The distal appendage (DAP) of the centriole is a pinwheel-like structure proposed to anchor cilia to the plasma membrane. Using quantitative proteomics and superresolution microscopy, Tanos et al. identify novel DAP components required for ciliogenesis. Loss of DAP proteins specifically blocks ciliogenesis at the membrane-docking step. This study reveals critical coordination between membrane docking and cilia initiation, allowing temporal and spatial regulation of ciliogenesis in response to cell cycle cues.
Keywords: centriole, cilia, distal appendages, transition fibers, membrane docking
Abstract
The distal appendages (DAPs) of centrioles have been proposed to anchor cilia to the plasma membrane, but their molecular composition, assembly, and exact function in ciliogenesis remain poorly understood. Using quantitative centrosome proteomics and superresolution microscopy, we identified five DAP components, including one previously described (CEP164), one partially characterized (CEP89 [ccdc123]), and three novel (CEP83 [ccdc41], SCLT1, and FBF1) DAP proteins. Analyses of DAP assembly revealed a hierarchy. CEP83 recruits both SCLT1 and CEP89 to centrioles. Subsequent recruitment of FBF1 and CEP164 is independent of CEP89 but mediated by SCLT1. All five DAP components are essential for ciliogenesis; loss of CEP83 specifically blocks centriole-to-membrane docking. Undocked centrioles fail to recruit TTBK2 or release CP110, the two earliest modifications found on centrioles prior to cilia assembly, revealing centriole-to-membrane docking as a temporal and spatial cue promoting cilia initiation.
The primary cilium is a microtubule (MT)-based organelle projecting from the plasma membrane of nearly every cell type in the human body and functions as a sensory hub for the transduction of diverse signals in the environment (Eggenschwiler and Anderson 2007). The cilium core, or axoneme, is composed of nine doublet MTs nucleated directly from a mature centriole known as the basal body. In addition to being a template for axoneme formation, basal bodies also serve as an anchor that docks the base of cilia to the plasma membrane (Anderson 1972). Electron microscopy (EM) analyses have suggested that docking is mediated by a pinwheel-like structure at centriole distal ends (Anderson 1972), described as the alar sheets, transition fibers, or distal appendages (DAPs). The DAPs project radially from the distal end of basal bodies to the membrane, marking the border of the plasma membrane and the ciliary membrane. Moreover, DAPs are also thought to be part of the ciliary gate or “ciliary pore complex” (Rosenbaum and Witman 2002; Reiter et al. 2012), working together with the septin ring barrier (Hu et al. 2010), nucleoporins (Kee et al. 2012), and the ciliary transition zone (TZ) (Chih et al. 2012) to allow selective targeting and sorting of proteins to and from the ciliary compartment. EM analyses of vertebrate cycling cells have revealed that newborn centrioles take about one and a half cell cycles to acquire appendages (a process known as centriole maturation), and only mature centrioles can support ciliogenesis (Vorobjev and Chentsov Yu 1982). As a result, cells in the beginning of the cell cycle inherit one appendage-equipped centriole (mother) that can grow a cilium and one immature centriole (daughter) that cannot (Vorobjev and Chentsov Yu 1982). Despite the importance of DAPs in ciliogenesis (Vorobjev and Chentsov Yu 1982; Graser et al. 2007), the molecular composition, assembly, and exact function of the DAP remain largely unclear.
Results and Discussion
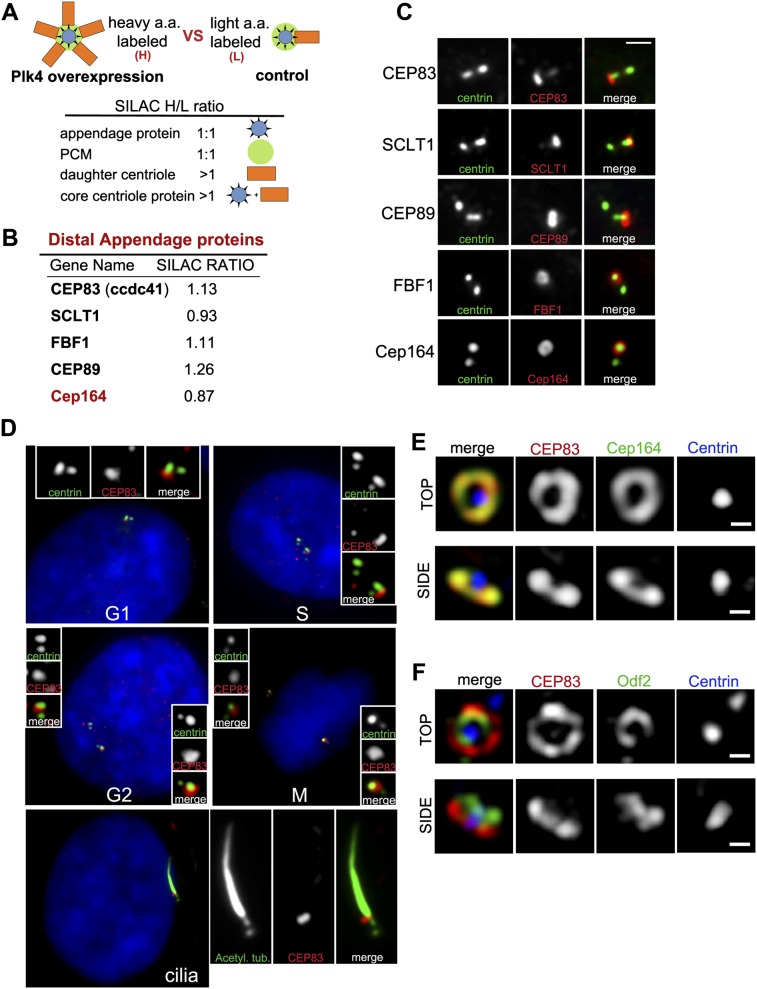
