pRB and its Drosophila ortholog, RBF1, are known cell cycle regulators. Nicolay et al. now uncover a role for RB proteins in glutamine metabolism. rbf1 mutant flies are sensitive to fasting. Metabolic profiling reveals that RBF1 loss causes major changes in glutathione synthesis and nucleotide metabolism. Similar metabolic effects are observed in human cancer cells upon pRB loss. This study reveals a striking metabolic vulnerability caused by RB protein inactivation that could be exploited to target cancer cells.
Keywords: RBF, pRB, Drosophila, metabolism
Abstract
Inactivation of the retinoblastoma tumor suppressor (pRB) alters the expression of a myriad of genes. To understand the altered cellular environment that these changes create, we took advantage of the Drosophila model system and used targeted liquid chromatography tandem mass spectrometry (LC-MS/MS) to profile the metabolic changes that occur when RBF1, the fly ortholog of pRB, is removed. We show that RBF1-depleted tissues and larvae are sensitive to fasting. Depletion of RBF1 causes major changes in nucleotide synthesis and glutathione metabolism. Under fasting conditions, these changes interconnect, and the increased replication demand of RBF1-depleted larvae is associated with the depletion of glutathione pools. In vivo 13C isotopic tracer analysis shows that RBF1-depleted larvae increase the flux of glutamine toward glutathione synthesis, presumably to minimize oxidative stress. Concordantly, H2O2 preferentially promoted apoptosis in RBF1-depleted tissues, and the sensitivity of RBF1-depleted animals to fasting was specifically suppressed by either a glutamine supplement or the antioxidant N-acetyl-cysteine. Effects of pRB activation/inactivation on glutamine catabolism were also detected in human cell lines. These results show that the inactivation of RB proteins causes metabolic reprogramming and that these consequences of RBF/RB function are present in both flies and human cell lines.
There is considerable evidence that the protein product of the retinoblastoma susceptibility gene (pRB) regulates gene expression. In most but not all chromatin contexts, pRB functions as a corepressor of transcription (for review, see Longworth and Dyson 2010). pRB is a promiscuous protein with >200 documented binding partners (Morris and Dyson 2001), but its best known activity is its role in the regulation of E2F-dependent transcription. Although E2F-binding sites are found in the promoters of genes with functions in many biological processes, the best-studied targets of E2F regulation are genes that function during cell cycle progression. When it is in an active conformation, pRB antagonizes E2F and suppresses the transcription of a suite of genes that are needed for cell proliferation. The inactivation of pRB is thought to remove this “brake” and places cells in a state that is permissive for cell division (Blais and Dynlacht 2004; van den Heuvel and Dyson 2008).
Given the widespread changes in chromatin marks in RB1 mutant cells (Zhang et al. 2012) and the extensive changes affecting programs in gene expression (Black et al. 2003), the loss of pRB clearly impacts multiple cellular processes. While it is known that the functional inactivation of pRB causes major changes, beyond the fact that these cells are more easily driven into the cell cycle, we currently understand very little about the range of cellular processes that are altered when pRB is lost. A major initiative in cancer research is the idea of personalized medicine and the use of treatments that are tailored to the unique properties of each tumor (Haber et al. 2011; Arteaga and Baselga 2012). This concept relies on the idea that changes in specific oncogenes/tumor suppressor genes generate unique dependencies that can be exploited. From a standpoint of cancer therapy, it would be very appealing to know how to selectively target premalignant tumor cells that lack active pRB. To date, however, there is no general strategy for targeting pRB-deficient cells. Without more information about the changes that occur when pRB is inactivated, it is difficult to predict which types of cellular processes should be targeted in order to distinguish pRB-deficient cells from normal cells.
Here we took advantage of the streamlined Rb pathway present in Drosophila. Mammalian cells contain three pRB family members (pRB, p107, and p130) that have partially overlapping functions. Although flies have two pRB-like proteins (RBF1 and RBF2), genetic studies indicate that RBF1 is the major player in most cell types (Stevaux et al. 2002; Dimova et al. 2003). RBF1 has several properties that resemble pRB: Like pRB, RBF1 antagonizes the ability of activators to promote the expression of genes sufficient for S-phase entry (Du et al. 1996; Du and Dyson 1999). In addition, pRB associates with repressor E2Fs and is an essential element of repressor complexes (Korenjak et al. 2004). Also like pRB, most of the literature on RBF1 centers on its ability to limit dE2F-dependent proliferation. However, RBF1 has recently been shown to possess additional functions in the immune response (Longworth et al. 2012), in cell differentiation (Lee et al. 2010, 2012; Nicolay et al. 2010; Popova et al. 2012), and in chromatin organization (Longworth et al. 2008; Manning et al. 2010). Thus, like its mammalian counterpart, the loss of RBF1 has ramifications that extend beyond the cell cycle.
Using this model system, we looked for ways to selectively induce apoptosis in rbf1 mutant cells. We found that RBF1-depleted cells and rbf1 mutant animals have a heightened sensitivity to fasting. To understand this phenotype, we performed metabolic profiling of control and RBF1-depleted larvae under both fed and fasted conditions to identify the metabolic changes that occur when RBF1 is removed. The results show that the loss of RBF1 leads to major changes in nucleotide synthesis and in glutathione production. Both of these processes depend on a supply of glutamine. We show that glutamine catabolism is altered when RBF1 is depleted and that a glutamine supplement is sufficient to suppress the sensitivity of RBF1-depleted animals. Under fasting conditions, RBF1-depleted larvae struggle to sustain sufficient glutathione production to both maintain red/ox balance and to synthesize deoxynucleotides (dNTPs). This places RBF1-depleted cells in an oxidized state where they are hypersensitive to DNA damage. Interestingly, pRB-dependent changes in glutamine flux were also detected in human cell lines, indicating that some of the metabolic consequences of RBF1 inactivation are conserved from flies to humans.
Results
RBF mutant cells are sensitive to fasting conditions
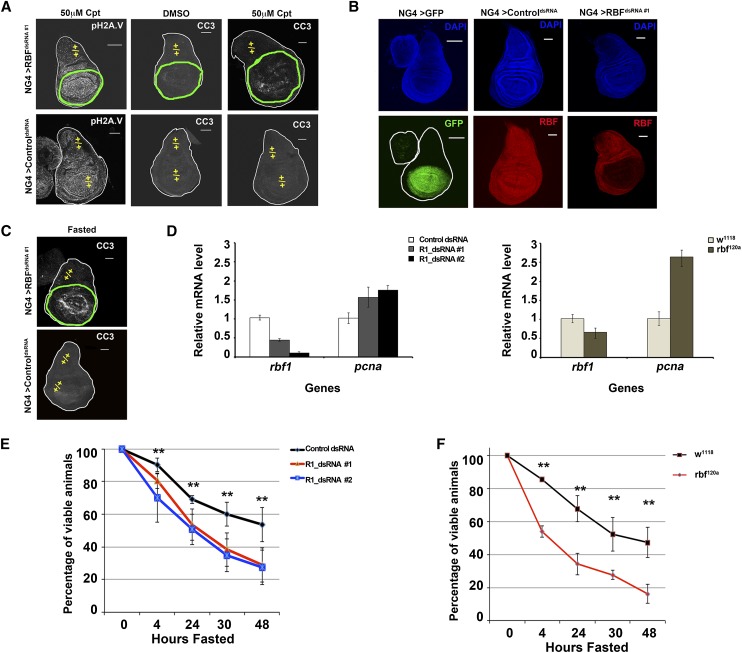
When compared side by side in mosaic imaginal discs, rbf1 mutant cells are more prone to DNA damage-induced apoptosis than adjacent wild-type tissue (Moon et al. 2008). This is illustrated in Figure 1A using wing discs in which Nubbin-Gal4 (NG4) was used to drive the expression of UAS-RBFdsRNA #1 (R1dsRNA #1) during development. Depletion of RBF1 protein was confirmed by immunostaining (Fig. 1B), and camptothecin (Cpt) was used to induce DNA breaks. A range of Cpt concentrations were tested, and the discs shown in Figure 1A were treated with a concentration (50 μM) that gave similar levels of DNA damage in both wild-type and RBF1-depleted tissue, as determined by phosphorylated Histone 2A.V (pH2AV) staining (Fig. 1A). Despite high levels of DNA damage throughout the disc, cleaved caspase-3 (CC3) was detected specifically in the portion of the disc containing RBF1-depleted cells (Fig. 1A). This observation is consistent with previous studies in Drosophila and in mammalian cells showing that the inactivation of RBF1 or pRB enhances cellular sensitivity to DNA-damaging agents (Zagorski et al. 2007; Liu et al. 2009).
