The Mediator complex regulates gene expression by linking transcription factors with RNA Pol II. These interactions are controlled by CDK8, yet the mechanisms regulating CDK8–Mediator association are largely unknown. Davis et al. find that Fbw7, a tumor suppressor and ubiquitin ligase, binds CDK8–Mediator and targets Mediator subunits MED13/13L for degradation. Fbw7 loss increases CDK8–Mediator association in proliferating cells, suggesting the possibility that Mediator and CDK8 contribute to Fbw7-associated tumorigenesis.
Keywords: Fbw7, MED13/13L, CDK8, SCF, Mediator
Abstract
The Mediator complex is an essential transcription regulator that bridges transcription factors with RNA polymerase II. This interaction is controlled by dynamic interactions between Mediator and the CDK8 module, but the mechanisms governing CDK8 module–Mediator association remain poorly understood. We show that Fbw7, a tumor suppressor and ubiquitin ligase, binds to CDK8–Mediator and targets MED13/13L for degradation. MED13/13L physically link the CDK8 module to Mediator, and Fbw7 loss increases CDK8 module–Mediator association. Our work reveals a novel mechanism regulating CDK8 module–Mediator association and suggests an expanded role for Fbw7 in transcriptional control and an unanticipated relationship with the CDK8 oncogene.
Fbw7 is the substrate recognition component of an SCF (Skp–Cullin–F-box) ubiquitin ligase that targets phosphorylated substrates for ubiquitylation and proteasomal degradation (Nakayama and Nakayama 2006; Tan et al. 2008; Welcker and Clurman 2008; Crusio et al. 2010). Substrates bind to Fbw7 through a conserved phosphodegron (CPD), ΦxΦΦΦ(T/S)PPx(T/S/E/D), where Φ represents hydrophobic residues and T/S are phosphoserine or phosphothreonine (Nash et al. 2001; Welcker and Clurman 2008). Phosphorylated CPDs make up to eight contacts with the Fbw7 substrate-binding domain, which includes three critical arginines that are required for substrate degradation (Orlicky et al. 2003; Hao et al. 2007). Importantly, Fbw7 is a tumor suppressor that is commonly mutated in cancers (Akhoondi et al. 2007) and degrades numerous oncogenic transcription factors (e.g., Myc, Notch, and Jun) as well as other proteins that contribute to carcinogenesis (e.g., Cyclin E and Mcl1) (Welcker and Clurman 2008; Inuzuka et al. 2011; Wertz et al. 2011). Fbw7 mutations are thought to drive tumorigenesis through inappropriate activity of its oncogenic substrates, and the causality of Fbw7 mutations in carcinogenesis has been demonstrated in murine models (Mao et al. 2004; Onoyama et al. 2007; Thompson et al. 2008; Sancho et al. 2010; Grim et al. 2012).
As a genome-wide regulator of transcription, the 26-subunit Mediator complex regulates gene expression by physically linking transcription factors to the general transcription machinery, including RNA polymerase II (Pol II) (Malik and Roeder 2005). Mammalian Mediator exists predominantly in two forms that are distinguished by the presence or absence of the CDK8 module. The CDK8 module reversibly associates with Mediator to regulate the Mediator–Pol II interaction (Elmlund et al. 2006; Knuesel et al. 2009) and controls transcription initiation and reinitiation. Additionally, CDK8–Mediator regulates transcription via chromatin modification and physical or functional interactions with elongation factors (Meyer et al. 2008; Donner et al. 2010; Ebmeier and Taatjes 2010). Thus, the CDK8 module plays multiple distinct roles in regulating Mediator function and transcription. Of note, the CDK8 module contains four subunits (CDK8, CCNC, MED12, and MED13) (Taatjes 2010) and associates with Mediator through contacts between MED13 and the Mediator core (Knuesel et al. 2009). Our present understanding of the mechanisms that regulate CDK8 module–Mediator interaction is limited to a study demonstrating that PARP-1 is required in a catalytically independent manner for CDK8 module–Mediator dissociation in the specific context of retinoic acid-regulated gene expression (Pavri et al. 2005). However, the general mechanisms that determine the amount of CDK8 module bound to Mediator are largely undefined.
Here, we demonstrate that the ubiquitously expressed SCFFbw7 ubiquitin ligase regulates CDK8 module–Mediator interactions and targets MED13 and MED13L for proteasomal degradation. Because Fbw7 loss increases the amount of CDK8 module bound to Mediator in proliferating cells, Fbw7 likely represents a general mechanism for controlling Mediator activity. Our results also suggest an expanded role for Fbw7 in transcriptional control that extends beyond its known substrates and raise the possibility that Mediator and CDK8 activity contributes to Fbw7-associated tumorigenesis.
