Abstract
Background
Aberrant DNA hypermethylation has been implicated as a component of an epigenetic mechanism that silences genes in cancers.
Methods
We performed a genome-wide search to identify differentially methylated loci between 26 tumor and adjacent non-tumor paired tissues from same lung cancer patients using restriction landmark genomic scanning (RLGS) analysis. Among 229 loci which were hypermethylated in lung tumors as compared to adjacent non-tumor tissues, solute carrier family 5, member 8 (SLC5A8) was one of the hypermethylated genes, and known as a tumor suppressor gene which is silenced by epigenetic changes in various tumors. We investigated the significance of DNA methylation in SLC5A8 expression in lung cancer cell lines, and 23 paired tumor and adjacent non-tumor lung tissues by reverse transcription-PCR (RT-PCR), quantitative methylation specific PCR (QMSP) and bisulfite modified DNA sequencing analyses.
Results
Reduced or lost expression of SLC5A8 was observed in 39.1% (9/23) of the tumor tissues as compared with paired adjacent non-tumor tissues. Bisulfite sequencing results of lung cancer cell lines and tissues which did not express SLC5A8 showed a densely methylated promoter region of SLC5A8. SLC5A8 was reactivated by treatment with DNA methyltransferase inhibitor, 5-Aza and/or HDAC inhibitor, trichostatin A (TSA) in lung cancer cell lines, which did not express SLC5A8. Hypermethylation was detected at the promoter region of SLC5A8 in primary lung tumor tissues as compared with adjacent non-tumor tissues (14/23, 60.9%).
Conclusion
These results suggest that DNA methylation in the SLC5A8 promoter region may suppress the expression of SLC5A8 in lung tumor.
Keywords: Lung cancer, Gene silencing, SLC5A8, Tumor suppressor gene, DNA methylation, Restriction landmark genomic scanning
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths in the United States. The poor survival is due to its advanced stage at the time of diagnosis. Over 50% of patients were found to have distant metastatic lesion at the time of diagnosis and only 15% of patients had localized disease of whom the survival could be expected 5 years with potentially-curative resection [1]. Recently, the National Lung Screening Trial has shown low-dose helical computed tomography to reduce lung cancer mortality [2], however there are still uncertainty to screen general population.
DNA methylation of promoter regions in tumor suppressor genes is considered as one of the contributing factors in carcinogenesis. The methylation status of the promoter region in many genes has been assessed in lung tumors. Several genes are known to be hypermethylated and are reported to be of unfavorable prognostic relevance in primary lung tumor [3]. Furthermore, promoter methylations in these genes were found in the early stage of carcinogenesis and remained during progression [4].
Solute carrier family 5 (iodide transporter) (SLC5) is a solute-linked carrier gene family that contains 12 sodium-coupled transporters for several chemicals [5]. Several studies reported that SLC5A8 is a sodium-coupled transporter for nicotinate and its structural analogs [6], lactate [7], pyroglutamate [8], short-chain fatty acids (butyrate) [9] and pyruvate which are known as histone deacetylase (HDAC) inhibitors [10–14].
SLC5A8 was first identified as a differentially methylated gene by restriction landmark genome scanning (RLGS) which provides a global analysis of methylation events in colon cancer cell lines [15]. Since then, increasing evidences suggest that gene silencing of SLC5A8 may contribute to the carcinogenesis and progression of tumors. Therefore, SLC5A8 has been suggested as a tumor suppressor and promoter methylation and gene-silencing in various tumors has been detected [4, 15–27].
In the present study, we report a differential methylation profile of lung tumor tissues, characterize the methylation status of SLC5A8 and investigate the role of epigenetic changes in the promoter region of SLC5A8 in its expression in lung cancer cell lines and lung tumor tissues.
MATERIALS AND METHODS
Tissue collection
Paired lung tumor and adjacent non-tumor tissues from 26 lung cancer patients were used for RLGS analysis. These patients were operated at the Moffitt Cancer Center (Tampa, FL) with primary adenocarcinoma (n=14), squamous cell carcinoma (n=9) and large cell carcinoma (n=3). The patients included 25 Caucasians and 1 African American, 18 males and 8 females ranging in age from 51 to 87 years (Table 1). For expression and DNA methylation analyses, 23 pairs of lung tissues which were analyzed in RLGS assay, were used. Three paired lung tumor and adjacent non-tumor tissues were excluded, because there were not enough samples left for analysis.