Figure 1.
Loss of the RBF1 protein leads to hypersensitivity to DNA damage and energetic stress. (A–C) Images shown are third instar wing discs. Bar, 100 μm. Genotypes are described in the Materials and Methods. The domain where the RBF1 protein has been depleted (green outline) or remains (yellow +/+) is designated. (A) Wing discs were treated with 50 μM Cpt or 50 μM DMSO as a control. DNA damage foci shown by pH2AV (grayscale). Apoptosis following Cpt challenge shown by CC3 (grayscale). (B) The nubbin-Gal4 expression domain is identified by the expression of a UAS-gfp (NG4 > GFP) (green). The RBF1 protein is efficiently depleted by “RBFdsRNA #1” (red), while ControldsRNA has no affect. Gross tissue architecture and like focal planes are indicated by DAPI (blue). (C) Fasting leads to apoptosis specifically in RBF1-depleted tissue, as indicated by CC3 (grayscale). (D) Error bars represent the standard deviation from the mean. mRNA levels were measured by quantitative real-time PCR (qPCR). RBF1 mRNA is efficiently depleted by both targeted dsRNAs. Depletion of RBF1 leads to increased levels of the traditional RBF1/dE2F target gene pcna. (E,F) Error bars represent the confidence interval (C.I.) of 95%. Mid-second instar larvae were fasted over 48 h. Reduced RBF1 protein leads to a statistically significant hypersensitivity to fasting when compared with appropriate control animals (at time points between 4 and 48 h the P-value < 0.01).
We used this experimental system to examine the effects of various conditions on RBF1-depleted cells. Unexpectedly, we found that fasting larvae for 4 h (see the Supplemental Material for details) also preferentially affected RBF1-depleted cells and resulted in CC3-stained cells specifically in the RBF1-depleted portion of the wing disc (Fig. 1C). Similar results were obtained using an alternative UAS-RBFdsRNA allele that targeted different sequences within RBF1 and when a different Gal4 driver was used to deplete RBF1 from wing discs in a different pattern (data not shown). Expression of controldsRNA had no effect on the expression of the RBF1 protein (Fig. 1B) and had no effect on CC3 staining in discs treated with Cpt (Fig. 1A) or following fasting (Fig. 1C).
These observations suggest that RBF1-depleted cells have a heightened sensitivity to the energetic stress caused by fasting conditions. Null alleles of rbf1 are lethal at the early larval stages; therefore, we tested this idea further using UAS-RBFdsRNA alleles to reduce the level of RBF1. Since actin-Gal4-driven expression of UAS-RBFdsRNA caused larval lethality, we used Armadillo-Gal4 (ArmG4), a weaker driver that is expressed ubiquitously and continuously during larval development (Flybase, http://flybase.org), to direct the expression of UAS-RBFdsRNA. ArmG4 expression of a control dsRNA had no effect on larval viability or on rates of eclosion. On standard growth medium, ArmG4-directed expression of the RBF dsRNAs had no effect on larval viability or developmental timing prior to 120 h after larval hatching (ALH). At later stages, ArmG4-driven expression of RBFdsRNA#1 led to 100% eclosion, but ArmG4-driven expression of RBFdsRNA#2 led to a rate of eclosion of only ∼30%. Consistent with this difference, we found that ArmG4; RBFdsRNA#2 gave a stronger knockdown of RBF1 than ArmG4; RBFdsRNA#1 (Fig. 1D).
To test the effects of fasting, larvae were shifted from a nutrient-rich food source between 40 and 48 h ALH and placed on a diet consisting only of PBS in order to maintain hydration. This time point was chosen because wild-type larvae pass through a transition point at ∼70 h ALH, and larvae fasted prior to this stage die before pupariation, while larvae fasted after this time will eclose as runted adults. After 4 h of fasting, we observed a statistically significant decrease in animal viability in ArmG4; RBFdsRNA#1 and ArmG4; RBFdsRNA#2 larvae compared with control larvae (Fig. 1E). An even greater sensitivity was observed in RBF1-depleted larvae after 24 h of fasting. The sensitivity of RBFdsRNA#2 was, in general, more severe (Fig. 1E), but these effects were observed with both RBF1 hairpins. Animals hemizygous for rbf120a, a viable hypomorphic allele, showed a similar statistically significant increase in sensitivity to fasting compared with the appropriate background control, which was evident after 4 h of fasting and seen at each time point examined (Fig. 1F). Taken together, these results indicate that a reduction in the level of RBF1 impacts the ability of cells or larvae to survive under energetic stress.
Fasted RBF1 mutants have altered levels of cellular metabolites
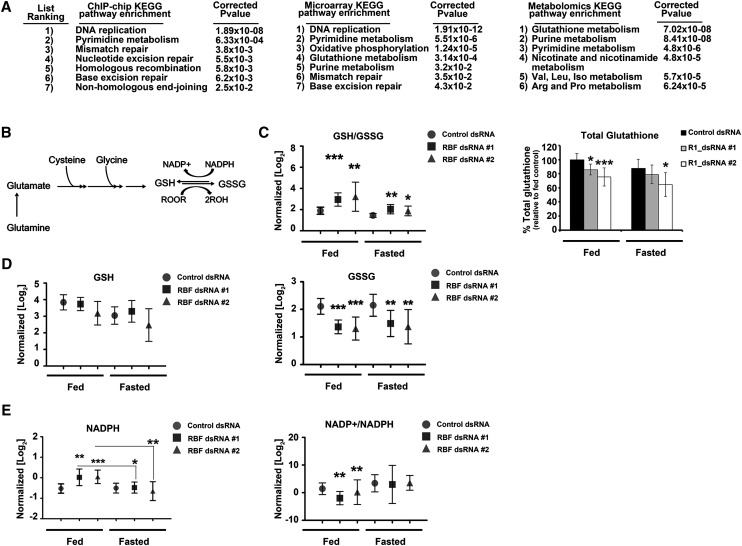
To obtain a general perspective on the changes likely to result from the loss of RBF1, we performed Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway analysis on the transcriptional targets of RBF1. Analysis of genes whose promoters were directly bound by RBF1 in third instar wild-type larvae (Korenjak et al. 2012) showed the strongest enrichment for genes with functions in DNA replication; pyrimidine metabolism and DNA repair were the next most significant categories (Fig. 2A). A similar analysis of the transcriptional changes in rbf1 mutant larvae (Longworth et al. 2012) showed that genes in these categories were also up-regulated in the mutants, consistent with the loss of a transcriptional repressor (Fig. 2A). Interestingly, the transcriptional profiles also revealed highly significant changes in pathways that were not strongly enriched in the chromatin immunoprecipitation (ChIP) data. Most notably, we observed changes in the expression of genes with functions in oxidative phosphorylation and glutathione metabolism (Fig. 2A). The differences between these lists raised the possibility that the altered gene expression profiles of rbf1 mutants include changes in direct targets of RBF1 and changes that occur in response to the altered rbf1 mutant state.
Figure 2.
RBF1 depletion leads to global metabolic changes and increased cellular oxidation. (A) KEGG pathway enrichment was performed on genes bound by RBF1 in wild-type third instar larvae (Korenjak et al. 2012), genes up-regulated in RBF1−/− third instar larvae (Longworth et al. 2012), and altered metabolites as a response to RBF1 depletion under fasting. The top seven pathways enriched from each analysis are shown. (B) Schematic of the flow of glutamine catabolism toward glutathione synthesis. (C–E) Normalized levels (see the Supplemental Material for details) of metabolite pools from fed and fasted control or RBF1-depleted third instar larvae. Error bars represent the C.I. of 95%. Statistical significance is designated as P-values as follows: (*) P < 0.06; (**) P < 0.02; (***) P < 0.001. Statistical differences are between dffects from RBF1 depletion compared with control animals within the two metabolic states unless designated otherwise.