Results and Discussion
Fbw7 interacts with Mediator
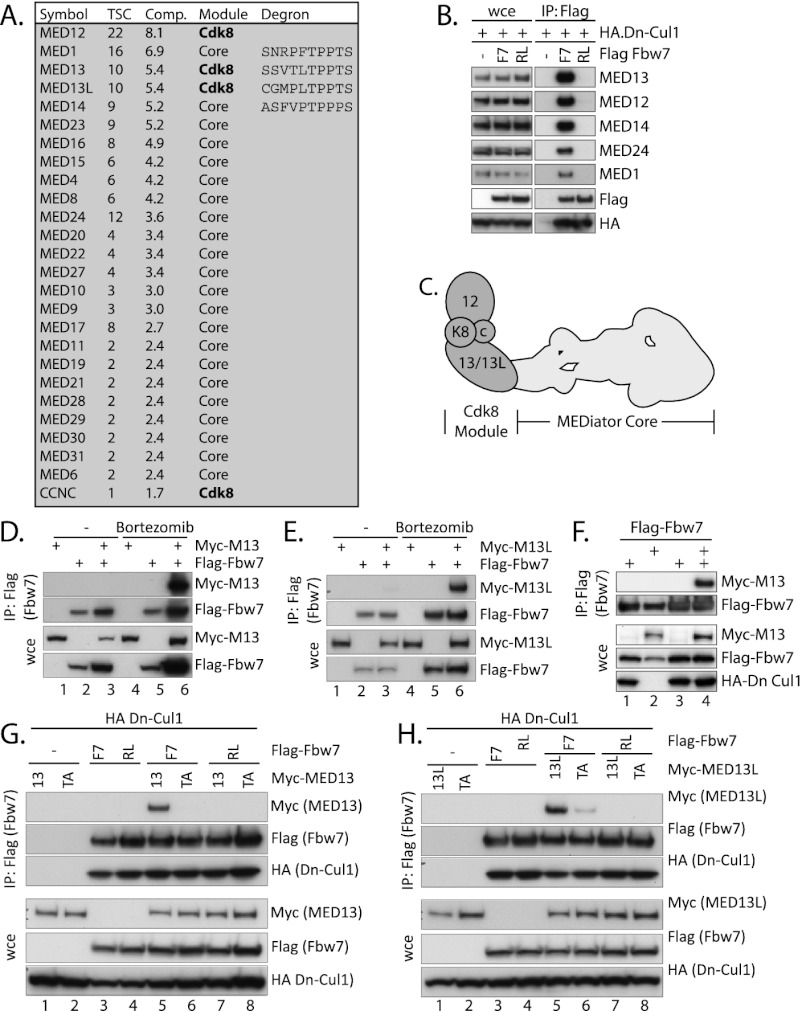
We used an affinity purification and mass spectrometry (MS)-based approach to identify Fbw7 substrates. To help identify candidate substrates, we compared proteins that bound to wild-type Fbw7 with those that bound an Fbw7 mutant in which one of the arginines (R505) required for substrate binding was converted to leucine (Fbw7 RL). Since substrates should bind to wild-type Fbw7 but not Fbw7 RL, we focused on proteins that specifically bound to wild-type Fbw7. In this manner, we identified 72 proteins (Supplemental Table 1), including known substrates, such as KLF5, Myc, Notch1, Notch2, and SREBP-2 (Welcker and Clurman 2008; Liu et al. 2010). We then grouped candidates into known functional complexes and surprisingly found that 26 of the 72 proteins belonged to one complex: Mediator (Fig. 1A).
Figure 1.
MED13/13L bind Fbw7 as canonical substrates. (A) Mediator subunits that associated with wild-type (wt) Fbw7 but not Fbw7 RL. (Comp) CompPASS specificity; (TSC) average total spectral counts of all peptides in duplicate runs. Optimal Fbw7 degrons are indicated (bold). (B) Mediator binds Fbw7. 293A cells were cotransfected with Flag-Fbw7 or Fbw7 RL and HA-DN-Cul1 to inhibit SCFFbw7-mediated degradation. Endogenous Mediator subunits that coprecipitated with Fbw7 are shown. (WCE) Whole-cell extract; (IP) immunoprecipitation. (C) Schematic of the CDK8-bound Mediator complex. (Dark gray) CDK8 module; (light gray) Mediator core. (D,E) Stable interactions between MED13 or MED13L and Fbw7 require proteasomal inhibition. Cells were cotransfected with Flag-Fbw7 and/or myc-MED13/13L and treated with the proteasome inhibitor bortezomib. MED13 (D) or MED13L (E) only coprecipitated after bortezomib treatment (lane 6). (F) DN-Cul1 allows stable MED13-Fbw7 binding. 293 cells were transfected with Flag-Fbw7, HA-DN-Cul1, and myc-MED13 as indicated, and the amount of MED13 bound to immunoprecipitated Fbw7 is shown. (G,H) Mutating the MED13/13L CPDs (TA) or CPD-interacting residues in Fbw7 (RL) abrogates the Fbw7-MED13/13L interaction. Cells were transfected as indicated with wild-type or T326A CPD mutants of MED13/13L and/or wild-type Fbw7 or Fbw7 RL as well as DN-Cul1.
The mammalian CDK8 module–Mediator complex contains 29 subunits (Sato et al. 2004). Interestingly, we detected multiple components of the CDK8 module (MED12, MED13, MED13L, and CCNC) but not MED26, whose binding to Mediator is mutually exclusive with the CDK8 module. This suggested that Fbw7 preferentially interacts with the CDK8–Mediator complex as opposed to the Mediator core. To validate this interaction, we transfected HEK 293A cells (293As) with Flag-tagged Fbw7 or Fbw7 RL and assayed the ability of Fbw7 to bind endogenous Mediator components (Fig. 1B). Cells were cotransfected with dominant-negative Cullin 1 (DN-Cul1) to inhibit substrate ubiquitylation by SCFFbw7 and permit stable Fbw7–substrate interactions. Consistent with our MS data, MED1, MED12, MED13, MED14, and MED24 associated with wild-type Fbw7 but not with Fbw7 RL (Fig. 1B).
Because most Mediator subunits likely interact with Fbw7 indirectly through one to several subunits that bind directly to Fbw7, we examined Mediator subunits for CPDs to identify likely substrates. MED1, MED13, MED13L, and MED14 contain optimal CPDs (Fig. 1A), whereas other subunits contain suboptimal CPDs (Supplemental Table 2). Two of the subunits with optimal CPDs, MED13/13L, are components of the CDK8 module (the MED13 and MED13L peptides detected by MS are shown in Supplemental Table 3). MED13 physically links the CDK8 module to the Mediator core (Fig. 1C; Knuesel et al. 2009), and it is likely that the less-characterized MED13L has similar functions. We hypothesized that Fbw7-dependent degradation of MED13/13L might disrupt the CDK8 module–Mediator complex and therefore determined whether MED13/13L were Fbw7 substrates.