To better understand the function and assembly of DAPs, we sought to systematically identify DAP-associated proteins. Since only the mother centriole in a centrosome acquires appendages (Fig. 1), an increase in the number of daughter centrioles within a centrosome would not change the total amount of appendage proteins in that centrosome, although other centriolar proteins would increase proportionally. Such differences can be detected and analyzed by SILAC (stable isotope labeling by amino acids in cell culture) mass spectrometry (Ong et al. 2002) and were used to quantitatively differentiate appendage proteins from other core centriolar components (Fig. 1A). In brief, the following two types of centrosomes were isolated from S-phase arrested cells: (1) wild-type centrosomes containing one mother centriole that is engaged with one newly formed daughter centriole and (2) Plk4-induced “rosette” centrosomes (Habedanck et al. 2005) containing one mother centriole engaged with four to six newly formed daughter centrioles. In both samples, only mother centrioles carry appendages. Thus, the amount of any appendage protein should be the same between rosette and wild-type centrosomes, showing a 1:1 ratio in the SILAC analysis (Fig. 1A). Conversely, higher amounts of core centriolar proteins should associate with rosette centrosomes, showing ratios >1. Importantly, as nonspecific proteins copurified with centrosomes also show a ratio close to 1, only those that are present in the published centrosome proteome (Jakobsen et al. 2011) were included in the subsequent analysis and considered as potential appendage-associated proteins.
Figure 1.
Identification of novel DAP proteins. (A) Schematic of SILAC experiment. DAP proteins belong to the group of proteins showing a 1:1 ratio. (B) Summary of the screen for DAP proteins. The SILAC ratio (H/L) is indicated. (C) RPE-1 cells in G1 were stained with antibodies to the indicated DAP proteins (red) or to centrin (green). Note the asymmetrical localization of DAP proteins. Bar, 1 μm. (D) Subcellular distribution of CEP83. RPE-1 cells stably expressing GFP-centrin 2 (green) were stained with an antibody to CEP83 (red) and DAPI (blue). Cell cycle stages are indicated; note that CEP83 is incorporated into centrioles during G2. (Bottom panel) In ciliated cells where primary cilia are marked with antibodies against acetylated α-tubulin (green), CEP83 localizes to the ciliary base. (E,F) RPE-1 cells stably expressing HA-tagged CEP83 were stained with antibodies to HA (red), Cep164 (green) (E) or Odf2 (green) (F) and centrin (blue) and examined with an OMX superresolution microscope (Applied Precision, Inc.). Note that CEP83 colocalizes with the DAP protein Cep164 both in top view (top panels) and side view (bottom panels) but does not colocalize with the SAP protein Odf2. Small bar, 200 nm.
Candidate DAP proteins were examined for their subcellular localization by fluorescence light microscopy and superresolution microscopy (OMX, DeltaVision). Five DAP components have been identified so far, including three unknown (CEP83 [ccdc41], SCLT1, and FBF1), one partially characterized (CEP89 [ccdc123]) (Sillibourne et al. 2011), and one previously described (CEP164) (Graser et al. 2007). These DAP proteins are evolutionarily conserved in some ciliated organisms but appear to be lost or highly divergent in others (Supplemental Table S1). The localization pattern of new DAP components during the cell cycle matches exactly with that of CEP164, which is present at one mature centriole in G1 and S phases but two in G2 and M phases (Fig. 1C; Supplemental Fig. S1A,B). Similar to the previously reported CEP89 (Sillibourne et al. 2011), superresolution microscopy revealed that CEP83, SCLT1, and FBF1 colocalize with the known DAP component CEP164 (Graser et al. 2007) but not with the subdistal appendage (SAP) marker ODF2 (Fig. 1E,F; Supplemental Fig. S1C–F; Ishikawa et al. 2005), suggesting that CEP83, SCLT1, CEP89, FBF1, and CEP164 form part of the core structure of DAPs.