These lists illustrate some of the potential caveats of the ChIP and expression data sets: Not all genes bound by RBF1 are deregulated in rbf1 mutant animals, the expression changes that do occur will include both direct and indirect effects, and only some of the gene expression changes that occur in rbf1 mutants will impact gene function. Therefore, we looked for an alternative approach to identify the changes responsible for the sensitivity to energetic stress when RBF1 is removed. We used targeted liquid chromatography tandem mass spectrometry (LC-MS/MS) to generate metabolite profiles for control and RBF1-depleted larvae in both fed and fasted conditions. Polar metabolites with coverage across all major metabolic pathways (including the pentose phosphate pathway, glycolysis, TCA cycle, urea cycle, nucleotide biosynthesis, and others) were examined to obtain a global view of the metabolic state (Yuan et al. 2012). We performed KEGG pathway enrichment analysis on the 219 (out of 258) metabolites that gave robust and reproducible peaks and weighted (Xia and Wishart 2011) the changes for the greatest effect in fasted, RBF1-depleted larvae (see the Materials and Methods for details). The results were remarkably consistent with the analysis of the ChIP–chip and gene expression data sets; indeed, many of the proteins that directly regulate the altered metabolites were previously found to be RBF1 transcriptional targets (Supplemental Tables 1–3; Korenjak et al. 2012). Therefore, this integrated analysis enabled us to focus on metabolic pathways that were functionally altered by the loss of RBF1. The three most statistically significant metabolic changes evident in RBF1-depleted larvae showed a strong enrichment for pathways involved in glutathione metabolism, purine metabolism, and pyrimidine metabolism (Fig. 2A). These changes are described in more detail below, and subsequent experiments suggest that these differences are important for the sensitivity of RBF1-deficient animals to fasting.
RBF1-depleted larvae require increased generation of reduced glutathione
Glutathione is a key metabolite that is used by glutathione peroxidase to reduce the accumulation of damaging reactive oxygen species (ROS), such as H2O2. To be able to perform this function, it is critical for the cell to maintain a pool of glutathione in the reduced state (GSH). NADPH is required for GSH regeneration from oxidized glutathione (GSSG). To maintain high levels of GSH, a cell must continually undergo de novo glutathione synthesis and/or maintain high levels of NADPH (Fig. 2B).
The depletion of RBF1 lead to a significant increase in the GSH/GSSG ratio compared with the control animals, and this change was evident when the RBF1-depleted larvae were either fed or fasted (Fig. 2C). Interestingly, no difference in the amount of GSH was detected between RBF1-depleted and control animals; rather, the increased GSH/GSSG ratio reflected a significantly smaller pool of GSSG (in both fed and fasted larvae) (Fig. 2D). Thus, the total pool of glutathione (GSH + GSSG) was decreased in RBF1-deficient larvae (Fig. 2C). We inferred that in RBF1-depleted larvae, the reduction in GSSG was due to an increased need for GSH regeneration, as NADPH levels were significantly increased when the larvae were fed (Fig. 2E). Conversely, NADPH levels dropped significantly when RBF1-depleted animals were fasted, while no change was detected in the control animals (Fig. 2E). This drop in total NADPH correlated with a decreased GSH/GSSG ratio in fasted RBF1-depleted animals (Fig. 2C).
Potentially, the lower level of glutathione in fasted RBF1-depleted larvae might result from reduced glutathione synthesis. Glutathione is synthesized from glutamate, cysteine, and glycine in a stepwise progression by two enzymes. The first, and rate-limiting, enzyme of glutathione synthesis is glutamate–cysteine ligase (GCL), and the second is glutathione synthetase (GS). Consistent with the reduced levels of glutathione, levels of both GCL and GS mRNA were lower in both fed and fasted RBF1-depleted animals (Supplemental Fig. 1). ChIP experiments showed that RBF1 bound the promoter of the GCL gene (GCLC); however, it was not detected at the GS promoter (Supplemental Fig. 1D), suggesting that RBF1 has both direct and indirect effects on the expression of enzymes important for glutathione synthesis. RBF1 is most commonly described as a repressor of gene transcription; however, there are many loci, like GCLC and GS, whose expression decreases in rbf1 mutants (Longworth et al. 2012).
Taken together, these observations indicate that RBF1-depleted larvae have less total glutathione than control larvae in both the fed and fasted states and decreased expression of GCL and GS, enzymes that are key for glutathione synthesis. The changes in GSH/GSSG ratio and NADPH levels suggest that RBF1-depleted animals have an increased need for GSH regeneration. These conflicts of supply and demand suggest that the RBF1-depleted larvae struggle to maintain a reduced state during fasting.
RBF1-depleted animals have an increased demand on nucleotide biosynthesis
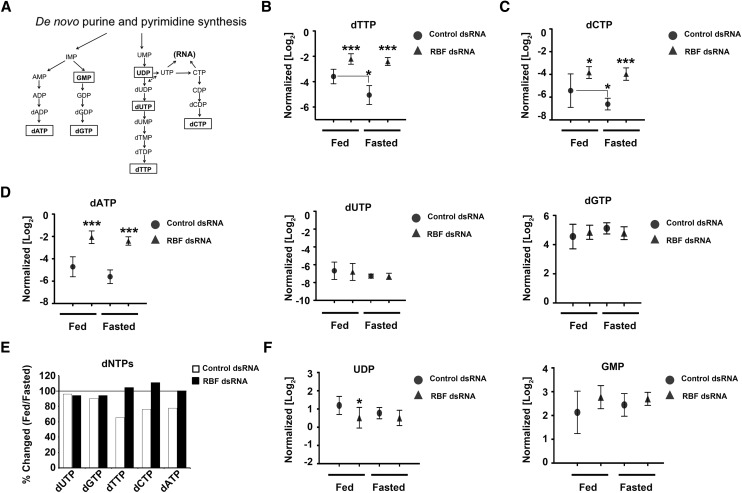
The loss of RBF1 led to an increase in the levels of each dNTP in both fed and fasted larvae (Fig. 3B–E). This was not unexpected, given that the loss of RBF1 is known to increase the proportion of cells undergoing DNA replication. However, two surprising features of this data were that RBF1-deficient larvae maintain their elevated dNTP pools after being fasted for 24 h and that dNTP levels were increased without an increase in dNTP precursor products (Fig. 3B–F). In contrast, control animals exhibited reduced levels of dNTPs after fasting, as would be predicted if replication slowed as carbon and nitrogen become limiting (Fig. 3B–E).
Figure 3.
RBF1 depletion allows sustained levels of dNTPs during fasting. (A) Schematic of de novo dNTP synthesis. (B–D,F) Normalized levels of ribonucleotide and dexoynucleotide pools from fed and fasted control or RBF1-depleted third instar larvae. Error bars represent the C.I. of 95%. Statistical significance is designated as P-values as follows: (*) P < 0.06; (***) P < 0.001. Statistical differences are between effects from RBF1 depletion compared with control animals within the two metabolic states unless designated otherwise. (E) Ratios of dNTP pools between fed and fasted conditions.
dNTPs can be produced from de novo nucleotide synthesis (Fig. 3A) or through nucleotide salvage pathways (Fig. 4C). To understand how RBF1-depleted larvae increased the level of dNTPs and then sustained these elevated levels of dNTPs, we first examined the contribution of de novo nucleotide synthesis to dNTP pools of RBF1-depleted larvae.
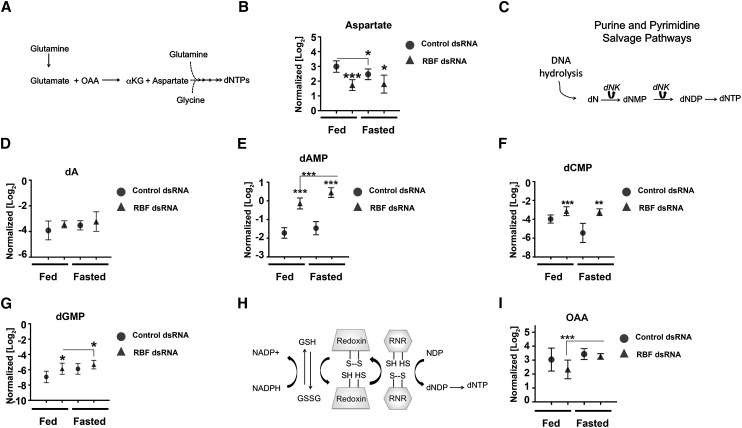
Figure 4.
