1 snRNA variant genes and pseudogenes can be found in the human genome (Kyriakopoulou et al. 2006). A recent study reports the expression of a subset of these U1 variants (O'Reilly et al. 2012), but their involvement in splicing is not clear.
Most importantly, all of these studies suggest that there is functional redundancy in the recognition of U2-type 5′ss by U1 base-pairing and other mechanisms, be it other components of the U1 snRNP, U6, or other factors. Such alternative mechanisms could enhance fidelity and provide a platform for regulation of 5′ss recognition. The role of U1 and/or other factors in early 5′ss recognition could be seen as just a mark on the substrate that—along with other marks—triggers assembly of spliceosomal complexes and then splicing catalysis. As part of active spliceosomal complexes, U6 determines the final transesterification site in a narrow but flexible sequence window centered at the site of U1 binding. Below, we describe how 5′ss recognition by U1 is influenced by proteins bound at nearby sequences and how a U1-tagged 5′ss becomes committed to splicing.
Extrinsic factors affecting 5′ss choices
SR proteins bound to exon sequences favor the nearest 5′ss downstream
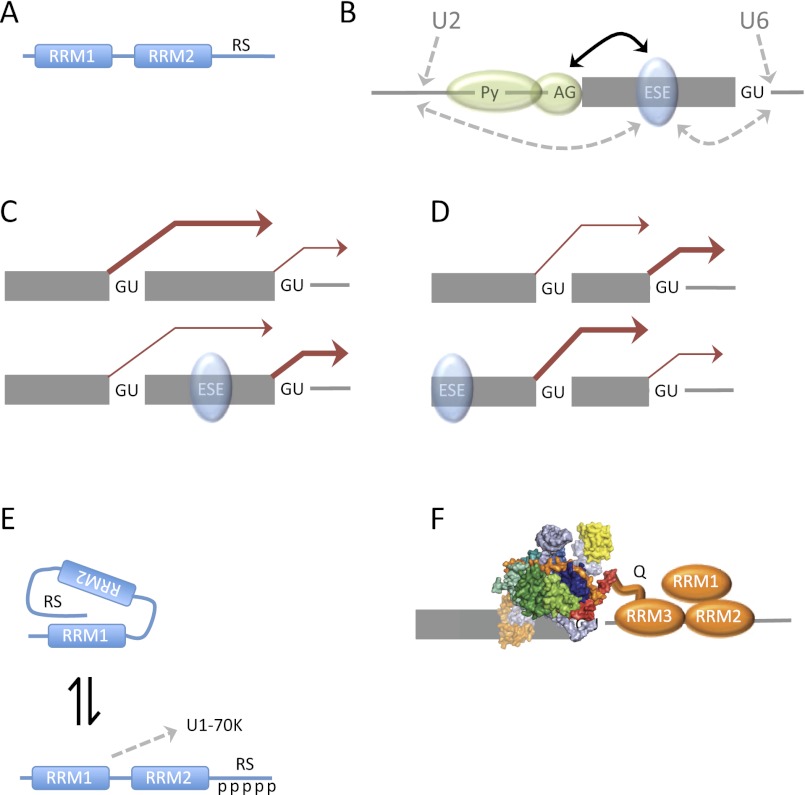
SR proteins comprise one or two RNA recognition motif (RRM)-type RNA-binding domains and a C-terminal RS domain, a region rich in arginine and serine (mostly as RS dipeptides) (Fig. 3A). They are involved in splicing, nuclear export of mRNA, the control of translation, and nonsense-mediated decay (for reviews, see Bourgeois et al. 2004; Long and Caceres 2009; Shepard and Hertel 2009; Zhong et al. 2009). A well-known activity of SR proteins is to stimulate the inclusion of exons with weak splice sites. This appears to involve binding to exonic splicing enhancer sequences (ESEs) in the exon, followed by direct (looping) or indirect interactions of the RS domain with either the RS domain of U2AF35 at the 3′ss (Lavigueur et al. 1993; Tian and Maniatis 1993; Wu and Maniatis 1993; Staknis and Reed 1994; Wang et al. 1995) or RNA duplexes formed by U2 and U6 snRNAs at the branchpoint and 5′ss (Fig. 3B; Shen and Green 2006). An ESE or an artificial tethering sequence for specific SR proteins (SRSF1, SRSF2, or SRSF7) between alternative 5′ss shifts splicing to the intron-proximal 5′ss (Fig. 3C; Bourgeois et al. 1999; Gabut et al. 2005; Spena et al. 2006; Wang et al. 2006; Erkelenz et al. 2013). This is likely to be the result of direct activation of the nearest 5′ss to the 3′ side, since in one case, a cryptic 5′ss is activated by a mutation creating an ESE (Gabut et al. 2005). However, simple looping-type models might not apply, as a central SR protein appears to be equally likely to make contacts with 5′ss on either side. An alternative possibility is that use of the intron-distal 5′ss is inhibited because this would place an SR-binding site in the intron (Ibrahim et al. 2005; Erkelenz et al. 2013). An observation that might support some form of local activation is that an ESE at the 5′ end of a pre-mRNA with identical alternative 5′ss shifts splicing from the intron-proximal to the intron-distal site, closer to the ESE (Fig. 3D; Lewis et al. 2012).
Figure 3.
Proteins that generally activate splicing on the basis of position or sequence of the sites. (A) Diagram of an SR protein. There are one or two RRM-type RNA-binding domains, which may also mediate interprotein contacts, and a C-terminal RS domain, rich in RS or SR dipeptides. SRSF1 has two RRMs with an intervening linker comprising arginine-rich sequences flanking a central block of nine consecutive glycines. This linker plays an important role in binding to ESEs (Cho et al. 2011a). (B) Some mechanisms by which RS domains bound to an ESE might stimulate exon inclusion. (Solid arrow) Interactions in early complexes between the RS domains of the SR protein (blue) and U2AF35 (binds the 3′ss AG; small green oval) become critical if the polypyrimidine tract and U2AF65 binding (larger green oval) are weak; (dotted arrows) interactions in later complexes with the duplexes formed by base-pairing of U2 or U6 snRNAs with pre-mRNA. (C) SR proteins generally promote a shift in splicing toward the intron-proximal 5′ss when either the concentrations of some SR proteins are raised (as with SRSF1) or an ESE is inserted between the alternative 5′ss (as shown). The relative levels of use of the two 5′ss are shown by the thickness of the brown arrows. (D) Effect of an ESE to the 5′ side of alternative 5′ss. (E) Effects of phosphorylation of the RS domain of SRSF1 (Cho et al. 2011b). A hypophosphorylated RS domain interacts with the RRMs and prevents them from binding to the RRM of U1-70K. (F) Binding of TIA-1 RRM2 and RRM3 to interrupted U tracts just 3′ of the 5′ss leaves RRM1 free to augment the interactions of the glutamine-rich domain (Q) with U1-C (Bauer et al. 2012).
SRSF1 shifts splicing to intron-proximal 5′ss without requiring an RS domain
SRSF1 is the most-studied SR protein. It was isolated originally as both a factor restoring splicing to inactive S100 extracts (SF2) (Krainer et al. 1990, 1991) and a factor affecting 5′ss selection in HEK293 cell extracts (ASF) (Ge and Manley 1990; Ge et al. 1991). It has two RRMs and an RS domain. The RS domain is not always required for splicing activation (Zhu and Krainer 2000; Shaw et al. 2007), and neither RRM1 nor the RS domain is essential for 5′ss switching activity (Caceres and Krainer 1993; Zuo and Manley 1993; Wang and Manley 1995; Caceres et al. 1997; van Der Houven Van Oordt et al. 2000). Removal of RRM2, however, results in a change in the pattern of 5′ss use (Caceres et al. 1997; van Der Houven Van Oordt et al. 2000).