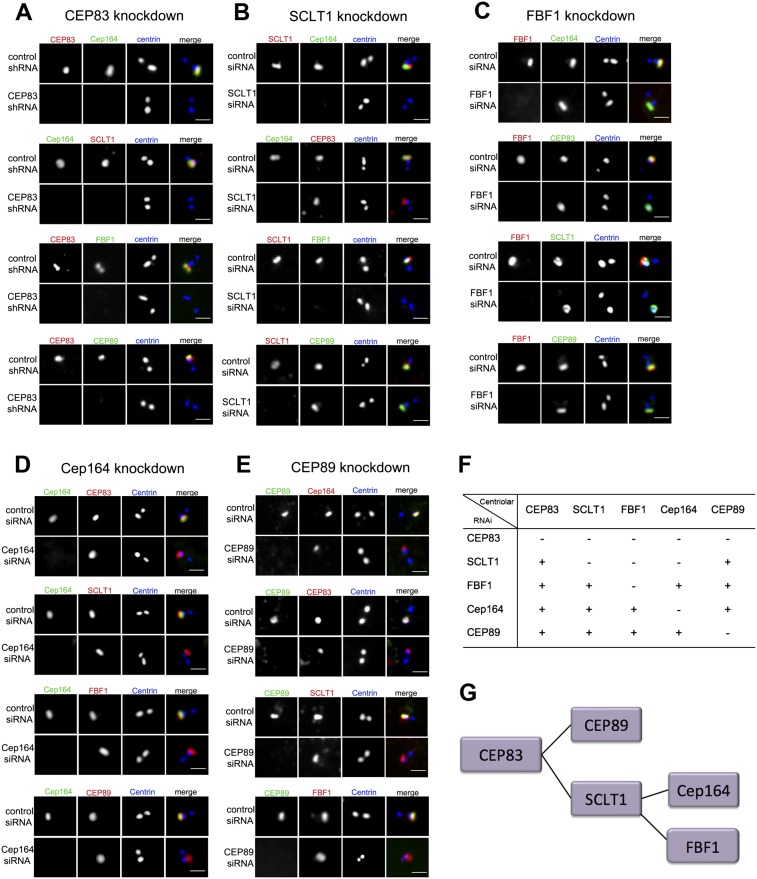
To explore DAP assembly, individual DAP components were depleted from cells by RNAi, and the localization of others was examined. Loss of CEP83 disrupts the centriolar recruitment of all other DAP components (Fig. 2A) without affecting their expression levels (Supplemental Fig. S2G). In contrast, CEP83 can localize to centriole distal ends in the absence of any other DAP proteins (Fig. 2B–E), suggesting that CEP83 is required early. The recruitments of CEP89 and SCLT1 depend on CEP83 but are independent of each other and of FBF1 or CEP164 (Fig. 2A,B,E), revealing a hierarchy of assembly downstream from CEP83. Interestingly, both FBF1 and CEP164 localize to centrioles in a manner depending on the presence of SCLT1 (Fig. 2B) but not CEP89 (Fig. 2E) or each other (Fig. 2C,D), revealing another branch downstream from SCLT1. Taken together, a tree of DAP assembly pathway is established (Fig. 2F,G) in which CEP83 is at the root, branching out through CEP89 and SCLT1, followed by another branch downstream from SCLT1 via FBF1 and CEP164.
Figure 2.
The DAP assembly pathway. (A) RPE1 cells were retrovirally transduced with control shRNA or a CEP83 targeting shRNA as indicated and stained with antibodies to CEP83 (red) and to the other DAPs identified (green) (see Fig. 1A). Centrioles are marked with centrin (blue). (First panel) Note that CEP83 is required for the centriolar localization of the known DAP protein Cep164 as well as all of the other DAP components. The second panel from the top shows that SCLT1 is absent from CEP83-depleted centrioles marked by the absence of Cep164. Bar, 1 μm. (B) Cells transfected with control siRNA or SCLT1 siRNA were stained with antibodies to SCLT1 (red) and to the other DAP proteins identified (green). Centrioles are marked with centrin (blue). Note that Cep164 is absent from SCLT1-depleted centrioles (first panel), whereas CEP83 is still present (second panel). Bar, 1 μm. (C) Cells transfected with control siRNA or FBF1 siRNA were stained with antibodies to FBF1 (red) and to all other DAP components (green). Centrioles are marked with centrin (blue). Bar, 1 μm. (D) Cells transfected with control siRNA or Cep164 siRNA were stained with antibodies to Cep164 (green) and to the other DAP proteins (red). Centrioles are marked with centrin (blue). Bar, 1 μm. (E) Cells transfected with control siRNA or CEP89 siRNA were stained with antibodies to CEP89 (green) and to the other DAP proteins (red). Centrioles are marked with centrin (blue). Bar, 1 μm. (F) A table showing the interdependency of centriolar localization for the proteins identified. (G) Assembly pathway of centriolar DAPs.
A previous EM study reported that DAPs are absent when the SAP protein ODF2 is depleted, suggesting that ODF2 may mediate DAP assembly (Ishikawa et al. 2005). To test this hypothesis, ODF2 was depleted from cells, and the localization of DAP components was examined. Surprisingly, while depletion of ODF2 abolished the recruitment of other SAP components such as centriolin, it had no effect on the assembly of DAP components (Supplemental Fig. S2A–D). Similarly, depletion of CEP83, while disrupting DAPs, had no effect on the localization of ODF2 or centriolin (Supplemental Fig. S2E,F), suggesting that the assemblies of SAPs and DAPs are largely independent of each other.