RBF1-depleted animals increase levels of nucleotide salvage pathway metabolites to maintain dNTP pools. Metabolite pools from fed and fasted animals from fed and fasted control or RBF1-depleted third instar larvae. Error bars represent the C.I. of 95%. Statistical significance is designated as P-values as follows: (*) P < 0.06; (**) P < 0.02; (***) P < 0.001. Statistical differences are between effects from RBF1 depletion compared with control animals within the two metabolic states unless designated otherwise. (A) Schematic of glutamine catabolism to dNTPs. (αKG) α-ketoglutarate. (B) Normalized aspartate pools. (C) Simplified schematic of the nucleotide salvage pathway from DNA hydrolysis. (dN) Deoxynucleotides; (dNK) deoxynucleotide kinase; (dNMP) deoxynucleotide monophosphate; (dNDP) deoxynucleotide diphosphate; (dNTP) deoxynucleotide triphosphate. (D) Normalized deoxyadenosine (dA) pools. (E–G) Normalized deoxynucleotide monophosphate pools. (H) Schematic of how the RNR enzyme complex generates deoxynucleotides from ribonucleotides. (I) Normalized OAA pools.
De novo nucleotide synthesis requires the contribution of carbon and nitrogen from glutamine, glutamate, aspartate, and glycine (Fig. 4A). The levels of glutamine, glutamate, and glycine were unaltered between genotypes in both fed and fasted larvae (Supplemental Fig. 2A–C). However, the pool of aspartate was significantly reduced in RBF1-depleted animals (Fig. 4B). Assuming that the pools were at a steady state, this reduction in aspartate, coupled with an increase in dNTP levels, suggests that the RBF1-depleted animals may have a reduced capacity for de novo nucleotide synthesis.
To assess the contribution of the nucleotide salvage pathways for dNTP production, we examined levels of nucleotide intermediates that are used by these pathways following DNA hydrolysis (Fig. 4C). Fed RBF1-depleted larvae had significantly elevated levels of metabolites from both pyrimidine and purine catabolism (Fig. 4D–G), suggesting that under nutrient-rich conditions, RBF1-depleted larvae preferentially use the nucleotide salvage pathways for dNTP production. When control animals were fasted, we observed that they also increased levels of nucleotide salvage pathway metabolites (Fig. 4D–G). The increased use of nucleotide salvage pathway metabolites in fasted control animals was associated with a 20% reduction in aspartate pools (Fig. 4B) and a general decrease in dNTP pools (Fig. 3B–E). Strikingly, and in contrast, no statistically significant change was evident in aspartate levels (Fig. 4B) or in dNTP pools (Fig. 3B–E) when RBF1-deficient larvae were fasted.
From these data, we infer that RBF1-deficient larvae use nucleotide salvage pathways to increase dNTP levels when they are fed. However, when these animals are starved, they are unable to further increase production through the salvage pathways and are, presumably, increasingly dependent on de novo nucleotide synthesis to maintain the elevated dNTP levels.
Glutamate is critical to maintain nucleotide and glutathione pools
Although we initially examined the changes in nucleotide and glutathione metabolism in RBF1-depleted larvae separately from one another, these changes are likely to interconnect. In addition to its role in reducing oxidative stress, glutathione also has important functions in protein thiol-disulfide balance. One example of this is the need for the ribonucleotide reductase (RNR) enzyme complex to be reduced to generate dNTPs from ribonucleosides (Fig. 4H). As a result, a change in glutathione metabolism has consequences for both oxidative stress protection and for replication. This relationship becomes especially important in the context of RBF1-depleted larvae as the levels of RNR increase, while levels of both GCl and GS decrease (Supplemental Fig. 1A,B).
From this, we surmised that glutathione would be an especially precious commodity in RBF1-depleted larvae and that metabolites, such as glutamate, that are required to both decrease oxidative stress and increase dNTP production would be especially important in these animals. To look for evidence of this, we examined the levels of oxaloacetate (OAA) in the RBF1-depleted larvae. Together with glutamate, OAA produces aspartate (Fig. 4A). The level of OAA was significantly reduced in fed RBF1-depleted animals compared with control animals (Fig. 4I). Reduced OAA would limit the catabolism of glutamate that provides carbon and nitrogen for nucleotide synthesis and would allow more glutamate to be shunted toward glutathione production. In contrast, in fasted RBF1-deficient larvae, OAA was increased by ∼50% relative to fed RBF1-deficient larvae, whereas no change was seen between fed and fasted control animals (Fig. 4I). This increase in OAA correlated with a sustained level of aspartate (Fig. 4B) and dNTPs (Fig. 3B–E), and the apparent increase in glutamate catabolism toward aspartate and dNTPs was associated with a general decrease in total glutathione (Fig. 2C–D). Together, these data suggest that glutamine catabolism is rate-limiting in fasted RBF1-deficient larvae.
To directly test the idea that insufficient flux from glutamine contributes to the sensitivity of RBF1-depleted larvae to fasting, we tested whether a glutamine supplement would be sufficient to rescue this phenotype. Animals were allowed to develop on a nutrient-rich diet for 40–48 h ALH prior to being shifted to either PBS or PBS + 25 mM L-glutamine, which is ∼10%–15% of the total glutamine detected in the normal food (data not shown). The addition of glutamine to the PBS had no significant impact on the overall viability of control animals. Remarkably, the addition of glutamine completely rescued the sensitivity of RBF1-depleted larvae to fasting (Fig. 5A), whereas the addition of glycine failed to rescue the sensitivity of RBF1-depleted larvae to fasting (Supplemental Fig. 3A). Glycine provides carbon and nitrogen for glutathione and nucleotide synthesis but inputs downstream from the entry of glutamine in both of these synthetic processes. The differential effects of glycine and glutamine in these assays suggests that RBF1 mutants have a particular need for glutamine.
Figure 5.
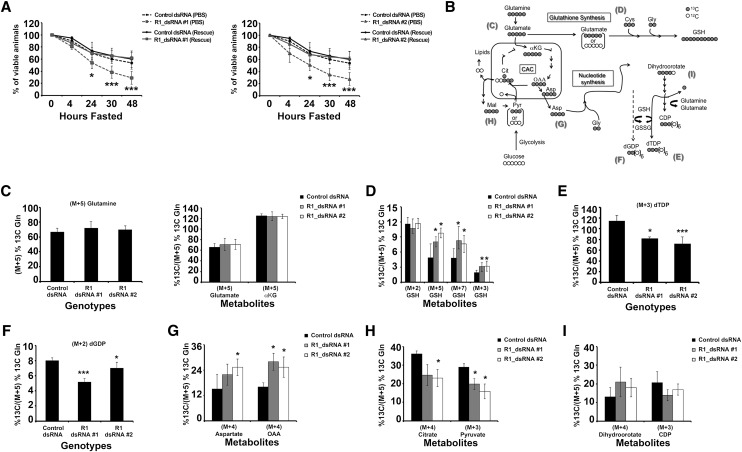
RBF1-depleted animals' sensitivity to fasting is rescued by glutamine supplementation. Genotypes of animals used are described in the Materials and Methods. Error bars represent the C.I. of 95%. Statistical significance is designated as P-values as follows: (*) P < 0.06; (***) P < 0.001. Statistical differences are between effects from RBF1 depletion compared with control animals. (A) Mid-second instar larvae were fasted or fed only 25 mM L-glutamine over the course of 48 h. (B) Schematic of [U-13C]-glutamine (gray circles) metabolism depicts the expected amount of 13C labeling after glutamine is metabolized through the citric acid cycle (CAC). Molecular symmetry and cellular compartments are not depicted. Unlabeled carbon (12C) is shown by open circles. Molecular symmetry and cellular compartments are not depicted. (αKG) α-Ketoglutarate; (Cit) citrate; (Asp) aspartate; (Mal) malate; (Pyr) pyruvate; (Gly) glycine; (Cys) cysteine; (CDP) cytodine diphosphate; (dTDP) thymidine diphosphate; (dGDP) deoxyguanosine diphosphate. (C–I) Enrichment of the [U-13C5]-glutamine tracer in metabolites associated with glutathione synthesis, the CAC, and nucleotide synthesis. All percentages are relative to the total amount of [U-13C5]-glutamine tracer in each sample.