Table 1.
| Patient | Age | Sex | Race | Stage | Tumor type1 | Methylated2 |
|---|---|---|---|---|---|---|
| 1 | 82 | M | Caucasian | III | AD | 38 |
| 2 | 83 | M | Caucasian | II | AD | 1 |
| 3 | 87 | F | Caucasian | I | AD | 29 |
| 4 | 87 | M | Caucasian | I | AD | 47 |
| 5 | 74 | M | Caucasian | I | AD | 14 |
| 63 | 74 | M | Caucasian | I | AD | 9 |
| 7 | 55 | F | Caucasian | III | AD | 29 |
| 8 | 63 | M | Caucasian | II | AD | 23 |
| 9 | 51 | M | Caucasian | II | AD | 7 |
| 10 | 73 | F | Caucasian | I | AD | 13 |
| 11 | 76 | M | Caucasian | I | AD | 5 |
| 12 | 83 | M | Caucasian | I | AD | 7 |
| 13 | 79 | F | Caucasian | I | AD | 25 |
| 143 | 60 | F | African American | I | AD | 7 |
| 15 | 64 | M | Caucasian | I | SCC | 19 |
| 16 | 85 | F | Caucasian | I | SCC | 14 |
| 17 | 69 | F | Caucasian | II | SCC | 9 |
| 18 | 80 | M | Caucasian | I | SCC | 5 |
| 193 | 64 | M | Caucasian | III | SCC | 6 |
| 20 | 66 | M | Caucasian | I | SCC | 40 |
| 21 | 78 | F | Caucasian | I | SCC | 16 |
| 22 | 72 | M | Caucasian | I | SCC | 21 |
| 23 | 78 | M | Caucasian | II | SCC | 5 |
| 24 | 70 | M | Caucasian | I | LC | 38 |
| 25 | 74 | M | Caucasian | III | LC | 5 |
| 26 | 58 | M | Caucasian | III | LC | 6 |
AD: Adenocarcinoma, SCC: Squamous cell carcinoma, LC: Large cell carcinoma
Number of hypermethylated loci identified by RLGS
Samples were excluded from QMSP
All tissues were collected under a protocol approved by the Institutional Review Board of the Moffitt Cancer Center and the University of South Florida. All adjacent non-tumor specimens were harvested freshly from morphologically normal appearing tissue located at least 3 cm from the tumor margin. A surgical pathologist performed histological evaluation of the tumor and non-tumor tissues. All tissues were snap frozen in liquid nitrogen and stored at −80°C.
Restriction landmark genome scanning (RLGS) analysis
After extracting DNA from the lung tissues, the RLGS technique, a two-dimensional gel electrophoresis system that uses radiolabeled restriction endonuclease sites to create “landmarks” seen on an autoradiograph, was used as previously described [28–30]. The tumor and normal gels of each patient were either visually compared or quantified by densitometry at each of ~2,000 sites. Differential methylation was detected by either the absence or decrease in signal intensity (> 50%).
RNA extraction from cell lines and tissues
RNA was extracted from 22 lung cancer cell lines, which were obtained from the American Type Culture Collection (Manassas, VA), and 23 paired lung tumors/adjacent non-tumors tissues with the Qiagen RNA isolation kit (Austin, TX) according to the manufacturer’s protocol.
Semi-quantitative and quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR)
We investigated the expression of SLC5A8 in lung cancer cell lines, and paired tumor and adjacent non-tumor tissues from lung cancer patients by a semi-quantitative and quantitative real-time RT-PCR. Semi-quantitative PCR was performed with primers selected from exon 1 and exon 2 (cttcatgtcagccgtcactg and caatgaagaggactgttccac, respectively). The resulting amplified DNA was analyzed by electrophoresis on an 8% PAGE gel as previously described [22].
Quantitative real-time PCR was performed using the ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA), according to the manufacturer’s protocol. Taqman Gene Expression Assay including 2 primers and a probe was used for the quantification of SLC5A8, and β-actin was used as reference gene as previously described [24]. Tumor down-regulation was defined as less than 0.5 ratio (raw) or 0.75 (log) of the expression of normal tissue.