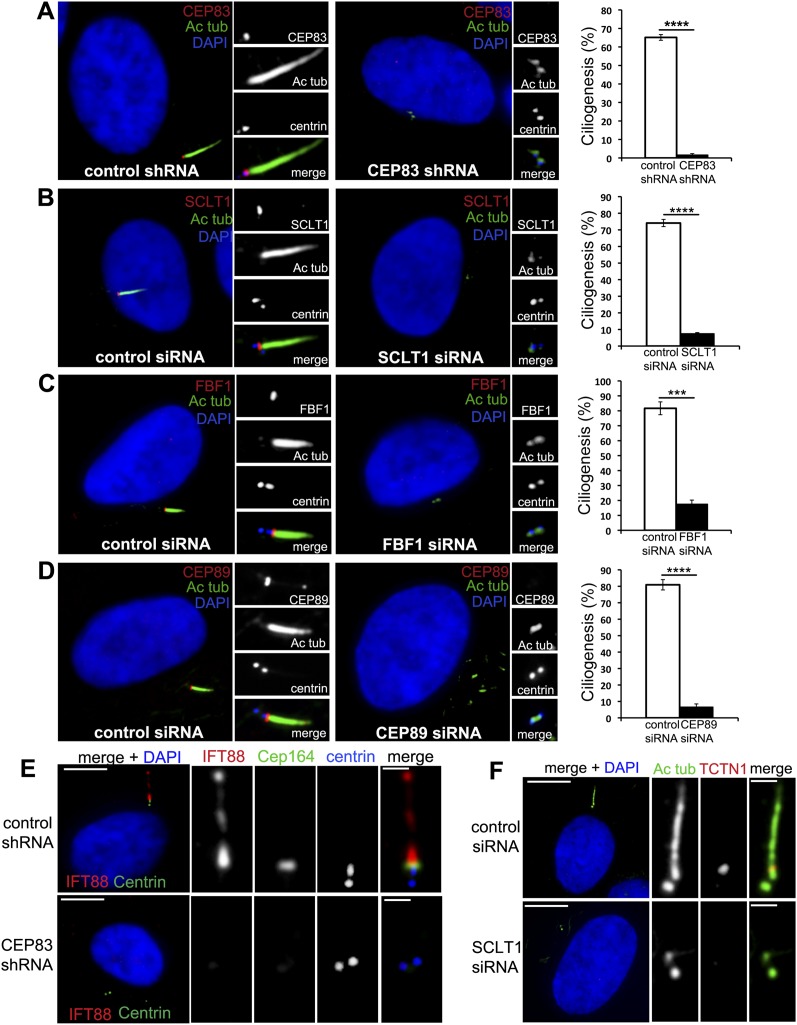
To determine the function of the DAPs, DAP proteins were individually depleted from cells by RNAi. It is known that CEP164 is essential for ciliogenesis (Graser et al. 2007), suggesting that any component acting upstream of CEP164 in DAP assembly should also be essential for ciliogenesis. Indeed, depletion of either CEP83 or SCLT1 in RPE1 cells, which disrupted CEP164 localization, completely blocked cilia formation (Fig. 3A,B). Interestingly, ciliogenesis also failed in CEP89-depleted cells (Fig. 3D) where the SCLT1 branch was intact at centrioles. Similar ciliogenesis defects were seen in cells depleted of FBF1 (Fig. 3C), a component parallel to CEP164 in the assembly pathway, indicating that all DAP components identified here play a critical role in ciliogenesis (Fig. 3). Note that these ciliogenesis defects could be rescued by expressing a RNAi-resistant form of DAP cDNA (Supplemental Fig. S3; data not shown), indicating that the phenotype is specific to loss of DAP components. Additionally, TZ-associated components that are normally recruited to centrioles before axoneme extension, including IFT88 and Tctn1, were all absent from DAP-defective centrioles (Fig. 3E,F; Supplemental Fig. S3), suggesting that loss of DAPs blocks ciliogenesis at a stage prior to TZ assembly.
Figure 3.
All DAP components are required for ciliogenesis. (A) RPE-1 cells stably expressing centrin-GFP (blue) were retrovirally transduced with control shRNA (left panel) or CEP83 shRNA (right panel), grown in serum-free medium for 48 h, and then fixed and stained with antibodies to CEP83 (red) and acetylated tubulin (green). DNA staining with DAPI is shown in blue. A quantification of this result is shown in the graph on the right (>300 cells). (B–D) RPE-1 cells stably expressing centrin-GFP (blue) were transfected with control siRNA (left panels) or SCLT1 (B), FBF1 (C), or CEP89 siRNA (D) (right panels), grown in serum-free medium for 48 h, and then fixed and stained with antibodies to SCLT1 (B), FBF1 (C), or CEP89 (red) and acetylated tubulin (green). DNA staining with DAPI is shown in blue. A graph with the quantification of each result is shown (>300 cells). (E) RPE-1 cells retrovirally transduced with control shRNA (top panels) or CEP83 shRNA (bottom panels) and stained with antibodies against IFT88 (red) or Cep164 (green) and centrin (blue). Note that in RPE-1 control cells, IFT88 localizes to both the basal bodies and ciliary axoneme. These localizations are lost in CEP83-depleted cells (marked by the absence of Cep164). (F) RPE-1 cells were transfected with control siRNA (top panels) or SCLT1 siRNA (bottom panels) and stained with antibodies to acetylated tubulin (green) or TCTN1 (red). DNA staining with DAPI is shown in blue. Large bar, 5 μm; small bar, 1 μm.