RBF1-depleted larvae shunt glutamine to glutathione under energetic stress
To understand the mechanism underlying the glutamine rescue, we replaced the unlabeled 25 mM L-glutamine with 25 mM [U-13C5]-glutamine. The 13C label made it possible to examine the metabolic fates of glutamine and identify differences between the control and RBF1-depleted animals (Fig. 5B). Polar metabolites were extracted from larvae following 24 h of labeling, and 13C incorporation was detected and quantified by gas chromatography mass spectrometry (GC-MS) or LC-MS/MS (see the Supplemental Material for details). This time point was used because it was the earliest time that the glutamine supplement gave a statistically significant rescue of viability in RBF1-depleted larvae. Importantly, after 24 h of labeling, no statistical difference was found between genotypes in the 13C enrichment of fully labeled (M+5) glutamine (Fig. 5C). As additional controls, we observed no difference between genotypes in the overall levels of glutamine (Supplemental Fig. 2B), and glutaminase activity also appeared to be unaltered, since no difference was evident in the 13C enrichment of glutamate or α-ketoglutarate (Fig. 5C).
First, we examined the contribution of 13C-glutamine to glutathione synthesis. Glutathione is synthesized in a stepwise process that first combines glutamate with cysteine, and then glycine is combined to the glutamate–cysteine intermediate. While no difference was seen between control and RBF1-depleted animals in 13C labeling in glycine (M+2), a significant increase in 13C enrichment in the cysteine precursor carbon serine (M+3) was detected (Supplemental Fig. 3B). Strikingly, RBF1-deficient larvae showed a greater 13C enrichment of M+3 (from cysteine), M+5 (from either fully labeled glutamate or the combined contribution of fully labeled cysteine and glycine), and M+7 glutathione (Fig. 5D). These data indicate that the amount of glutathione derived from glutamine was increased in RBF1-depleted larvae.
In contrast, when we next examined the contribution of glutamine catabolism toward nucleotide production, we found reduced 13C labeling in both thymidine diphosphate (dTDP) and deoxyguanosine diphosphate (dGDP) (Fig. 5E–F). To understand this difference, we then examined the intermediate stages of nucleotide synthesis. Aspartate supplies the backbone of pyrimidine rings during de novo synthesis, and strikingly, 13C enrichment was significantly increased in fully labeled (M+4) aspartate in the RBF1-deficient animals (Fig. 5G). In agreement with the analysis of the metabolite pools (Fig. 4B,I), we saw an increase in fully labeled (M+4) OAA in RBF1-deficient larvae when compared with control larvae (Fig. 5G). However, the increased flow of glutamine to aspartate was specific, as we found reduced labeling in both M+4 citrate and M+3 pyruvate (Fig. 5H). This suggests that the flow of glutaminolysis to aspartate is via the TCA cycle/malate–aspartate shuttle. Also, because M+3 pyruvate would be generated by malic enzyme to produce NADPH, these data suggest that malic enzyme activity may be reduced in RBF1-depleted animals.
When we followed the fate of the glutamine-derived aspartate through pyrimidine ring synthesis, we saw little or no change across genotypes in carbamoyl aspartate (Supplemental Fig. 3C), the initial product formed in pyrimidine synthesis derived from aspartate. However, the RBF1-depleted animals had reduced labeling in cytodine diphosphate (CDP) (Fig. 5I). CDP is synthesized from cytodine triphosphate (CTP) and represents activity from the last committed step in pyrimidine synthesis (Fig. 3A). CTP is generated by the addition of an amino group, donated by glutamine, to UTP. It is likely, therefore, that the reduced labeling in CDP as well as dTDP and dGDP in these experiments (Fig. 5) is due to insufficient levels of glutamine rather than a general aversion of de novo synthesis.
Glutamine appears to be a precious commodity in RBF1-depleted larvae, and these experiments reveal how it is used in animals where a limited quantity is sufficient to promote survival. In this context, glutamine flows preferentially toward production of GSH rather than toward synthesis of dNTPs.
Oxidative stress promotes DNA damage and apoptosis in RBF1-depleted tissue
Although RBF1-depleted larvae likely have an increased requirement for reducing potential in protein thiol-disulfide reactions for nucleotide synthesis, the 13C-labeling experiments suggest that the synthesis of GSH for protection against oxidative stress is the highest priority. If this interpretation is correct, then RBF1-deficient larvae would be predicted to be highly sensitive to oxidative stress, and decreased oxidative stress might rescue the sensitivity of RBF1-deficient larvae to fasting. Moreover, if RBF1-depleted cells drive glutamate toward glutathione synthesis, then the lower availability of glutamate for nucleotide production might lead to replication errors and elevated DNA damage.
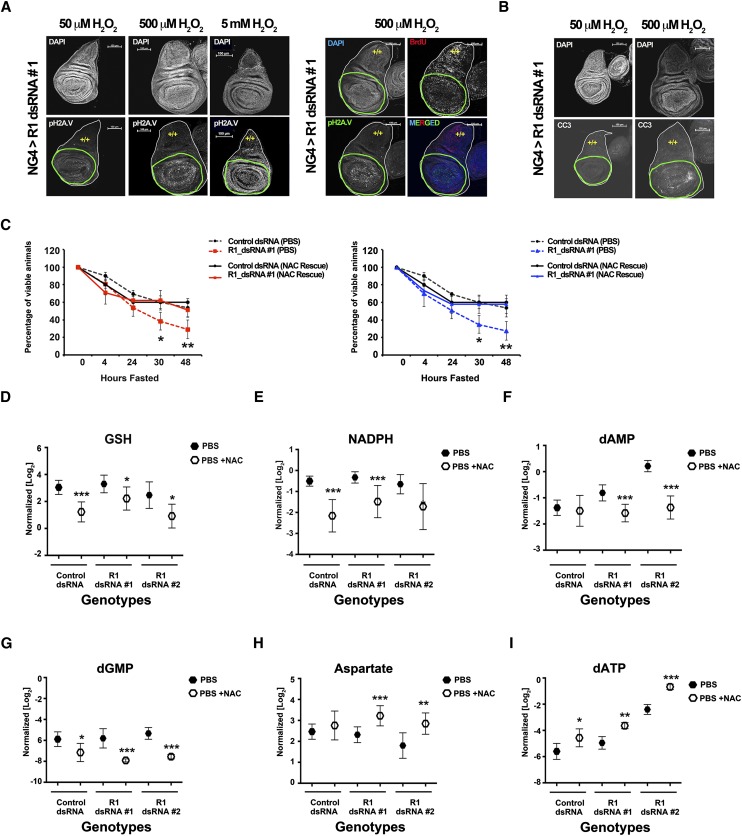
To test these predictions, larval wing discs from NG4; R1dsRNA#1 animals were dissected and treated with H2O2. Discs were then examined for DNA damage foci and CC3 formation by immunofluorescence. H2O2 specifically induced DNA damage foci (stained with pH2AV antibodies) in RBF1-depleted tissue but not in neighboring wild-type tissue (Fig. 6A; Supplemental Fig. 6A). This consequence was dose-dependent, and increased DNA damage was detected throughout the disc with a 100-fold increase in H2O2 (Fig. 6A; Supplemental Fig. 6A). Strikingly, RBF1-depleted cells continued to replicate DNA in the presence of DNA damage (Fig. 6A; Supplemental Fig. 6A). In addition to increased DNA damage, RBF1-depleted tissue also had increased formation of CC3 compared with wild-type tissue upon exposure to H2O2 (Fig. 6B; Supplemental Fig. 6B). This effect was also dose-dependent, as no CC3 formation was detected following treatment with the lowest H2O2 concentration (Fig. 6B; Supplemental Fig. 6B). These results indicate that RBF1-depleted tissue is hypersensitive to increased oxidation and that increased oxidation selectively causes DNA damage in RBF1-depleted cells.