Genomic DNA extraction and sodium bisulfite modified sequencing
Genomic DNA samples were extracted from selected cells and tissues using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. Sodium bisulfite conversion of genomic DNA was done as described [31] using EZ DNA Methylation Kit (Zymo Research, Irvine, CA). Bisulfite modified sequencing was performed to determine the density of methylation in the SLC5A8 promoter. Primers for bisulfite sequencing were selected using the Methprimer software program, which identifies the location and structure of CpG islands of SLC5A8 (www.urogene.org/methprimer). A 426-base pair (bp) region of the SLC5A8 promoter from −278 to +148 (+1 was the translation start site) encompassing 45 CpG sites were amplified by bisulfite modified specific primers (sense: gaggttttatatttgggtttgagg, anti-sense: cccatcaaaaaatccttaaaaatcta) (Supplementary figure 1). The methylation level of each CpG site on SLC5A8 was estimated by comparing the height of the cytosine peak with the height of the thymine peak on the chromatogram as previously described [32].
Quantitative methylation specific PCR (QMSP) assay for SLC5A8
The bisulfite-modified DNA samples were used as a templates for fluorescence-based QMSP for SLC5A8, as previously described with modification [15]. In brief, primers (sense: tcgaacgtatttcgaggc, anti-sense: acaacgaatcgattttccg), and a labeled probe (6FAM-caacgacgaatacaaaaacgactaccaac-BHQ-2) were designed to specifically amplify the bisulfite-converted sequence of SLC5A8. The ratios between the expression levels of SLC5A8 and the reference gene, β-actin were used as a measure for representing the relative level of methylation of promoter DNA in the tissues (SLC5A8/β-actin X 1000). Bisulfite-modified DNA was amplified by PCR with the following reaction conditions: 1 X buffer, 600 nM of each primer, 200 nM of probes, 0.2 mM of dNTPs, 0.5 U of Taq enzyme and 20 ng of bisulphite-modified DNA in a final volume of 20 μl. The condition of the QMSP was as follows: 95°C for 10 min for denaturation, 55 cycles of amplification (95°C, 30 s; 60°C, 30 s; 72°C, 30 s) with a final elongation step of 10 min at 72°C. Each plate contained tissue samples and water blanks, as well as positive and negative controls. Genomic DNA extracted from a human prostate cancer cell line (DU145) cells with bisulfite treatment served as a positive control for QMSP. Serial dilutions of this DNA were used for constructing the calibration curves on each plate as previously described [22]. The more than 3 fold ratio of methylation between tumor and normal tissue was considered as hypermethylated.
Treatment of DNA methyltransferase inhibitor, 5-Aza and HDAC inhibitor, trichostatin A (TSA)
Lung cancer cell lines were treated with either 5-Aza or TSA (Sigma, St. Louis, MO), which are commonly used to determine whether DNA methylation or histone modification influence gene expressions. Briefly, cells were split to low density (1 × 106 cells/T-75 flask) 24 hours before treatments. Stock solutions of 5-Aza and TSA were dissolved in phosphate-buffered saline and 100% ethanol, respectively. Cells were treated with 2 mM 5-Aza for 5 days or 300 nM TSA for 24 hours. We also mock treated cells with the same volume of phosphate-buffered saline or ethanol as negative controls. The medium was changed at 24 hours after addition of 5-Aza. RNA was extracted after the treatment for RT-PCR analysis. The cDNA samples were used for quantification of SLC5A8 expression using the above described primers. β-actin was co-amplified for normalization of the SLC5A8 expression.
Statistical methods
Student’s t-test was used to compare levels of expression or methylation between tumor and adjacent non-tumor tissues. Reported p values are two sided. P value < 0.05 was considered statistically significant. SAS version 9 software (SAS institute, Cary, NC) was used for all statistical analyses in this study.
RESULTS
RLGS analysis in lung cancer patient samples
Differential methylation profiles were obtained from the DNA samples of lung tumor and adjacent non-tumor tissues with RLGS analyses from 26 lung cancer patients. For each patient, the non-tumor tissue profile was compared to the tumor profile. Differential methylation was detected either by the absence or decrease in signal intensity. Using the surrounding single copy spots as an internal control, we could visually estimate the loss of intensity of a locus or quantify it by densitometry. We could reliably detect loss of spot intensity equal to 30–40% of methylation [28].