The first insight into the mechanism by which SRSF1 switches 5′ss use came when it was shown to enhance the formation of U1-dependent complexes at 5′ss (Eperon et al. 1993). Interestingly, this effect is not restricted to the intron-proximal 5′ss to which splicing shifts (Eperon et al. 1993). Similar results were found with other SR proteins, such as SRSF2 (Tarn and Steitz 1994; Zahler and Roth 1995), although SRSF5 has been reported to mediate selective U1 binding (Zahler and Roth 1995). Possible mechanisms by which an indiscriminate enhancement of U1 snRNP binding can switch splicing to an intron-proximal 5′ss use are discussed below in “How Does 5′ss Recognition Turn into Selection.” The relevance of enhanced U1 snRNP binding to splice site selection was called into question (Valcarcel and Green 1996) when it was shown that, unlike 5′ss switching, it involved the RS domain of SRSF1, which appeared to interact with a similar domain on the U1 snRNP 70-kDa subunit (U1-70K) (Kohtz et al. 1994; Jamison et al. 1995). However, the interaction between the two RS domains could be explained by bridging through contaminating RNA (Xiao and Manley 1998). Moreover, other assays showed that the RS domain was not required for the enhancement of U1 snRNP binding (Eperon et al. 2000). Recent results showed that the interaction between SRSF1 and U1-70K can be mediated by the RRMs of the two proteins, and the RS domain interferes with this by binding intramolecularly if it is hypophosphorylated. This suggests that phosphorylation of the RS domain is a switch that exposes the RRMs for interaction with the U1 snRNP, resulting in enhanced U1 snRNP binding to 5′ss (Fig. 3E; Cho et al. 2011b).
It remains unclear whether either U1 snRNP or SRSF1 binds the pre-mRNA first and also whether specific binding sites for SRSF1 have to be present. Sequences other than the 5′ss are not required for formation of a U1-snRNP/SRSF1/5′ss complex (Jamison et al. 1995; Zahler and Roth 1995). However, ultraviolalet cross-linking and immunoprecipitation (CLIP) analysis in HEK293T cells confirmed that SRSF1 binds to sequences with a loose consensus of GAAGARR (Sanford et al. 2009), fitting previous results based on selection in vitro (Tacke and Manley 1995; Liu et al. 1998). These sequence motifs are enriched in exons within ∼200 nt of splice sites, peaking at 20–40 nt from the sites. Binding involves cooperation between the RRMs and the intervening linker (Cho et al. 2011a).
Proteins that bind intron sequences and activate splicing by recruiting U1 snRNPs
Other proteins modulating splicing act more simply, by modulating the affinity of binding by U1 snRNPs only at recognizable sites. In yeast, the selection of weak alternative 5′ss can be modulated by the presence of a U-rich tract just downstream from the 5′ss. This tract is bound by Nam8p, which is a component of the U1 snRNP in yeast (Puig et al. 1999). Although there is no equivalent stable component of mammalian U1 snRNP, the existence of similar tracts downstream from human 5′ss led to the identification of TIA-1, a homolog of Nam8p, and a close relative named TIA-R. TIA-1 has a number of functions in the cytoplasm, where it is involved in translational repression, but in the nucleus, it has been shown to direct splicing to specific 5′ss in which a U-rich tract begins 5–9 nt downstream (Del Gatto-Konczak et al. 2000; Forch et al. 2000). Binding is synergistic: TIA-1 facilitates binding by U1 snRNP to the adjacent 5′ss (Forch et al. 2000), and the 5′ss and U1 snRNP enhance the binding of TIA-1 (Del Gatto-Konczak et al. 2000). TIA-1 binds directly to the N-terminal part of the U1-C polypeptide (Forch et al. 2002). TIA-1 consists of three RRMs and a glutamine-rich C-terminal sequence. The primary contact with U1-C is mediated by the glutamine-rich sequence, but the interaction is strengthened by RRM1 (Forch et al. 2002). A recent structural study suggested that high-affinity binding to polyU involves all three RRMs but that interruptions to the continuity of U-tracts, as in natural pre-mRNA, prevent the binding of RRM1 to RNA, liberating it to reinforce the interaction with U1-C (Fig. 3F; Bauer et al. 2012).
Another important sequence element is the G-triplet. Although these triplets were first identified as elements characteristic of short mammalian introns (McCullough and Berget 1997), they occur widely and are found preferentially toward the 5′ end of an intron, being most frequent at only 20–30 nt from the 5′ss (Xiao et al. 2009). When inserted between alternative 5′ss, G-triplets stimulate use of the upstream site (McCullough and Berget 1997). These properties suggested that the triplets stimulate the use of an adjacent upstream 5′ss. It was initially suggested that the motifs were themselves bound by U1 snRNPs, base-paired via nucleotides 8–10 of U1 snRNA (McCullough and Berget 2000). However, G-triplets can be bound by hnRNP H family proteins (Caputi and Zahler 2001; Dominguez et al. 2010), and there is evidence that these may play a role in recruiting U1 snRNPs (Wang and Cambi 2009). A more detailed analysis suggests that SR proteins are involved also and that the outcome depends on the nature of the 5′ss, distance from the sites, and, perhaps, the ability of G-runs to form quadruplex structures (Xiao et al. 2009; Wang et al. 2011).
Proteins that inhibit 5′ss by stabilizing U1 snRNP binding
The ability to stabilize U1 snRNP binding is not confined to proteins that activate splicing: A number of examples have been discovered in which U1 snRNP is part of an unproductive complex either at pseudo or bona fide 5′ss. A common feature is that the binding site for the protein is juxtaposed to the 5′ss, although spliceosome assembly may be stalled at different stages. Proteins that inhibit splicing when their binding site is in the exon include HMGA1A, which interacts with the U1-70K polypeptide of U1 snRNP (Ohe and Mayeda 2010); TIA-1 (Erkelenz et al. 2013); and hnRNP A1 (Yu et al. 2008). A systematic screen identified several other sequence classes that inhibit splicing when inserted at −7 relative to the 5′ss, although cognate proteins could not be identified (Yu et al. 2008). The nature of the inhibition has not yet been defined, although it appears to operate at an early stage after U1 snRNP binding, preventing either formation of complex E (Ohe and Mayeda 2010) or progression to complex A (Erkelenz et al. 2013). Other proteins that inhibit when bound within an exon not only act on U1 snRNP but stabilize an exon-defining prespliceosomal complex containing U1 and U2 snRNPs, preventing the components from forming cross-intron interactions (House and Lynch 2006; Bonnal et al. 2008).