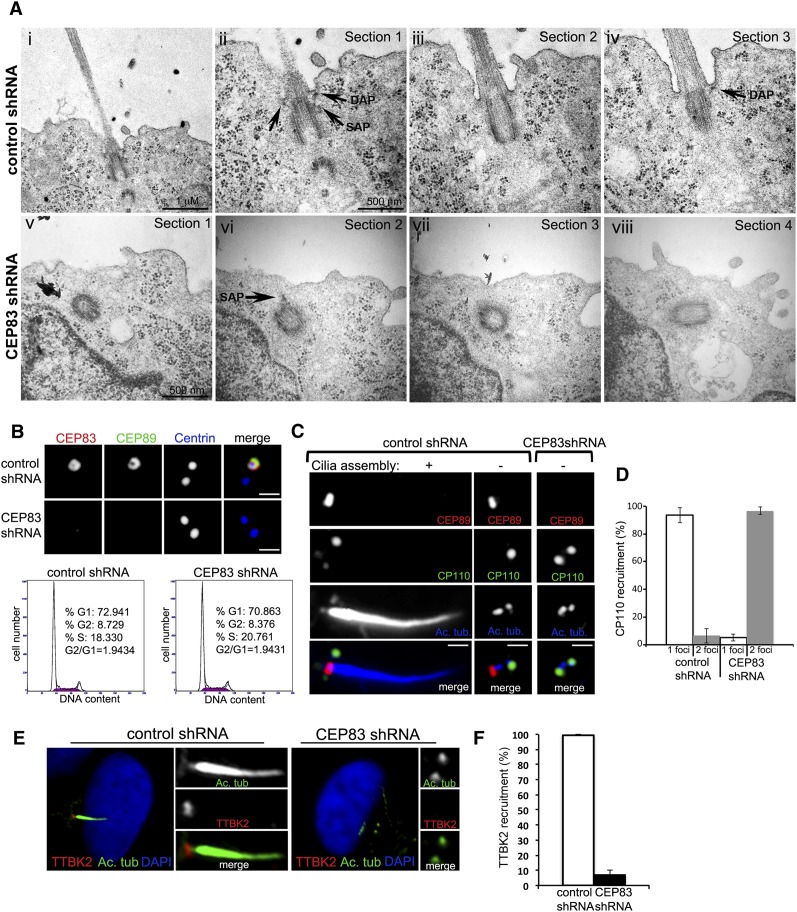
To determine whether DAPs are required for centriole-to-membrane docking, EM was used to examine fully polarized IMCD3 cells in which cilia normally form at the apical plasma membrane. Polarized IMCD3 cells were fixed and sectioned specifically at the plane in parallel to the apical–basal axis. As expected, nearly all centrioles in control IMCD3 cells docked to the apical membrane, aligned longitudinally to the apical–basal axis, and supported cilia formation (Fig. 4A; Supplemental Fig. S4). In contrast, in CEP83-depleted cells, while centrioles were able to position near the apical cortex (Fig. 4A; Supplemental Figs. S4, S5), they failed to interact with the membrane and aligned randomly with respect to the apical–basal axis, showing a small gap between the DAP-defective centriole and the plasma membrane (Fig. 4A; Supplemental Fig. S4). Moreover, no ciliary buds were seen growing from these undocked, CEP83-depleted centrioles (Fig. 4A; Supplemental Fig. S4). Note that IMCD3 cells depleted of CEP83 showed no detectable change in the distribution of the basolateral protein E-Cadherin, tight junction marker ZO-1, or apical F-actin (Supplemental Fig. S5), indicating that apical–basal polarity is unaltered. Together, these results indicate that the DAPs are specifically required for centriole-to-membrane docking.
Figure 4.