We found that 229 RLGS fragments showed hypermethylation (spots present at least one patient’s normal tissue profile that was absent or decreased in intensity from that patient’s tumor profile). Among these 229 loci, 108 different loci were found in only one tumor/normal pair out of all 26 subjects. Sixty eight loci were detected in 2 tumor/normal pairs, 31 loci in 3 pairs, 15 loci in 4 pairs, 5 loci in 5 pairs, and 2 loci were hypermethylated in 8 pairs, respectively. These RLGS fragment losses are indicative of hypermethylation of those loci. Supplementary table 1 shows the degree of hypermethylation in different histological subtypes of lung cancer. The percentage of hypermethylated RLGS fragments ranges from 0.05% to 2.34% among tumors. The frequencies of locus methylation by tumor cell type are followed: 186 of the 229 loci were methylated in adenocarcinoma, 109 in squamous cell carcinoma and 45 in large cell carcinoma (Supplementary figure 2). Among the 229 hypermethylated loci, information of 109 hypermethylated loci was available from previous studies [33–35]. Among these 109 sites, we found annotated genes for 69 loci. Information for chromosomal location, CpG island traits, and the genomic context were assessed (supplementary table 2). For the 69 annotated genes, most of the CpG islands from which the RLGS fragment arises were found in the promoter region at the 5′ end of the gene and were not previously reported to be hypermethylated in lung cancer (supplementary table 2). Hypermethylation sites identified by RLGS were found at loci on chromosomes 2p, 2q, 3p, 4q, 5p, 6p, 10p, 17p, 17q, 18p, 18q, and 21q sites, where loss of heterozygosity has been frequently reported in lung cancer [36–39]. This analysis revealed that 15.3% (4 of 26) of the lung tumors show differential methylation in SLC5A8 (supplementary table 2).
Expression of SLC5A8 in lung cancer cell lines, tumor and adjacent non-tumor tissues
Semi-quantitative RT-PCR was used to characterize SLC5A8 expression in 8 pared tumor and adjacent non-tumor tissues (Fig. 1A) and in 22 lung cancer cell lines. The SLC5A8 was not expressed in most lung cancer cell lines (21/22) except H69 cell line (Supplementary figure 3). In addition, SLC5A8 expression in tumor tissues was reduced as compared with that in the adjacent non-tumor tissues. Quantitative real-time RT-PCR showed that the SLC5A8 was well expressed in adjacent non-tumor lung tissues, while 39.1% (9/23) of tumor tissues were either silenced or reduced (Fig. 1B). Among these 9 down–regulated tumors, 5 tumors (55.6%, 5/9) were hypermethylated and one sample (11.1%, 1/9) was hypomethylated. Conversely, 2 out of 6 over-expressed tumors were hypomethylated and hypermethylated, respectively. Different methylation status of each samples were indicated in Fig. 1B.
Fig. 1.
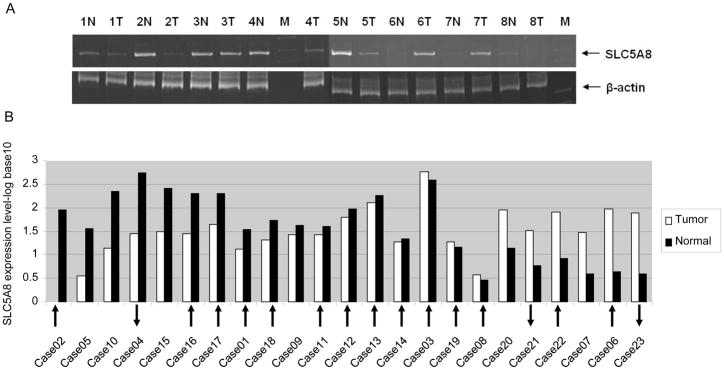
SLC5A8 expression from RT-PCR analysis. (A) Representative semi-quantitative RT-PCR results of SLC5A8 transcripts from RNA isolated from 8 pairs of tumor (T) and adjacent non-tumor (N) tissues. M: DNA marker, Lanes 1~16: RNA from 8 tumor (T) and adjacent non-tumor tissue (N) pairs. (B) Quantitative real-time RT-PCR results of SLC5A8 transcripts from RNA isolated from 23 pairs of tumor and adjacent non-tumor tissues. 9 out of 23 tumor tissues were significantly down-regulated. Arrows indicate the status of methylation of tumor as compared with adjacent non-tumor tissues. Direction of arrow indicates either hypermethylation or hypomethylation.