MED13/13L bind to Fbw7-like canonical substrates
Based on their putative CPDs centered on Thr 326 (T326) (Supplemental Fig. 1), we determined whether MED13 and MED13L bound to Fbw7 like other substrates. First, we asked whether stable interactions required binding to be uncoupled from degradation. We immunoprecipitated Fbw7 from 239As transfected with MED13/13L and/or Fbw7 and determined the amount of bound MED13/13L in cells treated with or without the proteasome inhibitor bortezomib (Fig. 1D,E, cf. lanes 3 and 6). MED13/13L only coprecipitated with Fbw7 after proteasomal inhibition. We also used DN-Cul to inhibit proteolysis by preventing SCF function, and this also allowed stable MED13–Fbw7 binding (Fig. 1F). We next determined whether the MED13/13L CPD was required for Fbw7 binding by mutating T326 to alanine (T326A) and disabling SCFFbw7 with DN-Cul1. We found that while wild-type MED13 coprecipitated with wild-type Fbw7, MED13 T326A did not (Fig. 1F). An analogous MED13L T326A mutation similarly reduced MED13L–Fbw7 binding (Fig. 1G). We then asked whether MED13/13L binding requires an intact Fbw7 substrate-binding domain and, consistent with our MS data, found that Fbw7 RL did not bind to MED13/13L (Fig. 1F,G). Finally, MED13 T326 phosphorylation in vivo has been described in proteomic studies (Daub et al. 2008; Oppermann et al. 2009), and we determined whether MED13/13L are phosphorylated at T326. Because the T326 CPD is highly similar to the Cyclin E T380 CPD (Supplemental Fig. 2A), we tested whether a phospho-T380-specific cyclin E antibody recognized MED13/13L (Supplemental Fig. 2B). Anti-pT380 cyclin E antibody detected both MED13 and MED13 but not the corresponding T326A mutations (Supplemental Fig. 2B), and this recognition was inhibited by phosphatase treatment (Supplemental Fig. 2C). The anti-pT380 cyclin E antibody thus recognizes T326 phosphorylated MED13/13L. In sum, these data show that MED13/13L–Fbw7 binding requires the T326 CPDs, an intact Fbw7 substrate-binding interface, and disabled SCF-mediated proteolysis; all of these features are exhibited by Fbw7 substrates.
Fbw7 promotes MED13/13L degradation and ubiquitylation
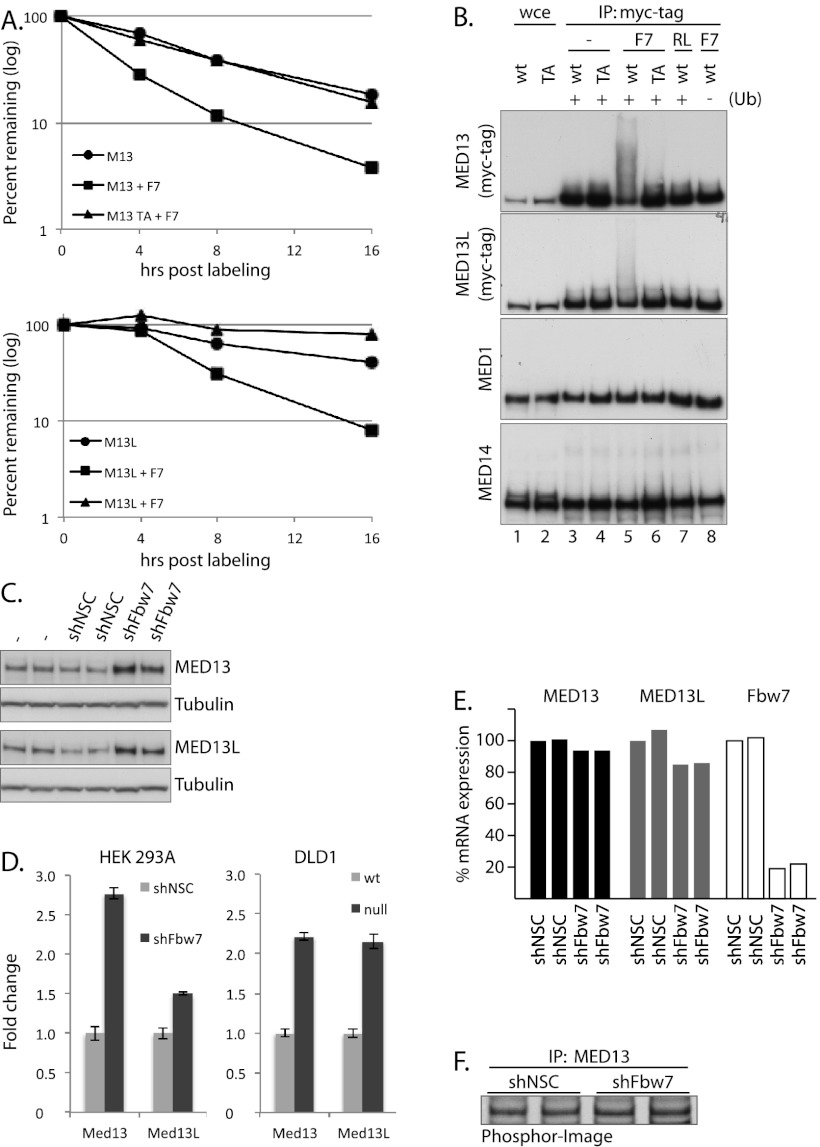
To determine whether Fbw7 regulates MED13/13L stability, we expressed myc-tagged wild-type or T326A mutants of MED13/13L with or without Fbw7 and conducted metabolic pulse-chase experiments (Fig. 2A; Supplemental Fig. 3). Fbw7 coexpression increased MED13/13L turnover, which was prevented by mutating the MED13/13L T326 CPD. Fbw7 decreased the half-life of MED13 from 6.0 h to 3.0 h, whereas the half-life of MED13 T326A coexpressed with Fbw7 was 6.0 h (Fig. 2A). Similarly, Fbw7 coexpression decreased the half-life of MED13L from 12 h to 4 h, whereas MED13L T326A exhibited a half-life too long to be measured in this experiment (Fig. 2A; Supplemental Fig. 3). Thus, Fbw7 overexpression drives MED13/13L turnover in a CPD-specific manner.