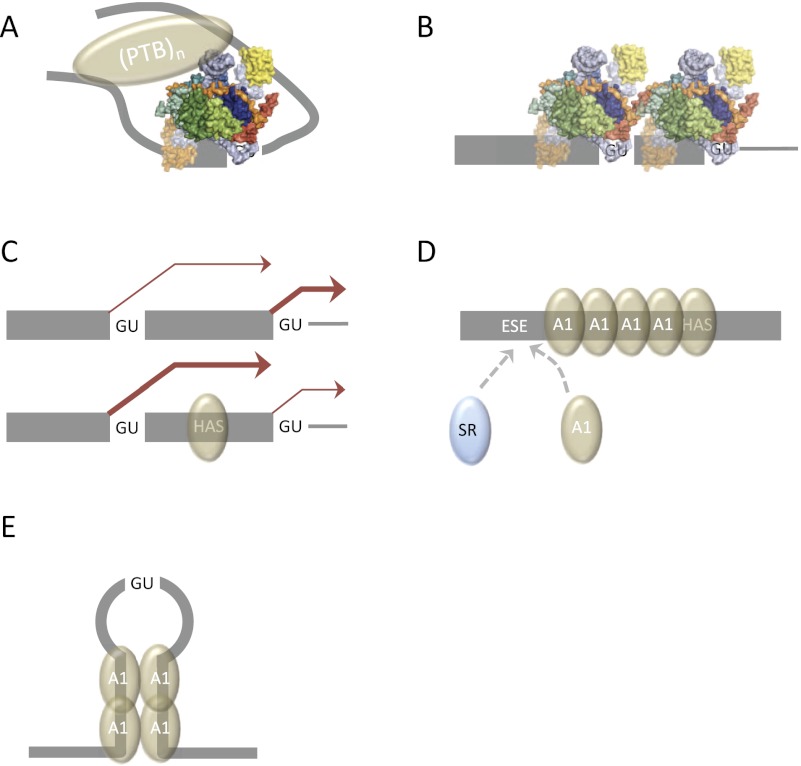
Proteins can also form unproductive complexes when juxtaposed to the 5′ss on the intron side. Polypyrimidine tract-binding protein (PTB) binds to pyrimidine-rich tracts flanking the N1 exon of Src in nonneuronal cells, preventing progression beyond the prespliceosomal A complex (Sharma et al. 2008). The explanation for this appears to lie in the ability of RRM1 and RRM2 to bind to stem–loop 4 of U1 snRNA, which is exposed in the snRNP. This may lock the U1 snRNA in a conformation that blocks further interactions (Fig. 4A; Sharma et al. 2011). SRSF7 is also inhibitory when bound in the intron, where it prevents progression beyond complex E (Erkelenz et al. 2013), and several other classes of sequence are inhibitory when inserted at +11 relative to the 5′ss, including a sequence bound by hnRNP A1 (Yu et al. 2008). Interestingly, it has been suggested on the basis of tests with several proteins that it is a general property of SR and hnRNP proteins that they stimulate splicing when bound close to the 5′ss on the exonic or intronic sides, respectively, and inhibit splicing when bound on the opposite side (Erkelenz et al. 2013). hnRNP A1 may be an exception, since it inhibits when bound to a high-affinity site on either side (Yu et al. 2008). Whereas HMGA1A and PTB have specific mechanisms for inhibition, it may be that proteins generally considered to activate splicing act as repressors when juxtaposed to U1 snRNP on the “wrong” side because they interfere sterically with the interactions made by U1 snRNP shortly after its binding. The U1 snRNP extends its interactions with the pre-mRNA such that ∼20 nt on either side of the 5′ss are protected against nucleases (Chabot and Steitz 1987), although the molecular basis and roles of this extended footprint are unknown.
Figure 4.
Inhibition of 5′ss. (A) Stabilization of U1 snRNP binding to the 5′ss by PTB, including interactions with stem–loop 4 (Sharma et al. 2011), prevents progression beyond complex A. The number and distribution of PTB molecules bound to the flanking intron sequences are not known, and hence the PTB molecules are represented by a single oval. (B) Simultaneous occupancy by two U1 snRNPs of 5′ss in close proximity prevents the development of extended interactions associated with spliceosome assembly. (C) hnRNP A1 (brown) promotes a shift in splicing toward the intron-distal 5′ss when either the protein concentration is raised or a high-affinity site is inserted between the alternative 5′ss (as shown). (D) Model for nucleation of cooperative binding of hnRNP A1 from a high-affinity site. At the ESE, hnRNP A1 association may block the binding of some SR proteins (SRSF2), whereas SRSF1 binding will terminate propagation (Zhu et al. 2001). (E) Model for looping interactions of hnRNP A1 that prevent use of a 5′ss.
Steric interference with the development of interactions by U1 snRNP may account also for the drastic inhibition seen when two strong 5′ss are in close proximity. It has been shown by both ribonuclease protection (Nelson and Green 1988; Eperon et al. 1993) and single-molecule experiments (Hodson et al. 2012) that the 5′ss are both occupied by U1 snRNPs. It is likely that the two snRNPs mutually block further interactions (Fig. 4B).
Effects and mechanisms of competition
A protein purified as an antagonist to the effects of SRSF1 on 5′ss selection turned out to be hnRNP A1 (Mayeda and Krainer 1992). This protein shifts 5′ss preferences to the intron-distal site and favors exon skipping rather than inclusion (Mayeda and Krainer 1992; Mayeda et al. 1993; Yang et al. 1994). The protein has two RRMs and a C-terminal glycine-rich domain that promotes cooperative binding and interactions with other proteins (CasasFinet et al. 1993; Cartegni et al. 1996). Like SRSF1, hnRNP A1 appears to affect 5′ss selection with no absolute requirement for a high-affinity binding site and acts indiscriminately on U1 snRNP binding at candidate 5′ss—in this case, to reduce U1 snRNP binding (Eperon et al. 2000). Rather than interacting directly with U1 snRNPs or the U1–pre-mRNA complex, hnRNP A1 appears to act by competition with SR proteins and U1 snRNPs for binding to pre-mRNA (Eperon et al. 2000; Zhu et al. 2001). The presence of a high-affinity binding site in an exon promotes skipping (Caputi et al. 1999; Del Gatto-Konczak et al. 1999), and when it is placed between alternative 5′ss, the glycine-rich domain promotes a shift to the distal 5′ss (Fig. 4C; Eperon et al. 2000; Wang et al. 2006). High-affinity sites nucleate cooperative binding of pure hnRNP A1 that can displace other proteins as it spreads along the RNA, a process that is more efficient between two high-affinity sites (Fig. 4D; Zhu et al. 2001; Okunola and Krainer 2009). However, there is also evidence that the presence of multiple sites flanking a 5′ss can repress it by the formation of looping interactions mediated by the glycine-rich domain (Blanchette and Chabot 1999; Nasim et al. 2002), which may also involve interactions with hnRNP H (Fig. 4E; Fisette et al. 2010). The formation of loops does not prevent binding of U1 snRNPs but prevents further spliceosome assembly for unknown reasons (Nasim et al. 2002). It remains to be seen whether these two models for the effects of high-affinity sites indicate that there are different mechanisms or that there is an undiscovered unifying mechanism. One of the major difficulties in investigating the mechanisms of action of hnRNP A1, as with SRSF1, is that there are likely to be numerous low-affinity sites.
How does 5′ss recognition turn into selection?
The 9000 different bona fide human 5′ss sequences in the −3 to +6 region (Roca et al. 2012) represent over half of all possible sequences, assuming an absolute requirement for a GU at +1/+2. This means that the consensus region restricts the number of GU motifs in a sequence that could be potential 5′ss by only a factor of two. This range of possible sites is so broad that most genes will comprise many more pseudo-5′ss than actual 5′ss. We described above the evidence that most sites are recognized by U1 snRNP, even if this requires base-pairing to accommodate altered registers and bulges. We summarized the evidence that indicates that in most cases, those factors that modulate 5′ss usage act via a U1 snRNP by either stabilizing its interactions at a 5′ss, competing for binding, or binding adjacent to the U1 snRNP and stabilizing it in an inactive conformation. This leaves the focus of our attention on the question of how, if U1 snRNP recognizes so many sites, it can also be involved in selecting them. To put the same question differently: If U1 snRNPs mark the 5′ss to be used, then how does the mechanism ensure that only one U1 snRNP marks each intron?