Treatments of 5-Aza and/or TSA restore SLC5A8 expression
To investigate a relationship between epigenetic changes and expression of SLC5A8, three lung cancer cell lines (H322, Calu3, and A549) were treated with a DNA methyltransferase inhibitor, 5-Aza, and/or a HDAC inhibitor, TSA. SLC5A8 mRNA expression was compared between the inhibitor-treated and untreated cell lines using semi-quantitative RT-PCR. Among three lung cancer cell lines which SLC5A8 expression was not detected, all three cell lines were reactivated by TSA, while only Calu3 was reactivated by 5-Aza treatment. These results imply that the SLC5A8 is suppressed by DNA methylation and/or histone modification (Fig. 2).
Fig. 2.
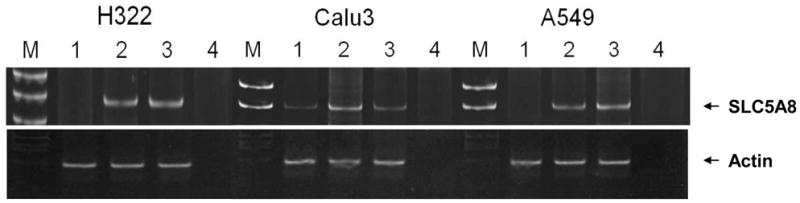
RT-PCR results of SLC5A8 transcripts from RNA isolated from lung cancer cell lines with either 5-Aza or TSA treatment. Lane 1: 5-Aza treatment, Lane 2: TSA treatment, Lane 3: 5-Aza and TSA treatment, Lane 4: negative control.
Quantitative methylation specific PCR (QMSP) assay and bisulfite sequencing of the promoter region of SLC5A8
QMSP was performed to evaluate the methylation status of SLC5A8 promoter in lung tissues (Fig. 3). Hypermethylation at the promoter region of SLC5A8 was primarily detected in lung tumor tissues as compared with that in adjacent non-tumor tissues (14/23, 60.9%). Average of log-transformed methylation levels, log [(SLC5A8/β-actin) × 1000] in the tumor are significantly higher than those in the adjacent non-tumor tissues (−1.16 vs. −2.09, p<0.01). The distribution of methylation levels and pair-wise comparisons are graphically illustrated in Fig. 3.
Fig. 3.
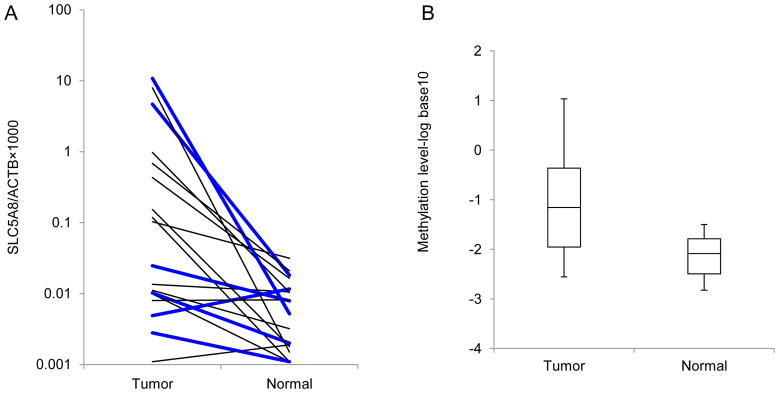
Results of QMSP analysis. (A) Methylation levels of SLC5A8 in tumor and adjacent non-tumor tissues from lung cancer patients were measured by QMSP. Calculation of SLC5A8/β-actin ratios was based on the fluorescence emission intensity values for both SLC5A8 and β-actin obtained by quantitative real-time PCR analysis. The relative amount of methylated promoter DNA was higher in tumor compared with adjacent non-tumor tissues (14/23, 60.9%). Results of 5 pairs were excluded because methylation levels of one of these pairs were below the limit of detection. Bold lines represent low SLC5A8 expression in tumors. (B) Distribution of methylation level in two groups is presented. Average methylation level (log10) is −1.16 and −2.09 in tumor and adjacent non-tumor tissues, respectively.