Figure 2.
MED13 and MED13L are Fbw7 substrates. (A) Fbw7 increases MED13/13L turnover. 293 cells were cotransfected with myc-MED13/13L or MED13/13L T326A and Flag-Fbw7, labeled with 35S-Met, and chased with cold methionine as indicated. Turnover of labeled MED13/13L was assayed by phosphor-imaging of immunoprecipitated MED13, and the graphs show averages of two independent experiments. Entire gels are shown in Supplemental Figure 3. (B) Fbw7 specifically ubiquitylates MED13/13L in vitro. Myc-MED13/13L (wild-type [wt]) or myc-MED13/13L CPD mutants (TA) were immunoprecipitated and subjected to in vitro ubiquitylation assays containing recombinant wild-type Fbw7 (F7) or Fbw7 RL (RL) and other ubiquitylation components (E1, Cul1, E2, and ubiquitin). Controls were performed without Fbw7 (lanes 3,4) or ubiquitin (Ub) (lane 8). Membranes were reprobed to detect MED1 and MED14. (C) shRNA-mediated knockdown of Fbw7 increases endogenous MED13 and MED13L abundance. Cells were untreated or transduced with nonsilencing control shRNA (shNSC) or an Fbw7-specific shRNA (shFbw7). Two biological replicates are shown. (D) Quantitation of MED13/13L abundance in 293 cells expressing sh-Fbw7 or control shRNA (two biological replicates) (Fig 3A) or Fbw7-null DLD1 cells and controls (three biological replicates) (Fig. 3A). Bands were quantified with an ImageQuant Bioanalyzer and ImageQuant TL. (E) Knockdown of Fbw7 does not increase MED13 or MED13L transcription. MED13/13L mRNA was quantified by real-time PCR (normalized to GAPDH). (F) Knockdown of Fbw7 does not increase MED13 translation. Cells were pulse-labeled with 35S-Met, and endogenous MED13 was immunoprecipitated.
Because changes in protein stability in vivo could reflect indirect effects of Fbw7, we determined whether SCFFbw7 directly ubiquitylates MED13/13L in vitro. Wild-type or T326A MED13/13L were immunoprecipitated from transfected cells and subjected to in vitro ubiquitylation reactions using purified components. Wild-type Fbw7 but not the Fbw7 RL substrate-binding mutant ubiquitylated wild-type MED13/13L (Fig. 2B), as demonstrated by the ubiquitin-dependent formation of high-molecular-weight species and the corresponding reduction in unmodified MED13/13L. In contrast, MED13/13L T326A was not ubiquitylated, consistent with its inability to bind Fbw7. Thus, MED13/13L are directly ubiquitylated by SCFFbw7 in vitro, and, like the coprecipitation and turnover experiments, this requires intact CPDs and a wild-type Fbw7 substrate-binding region.
In addition to MED13/13L, MED1 and MED14 also contain optimal CPDs (Fig. 1A). To test whether they might be Fbw7 substrates, membranes used in assaying the in vitro ubiquitylation of MED13/13L were stripped and reprobed for MED1 and MED14 contained within the MED13 immunoprecipitates (Fig. 2B). Neither MED1 nor MED14 was ubiquitylated by Fbw7, suggesting that Fbw7 specifically ubiquitylates MED13/13L and not simply all Mediator components with CPD motifs.
Endogenous Fbw7 regulates endogenous MED13/13L abundance
We next determined the effects of Fbw7 loss on endogenous MED13/13L abundance using stable expression of an Fbw7-specific shRNA (Fig. 2C,D). Fbw7 knockdown increased endogenous MED13/13L protein abundance, as compared with controls expressing no or nonspecific shRNA (Figs. 2C,D, 3A). To eliminate possible off-target shRNA effects, we also examined MED13/13L in DLD1 colon carcinoma cells in which Fbw7 was inactivated by gene targeting (Rajagopalan et al. 2004) and found that MED13/13L abundance was increased in Fbw7 knockout cells, compared with isogenic controls (Figs. 2D, 3A). Additionally, transient Fbw7 silencing by siRNA transfection increased MED13/13L and pT326-Med13/13L abundance in other cell types, which exhibited cell type-specific differences with respect to the extent of MED13 and MED13L regulation (Supplemental Fig. 4). Because we were unable to measure endogenous MED13/13L half-life for technical reasons, we determined whether Fbw7 loss affected MED13/13L synthesis by examining their mRNA abundance by quantitative PCR (qPCR) (Fig. 2E) and MED13 translation rates by 35S-Met pulse labeling (Fig. 2F). Neither was impacted by Fbw7 loss; thus, the increased abundance of endogenous MED13/13L likely reflects changes in protein stability.