Coordination of centriole-to-membrane docking and cilia initiation. (A) IMCD3 cells were retrovirally transduced with either a luciferase control shRNA (panels i–iv) or a CEP83 targeting shRNA (panels v–viii). Three days after infection, cells were plated on top of polystyrene transwell filters and allowed to polarize and form a monolayer for 72 h. Filters were then fixed and processed for transmission EM. (Panel i) Thin section showing a centriole pair and cilia in IMCD3 control cells (25K). Bar, 1 μm. (Panel ii) Centriole pair in panel i at a higher magnification (50K). Note the mother centriole docked to the plasma membrane and daughter centriole in cross-section. DAPs and SAPs can be clearly seen (arrows, indicated). Bar, 500 nm. (Panel iii) Consecutive section showing the same centriole pair as in panel ii and the beginning of the ciliary axoneme. (Panel iv) Consecutive section showing the same centriole pair as in panel iii. Note that the tip of a distal appendage points to the site of attachment (arrow). (Panel v) Mother centriole of a pair in an IMCD3 cell depleted of CEP83 (50K). Bar, 500 nm. (Panel vi) Consecutive section of centriole in panel v. Note that the centriole is close to the apical cortex but has not docked; one subdistal appendage can be seen (arrow). (Panel vii) Continuous section. (Panel viii) Continuous section showing the other centriole of the pair. (B) RPE-1 cells stably expressing centrin-GFP (blue) were retrovirally transduced with control shRNA (top panels) or CEP83 shRNA (bottom panels), serum-starved for 48 h, and stained with antibodies to CEP83 and CEP89. Note that in RPE-1 cells, CEP83 is required for CEP89 localization to centrioles. Bar, 1 μm. The bottom panels show the cell cycle profile of control RPE-1 cells (left) and CEP83-depleted cells (right). (C) RPE-1 cells transduced with control shRNA or CEP83 shRNA were serum-starved for 48 h, fixed, and stained for CEP89 (red), CP110 (green), and acetylated tubulin (blue). Note that CP110 removal normally occurs at mature centrioles before cilia assembly initiates but fails in CEP83-depleted centrioles. Bar, 1 μm. (D) Quantification of CP110 recruitment. (E) RPE-1 cells transduced with control shRNA or CEP83 shRNA were serum-depleted for 48 h, fixed, and stained for TTKB2 (red) and acetylated tubulin (green). Note that in control cells, TTKB2 localizes to the centriole that supports cilia formation, whereas in CEP83-depleted cells, the recruitment of TTKB2 fails. (F) Quantification of TTBK2 recruitment.
Last, we examined the relationship between cilia initiation and centriole-to-membrane docking. The removal of CP110 from the distal end of mother centrioles (but not daughter) is a prerequisite for enabling the outgrowth of the axoneme (Tsang et al. 2008) and appears to be one of the earliest steps that initiate ciliogenesis. However, how CP110 removal is temporally and spatially regulated remains unclear. Intriguingly, CP110 removal failed completely in undocked, DAP-defective centrioles (Fig. 4C,D), even though these centrioles were exposed to cell cycle signals that induce ciliogenesis by serum starvation (Fig. 4B). Moreover, a recent report found that targeting of the kinase TTBK2 to mother centrioles upon serum starvation is required to promote CP110 removal and ciliogenesis (Goetz et al. 2012). Again, we found that TTBK2 cannot be targeted to undocked centrioles upon serum starvation in both RPE-1 cells (Fig. 4E,F) and murine IMCD3 cells (data not shown). These data suggest that centriole-to-membrane docking mediated by the DAP may serve as an instructive signal that temporally and spatially regulates cilia initiation.
Conclusion
Ciliogenesis in vertebrate cells follows a series of ordered steps. The interaction between centriole distal ends and membrane vesicles marks the first morphological event during ciliogenesis, followed by the outgrowth of ciliary MTs from centrioles, assembly of the TZ, and finally, elongation of the axoneme or cilium. The DAP localizes to the site of centriole-to-membrane docking and has been proposed to function at this initial step. Through centrosome proteomics and subcellular imaging, we uncovered molecular components of the DAP and established a DAP assembly pathway in human cells. Functional studies showed that DAPs are essential for ciliogenesis and that loss of DAPs specifically blocks ciliogenesis at the step of centriole-to-membrane docking. Undocked centrioles lack some earliest signs of undergoing cilia assembly, even when cells are exposed to the cell cycle signals that promote ciliogenesis. Both TTBK2 recruitment and CP110 removal, two of the earliest modifications found on centrioles prior to cilia assembly, fail to occur on undocked centrioles, suggesting that without DAPs, cilia assembly does not initiate. This reveals a critical coordination between membrane docking and cilia initiation, allowing temporal and spatial regulation of ciliogenesis in response to cell cycle signals. It remains to be determined how the DAPs interact with the membrane. Interestingly, the DAP component SCLT1 has been shown to interact with the membrane coat protein clathrin (Liu et al. 2005), raising a possibility that centrioles may directly associate with clathrin-coated vesicles via SCLT1. Future determination of the mechanisms by which DAPs mediate centriole-to-membrane docking and promote the removal of CP110 will provide a fundamental understanding of the biogenesis of this remarkable organelle.
Material and methods
Cell culture
Retinal pigment epithelial cells (RPE-1) and murine inner medullary collecting duct (IMCD3) cells were cultured in DME/F-12 (1:1) medium supplemented with 10% FBS and 1% penicillin–streptomycin. 293T, HeLa, and U2OS cells were cultured in DMEM with 10% FBS and 1% penicillin–streptomycin.