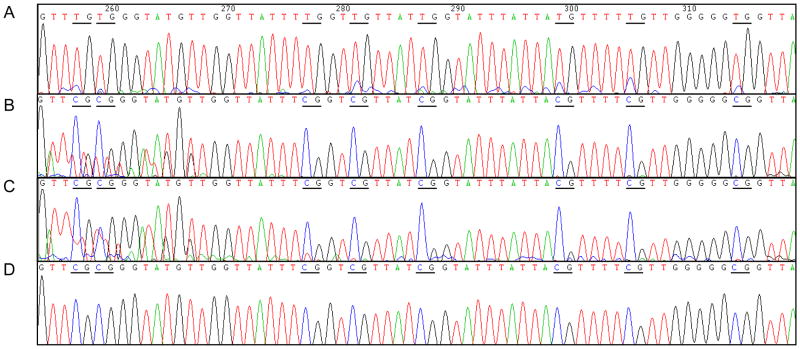
The extent of methylation in the SLC5A8 promoter region was also assessed in lung cancer cell lines and tumor/adjacent non-tumor tissues by bisulfite modified sequencing. Representative sequencing results are presented in Fig. 4. In the SLC5A8-silenced A549 lung cancer cell line and tumor tissues, most of CpG sites were methylated while the CpG sites were unmethylated in the adjacent non-tumor tissues.
Fig. 4.
Representative sequencing results demonstrate methylation status in the promoter region of SLC5A8 in DNA from adjacent non-tumor tissue, tumor tissue, lung cancer cell line, and DU145 cell. (A) Bisulfite-modified DNA sequence of SLC5A8 expressing adjacent non-tumor tissue. (B) Methylation of CpG sites in tumor tissue (C) Methylation of CpG sites in A549 lung cancer cell line. (D) Methylation of DU145 prostate cancer cell line was assessed as a positive control.
DISCUSSION
Among hypermethylated genes identified, SLC5A8 was suggested as a tumor suppressor gene. As previous studies described in other cancer sites [15, 16, 18–24, 27, 40], we observed DNA methylation in the promoter region of SLC5A8 and its association with gene silencing of SLC5A8 in lung cancer cell lines and tumor tissues. Among nine SLC5A8 down-regulated tumor tissues, five (56%) of them were hypermethylated, while two out of six high SLC5A8 expression group (33%) were hypermethylated (p>0.05). In addition, the SLC5A8 expression could be restored by treatment with the demethylating agent, 5-Aza (Calu3); HDAC inhibitor, TSA (H322, Calu3 and A549) or both inhibitors. Therefore, epigenetic gene silencing of SLC5A8, which is reflected by aberrant SLC5A8 methylation and/or histone modification, could represent a principal mechanism for inactivating this gene in lung tumor.
Since the SLC5A8 was firstly proposed as a tumor suppressor gene in colon cancer, many studies have reported methylated SLC5A8 in different cancer sites, such as brain, thyroid, gastric, colon, prostate, pancreas, breast and head & neck [11, 15, 16, 18–24] based upon QMSP which detect hypermethylated CpG sites responsible for SLC5A8 down-regulation. However, data regarding the relationship between DNA methylation and expression of SLC5A8 in lung tumor were not reported yet. Therefore, our present findings demonstrate the first evidence for epigenetic regulation of SLC5A8 in lung cancer, providing new insights into the role of SLC5A8 in tumorigenesis.
Li et al. [15] reported that 59% of colon tumors were methylated while only 5% of normal tissues were methylated. Moreover, DNA methylation in SLC5A8 appears an early event in carcinogenesis. Two independent studies reported that 60~80% of precursor lesions were methylated in SLC5A8 [4, 15]. Expression of SLC5A8 is also associated with survival of colon cancer patients. Paroder et al. [21] reported that expression of SLC5A8 in colon samples from colorectal cancer patients correlates with survival.