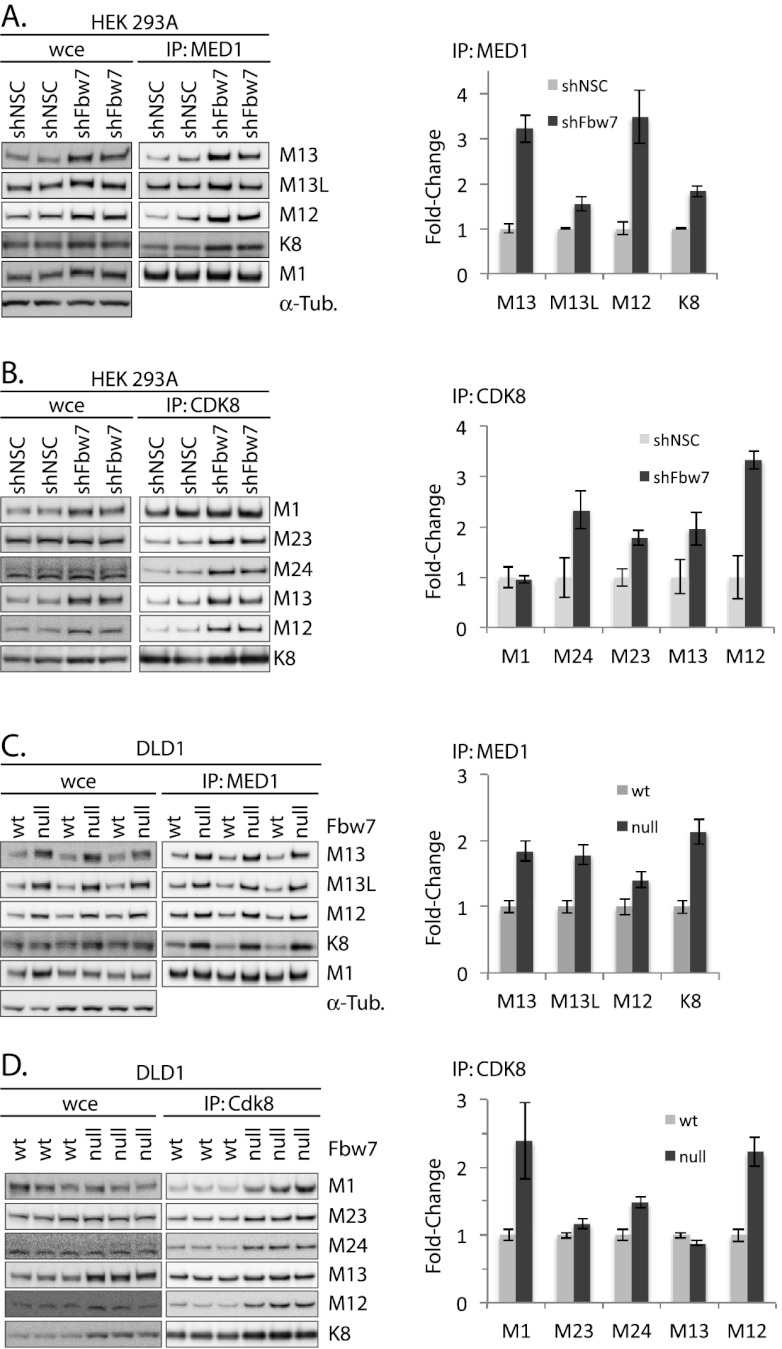
Figure 3.
Fbw7 loss increases CDK8 module–Mediator association. (A) Fbw7 was silenced by shRNA in 293As as shown in Figure 2. The amount of endogenous CDK8 module–Mediator association was determined by immunoprecipitation with the Mediator core component MED1 and Western analyses of the indicated proteins. Four independent biological replicates were quantified with an ImageQuant Bioanalyzer and ImageQuant TL. Values were normalized to the amount of immunoprecipitated MED1 in each sample, and error bars show ±standard error of the mean (SEM). (B) Fbw7-dependent changes in CDK8 module–Mediator association were examined as in A, except CDK8 was immunoprecipitated. The normalized amounts of CDK8-bound core Mediator and CDK8 module subunits are shown (two biological replicates). (C,D) Lysates from wild-type and Fbw7-null DLD1 cells were analyzed as shown in A and B. The unnormalized quantified data used for this figure are shown in Supplemental Figure 5.
Fbw7 regulates CDK8 module–Mediator association
A fundamental role for MED13 (and likely MED13L) is to bridge the CDK8 module to the Mediator complex (Knuesel et al. 2009), suggesting that Fbw7 loss might increase the association of CDK8 module components with Mediator. To test this, we first immunoprecipitated Mediator via a core component, MED1, and determined whether Fbw7 loss changed the amount of CDK8 module bound to Mediator. We normalized the abundance of bound proteins to the amount of MED1 in the immunoprecipitates and found that Fbw7 loss increased Mediator-associated MED13, MED13L, MED12, and CDK8 in 239As expressing Fbw7 shRNA (Fig. 3A) and in Fbw7-null DLD1s (Fig. 3C). The unnormalized data from these immunoprecipitates are shown in Supplemental Figure 5. Thus, Fbw7 loss increased the association of CDK8 module subunits bound to Mediator as measured through the coimmunoprecipitation with the Mediator core protein Med1.