U1 snRNPs may bind independently to multiple candidate 5′ss
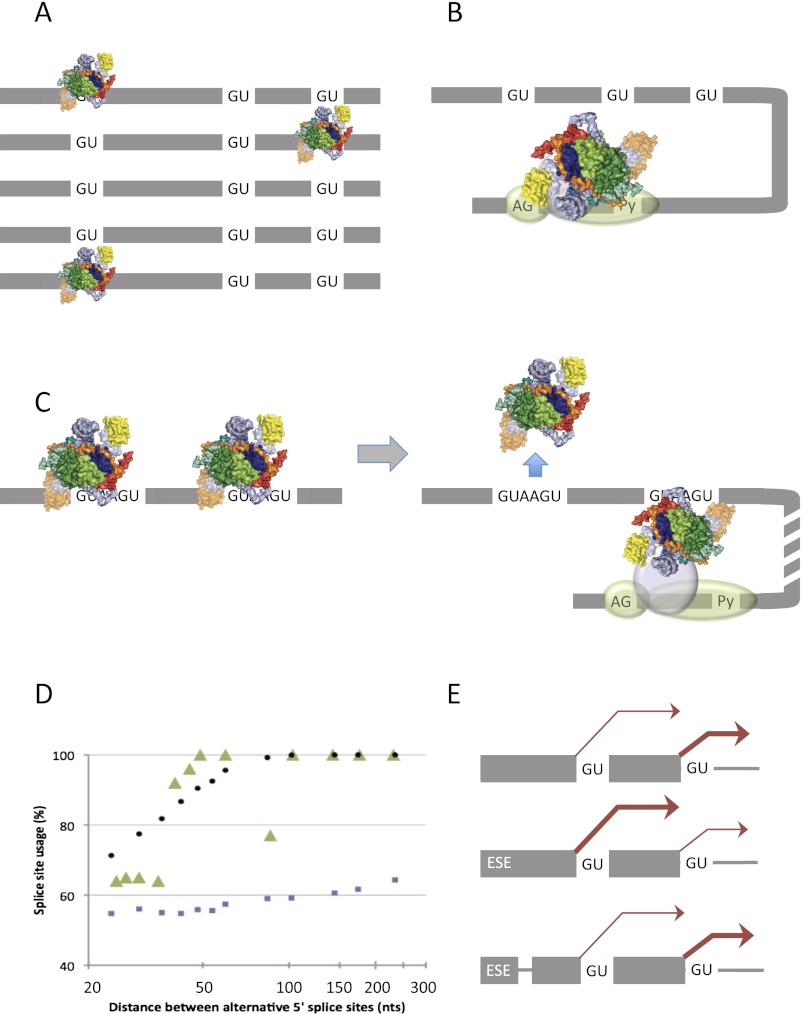
If the main determinant of 5′ss use is the affinity of interactions with U1 snRNP, we need to ask how these affinities become apparent to the splicing apparatus. The answer will depend on how U1 snRNP binds the pre-mRNA. Multiple U1 snRNPs might interact independently with all candidate sequences (Fig. 5A), or a single U1 snRNP might be recruited by, for example, components at the 3′ss and then sample all local candidates (Fig. 5B). The latter model fits well with what is known of complex E. This is an ATP-independent complex that is presumed to equate to the first complex that assembles on a pre-mRNA under normal conditions (Michaud and Reed 1991), although it has only been shown to be a prerequisite for in vitro spliceosome assembly on one substrate (Jamison et al. 1992). It contains U1 snRNP, weakly associated U2 snRNP, and other proteins, including SRSF1 (Staknis and Reed 1994; Makarov et al. 2012). Importantly, it is committed to splicing (Michaud and Reed 1991; Jamison et al. 1992), and there is a functional interaction in assembly between the 5′ss and 3′ss (Michaud and Reed 1993). Hydroxy radical probes tethered near either the 3′ss or the 5′ end of U2 snRNA were used to show that the 5′ss and 3′ss are in close proximity (Kent and MacMillan 2002; Donmez et al. 2007). Interestingly, the addition of Drosophila SR-related proteins (Tra/Tra2) promoted a switch between alternative 5′ss of fruitless pre-mRNA even after complex E had formed, whereas it did not do so after assembly of the first ATP-dependent complex A (Kotlajich et al. 2009). Hence, candidate 5′ss may still be being explored by the bound U1 snRNP. Complex E is therefore consistent with the model in which a single U1 snRNP is recruited by 3′ss components. However, this model is contradicted by experiments showing that alternative strong 5′ss can be protected concurrently against ribonuclease digestion (Nelson and Green 1988; Eperon et al. 1993) and, more clearly, by single-molecule experiments showing that two U1 snRNPs are associated with most molecules of pre-mRNA containing two strong 5′ss in the absence of ATP, whereas only one is bound in complex A (Hodson et al. 2012). These results suggest that U1 snRNPs can interact independently with the candidate 5′ss and that selection is associated with the dissociation of the surplus U1 snRNPs (Fig. 5C).
Figure 5.
Models for the effects of affinity for U1 snRNP and 5′ss position upon selection. (A,B) Possible mechanisms by which affinity determines outcome. The candidate 5′ss have low affinity, and the level of occupancy is low such that there is no multiple occupancy and the distribution of U1 snRNPs reflects the affinities for the 5′ss. (A) Independent binding by U1 snRNPs. (B) Recruitment of a single U1 snRNP by components assembled, for example, at the 3′ss and its rapid interactions with the 5′ss result in a distribution of occupancy that reflects the affinities. (C) With high-affinity sites, binding is saturating and there is multiple occupancy (consistent with the model in A) in complex E conditions; during formation of complex A, the intron-proximal U1 snRNP is selected, and the distal snRNP is displaced (Hodson et al. 2012). (D) The effects of varying the distance between two alternative 5′ss suggest that the preference for an intron-proximal site in C is not accounted for by 3D diffusion of free RNA. (Green triangles) Experimental data from Cunningham et al. (1991); (purple squares) outcome expected from simulation with substrate as free RNA (from a distribution of possible conformations of a homogeneous random coil modeled as a freely jointed chain); (black circles) outcomes expected if the sequence between the 5′ss is rigid. From Hodson et al. (2012), by permission of Oxford University Press. (E) Insertion of a non-RNA linker between an ESE and the alternative 5′ss blocks its effects, arguing against a looping model (Lewis et al. 2012).
Selection by affinity may require low levels of occupancy by U1 snRNPs
The affinity of binding determines the fraction of molecules in which a particular 5′ss is bound by U1 snRNP, which is equivalent to the probability that a particular site on a molecule is occupied at any given time. If the affinities are so low that only one 5′ss (or none) is occupied on any given molecule of pre-mRNA at the time when selection takes place, then affinity-based selection would become merely a matter of selecting whichever U1 snRNP is present: The probabilities that the sites are occupied will determine the relative use of the possible 5′ss. Clearly, such a model would not work if more than one site were bound per molecule of pre-mRNA.
There are several indications of the likely range of lifetimes of U1:pre-mRNA complexes. The lifetime of a complex between pure U1 snRNP and a consensus 5′ss is ∼10 min (Eperon et al. 2000). If base-pairing limits the dissociation rate, then the lifetimes at more typical 5′ss would be shorter by two or three orders of magnitude. In vivo measurements suggest that the lifetime of bound U1 snRNP averages <1 sec (Huranova et al. 2010). In active yeast extracts with a substrate containing a highly conserved 5′ss, the lifetime of the complex formed between uncommitted U1 snRNP and pre-mRNA was estimated by single-molecule methods at ∼0.1 min (Hoskins et al. 2011). Given that transcription of a mammalian intron would generally take at least a few minutes, if not hours, a situation approaching equilibrium between the candidate 5′ss and U1 snRNPs might be established before the point is reached at which the 5′ss is selected.
The process of selection itself is unknown. If the 5′ss and 3′ss have been brought into close proximity in complex E, but the 5′ss to be used is still negotiable, then selection must involve more than simply forming contacts between the U1 snRNP and 3′ss components. It is associated with formation of complex A and ATP hydrolysis (Kotlajich et al. 2009) and may include the process by which surplus U1 snRNPs are irreversibly displaced if there are multiple strong 5′ss (Hodson et al. 2012). A candidate component for such a role is DDX46, a DEAD-box helicase that interacts with U1 and U2 snRNPs (Xu et al. 2004). Its yeast homolog, Prp5, is a spliceosomal protein that interacts with U1-A (Shao et al. 2012) and has been proposed to recruit U2 snRNP and incorporate it stably into spliceosomes after ATP hydrolysis (Kosowski et al. 2009).