SILAC labeling and mass spectrometry
HeLa cells with Plk4 expression under a tetracycline-inducible promoter were used in this experiment. Cells were metabolically labeled with either normal arginine and lysine or heavier isotopic variants of the two amino acids (L-lysine 2HCL [U-13C6] and L-arginine HCL [U-13C6 and U-N15N4]) as previously described (Ong et al. 2002). SILAC medium was purchased from Invitrogen and prepared according to the manufacturer's recommendations. After labeling, cells in both populations were arrested in S phase by aphidicolin (Sigma), but cells fed with heavy amino acids were additionally induced to overexpress Plk4 and form rosette centrosomes. The two populations of cells were mixed and processed for centrosome purification using the sucrose gradient centrifugation as described (Tsou and Stearns 2006). Centrosomes or SILAC-labeled protein mixtures were run by SDS-PAGE, and 12 gel sections were excised and digested overnight at pH 8.0 with modified sequencing grade trypsin (Promega Corp.). Peptide mixtures were eluted, and each gel section was analyzed separately by microcapillary liquid chromatography tandem mass spectrometry (LC-MS/MS) using the EASY-nLC nanoflow HPLC (Thermo Fisher Scientific) with a 75-μm inner diameter × 15-cm length Picofrit capillary column (New Objective, Inc.) self-packed with 5 μm Magic C18 resin (Michrom Bioresources) coupled to a hybrid LTQ Orbitrap XL-ETD mass spectrometer (Thermo Fisher Scientific). MS/MS fragmentation spectra were searched for protein identification using the Andromeda search engine (http://www.andromeda-search.org) against the reversed and concatenated IPI_HUMAN protein database (version 3.87) (http://www.ebi.ac.uk/IPI/IPIhuman.html). One unique peptide was required for high-confidence protein identifications, and a minimum ratio count of two peptides (one unique and one razor) was required for SILAC ratio determination. Normalized SILAC ratios (H/L) were used for subsequent analysis.
siRNA, shRNA, and stable DNA expression
RNAi-mediated knockdown of SCLT1, FBF1, CEP89, and Cep164 in RPE-1 cells was carried out using siGenome SMARTpool siRNAs (Dharmacon). siRNA oligos were delivered using Lipofectamine RNAiMAX (Invitrogen). Odf2 depletion was carried out using a lentiviral shRNA (Sigma-Aldrich, NM_002540). CEP83 RNAi was carried out using a combination of three different lentiviral shRNAs with the following targeting sequences: 1 (CAAAGAAGAAGGTTATCAA) (Open Biosystems), 2 (GCATAAAGCTGAACGAGAAAT), and 3 (AGGCTGAAGTAGCGGAATTAA) (Sigma-Aldrich) infected sequentially.
Human CEP83 was PCR-amplified from a HeLa cDNA library (Invitrogen) with primers containing BamHI sites (5′-AATGGATCCGCCATGGTTGTCAGCACATTTACCG-3′ and 5′-GGTGGATCCTCATTCTCCGGAAGATCCAA-3′) and cloned as a BamHI restriction fragment into pCDNA3. This BamH1 fragment was then shuttled into pLVX-tight puro (Takara Bio, Inc.) to generate pLVX-tight-HA-CEP83. To engineer inducible expression of CEP83, RPE1 cells stably expressing rtTA (generated through lentiviral transduction of rtTA-Advanced from Clontech) were stably infected with pLVX-tight-HA-CEP83, whereby CEP83 expression is under the control of a tetracycline-inducible promoter. CEP83 expression was induced with 1 μg/mL doxycycline for 48 h. All lentiviruses were generated by transient cotransfection of 293T cells with packaging and envelope vectors (Clontech) using the calcium phosphate precipitation method. Stable expressors were derived through antibiotic selection (5 μg/mL puromycin for RPE-1 cells, and 2 μg/mL for IMCD3).
Ciliogenesis experiments
RPE-1 or IMCD3 cells were plated in acid-washed poly-lysine-coated coverslips in 3.5-cm plates at 0.4 × 106 cells per well. Cells were allowed to attach for 24 h, then washed twice with serum-free medium, and left in serum-free medium for an additional 48 h. When grown in filters, IMCD3 cells were plated at 0.3 × 106 cells per filter in 1.2-cm, 0.4-μm pore size, transwell polycarbonate filters and allowed to polarize for 3 d.
Immunofluorescense, microscopy, and FACS
For centrosomal staining, cells were permeabilized for 2 min in PTEM buffer containing 20 mM PIPES (pH 6.8), 0.2% Triton X-100, 10 mM EGTA, and 1 mM MgCl2; fixed for 10 min with 4% PFA or cold methanol (depending on specific conditions for each antibody); and blocked with 3% (w/v) bovine serum albumin and 0.1% Triton X-100 in PBS for 30 min. Primary antibody incubation was done for 1 h in blocking solution, followed by five 1× PBS, 0.1% Triton X-100 washes. Secondary antibody incubation was carried out for 1 h followed by five washes and incubation with DAPI (Invitrogen) for 2 min to visualize DNA. Fluorescent images were acquired on an upright microscope (Axio imager, Carl Zeiss) equipped with 100× oil objectives, 1.4 NA, a camera (ORCA ER, Hamamatsu Photonics), and a computer loaded with image-processing software (Axiovision). Individual images were cropped and assembled into figures using Photoshop (CS5, Adobe). Confocal images were acquired with a Leica SP5 point scanner equipped with a 63× oil objective. Superresolution microscopy was carried out with the DeltaVision OMX 3D-SIM system (Applied Precision) using an Olympus 100×, 1.4 NA oil objective; 405 nm, 488 nm, and 593 nm laser illumination; and standard excitation and emission filter sets. Raw images were acquired using a 125-nm z-step size and then aligned and reconstructed in three dimensions with softWoRx 5.0.0 (Applied Precision) software. FACS analyses were performed by collecting ∼106 cells by trypsinization, washing twice with 1× PBS, fixing in cold methanol, and then resuspending in propidium iodide staining buffer supplemented with RNase and analyzed with a Becton-Dickinson FACSCalibur flow cytometer.