Figure 6.
RBF1-depleted animals' sensitivity to fasting is rescued by NAC supplementation. Genotypes of animals used are described in the Materials and Methods. (A,B) RBF1-depleted cells (green outline) show increased sensitivity to H2O2 when compared with wild-type tissue (yellow +/+). RBF1-depleted cells have increased DNA damage, shown by pH2AV (grayscale and green), but continue to proliferate, shown by BrdU (red). This increased damage leads to increased apoptosis, shown by CC3 (grayscale). Error bars represent the C.I. of 95%. Statistical significance is designated as P-values as follows: (*) P < 0.06; (**) P < 0.02; (***) P < 0.001. Statistical differences are between effects from RBF1 depletion compared with control animals. (C) Mid-second instar larvae were fasted or fed only 0.6 mM NAC over the course of 48 h. (D–I) Normalized metabolite pools from larvae fasted (closed hexagons) or fed 0.6 mM NAC (open hexagons) for 24 h.
Decreased oxidative stress rescues the sensitivity RBF1-depleted larvae to fasting and impacts nucleotide synthesis
To directly test the idea that oxidative stress is important for the sensitivity of RBF1-depleted larvae to fasting, we asked whether the strong antioxidant N-acetyl-cysteine (NAC) was sufficient to rescue this sensitivity. As before, control and RBF1-depleted animals were shifted from nutrient-rich food sources 40–48 h ALH and placed in either PBS or PBS + NAC. Viability comparisons showed that NAC treatment provided no benefit in control animals but completely rescued the increased sensitivity of RBF1-depleted animals to fasting (Fig. 6C).
To understand the basis for this rescue, we examined the effects of NAC treatment on glutathione and nucleotide synthesis. NAC treatment decreased the levels of GSH in all genotypes (Fig. 6D) presumably because of the reduced need to prevent oxidative stress. Consistent with this change in GSH production, NADPH levels significantly dropped in all genotypes as well (Fig. 6E). Hence, the levels of GSH and NADPH production in the RBF1-depleted larvae appear to be tightly associated with the need for GSH.
These changes suggested to us that more glutamate carbon would be available for de novo nucleotide synthesis and that more GSH would be available for dNTP formation. We hypothesized that this might decrease the use of nucleotide salvage pathways for nucleotide production. Indeed, NAC treatment dramatically reduced the levels of nucleotide salvage pathway intermediates in both control and RBF1-depleted larvae (Fig. 6F-G). This raises the possibility that one of the reasons why the nucleotide salvage pathways are heavily used in RBF1-depleted cells is to conserve GSH for oxidative stress protection.
If a decrease in oxidative stress does indeed release glutamate and glutathione available for use in nucleotide synthesis in RBF1-depleted larvae, then we would expect to see increased levels of aspartate and dNTPs from de novo synthesis in the presence of NAC. NAC treatment had no significant effect on aspartate pools in control animals but significantly increased the aspartate pools in the RBF1-depleted larvae (Fig. 6H), a change that correlated with a greater increase in OAA levels (Supplemental Fig. 3D). NAC treatment also increased the pools of dNTPs in both control and RBF1-depleted larvae, with the increase being more significant in RBF1-depleted animals (Fig. 6I). These observations show a clear link between the oxidative stress in RBF1-depleted cells and nucleotide synthesis.
Taken together, our results demonstrate that RBF1 regulates glutamate usage in both fed and fasted conditions. The increased demands for nucleotide and GSH synthesis that occur when RBF1 is depleted place strong pressure on the supply of glutamine carbons, and this becomes especially significant when RBF1-depleted animals are fasted.
pRB modulates glutamine catabolism in human tumor cell lines
To test whether the role of pRB family proteins in regulating glutamine usage is conserved in human cells, we used [U-13C5]-glutamine to trace the metabolic fate of glutamine in hTERT-RPE-1 (RPE) cells (a line of hTERT-immortalized retinal pigment epithelial cells) and looked for changes in flux following pRB knockdown (Supplemental Fig. 4A).
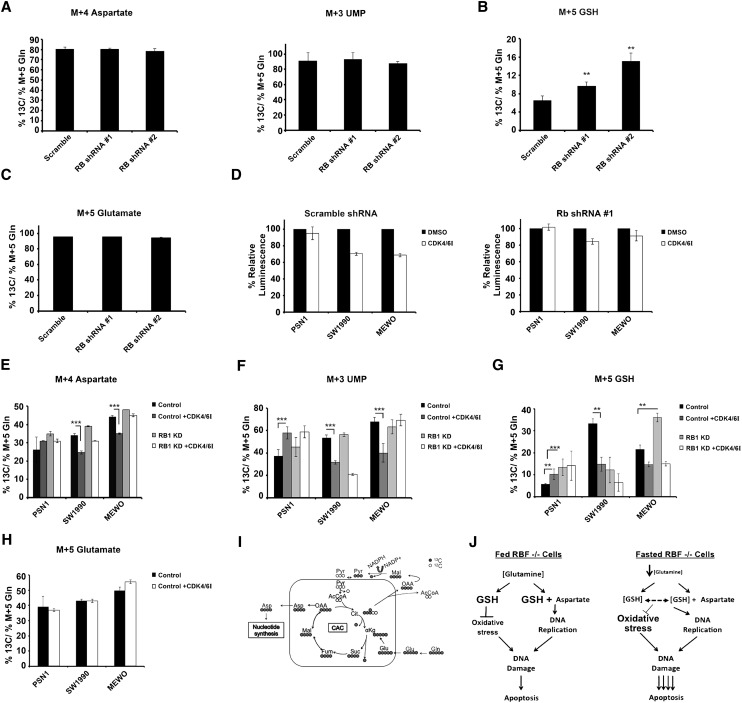
First, we examined the flow of glutamine into nucleotide precursors. Unlike RBF1-depleted flies, knockdown of pRB alone did not increase 13C labeling in either aspartate (M+4) or the nucleotide precursor UMP (M+3) (Fig. 7A). Interpretation of this is complicated by the fact that RPE cells express two pRB-related proteins, p107 and p130, with functions that partially overlap with pRB and that may also regulate nucleotide production. Second, we examined the impact of pRB depletion on the contribution of glutamine to glutathione synthesis. Strikingly, and as seen in RBF1-depleted flies, knockdown of pRB significantly increased 13C labeling in glutathione (GSH [M+5]) (Fig. 7B). Importantly, this increase was not due to increased 13C labeling in glutamate (Fig. 7C); rather, it represented a gross increase in the amount of GSH being generated from glutamine in pRB-depleted cells. Thus, the loss of pRB/RBF1 alters the usage of glutamine in both flies and humans.
Figure 7.
Alterations to glutamine catabolism following RBF1 depletion are partially conserved in human tumor cells. (A–C) RPE cells were fed the [U-13C5]-glutamine tracer in the presence or absence of a pRB shRNA. Error bars represent the C.I. of 95%. (D) The CellTiter-Glo assay (Promega) was used to measure cell number in human tumor cell lines over a 72-h period treated with or without the CDK4/6 inhibitor PD0332991 in the presence or absence of a pRB shRNA. The assays for each cell line were all done in parallel on the same plate but for visual representation are shown separated. Error bars represent the standard deviation from the mean. (A–C,E–H) Enrichment of the [U-13C5]-glutamine tracer in metabolites associated with glutathione synthesis, the CAC, and nucleotide synthesis. All percentages are relative to the total amount of [U-13C5]-glutamine tracer in each sample. In all panels except A–C, cells fed the [U-13C5]-glutamine tracer (see the Materials and Methods for details) were treated with or without the CDK4/6 inhibitor PD0332991 in the presence or absence of a pRB shRNA. Error bars represent the C.I. of 95%. Statistical significance is designated as P-values as follows: (**) P < 0.02; (***) P < 0.001. (I) Schematic of [U-13C]-glutamine (gray circles) metabolism depicts the expected amount of 13C labeling after glutamine is metabolized through the CAC. Molecular symmetry and cellular compartments are not depicted. Unlabeled carbon (12C) is shown by open circles. (αKG) α-Ketoglutarate; (Cit) citrate; (Asp) aspartate; (Mal) malate; (Pyr) pyruvate; (Suc) succinate; (Fum) fumarate; (Glu) glutamate; (Gln) glutamine; (AcCoA) acetyl-CoA. (J) Model of RBF1-depleted animals' hypersensitivity to energetic stress.