Higher levels of occupancy are associated with selection by position
In addition to selection by affinity, there is an additional method of selection based on the relative positions of candidate 5′ss. This is manifested as a strong preference for the intron-proximal 5′ss if the pre-mRNA contains two or more 5′ss with high affinity for U1 snRNP (Reed and Maniatis 1986; Cunningham et al. 1991; Yu et al. 2008; Hicks et al. 2010) or the concentration of some SR proteins, such as SRSF1, is increased (see ”Extrinsic Factors Affecting 5′ss Choices” above). With high-affinity candidate 5′ss, multiple sites on a molecule of pre-mRNA are occupied by U1 snRNPs at the same time (Eperon et al. 1993, 2000; Hodson et al. 2012), presumably because the higher affinities increase the independent chance that each site is occupied and therefore increase the proportion of molecules on which multiple sites are occupied. The same principle applies to the effects of SRSF1, which increases the level of binding by U1 snRNPs at intron-distal and intron-proximal 5′ss (Eperon et al. 1993). The result in either case is an increase in the proportion of pre-mRNA molecules bound by U1 snRNPs at more than one site simultaneously, in which case the basic condition for affinity-dependent selection is broken and there is a switch to position-dependent selection. hnRNP A1 has the opposite effect on splicing, and the likely explanation is that it reduces the proportion of molecules bound by multiple U1 snRNPs (Eperon et al. 2000).
It is not known why the intron-proximal 5′ss is favored when multiple sites are occupied by U1 snRNP. Simple explanations based on the relative probabilities that each 5′ss and its bound U1 snRNP will encounter the 3′ss by three-dimensional (3D) diffusion of an RNA chain (Yu et al. 2008) do not account for the extent of the preference or its dependence on the distance between the alternative 5′ss (Fig. 5D; Cunningham et al. 1991; Hodson et al. 2012). Interestingly, a closer approximation to the observed behavior is seen if the exon sequence is treated as a rigid body (Hodson et al. 2012), which is not unreasonable given the high density of associated proteins in exons (Beyer et al. 1981). The binding of U1 snRNP might recruit SR proteins, which in turn may nucleate the binding of further proteins to purine-rich sequences and other motifs characteristic of exons.
Enhancer sequences may not act by looping
The question of the adequacy of 3D diffusion or looping models was mentioned above when discussing the ability of enhancer sequences placed between alternative 5′ss to stimulate use of the intron-proximal 5′ss. A model in which SR proteins bound to ESEs make contacts by looping with splice site components has been invoked to explain the stimulation of U2AF binding at 3′ss as well as U1 snRNP at 5′ss (Fig. 3B; Lavigueur et al. 1993; Tian and Maniatis 1993; Wu and Maniatis 1993; Staknis and Reed 1994; Wang et al. 1995). However, it has been supported directly only by two observations: cross-linking of an exon-tethered RS domain to pre-mRNA at splice site regions (Shen and Green 2004, 2006) and the attenuation of stimulation by increasing distance from a target 3′ss (Graveley et al. 1998). Recently, looping was directly tested using an ESE at the 5′ end of pre-mRNA that stimulated the use of an upstream alternative 5′ss. Insertion of a flexible alkyl or PEG linker between the ESE and the rest of the pre-mRNA abolished its action, which is inconsistent with looping and implies that the elements have to be connected by RNA (Fig. 5E; Lewis et al. 2012). Therefore, it is possible that the enhancer affects protein binding along the RNA.
Implications for genetic diseases
We reviewed a wide range of mechanisms that dictate 5′ss selection and their implication in multiple types of alternative splicing events, which highlights the importance of research on 5′ss recognition. In addition, elucidating such mechanisms is highly relevant for human genetics. Around 10% of all disease-causing mutations affect either of the two splice sites (Krawczak et al. 2007), and this percentage increases to nearly 50% in the particular cases of NF1 and ATM genes, whose inactivation cause neurofibromatosis type 1 and ataxia telangiectasia, respectively (Teraoka et al. 1999; Ars et al. 2000). About half of such mutations affect 5′ss.
The two most important parameters of a splice site mutation are (1) the severity, which refers to the extent of reduction of correct splicing, and (2) the molecular consequence, which in humans can be, by order of frequency, skipping of the exon (in the case of internal exons), activation of cryptic splice sites, and intron retention. These two parameters often correlate with disease severity. Ab initio predictions of mutation severity by 5′ss scoring methods are largely accurate; i.e., the higher the difference in scores between the wild-type and mutant 5′ss, the more severe the mutation. Predicting the precise consequence of the mutation is far more difficult, although recent analyses have shown some progress (Wimmer et al. 2007; Divina et al. 2009).
The most deleterious mutations at a 5′ss are those affecting the nearly invariant GU dinucleotide, and this is reflected by the high frequency of mutations at these positions causing genetic diseases. For the remaining nine positions, the diagnosis can be more difficult, often relying on analyses of the mRNA from patients and of minigenes in vitro or in cultured cells. Most methods take into account the extent of conservation of the affected nucleotide, which correlates with the severity of the disruption. The effects of certain mutations are more complex, as is the case of +3A-to-G transitions causing genetic diseases, even though both A and G occur with almost equal frequency at this position in bona fide 5′ss. Such transitions are deleterious when the affected 5′ss has nonconsensus nucleotides at the adjacent positions +4 and +5, as proven by pairwise associations in genomic data sets and experimental analyses (Ohno et al. 1999; Madsen et al. 2006; Roca et al. 2008). In certain cases, the 5′ss scores do not reflect the effect of the mutation on 5′ss strength. For example, a 5′ss +5 A-to-G transition in the RARS2 gene causes pontocerebellar hypoplasia, and the deleterious effects of this mutation can be explained by considering the shifted 5′ss/U1 base-pairing register (Roca and Krainer 2009). Finally, a smaller group of disease-causing mutations create new 5′ss, termed de novo 5′ss, which are selected instead of the natural 5′ss. Recent reports investigated the particular sequence patterns surrounding cryptic and de novo 5′ss, which help to understand the selection of such aberrant 5′ss (Kralovicova and Vorechovsky 2007). Furthermore, promising approaches are being developed to rescue splicing defects, including 5′ss mutations not affecting positions +1 or +2. Such technologies are based on antisense oligonucleotides or larger RNA molecules that can affect splicing (Hammond and Wood 2011). These few examples illustrate that a better understanding of the mechanisms of 5′ss selection will likely improve the molecular diagnosis of 5′ss mutations and facilitate therapeutics development.
Finally, single-nucleotide polymorphisms (SNPs) can affect splicing signals, and >1000 SNPs in the human genome map to bona fide human 5′ss (Roca et al. 2008). Whereas most such variations do not substantially change the strength and use of the 5′ss, a fraction of them do affect splicing (Lu et al. 2012). Thus, progress in the field of 5′ss selection will also help in the identification of SNPs that affect splicing with potential phenotypic consequences.
Future perspectives
This review highlights a number of unsolved questions about 5′ss selection, some of which we reiterate here as a spur to future work. Even the apparently simple issue of 5′ss base-pairing potential to U1 snRNA (a short helix with a maximum of 11 bp) is not fully understood, as this interaction is very flexible, allowing for different registers, bulged nucleotides, and perhaps other subtle yet important modifiers of 5′ss strength. Also the limitations of nearest-neighbor parameters for modified nucleotides (Ψ) or bulges limits the inference of 5′ss strength based on the free energy of base-pairing to U1. Taking into account positions +7 and +8 as well as other base-pairing registers should improve existing 5′ss scoring tools. The contribution of proteins, such as U1C and hnRNPs, to 5′ss strength is also poorly understood. Likewise, U6 replaces U1 at the 5′ss in active spliceosomes, but its contribution to 5′ss selection has been shown in very few cases.