Antibodies and reagents
Rabbit polyclonal antibodies against CEP83, FBF1, SCLT1, and Odf2 were obtained from Sigma-Aldrich (1:200). Cep164 rabbit antibody was from Strategic Diagnotics (1:5000), and mouse Cep164 antibody (1:250) was a kind gift from Eva Lee (University of California at Irvine). Rabbit polyclonal antibody against IFT88 (1:500) was purchased from Protein-Tech. Mouse Odf2 (1:200) was from Novus Biologicals. Other antibodies used in this study include mouse anti-γ-tubulin (1:500; Santa Cruz Biotechnology, Inc.), mouse anti-acetylated-tubulin (1:2000; Sigma-Aldrich), mouse anti-α-tubulin (1:2000; Sigma-Aldrich), rabbit anti-ZO-1 (1:100; Invitrogen), and rat E-Cadherin (1:100; Invitrogen). All secondary antibodies—Alexa-Fluor 488, 594, and 680—were purchased from Molecular Probes (1:500; Invitrogen), except Dylight 405 (1:100; Jackson Immunoresearch). Phalloidin-rhodamine (1:100) was purchased from Invitrogen. Aphidicolin was purchased from Sigma-Aldrich. Rat antibodies against CEP89 (1–230 amino acids) and SCLT1 (1–224 amino acids) were generated by expressing the GST-tagged fragments in Escherichia coli, purifying the fusion proteins, and immunizing naive rats (Covance, Inc.).
Western blots
Cells were lysed in RIPA buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate) supplemented with 1 mM b-glycerophosphate, 2.5 mM sodium pyrophosphate, and Complete protease and phosphatase inhibitors (Roche). Lysates were sonicated and cleared by centrifugation.
EM
For ultrastructural analysis, IMCD3 cells grown on transwell filters as described above were washed with serum-free medium and then fixed in 4% paraformaldehyde and 2.5% glutaraldehyde with 0.1% tannic acid in 0.1 M sodium cacodylate buffer for 30 min at room temperature, post-fixed in 1% OsO4 in sodium cacodylate buffer for 30 min on ice, dehydrated in a graded series of ethanol, infiltrated with EPON812 resin (Electron Microcopy Sciences), and then embedded in the resin. Serial sections (∼90 nm thick) were cut on a microtome (Ultracut UC6, Leica) and stained with 1% uranyl acetate as well as 1% lead citrate. Samples were examined on a JEOL transmission electron microscope.
Acknowledgments
We thank Peter Satir and Birgit Satir (Albert Einstein College of Medicine) for their advice on this project. We thank Leslie Cummins (Albert Einstein College of Medicine), L. Cohen-Gould (Cornell University), N. Lampen (Memorial Sloan-Kettering Cancer Center [MSKCC]), and K. Uryu (Rockefeller University) for EM assistance; E. Lee (University of California at Irvine) for Cep164 antibody; and Y. Romin and S. Fujisawa (MSKCC-Molecular Cytology) for the assistance of confocal microscopy. We thank K. Anderson and S. Goetz at MSKCC for IMCD3 cells, A. Hall laboratory for reagents and helpful discussions, and A. Hall and C. Haynes at MSKCC for comments on the manuscript. We are grateful for A. North (Rockefeller University) for the use of superresolution microscopy (OMX, DeltaVision), supported by the award S10RR031855 from the National Center for Research Resources. This work was supported by the National Institutes of Health post-doctoral fellowship GM096558 (to B.E.T.) and grant GM088253 (to M.-F.B.T.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.207043.112.
References
- Anderson RG 1972. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol 54: 246–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS 2012. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol 14: 61–72 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV 2007. Cilia and developmental signaling. Annu Rev Cell Dev Biol 23: 345–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Liem KF Jr, Anderson KV 2012. The spinocerebellar ataxia-associated gene tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179: 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA 2005. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146 [DOI] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ 2010. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7: 517–524 [DOI] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. 2011. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J 30: 1520–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol 14: 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Cummins TR, Tyrrell L, Black JA, Waxman SG, Dib-Hajj SD 2005. CAP-1A is a novel linker that binds clathrin and the voltage-gated sodium channel Na(v)1.8. Mol Cell Neurosci 28: 636–649 [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Reiter JF, Blacque OE, Leroux MR 2012. The base of the cilium: Roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13: 608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB 2002. Intraflagellar transport. Nat Rev Mol Cell Biol 3: 813–825 [DOI] [PubMed] [Google Scholar]
- Sillibourne JE, Specht CG, Izeddin I, Hurbain I, Tran P, Triller A, Darzacq X, Dahan M, Bornens M 2011. Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton (Hoboken) 68: 619–627 [DOI] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD 2008. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell 15: 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442: 947–951 [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Yu S 1982. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol 93: 938–949 [DOI] [PMC free article] [PubMed] [Google Scholar]