We also performed reciprocal experiments in which we immunoprecipitated CDK8 and examined how Fbw7 loss affected the association of bound Mediator core subunits (MED1, MED23, and MED24) as well as other CDK8 module proteins (Fig. 3B,D). As expected, these studies also demonstrated an increase in CDK8 module–Mediator association upon Fbw7 loss. Additionally, Fbw7 loss increased CDK8-bound MED12 and MED13. This is consistent with the fact that CDK8 also exists as a dimer with CCNC (i.e., without MED12 and MED13) (Knuesel et al. 2009), and there is thus additional capacity to bind MED13 and MED12 and form complete Cdk8 modules. Importantly, because both shRNA-mediated knockdown of Fbw7 in 293As and somatic deletion of Fbw7 in DLD1s increased CDK8 module–Mediator association, these effects are Fbw7-dependent. In light of the fact that only 30% of cellular MED13 is not associated with Mediator (presumably as free CDK8 module), the magnitude of the increased CDK8 module–Mediator association observed upon Fbw7 loss appears quite significant (Knuesel et al. 2009).
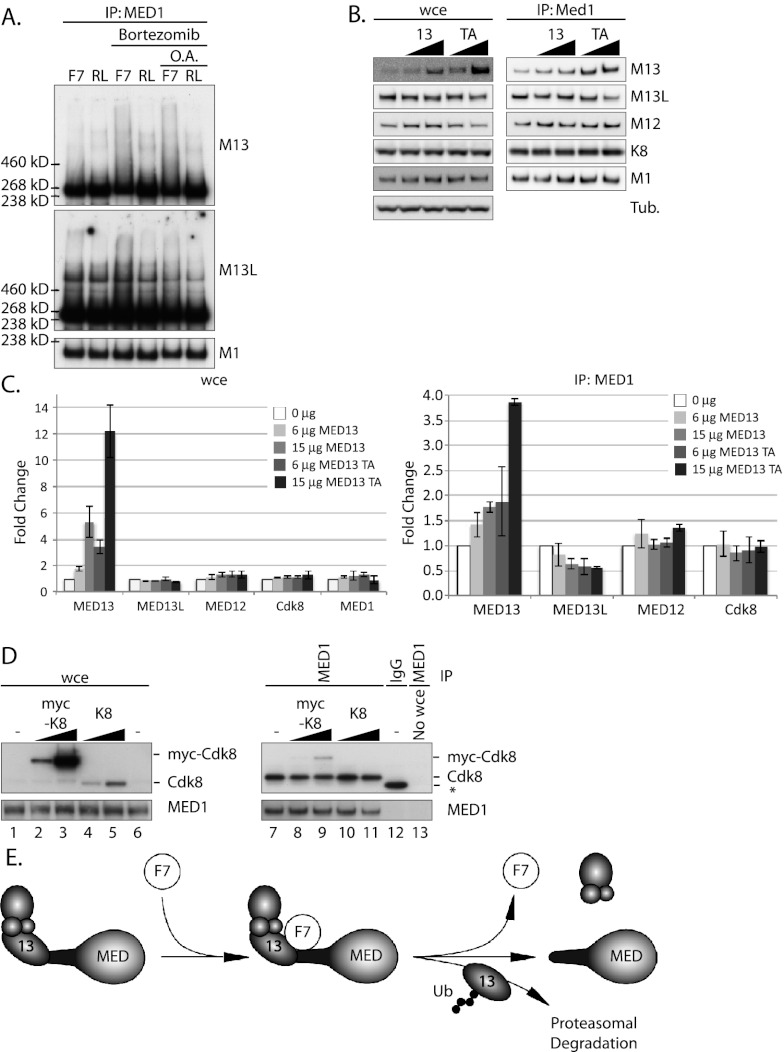
Because Fbw7 specifically ubiquitylates the proteins that link the CDK8 module to Mediator, we hypothesized that Fbw7 might disrupt the CDK8 module–Mediator association. If so, Fbw7 must be able to ubiquitylate Mediator-bound MED13/13L. Therefore, we immunoprecipitated Mediator via MED1 and conducted in vitro MED13/13L ubiquitylation assays (Fig. 4A). As shown in Figure 4A, wild-type Fbw7 did indeed ubiquitylate Mediator-bound MED13/13L, whereas Fbw7 RL did not. The amount of MED13/13L ubiquitylated in vitro was increased when MED13/13L was isolated from cells treated with bortezomib prior to lysis; this likely prevented degradation of the phosphorylated MED13/13L species required for Fbw7 binding. We also pretreated cells with okadaic acid in an effort to increase degron phosphorylation by inhibiting phosphatases, but this had no additional impact on MED13/13L ubiquitylation (Fig. 4A, lanes 5,6). Therefore, Fbw7 not only coprecipitates with the CDK8 module–Mediator complex, but also ubiquitylates Mediator-bound MED13/13L. These findings support the idea that Fbw7 can liberate the CDK8 module from Mediator by targeting MED13/13L for degradation.
Figure 4.
Fbw7 ubiquitylates Mediator-bound Med13/13L. (A) Fbw7 ubiquitylates Mediator-bound MED13. Mediator was immunoprecipitated via MED1 and subjected to in vitro ubiquitylation. Cells were pretreated with bortezomib (lanes 3,4) or bortezomib and okadaic acid (lanes 5,6) prior to lysis. (B,C) Overexpression of MED13 or MED13 TA does not increase CDK8 module–Mediator association. Increasing amounts of MED13 or MED13 TA were transfected into 239As. Mediator was immunoprecipitated via MED1, and CDK8 module–Mediator association was assayed via Western analysis. Three experiments were quantified as in Figure 3C and normalized against immunoprecipitated MED1. Error bars show the standard error of the mean. (B) A representative Western blot is shown. (D) Overexpression of CDK8 does not increase the amount of CDK8 bound to Mediator. Increasing amounts of myc-tagged or untagged CDK8 were transfected into 293As. Mediator was immunoprecipitated through MED1, and the positions of myc-tagged CDK8 and endogenous CDK8 are shown. Controls include immunoprecipitation with nonspecific IgG (lane 12) or no cell lysate (lane 13). (E) Model: Fbw7 binds and ubiquitylates Mediator-bound MED13/13L, resulting in their degradation and the dissociation of the remaining CDK8 module components from the Mediator core (Med).