The whole question of how the existence and perhaps recognition of many candidate 5′ss is turned into selection of just one is now an urgent issue for all areas of research into mammalian splicing. Processes such as transcription can restrict the number of accessible sites, as suggested in the “first come, first served” model (Kuhne et al. 1983), but the clear dependence of 5′ss selection on affinity, context, and the concentrations of factors shows that potential alternative 5′ss are exposed to recognition. The simplest explanation of affinity-based selection is that free U1 snRNPs are at first able to interact reversibly and independently with potential 5′ss, but it is difficult to reconcile this with what we know of the ATP-independent complex E. Is complex E a subsequent state in which one site has been selected, but events dependent on ATP hydrolysis have not yet removed surplus snRNPs or locked the U1 snRNP onto a specific site? Is the state of free but weak binding shown in Figure 5A followed by the state shown in Figure 5B?
The state of the pre-mRNA is hard to model at present. We referred to several proteins that affect 5′ss selection, mostly via interactions with U1 snRNP. However, very comprehensive experiments on exon inclusion involving analyses of sequence conservation (Goren et al. 2006; Friedman et al. 2008; Barash et al. 2010), selection from large pools (Ke et al. 2011), or systematic mutagenesis (Singh et al. 2004) indicate that most nucleotides in an exon could and in fact probably do affect splicing, presumably by influencing the binding of the 100 or so pre-mRNA-binding proteins. Does this mean that the exon (and perhaps flanking intron sequences) is smothered in proteins either before or after splice sites are selected? Unfortunately, much of the evidence for protein interactions depends on cross-linking and immunoprecipitation. While these are invaluable methods, they provide no indication as to whether all, some, or just a few pre-mRNAs in the reaction mixture are bound by a particular protein at a particular site. Moreover, they provide no information about stoichiometry. This requires single-molecule methods (Cherny et al. 2010). Hence, we do not yet know whether, in vivo or in vitro, U1 snRNP encounters accessible RNA or an RNA–protein complex that might limit binding. We described experiments that suggest that free RNA is an inappropriate model to account for the selection of intron-proximal strong sites or ESE-proximal sites. Do stably bound U1 snRNPs and ESEs trigger the propagation of proteins that alter the physical behavior of exons? Nothing at all is known yet about the flexibility and other physical properties of exons and introns, but we predict that they will behave very differently. Gross changes in physical properties might be one way of integrating the influences of numerous proteins and sites.
Acknowledgments
We are grateful to Chris Oubridge and Kiyoshi Nagai (Medical Research Council Laboratory of Molecular Biology, Cambridge, UK) for the space-filling model of U1 snRNP.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.209759.112.
References
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C 1986. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47: 555–565 [DOI] [PubMed] [Google Scholar]
- Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X 2000. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet 9: 237–247 [DOI] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Bauer WJ, Heath J, Jenkins JL, Kielkopf CL 2012. Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. J Mol Biol 415: 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA 1977. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci 74: 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer AL, Bouton AH, Miller OL Jr 1981. Correlation of hnRNP structure and nascent transcript cleavage. Cell 26: 155–165 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Chabot B 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J 18: 1939–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J 2008. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell 32: 81–95 [DOI] [PubMed] [Google Scholar]
- Bourgeois CF, Popielarz M, Hildwein G, Stevenin J 1999. Identification of a bidirectional splicing enhancer: Differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol Cell Biol 19: 7347–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stevenin J 2004. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 78: 37–88 [DOI] [PubMed] [Google Scholar]
- Brackenridge S, Wilkie AO, Screaton GR 2003. Efficient use of a ‘dead-end’ GA 5′ splice site in the human fibroblast growth factor receptor genes. EMBO J 22: 1620–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S 1991. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol 220: 49–65 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle M, De Conti L, Baralle D, Romano M, Ayala YM, Baralle FE 2004. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHβ genes. Nucleic Acids Res 32: 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB 1998. Modeling dependencies in pre-mRNA splicing signals. Comput Meth Mol Biol 8: 129–164 [Google Scholar]
- Caceres JF, Krainer AR 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J 12: 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol 138: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Zahler AM 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J Biol Chem 276: 43850–43859 [DOI] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J 18: 4060–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel I, Tal S, Vig I, Ast G 2004. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA 10: 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G 1996. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol 259: 337–348 [DOI] [PubMed] [Google Scholar]
- CasasFinet JR, Smith JJ, Kumar A, Kim JG, Wilson SH, Karpel RL 1993. Mammalian heterogeneous ribonucleoprotein A1 and its constituent domains. Nucleic acid interaction, structural stability and self-association. J Mol Biol 229: 873–889 [DOI] [PubMed] [Google Scholar]
- Chabot B, Steitz JA 1987. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol 7: 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny D, Gooding C, Eperon GE, Coelho MB, Bagshaw CR, Smith CW, Eperon IC 2010. Stoichiometry of a regulatory splicing complex revealed by single-molecule analyses. EMBO J 29: 2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Hoang A, Chakrabarti S, Huynh N, Huang DB, Ghosh G 2011a. The SRSF1 linker induces semi-conservative ESE binding by cooperating with the RRMs. Nucleic Acids Res 39: 9413–9421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G 2011b. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci 108: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8 [DOI] [PubMed] [Google Scholar]
- Cohen JB, Snow JE, Spencer SD, Levinson AD 1994. Suppression of mammalian 5′ splice-site defects by U1 small nuclear RNAs from a distance. Proc Natl Acad Sci 91: 10470–10474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, Sharp PA 1995. A U6 snRNA:pre-mRNA interaction can be rate-limiting for U1-independent splicing. Genes Dev 9: 2314–2323 [DOI] [PubMed] [Google Scholar]
- Crispino JD, Blencowe BJ, Sharp PA 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265: 1866–1869 [DOI] [PubMed] [Google Scholar]
- Crispino JD, Mermoud JE, Lamond AI, Sharp PA 1996. Cis-acting elements distinct from the 5′ splice site promote U1-independent pre-messenger RNA splicing. RNA 2: 664–673 [PMC free article] [PubMed] [Google Scholar]
- Cunningham SA, Else AJ, Potter B, Eperon IC 1991. Influences of separation and adjacent sequences on the use of alternative 5′ splice sites. J Mol Biol 217: 265–281 [DOI] [PubMed] [Google Scholar]
- Davis DR 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Conti L, Skoko N, Buratti E, Baralle M 2012. Complexities of 5′splice site definition: Implications in clinical analyses. RNA Biol 9: 911–923 [DOI] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol 19: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stevenin J, Breathnach R 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol 20: 6287–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divina P, Kvitkovicova A, Buratti E, Vorechovsky I 2009. Ab initio prediction of mutation-induced cryptic splice-site activation and exon skipping. Eur J Hum Genet 17: 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Fisette JF, Chabot B, Allain FH 2010. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol 17: 853–861 [DOI] [PubMed] [Google Scholar]
- Donahue CP, Muratore C, Wu JY, Kosik KS, Wolfe MS 2006. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J Biol Chem 281: 23302–23306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Kastner B, Will CL, Luhrmann R 2007. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell 25: 399–411 [DOI] [PubMed] [Google Scholar]
- Du H, Rosbash M 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–90 [DOI] [PubMed] [Google Scholar]
- Eperon LP, Estibeiro JP, Eperon IC 1986. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature 324: 280–282 [DOI] [PubMed] [Google Scholar]
- Eperon LP, Graham IR, Griffiths AD, Eperon IC 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: Is folding limited to a region behind the transcribing RNA polymerase? Cell 54: 393–401 [DOI] [PubMed] [Google Scholar]
- Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR 1993. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J 12: 3607–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Caceres JF, Hayward DG, Krainer AR 2000. Selection of alternative 5′ splice sites: Role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol 20: 8303–8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkelenz S, Mueller WF, Evans ME, Busch A, Schöneweiss K, Hertel KJ, Schaal H 2013. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 19: 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette JF, Toutant J, Dugre-Brisson S, Desgroseillers L, Chabot B 2010. hnRNP A1 and hnRNP H can collaborate to modulate 5′ splice site selection. RNA 16: 228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre- mRNA splicing. Mol Cell 6: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J 21: 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund M, Asang C, Kammler S, Konermann C, Krummheuer J, Hipp M, Meyer I, Gierling W, Theiss S, Preuss T, et al. 2003. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res 31: 6963–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund M, Hicks MJ, Konermann C, Otte M, Hertel KJ, Schaal H 2005. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res 33: 5112–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman BA, Stadler MB, Shomron N, Ding Y, Burge CB 2008. Ab initio identification of functionally interacting pairs of cis-regulatory elements. Genome Res 18: 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura K, Taniguchi I, Sakamoto H, Ohno M, Inoue K 2009. U1-independent pre-mRNA splicing contributes to the regulation of alternative splicing. Nucleic Acids Res 37: 1907–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Mine M, Marsac C, Brivet M, Tazi J, Soret J 2005. The SR protein SC35 is responsible for aberrant splicing of the E1α pyruvate dehydrogenase mRNA in a case of mental retardation with lactic acidosis. Mol Cell Biol 25: 3286–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Manley JL 1990. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell 62: 25–34 [DOI] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL 1991. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 66: 373–382 [DOI] [PubMed] [Google Scholar]
- Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G 2006. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol Cell 22: 769–781 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T 1998. A systematic analysis of the factors that determine the strength of pre- mRNA splicing enhancers. EMBO J 17: 6747–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KB, McLaughlin LW 1991. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry 30: 1795–1801 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Wood MJ 2011. Genetic therapies for RNA mis-splicing diseases. Trends Genet 27: 196–205 [DOI] [PubMed] [Google Scholar]
- Hartmann L, Theiss S, Niederacher D, Schaal H 2008. Diagnostics of pathogenic splicing mutations: Does bioinformatics cover all bases? Front Biosci 13: 3252–3272 [DOI] [PubMed] [Google Scholar]
- Hicks MJ, Mueller WF, Shepard PJ, Hertel KJ 2010. Competing upstream 5′ splice sites enhance the rate of proximal splicing. Mol Cell Biol 30: 1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson MJ, Hudson AJ, Cherny D, Eperon IC 2012. The transition in spliceosome assembly from complex E to complex A purges surplus U1 snRNPs from alternative splice sites. Nucleic Acids Res 40: 6850–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ 2011. Ordered and dynamic assembly of single spliceosomes. Science 331: 1289–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW 2006. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol 13: 937–944 [DOI] [PubMed] [Google Scholar]
- Huranova M, Ivani I, Benda A, Poser I, Brody Y, Hof M, Shav-Tal Y, Neugebauer KM, Stanek D 2010. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. J Cell Biol 191: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DY, Cohen JB 1996. U1 snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes Dev 10: 338–350 [DOI] [PubMed] [Google Scholar]
- Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T 2005. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci 102: 5002–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Crow A, Garcia-Blanco MA 1992. The spliceosome assembly pathway in mammalian extracts. Mol Cell Biol 12: 4279–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Pasman Z, Wang J, Will C, Luhrmann R, Manley JL, Garcia-Blanco MA 1995. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: Characterization of required elements. Nucleic Acids Res 23: 3260–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Yang Y, Zhang P 2011. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol 8: 450–457 [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S, Seraphin B 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039 [DOI] [PubMed] [Google Scholar]
- Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, Ju J, Chasin LA 2011. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res 21: 1360–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, MacMillan AM 2002. Early organization of pre-mRNA during spliceosome assembly. Nat Struct Biol 9: 576–581 [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368: 119–124 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA 1986. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46: 845–855 [DOI] [PubMed] [Google Scholar]
- Kosowski TR, Keys HR, Quan TK, Ruby SW 2009. DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle. RNA 15: 1345–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlajich MV, Crabb TL, Hertel KJ 2009. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in the A complex. Mol Cell Biol 29: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev 4: 1158–1171 [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G 1991. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66: 383–394 [DOI] [PubMed] [Google Scholar]
- Kralovicova J, Vorechovsky I 2007. Global control of aberrant splice-site activation by auxiliary splicing sequences: Evidence for a gradient in exon and intron definition. Nucleic Acids Res 35: 6399–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Keller W, Appel B, Luhrmann R 1984. The 5′ terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell 38: 299–307 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN 2007. Single base-pair substitutions in exon-intron junctions of human genes: Nature, distribution, and consequences for mRNA splicing. Hum Mutat 28: 150–158 [DOI] [PubMed] [Google Scholar]
- Kubota T, Roca X, Kimura T, Kokunai Y, Nishino I, Sakoda S, Krainer AR, Takahashi MP 2011. A mutation in a rare type of intron in a sodium-channel gene results in aberrant splicing and causes myotonia. Hum Mutat 32: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne T, Wieringa B, Reiser J, Weissmann C 1983. Evidence against a scanning model of RNA splicing. EMBO J 2: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou C, Larsson P, Liu L, Schuster J, Soderbom F, Kirsebom LA, Virtanen A 2006. U1-like snRNAs lacking complementarity to canonical 5′ splice sites. RNA 12: 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A, La Branche H, Kornblihtt AR, Chabot B 1993. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev 7: 2405–2417 [DOI] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA 1980. Are snRNPs involved in splicing? Nature 283: 220–224 [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C 1993. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science 262: 1982–1988 [DOI] [PubMed] [Google Scholar]
- Lewis H, Perrett AJ, Burley GA, Eperon IC 2012. An RNA splicing enhancer that does not act by looping. Angew Chem Int Ed Engl 51: 9800–9803 [DOI] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev 12: 1998–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF 2009. The SR protein family of splicing factors: Master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Lu ZX, Jiang P, Xing Y 2012. Genetic variation of pre-mRNA alternative splicing in human populations. Wiley Interdiscip Rev RNA 3: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M, Kjems J 2002. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA 8: 166–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen PP, Kibaek M, Roca X, Sachidanandam R, Krainer AR, Christensen E, Steiner RD, Gibson KM, Corydon TJ, Knudsen I, et al. 2006. Short/branched-chain acyl-CoA dehydrogenase deficiency due to an IVS3+3A>G mutation that causes exon skipping. Hum Genet 118: 680–690 [DOI] [PubMed] [Google Scholar]
- Makarov EM, Owen N, Bottrill A, Makarova OV 2012. Functional mammalian spliceosomal complex E contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucleic Acids Res 40: 2639–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68: 365–375 [DOI] [PubMed] [Google Scholar]
- Mayeda A, Helfman DM, Krainer AR 1993. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol 13: 2993–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough AJ, Berget SM 1997. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol 17: 4562–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough AJ, Berget SM 2000. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol Cell Biol 20: 9225–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Reed R 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev 5: 2534–2546 [DOI] [PubMed] [Google Scholar]
- Michaud S, Reed R 1993. A functional association between the 5′ and 3′ splice sites is established in the earliest prespliceosome complex (E) in mammals. Genes Dev 7: 1008–1020 [DOI] [PubMed] [Google Scholar]
- Montell C, Fisher EF, Caruthers MH, Berk AJ 1982. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature 295: 380–384 [DOI] [PubMed] [Google Scholar]
- Nasim FU, Hutchison S, Cordeau M, Chabot B 2002. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 8: 1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KK, Green MR 1988. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev 2: 319–329 [DOI] [PubMed] [Google Scholar]