In thyroid cancer studies, 26–63% of thyroid tumor tissues were methylated [19, 20, 41]. Thyroid cell lines, which do not express SLC5A8, were densely methylated and reactivated by 5-Aza [20]. However, DNA methylation in SLC5A8 may not be an early event in thyroid tumorigenesis. Schagdarsurengin et al. [41] observed no methylation in the follicular adenoma (0/10). Hu et al. [20] reported that aberrant methylation of SLC5A8 is associated with progression of thyroid cancer although these results were not confirmed by other investigators [41].
Ueno et al. [16] reported that 30% (23/71) of gastric tumor tissues and most gastric cell lines (10/12) are methylated. Among the methylated cell lines, SLC5A8 was not expressed in most of the methylated cell lines. Methylated cell lines, which did not express SLC5A8, were reactivated by 5-Aza. Similar results were reported in prostate and pancreatic tumor, thus demonstrating that frequent methylation and low expression of the SLC5A8 gene product is observed in tumor tissues across cancer sites [22, 24] (Table 2).
Table 2.
DNA methylation in the promoter region of SLC5A8 and expression in various cancer sites
| Samples | Methylation | Method | Low expression | Method | Reactivation by 5-Aza |
|---|---|---|---|---|---|
| Brain cell lines | 2/2 (100)1 | Sequencing2 | 2/2 (100) | RT-PCR | 2/2 (100) |
| Brain tumor | 28/40 (70) | RLGS | 13/13 (100) | RT-PCR | |
|
| |||||
| Breast tumor | 27/30 (90) | Tissue array | |||
| Breast cell lines (transformed) | 3/3 (100) | RT-PCR | 3/3 (100) | ||
| Breast cell lines (non-transformed) | 0/3 (0) | RT-PCR | |||
|
| |||||
| Colon cell lines | 16/31 (52) | MSP | 23/31 (74) | RT-PCR | 6/8 (75) |
| Colon tumor | 33/40 (83) | MSP | |||
| Colon tumor | 38/64 (59) | MSP | |||
| Colon normal | 3/64 (5) | MSP | |||
| Colon adenoma | 17/29 (59) | MSP | |||
|
| |||||
| Gastric cell lines | 10/12 (83) | COBRA3 | 6/7 (86) | RT-PCR | 3/7 (43) |
| Gastric tumor | 23/71 (30) | COBRA | |||
|
| |||||
| Head and Neck | 9/40 (23) | MassARRAY | 4/10 (40) | ||
|
| |||||
| Lung tumor | 1/5 (20) | RLGS | |||
|
| |||||
| Lung tumor | 14/23 (61) | QMSP | 9/23 (39) | RT-PCR | 1/3 (33) |
|
| |||||
| Pancreas cell lines | 3/3 (100) | Sequencing | 3/3 (100) | RT-PCR | 3/3 (100) |
| Pancreas tumor | 7/10 (70) | MSP | 10/10 (100) | RT-PCR | |
| Pancreas normal | 3/28 (11) | MSP | 17/28 (61) | RT-PCR | |
|
| |||||
| Prostate cell lines | 2/2 (100) | Sequencing | 2/2 (100) | RT-PCR | 2/2 (100) |
| Prostate tumor | 7/10 (70) | QMSP | 7/10 (70) | RT-PCR | |
|
| |||||
| Thyroid cell lines | 3/3 (100) | QMSP | 3/3 (100) | RT-PCR | 2/3 (67) |
| Thyroid tumor | 76/231 (33) | QMSP | |||
| Thyroid tumor-cf | 16/18 (89) | MSP | 16/18 (89) | RT-PCR | |
| Thyroid cell lines | 1/4 (25) | MSP | |||
| Thyroid tumor | 9/34 (26) | MSP | |||
Numbers in bracket denote percentage
Bisulphite modified sequencing
COBRA: combined bisulfite restriction analysis
The tumor suppressive role of SLC5A8 was suggested by Ganapathy et al. [17]. The SLC5A8 is a plasma membrane transporter belonging to the Na+-coupled transporter gene family. The transporter mediates Na+-dependent and electrophilic transport of monocarboxylates such as, short-chain fatty acids. Butyrate, one of the substrates for SLC5A8, is a histone deacetylase inhibitor and is known to induce apoptosis in a variety of tumors. SLC5A8 mediates the concentrative entry of butyrate into epithelial cells. Consequently, the transport function of SLC5A8 has the ability to influence the acetylation status of histones and hence gene expression in cells. The ability of SLC5A8 to deliver butyrate into epithelial cells most likely underlies the tumor suppressive role of this transporter [9].