Given the frequent inactivation of pRB in human tumors and the known dependency of many human tumor cells on glutamine, we next tested whether we could detect effects of pRB activation/inactivation on the catabolism of glutamine human tumor cell lines. For this purpose, we selected a panel of seven cancer cell lines (PSN1, SW1990, MEWO, T98G, BxPC3, HuPT3, and HT29) that retain an intact pRB gene but lack p16INK4, an upstream component of the pRB pathway. pRB is predicted to be functionally inactivated in these cells via increased phosphorylation by Cyclin D/CDK4/6 kinases. Incubation with Cyclin D/CDK4/6 kinase inhibitor PD0332991 (hereafter called “CDKi”) is expected to activate pRB in these cells. We used shRNAs to deplete pRB to assess whether any effects of CDKi on glutamine use in these cells were pRB-dependent.
To various degrees, the addition of CDKi reduced the proliferation of SW1990, MEWO, HuPT3, BxPC3, T98G, and HT29 cells (Fig. 7D; Supplemental Fig. 4B–D). Extensive cell line screens have shown that not all pRB+ cells respond to CDK4/6 inhibition, and for reasons that are not yet known, PSN1 cell proliferation is resistant to CDKi at the concentration tested (Fig. 7D). The knockdown experiments confirmed that in each line that reduced proliferation in response to CDKi, these effects were largely dependent on pRB (Fig. 7D; Supplemental Fig. 4B–D).
When we used [U-13C5]-glutamine to trace glutamine usage in this panel of cells, we observed that CDKi significantly reduced 13C labeling in aspartate (M+4) and the nucleotide precursor UMP (M+3) in each of the lines that slowed proliferation in response to CDKi (representative lines, SW1990 and MEWO, are shown in Fig. 7E,F). These effects were dependent on pRB, since knockdown of pRB restored the flow of glutamine to aspartate in the presence of the drug. These changes were consistent with the altered flux of glutamine to aspartate in RBF1-depleted larvae. Interestingly, in PSN1 cells, CDKi increased the 13C labeling in both aspartate and UMP (Fig. 7E–F), although, as seen in RPE cells, 13C labeling in these nucleotide precursors was not significantly changed following pRB knockdown alone (Fig. 7E–F). Together, these results suggest that pRB-mediated effects on cell proliferation correlate with reduced flow of glutamine toward nucleotide precursors.
The impact of pRB activity on glutamine flux toward glutathione varied between tumor cell lines (examples are shown in Fig. 7G). As in the RPE cells and RBF1-depleted flies, knockdown of pRB alone in PSN1 and MEWO cells led to increased 13C labeling of glutathione; but the opposite effect was found in SW1990 cells (Fig. 7G). Furthermore, the impact of CDKi on 13C labeling of glutathione appeared to be independent of pRB in SW1990 and MEWO cells but pRB-dependent in PSN1 cells (Fig. 7G). From this variety of responses, we infer that pRB activity can impact the flow of glutamine toward glutathione in tumor cell lines, but this effect varies considerably, and the impact of pRB presumably depends greatly on the different oncogenic landscape present in each of the tumor cell lines.
Together, these results confirm that pRB depletion causes metabolic changes and suggest that specific types of pRB-dependent metabolic changes may correlate with pRB-mediated suppression of cell proliferation. Our observations show that the CDKi PD0332991 can cause pRB-dependent reprogramming of glutamine metabolism in human cancer cells, but the specific nature of the changes varied in tumor cell lines. Perhaps more importantly, in nontransformed RPE cells, the depletion of pRB increased the flux of glutamine to glutathione. This effect resembled the changes seen in RBF1-deficient larvae, suggesting that pRB/RBF1 have a conserved ability to affect glutamine catabolism.
We propose that RBF1-deficient larvae under conditions of energetic stress (such as fasting) retain an increased demand for dNTP production. In the absence of RBF1, a reduced supply of glutamine causes a significant drop in glutathione levels, resulting in increased sensitivity to oxidative stress and to DNA damage. DNA repair requires the synthesis of more nucleotides for resectioning, increasing the demand for glutamine. This feed-forward loop further increases the likelihood of oxidative stress, DNA damage, and, ultimately, cell death (Fig. 7I).
Discussion
Numerous studies have assayed the “function” of RB proteins using transcription assays and have cataloged the changes in gene expression that occur when RB proteins are inactivated. Directly or indirectly, the loss of pRB or RBF affects the expression of a very large number of genes. The precise number varies in different reports from several hundred to more than a thousand. It is evident from these data that the altered properties of a RB/RBF1 mutant cell cannot be attributed to the misexpression of any single target. However, at the same time, very little is known about the net effects of all of these gene expression changes on the cell. In this study, we took a different approach to understanding the function of RBF1. Prompted by our finding that RBF1-depleted cells and rbf1 mutant animals are sensitive to fasting, we used metabolic profiling of RBF1-depleted larvae to identify metabolites that are significantly altered by the loss of RBF1. The results identify biological processes that are functionally affected by the loss of RBF1. Reassuringly, the results are consistent with previous ChIP–chip and gene expression studies yet provide a very different perspective on the changes that occur when RBF1 is lost. Importantly, the results highlight a metabolic vulnerability of the RBF1-depleted animals.
Our results show that loss of RBF1 leads to major changes in nucleotide metabolism and a significant change in the GSH/GSSG balance. RBF1-depleted larvae have greatly elevated nucleotide pools that are presumably important for the altered patterns of DNA replication. At the same time, these animals also have low levels of glutathione and are selectively rescued by glutamine supplementation. This combination of events means that glutamine is particularly important in the RBF1-deficient animals. Glutathione is synthesized from glutamine and is a key metabolite in the reduction of oxidative stress. In addition, elevated levels of DNA replication requires glutamine carbon and nitrogen to be used for dNTP synthesis. dNTP formation is initially catalyzed by the RNR enzyme complex, leading to the oxidation of RNR. Importantly, RNR must be constantly reduced in order to produce dNTPs from ribonucleosides. This protein thiol-disulfide cycle is balanced, in part, by the ability of a cell to produce sufficient glutathione.
These changes put RBF1-deficient cells and larvae into a “catch-22” situation when faced with glutamine-limited conditions: If the cell shunts glutamine toward dNTP production, it becomes vulnerable to oxidative stress; conversely, if the cell shunts glutamine away from dNTP synthesis, it risks an imbalance in dNTP pools and replication error-dependent DNA damage. In keeping with this model, RBF1-depleted cells are highly sensitive to H2O2, and the sensitivity of RBF1-depleted larvae to fasting can also be rescued by addition of the antioxidant NAC. The NAC rescue shows that glutamine is a fungible commodity in RBF1-depleted larvae, as NAC reduces the level of GSH and increases levels of dNTP synthesis.
[U-13C5]-glutamine-labeling experiments revealed that the flux of glutamine to both glutathione and aspartate (for nucleotide production) is increased in RBF1-depleted larvae. Strikingly, the role of RBF1 as a regulator of glutamine catabolism appears to be conserved by its human homolog, pRB. Following pRB knockdown in nontransformed RPE cells, we found increased 13C labeling in glutathione. However, when we performed similar experiments in a panel of tumor cell lines, we observed a more complex pattern of changes. These results suggest that pRB activation (or inactivation) can lead to a variety of metabolic changes and raise the possibility that the type of metabolic change that occurs may influence pRB's ability to suppress proliferation.