- Ohe K, Mayeda A 2010. HMGA1a trapping of U1 snRNP at an authentic 5′ splice site induces aberrant exon skipping in sporadic Alzheimer's disease. Mol Cell Biol 30: 2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K, Brengman JM, Felice KJ, Cornblath DR, Engel AG 1999. Congenital end-plate acetylcholinesterase deficiency caused by a nonsense mutation and an A→G splice-donor-site mutation at position +3 of the collagenlike-tail-subunit gene (COLQ): How does G at position +3 result in aberrant splicing? Am J Hum Genet 65: 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okunola HL, Krainer AR 2009. Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol Cell Biol 29: 5620–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S 2012. Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res doi: 10.1101/gr.142968.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Gottschalk A, Fabrizio P, Seraphin B 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev 13: 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M, Buratti E, Dassie E, Upadhyaya M, Baralle D 2009. Low U1 snRNP dependence at the NF1 exon 29 donor splice site. FEBS J 276: 2060–2073 [DOI] [PubMed] [Google Scholar]
- Reddy R, Henning D, Busch H 1981. Pseudouridine residues in the 5′-terminus of uridine-rich nuclear RNA I (U1 RNA). Biochem Biophys Res Commun 98: 1076–1083 [DOI] [PubMed] [Google Scholar]
- Reed R, Maniatis T 1986. A role for exon sequences and splice-site proximity in splice-site selection. Cell 46: 681–690 [DOI] [PubMed] [Google Scholar]
- Roca X, Krainer AR 2009. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat Struct Mol Biol 16: 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Olson AJ, Rao AR, Enerly E, Kristensen VN, Borresen-Dale AL, Andresen BS, Krainer AR, Sachidanandam R 2008. Features of 5′-splice-site efficiency derived from disease-causing mutations and comparative genomics. Genome Res 18: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Akerman M, Gaus H, Berdeja A, Bennett CF, Krainer AR 2012. Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev 26: 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan PK, Schneider TD 1995. Using information content and base frequencies to distinguish mutations from genetic polymorphisms in splice junction recognition sites. Hum Mutat 6: 74–76 [DOI] [PubMed] [Google Scholar]
- Rogers J, Wall R 1980. A mechanism for RNA splicing. Proc Natl Acad Sci 77: 1877–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahashi K, Masuda A, Matsuura T, Shinmi J, Zhang Z, Takeshima Y, Matsuo M, Sobue G, Ohno K 2007. In vitro and in silico analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5′ splice sites. Nucleic Acids Res 35: 5995–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y 2009. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19: 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P, Shapiro MB, Harris NL 1990. Splice junctions, branch point sites, and exons: Sequence statistics, identification, and applications to genome project. Methods Enzymol 183: 252–278 [DOI] [PubMed] [Google Scholar]
- Seraphin B, Kandels-Lewis S 1993. 3′ Splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell 73: 803–812 [DOI] [PubMed] [Google Scholar]
- Seraphin B, Kretzner L, Rosbash M 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J 7: 2533–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Kim HS, Cao Y, Xu YZ, Query CC 2012. A U1-U2 snRNP interaction network during intron definition. Mol Cell Biol 32: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P 1987. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res 15: 7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol 15: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Maris C, Allain FH, Black DL 2011. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol Cell 41: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SD, Chakrabarti S, Ghosh G, Krainer AR 2007. Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing. PLoS ONE 2: e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR 2004. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell 16: 363–373 [DOI] [PubMed] [Google Scholar]
- Shen H, Green MR 2006. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev 20: 1755–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ 2009. The SR protein family. Genome Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R 2006. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res 34: 3955–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano PG, Guthrie C 1988. 5′ splice site selection in yeast: Genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev 2: 1258–1267 [DOI] [PubMed] [Google Scholar]
- Siliciano PG, Jones MH, Guthrie C 1987. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science 237: 1484–1487 [DOI] [PubMed] [Google Scholar]
- Singh NN, Androphy EJ, Singh RN 2004. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA 10: 1291–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D 1985. Alternative splicing caused by RNA secondary structure. Cell 43: 667–676 [DOI] [PubMed] [Google Scholar]
- Solnick D, Lee SI 1987. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol 7: 3194–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena S, Tenchini ML, Buratti E 2006. Cryptic splice site usage in exon 7 of the human fibrinogen Bβ-chain gene is regulated by a naturally silent SF2/ASF binding site within this exon. RNA 12: 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staknis D, Reed R 1994. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol 14: 7670–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64 [DOI] [PubMed] [Google Scholar]
- Tacke R, Manley JL 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J 14: 3540–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev 8: 2704–2717 [DOI] [PubMed] [Google Scholar]
- Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA, et al. 1999. Splicing defects in the ataxia-telangiectasia gene, ATM: Underlying mutations and consequences. Am J Hum Genet 64: 1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T 1993. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 74: 105–114 [DOI] [PubMed] [Google Scholar]
- Treisman R, Orkin SH, Maniatis T 1983. Structural and functional defects in β-thalassemia. Prog Clin Biol Res 134: 99–121 [PubMed] [Google Scholar]
- Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D 2010. FragSeq: Transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods 7: 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S 2010. Role of the snRNAs in spliceosomal active site. RNA Biol 7: 345–353 [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Green MR 1996. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci 21: 296–301 [PubMed] [Google Scholar]
- van Der Houven Van Oordt W, Newton K, Screaton GR, Caceres JF 2000. Role of SR protein modular domains in alternative splicing specificity in vivo. Nucleic Acids Res 28: 4822–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Cambi F 2009. Heterogeneous nuclear ribonucleoproteins H and F regulate the proteolipid protein/DM20ratio by recruiting U1 small nuclear ribonucleoprotein through a complex array of G runs. J Biol Chem 284: 11194–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Manley JL 1995. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1: 335–346 [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hoffmann HM, Grabowski PJ 1995. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA 1: 21–35 [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xiao X, Van Nostrand E, Burge CB 2006. General and specific functions of exonic splicing silencers in splicing control. Mol Cell 23: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Mueller WF, Hertel KJ, Cambi F 2011. G Run-mediated recognition of proteolipid protein and DM20 5′ splice sites by U1 small nuclear RNA is regulated by context and proximity to the splice site. J Biol Chem 286: 4059–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA 1992. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science 257: 1918–1925 [DOI] [PubMed] [Google Scholar]
- Wieringa B, Meyer F, Reiser J, Weissmann C 1983. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit β-globin gene. Nature 301: 38–43 [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R 2005. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 386: 713–724 [DOI] [PubMed] [Google Scholar]
- Wimmer K, Roca X, Beiglbock H, Callens T, Etzler J, Rao AR, Krainer AR, Fonatsch C, Messiaen L 2007. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum Mutat 28: 599–612 [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75: 1061–1070 [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL 1998. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J 17: 6359–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB 2009. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol 16: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J 23: 376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bani MR, Lu SJ, Rowan S, BenDavid Y, Chabot B 1994. The A1 and A1(B) proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci 91: 6924–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 135: 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Roth MB 1995. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci 92: 2642–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Krainer AR 2000. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev 14: 3166–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mayeda A, Krainer AR 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell 8: 1351–1361 [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46: 827–835 [DOI] [PubMed] [Google Scholar]
- Zuo P, Manley JL 1993. Functional domains of the human splicing factor ASF/SF2. EMBO J 12: 4727–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]