In addition to regulating CDK8 module–Mediator association through ubiquitylating Mediator-bound MED13/13L, Fbw7 could also regulate the amount of CDK8 bound to Mediator by simply increasing Mediator-free MED13/13L abundance. We thus overexpressed MED13 or MED13 T326A and examined the amount of endogenous MED13, MED12, and CDK8 that coimmunoprecipitated with Mediator. As shown in Figure 4C, MED13 overexpression increased the amount of Mediator-bound MED13 but not the amount of Mediator-bound MED12 or CDK8. Thus, increased MED13 abundance is not sufficient to load additional CDK8 module components onto Mediator, and this is unlikely to be a major mechanism by which Fbw7 regulates CDK8–Mediator association. The failure of elevated MED13 to recruit other CDK8 module proteins to Mediator could reflect additional regulatory mechanisms that control assembly of the CDK8 module itself.
CDK8 is an oncogene that has been linked to colorectal carcinogenesis, and this may primarily involve its role in Mediator regulation (Firestein et al. 2008; Firestein and Hahn 2009). To examine how Fbw7 loss might impact CDK8 module–Mediator association, we overexpressed CDK8 and assayed its binding to Mediator (Fig. 4D). CDK8 overexpression did not increase the amount of CDK8 associated with Mediator. Thus, Fbw7 loss may modulate the impact of overexpressed CDK8 on Mediator activity in cancer, assuming that the oncogenic function of CDK8 involves its association with Mediator. The roles of Mediator and Fbw7 in CDK8-associated cancer thus require further study.
Fbw7's substrates include transcription factors that regulate complex gene networks (e.g., Myc and Notch) and play key roles in tumor suppression by Fbw7. Our finding that Fbw7 regulates the amount of CDK8 module bound to Mediator greatly expands Fbw7's potential impact on transcription in normal and cancer cells, especially given the genome-wide role of CDK8–Mediator in transcription and the role of CDK8 in tumorigenesis. Because Fbw7 and other SCF proteins are ubiquitously expressed, the signaling pathways that phosphorylate CPDs largely regulate the timing and context of substrate degradation. Although the specific pathways that stimulate MED13/13L CPD phosphorylation are unknown, all of our studies used asynchronously growing cells under normal growth conditions. Thus, our findings that Fbw7 regulates MED13/13L abundance and CDK8 module–Mediator association in these conditions suggest that Fbw7 is a general regulator of the CDK8 module–Mediator interaction.
Fbw7 degrades a network of oncogenic proteins, and defining specific roles for its individual substrates in tumor suppression remains challenging. Regulation of the CDK8–Mediator complex adds yet another layer of complexity to the Fbw7 substrate network and provides unanticipated connections between tumor suppression by Fbw7 and Mediator activity as well as a potential link between Fbw7 and oncogenesis associated with CDK8. In summary, our data support a model whereby Fbw7 targets Mediator-bound MED13/13L for ubiquitylation and degradation, thereby disrupting the CDK8 module–Mediator complex (Fig. 4E). As such, this defines the first mechanism regulating the reversible interaction between the CDK8 module and the Mediator complex.
Materials and methods
Cell culture
HEK 293A, HEK 293T, HeLa, U20S, HT1080, and DLD-1 cells were cultured in Dulbecco's modified essential medium containing L-glutamine, 4.5 g/L D-glucose, 10% FCS, and 1% penicillin/streptomycin. Fbw7-null DLD1 cells have been described previously (Rajagopalan et al. 2004). Transfection of plasmids or siRNA was performed with calcium phosphate or Lipofectamine RNAiMAX (Invitrogen). Cells were treated with 0.5 μM bortezomib for 10 h prior to lysis when indicated.
Plasmids and siRNA
MED13 and MED13L cDNA as well as Fbw7 shRNA constructs were from OpenBiosystems: MED13 (MHS1768-99865132), MED13L (MHS1768-9981314), and Fbw7 shRNA (clone V2LHS_202932). MED13/13L were subcloned into pCS2.MT (N-terminal myc tag) or pCS2 expression vectors. MED13/13L T326A were generated by site-directed mutagenesis. pFlagFbw7, pCS2.HA Dn-Cul1, and Fbw7-specific siRNA have been described previously (Welcker et al. 2003; Ye et al. 2004; van Drogen et al. 2006).
Antibodies
Antibodies from Bethyl Laboratories, Inc., were MED12 (A300-774A), MED13 (A301-278A), MED13L (A302-421A), MED14 (A301-044A), Med15 (A302-423A), and Med24 (301-472A). Antibodies from Santa Cruz Biotechnology, Inc., were CDK8 (sc-1521), Cyclin C (sc-7431), and MED1 (sc-5334). Antibodies from Sigma were Flag tag-specific antibody clone M2 and Tubulin-specific antibody clone DM1α. We used Covance HA tag-specific antibody clone HA-11 (MMS-101P) and in-house myc tag antibody 9E10.
Immunoprecipitations and Western blot analysis
Cells were lysed in 0.5% NP-40 buffer with 1 mM PMSF, 1 mM EDTA, 2 μg/μL aprotinin, 2 μg/μL leupeptin, 2 μg/μL pepstatin, 1 mM Na orthovanadate, and 50 mM NaF. Lysates were briefly sonicated. Immunoprecipitations were carried out overnight at 4°C. Lysates and immunoprecipitations were run on 3%–8% Tris-acetate gels (Invitrogen) and transferred to PVDF. Bands were visualized by ECL (made in-house) and either exposure to film or quantitation using an ImageQuant Bioanalyzer and ImageQuant TL. IgG light chain-specific secondary antibodies were used to visualize CDK8 after coimmunoprecipitation due to comigration with IgG heavy chain.