The evidence that this role of RBF1 is conserved in mammalian cells is consistent with the findings of several previous reports. Previous studies showed that pRB is needed to block cell proliferation following DNA damage (Harrington et al. 1998) and that the loss of pRB leads to an increased level of DNA damage in the absence of any exogenous damaging agent (Pickering and Kowalik 2006). More recently, it has been shown that human papillomavirus (HPV) E7-expressing cells that lack pRB function and with deregulated E2F have an imbalance in nucleotide pools that leads to stalled replication forks (Bester et al. 2011). Our findings are also in agreement with recent studies from the Frolov laboratory (AM Ambrus, ABMMK Islam, K Holmes, R Suckling, N-S Moon, N Lopez-Bigas, EV Benevolenskaya, and MV Frolov, in prep.) showing that dDP−/− cells have reduced mitochondrial activity. Taken together, these studies strongly support the emerging view that the E2F/RB pathway has significant impact on metabolic pathways. We note that the reduced mitochondrial activity observed in dDP−/− mutants by AM Ambrus, ABMMK Islam, K Holmes, R Suckling, N-S Moon, N Lopez-Bigas, EV Benevolenskaya, and MV Frolov will reduce the natural production of ROS stemming from oxidative phosphorylation. Consistent with this, rbf1−/−; dDP−/− cells were found to be protected from ionizing radiation (IR)-induced apoptosis (Moon et al. 2008). Decreased production of ROS may enable rbf1−/− cells to redirect the flow of glutamine toward dNTP synthesis and reduce the extent of DNA damage.
This work adds to the idea that metabolic profiling of tumor samples can enhance the use of gene expression signatures or genomic information to predict tumor cell sensitivities. This concept has also been documented in tumors driven by the expression of the c-Myc transcription factor (Yuneva et al. 2012). In that study, Yuneva et al. (2012) demonstrated that c-Myc overexpression in either the lung or liver led to distinct metabolic changes while retaining highly similar changes in gene expression of classic c-Myc target genes. Yuneva et al. (2012) further showed how these changes could be exploited in c-Myc-overexpressing, but not wild-type, cells. Thus, the use of metabolic profiling may one day help to identify sensitivities of tumor cells that are refractory to genotype-selected, targeted therapies and may help to pinpoint mechanisms of resistance.
In future studies, it may be possible to exploit these metabolic phenotypes to target tumor cells lacking functional pRB. The conserved role of pRB actively regulating glutamine catabolism opens the possibility that p16−/− or pRB−/− tumors may be sensitive to glutaminolysis inhibitors used in conjunction with DNA-damaging agents. Finally, we note that one of the implications of our observations is the likelihood that antioxidants will enhance the growth of some tumors. In agreement with this, it has recently been shown that AMPK activation can be helpful to tumor development. While at first pass this may appear counterintuitive, AMPK activation was shown to benefit a tumor by increasing the amount of reduced glutathione available to a tumor cell, thus reducing oxidative stress (Jeon et al. 2012). Given the emerging trend to use AMPK agonists such as metformin and AICAR to treat tumors, this is a point that clearly needs further investigation.
Materials and methods
Fly genetics
Genotypes assayed and described in all figures are UAS hairpins associated with “RBF dsRNA #1” [Vienna Drosophila RNAi Center (VDRC): GD4484(v10696)}, “RBF dsRNA #2” (Transgenic RNAi Project [TRiP]: HMS03004), “Control dsRNA” (TRiP: valium 20-Gal4.1), and “GFP” [UAS-GFP (II)].
In Figures 1, A–C, and 6, A and B, UAS hairpins were driven by nubbin-Gal4. In all other instances, UAS hairpins were driven by armG4.
w1118 and rbf120a stocks were also used. The w1118 and rbf120a stocks were backcrossed for five generations. Stocks were then re-established by selecting against genetic markers.
ChIP
ChIP was performed as previously described (Korenjak et al. 2012).
Antibodies
Antibodies used in this study included the following: mouse anti-RBF (DX5- 1:5), rabbit anti-CC3 (1:100; Cell Signaling), rabbit anit-pH2AV (1:500; Abnova), mouse anti-BrdU (1:100; Beckton Dickinson), Cy3/5 anti-mouse (1:500; Jackson ImmunoLaboratories), and Cy3/5 anti-rabbit (1:500; Jackson ImmunoLaboratories).
All other materials and methods can be found in the Supplemental Material.
Acknowledgments
We thank N. Bardeesy, L. Cantley, H. Coller, J. Lees, A. Manning, R. Mostoslavsky, A. Näär, and L. Zou for discussions and critical reading of the manuscript, and A. Ambrus and M. Frolov for sharing data prior to publication. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks and/or plasmid vectors used in this study, and the Vienna Drosophila RNAi Center (VDRC) for fly stocks. B.N.N. is supported by the NIH NRSA fellowship F32CA165856. P.A.G. is supported by a fellowship from the Foundation for Science and Technology (FCT), Portugal. This work was supported by the MGH Federal Share of Program Income under C06 CA059267 and by NIH R01 CA122591 (to O.I.). N.J.D. is the James and Shirley Curvey MGH Research Scholar.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.206227.112.
References
- Arteaga CL, Baselga J 2012. Impact of genomics on personalized cancer medicine. Clin Cancer Res 18: 612–618 [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black EP, Huang E, Dressman H, Rempel R, Laakso N, Asa SL, Ishida S, West M, Nevins JR 2003. Distinct gene expression phenotypes of cells lacking Rb and Rb family members. Cancer Res 63: 3716–3723 [PubMed] [Google Scholar]
- Blais A, Dynlacht BD 2004. Hitting their targets: An emerging picture of E2F and cell cycle control. Curr Opin Genet Dev 14: 527–532 [DOI] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Dyson N 1999. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J 18: 916–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Vidal M, Xie JE, Dyson N 1996. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev 10: 1206–1218 [DOI] [PubMed] [Google Scholar]
- Haber DA, Gray NS, Baselga J 2011. The evolving war on cancer. Cell 145: 19–24 [DOI] [PubMed] [Google Scholar]
- Harrington EA, Bruce JL, Harlow E, Dyson N 1998. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci 95: 11945–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N 2012. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 485: 661–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193 [DOI] [PubMed] [Google Scholar]
- Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ 2012. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol Cell Biol 32: 4375–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Ohno K, Voskoboynik Y, Ragusano L, Martinez A, Dimova DK 2010. Drosophila RB proteins repress differentiation-specific genes via two different mechanisms. Mol Cell Biol 30: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Ragusano L, Martinez A, Gill J, Dimova DK 2012. A dual role for the dREAM/MMB complex in the regulation of differentiation-specific E2F/RB target genes. Mol Cell Biol 32: 2110–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Knabb JR, Spike BT, Macleod KF 2009. Elevated poly-(ADP-ribose)-polymerase activity sensitizes retinoblastoma-deficient cells to DNA damage-induced necrosis. Mol Cancer Res 7: 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Dyson NJ 2010. pRb, a local chromatin organizer with global possibilities. Chromosoma 119: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Herr A, Ji JY, Dyson NJ 2008. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev 22: 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Walker JA, Anderssen E, Moon NS, Gladden A, Heck MM, Ramaswamy S, Dyson NJ 2012. A shared role for RBF1 and dCAP-D3 in the regulation of transcription with consequences for innate immunity. PLoS Genet 8: e1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Longworth MS, Dyson NJ 2010. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev 24: 1364–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon NS, Di Stefano L, Morris EJ, Patel R, White K, Dyson NJ 2008. E2F and p53 induce apoptosis independently during Drosophila development but intersect in the context of DNA damage. PLoS Genet 4: e1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ 2001. Retinoblastoma protein partners. Adv Cancer Res 82: 1–54 [DOI] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, Frolov MV 2010. Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet 6: e1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MT, Kowalik TF 2006. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene 25: 746–755 [DOI] [PubMed] [Google Scholar]
- Popova MK, He W, Korenjak M, Dyson NJ, Moon NS 2012. Rb deficiency during Drosophila eye development deregulates EMC, causing defects in the development of photoreceptors and cone cells. J Cell Sci 124: 4203–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J 21: 4927–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ 2008. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS 2011. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr Protoc Bioinformatics 34: 14.10.1–14.10.48 [DOI] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, Asara JM 2012. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 7: 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. 2012. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 15: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorski WA, Knudsen ES, Reed MF 2007. Retinoblastoma deficiency increases chemosensitivity in lung cancer. Cancer Res 67: 8264–8273 [DOI] [PubMed] [Google Scholar]
- Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X, Ulyanov A, Wu G, Wilson M, Wang J, et al. 2012. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]