Pulse label and pulse-chase experiments
Cells were transfected as indicated. Forty hours post-transfection, cells were washed with PBS; starved in Met-free, Cys-free medium for 20 min; and labeled with 250 μCi/mL 35S-Met for 25 min at 37°C. Cells were then washed and chased in starvation medium supplemented with 0.8 mg/mL cold Met and incubated at 37°C. Immunoprecipitated samples were electrophoresed, dried gels were analyzed with a PhosphorImager, and bands were quantified with ImageQuant TL. Half-lives were determined by averaging two independent experiments, graphing the signal intensities on a semilog plot, and using exponential regression to calculate the best-fit line.
In vitro ubiquitylation assays
Substrates were immunoprecipitated from 500 μg of protein lysate overnight, and beads were washed in lysis buffer. Reactions contained 10 ng/μL Fbw7/Skp1, 100 ng/μL Cul1/Rbx1, 80 ng/μL GST-E1, 150 ng/μL Ubch5, and 0.6 μg/μL HA-Ub (BostonBiochem, U-110) as well as 2 mM ATP, 10 mM MgCl2, and bortezomib. The reaction volume was 14 μL in 0.5% NP-40 supplemented with protease inhibitors and 1 mM DTT. Reactions were carried out for 1 h at room temperature with occasional agitation and were analyzed by Western blotting.
MS-based screen for novel substrates
Identification of novel Fbw7 substrates followed the methods of Sowa et al. (2009). HA-tagged Fbw7 or HA-Fbw7 RL was transfected into HEK 293A cells, cells were treated for 10 h with bortezomib, and Fbw7 was immunoprecipitated with anti-HA Agarose Conjugate (Sigma, A2095). Beads were washed three times in NP-40 lysis and three times in PBS, and bound complexes were eluted twice with 100 μg/mL HA peptide (Sigma, I2149) in PBS. Eluates were TCA-precipitated, trypsinized, and submitted for MS analysis. Each sample was run in duplicate on an LTQ-FT MS in the Fred Hutchinson Cancer Research Center Proteomics resource. Peptide predictions were made with X!Tandem and searched against both IPI Human version 3.69 and the pseudo-reverse IPI Human version 3.52. Predicted proteins and total spectral counts were used for CompPASS analysis (Sowa et al. 2009). Proteins with a CompPASS normalized D-score (NWD) >1.00 were pursued as specific interactors.
Acknowledgments
We thank Ning Zheng for providing purified proteins for ubiquitylation reactions, and Wade Harper and Matthew Sowa for ComPASS analyses. This work was supported by NIH grants CA084069 and CA102742 (to B.E.C.), CA127364 (to D.J.T.), and T32 CA080416 (to M.A.D.). B.E.C. is supported by the Burroughs Welcome Founation.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.207720.112.
References
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. 2007. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res 67: 9006–9012 [DOI] [PubMed] [Google Scholar]
- Crusio KM, King B, Reavie LB, Aifantis I 2010. The ubiquitous nature of cancer: The role of the SCF(Fbw7) complex in development and transformation. Oncogene 29: 4865–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 31: 438–448 [DOI] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 17: 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeier CC, Taatjes DJ 2010. Activator–Mediator binding regulates Mediator–cofactor interactions. Proc Natl Acad Sci 107: 11283–11288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM 2006. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci 103: 15788–15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Hahn WC 2009. Revving the Throttle on an oncogene: CDK8 takes the driver seat. Cancer Res 69: 7899–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. 2008. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 455: 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J, Delrow JJ, Small T, Grady WM, Nakayama KI, et al. 2012. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol Cell Biol 32: 2160–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP 2007. Structure of a Fbw7–Skp1–cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 26: 131–143 [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. 2011. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471: 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ 2009. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev 23: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, Wang Y, Wang W, Wang R, Wang Q, et al. 2010. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem 285: 18858–18867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30: 256–263 [DOI] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A 2004. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432: 775–779 [DOI] [PubMed] [Google Scholar]
- Meyer KD, Donner AJ, Knuesel MT, York AG, Espinosa JM, Taatjes DJ 2008. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J 27: 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K 2006. Ubiquitin ligases: Cell-cycle control and cancer. Nat Rev Cancer 6: 369–381 [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, Nakayama KI 2007. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med 204: 2875–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H 2009. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics 8: 1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256 [DOI] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell 18: 83–96 [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C 2004. Inactivation of hCDC4 can cause chromosomal instability. Nature 428: 77–81 [DOI] [PubMed] [Google Scholar]
- Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A 2010. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 139: 929–941 [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. 2004. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 14: 685–691 [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ 2010. The human Mediator complex: A versatile, genome-wide regulator of transcription. Trends Biochem Sci 35: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Sangfelt O, Spruck C 2008. The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett 271: 1–12 [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I 2008. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med 205: 1395–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, Sangfelt O, Malyukova A, Matskova L, Yeh E, Means AR, Reed SI 2006. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol Cell 23: 37–48 [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE 2008. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8: 83–93 [DOI] [PubMed] [Google Scholar]
- Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, Clurman BE, Roberts JM 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell 12: 381–392 [DOI] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. 2011. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471: 110–114 [DOI] [PubMed] [Google Scholar]
- Ye X, Nalepa G, Welcker M, Kessler BM, Spooner E, Qin J, Elledge SJ, Clurman BE, Harper JW 2004. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem 279: 50110–50119 [DOI] [PubMed] [Google Scholar]