As we demonstrated, expression of SLC5A8 was reactivated by 5-Aza and/or TSA, which are used in clinical trials for various human cancers. Rationale for this approach is that this agent reactivates the defective expression of tumor suppressor genes. Previous studies reported that over-expression of SLC5A8 induced apoptosis in breast [11], and colon cancer cell lines [42]. Our data support that such a strategy might be a potential therapy for lung cancer.
In conclusion, the present study suggests that SLC5A8 plays an important role as a tumor suppressor in lung tumor and the reduced expression of SLC5A8 by promoter methylation could become a major molecular target for the diagnosis and treatment of lung cancer.
Supplementary Material
Acknowledgments
This work was supported in part by US National Institutes of Health by grant R03CA114713 (PI: Park, J). We are grateful to the Molecular Biology Core facility and Tissue Procurement facilities at Moffitt Cancer Center for excellent technical assistance.
Funding: This research was supported in part by the National Cancer Institute grant R03CA114713 (PI: Park, J).
Abbreviations
- 5-Aza
5-azacytidine
- HDAC
Histone deacetylase
- QMSP
Quantitative methylation specific PCR
- RLGS
Restriction landmark genomic scanning
- RT-PCR
Reverse transcription-polymerase chain reaction
- iodide transporter
Solute carrier family 5
- SLC5A8
member 8
- TSA
Trichostatin A
Footnotes
Conflicts of interest statement
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heller G, Zielinski CC, Zochbauer-Muller S. Lung cancer: from single-gene methylation to methylome profiling. Cancer Metastasis Rev. 2010;29:95–107. doi: 10.1007/s10555-010-9203-x. [DOI] [PubMed] [Google Scholar]
- 4.Dong SM, Lee EJ, Jeon ES, Park CK, Kim KM. Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol. 2005;18:170–8. doi: 10.1038/modpathol.3800261. [DOI] [PubMed] [Google Scholar]
- 5.Srinivas SR, Gopal E, Zhuang L, Itagaki S, Martin PM, Fei YJ, et al. Cloning and functional identification of slc5a12 as a sodium-coupled low-affinity transporter for monocarboxylates (SMCT2) Biochem J. 2005;392:655–64. doi: 10.1042/BJ20050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopal E, Miyauchi S, Martin PM, Ananth S, Roon P, Smith SB, et al. Transport of nicotinate and structurally related compounds by human SMCT1 (SLC5A8) and its relevance to drug transport in the mammalian intestinal tract. Pharm Res. 2007;24:575–84. doi: 10.1007/s11095-006-9176-1. [DOI] [PubMed] [Google Scholar]
- 7.Martin PM, Gopal E, Ananth S, Zhuang L, Itagaki S, Prasad BM, et al. Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ketone bodies in the brain. J Neurochem. 2006;98:279–88. doi: 10.1111/j.1471-4159.2006.03878.x. [DOI] [PubMed] [Google Scholar]
- 8.Miyauchi S, Gopal E, Babu E, Srinivas SR, Kubo Y, Umapathy NS, et al. Sodium-coupled electrogenic transport of pyroglutamate (5-oxoproline) via SLC5A8, a monocarboxylate transporter. Biochim Biophys Acta. 2010;1798:1164–71. doi: 10.1016/j.bbamem.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Thangaraju M, Cresci G, Itagaki S, Mellinger J, Browning DD, Berger FG, et al. Sodium-coupled transport of the short chain fatty acid butyrate by SLC5A8 and its relevance to colon cancer. J Gastrointest Surg. 2008;12:1773–81. doi: 10.1007/s11605-008-0573-0. discussion 81–2. [DOI] [PubMed] [Google Scholar]
- 10.Gupta N, Martin PM, Prasad PD, Ganapathy V. SLC5A8 (SMCT1)-mediated transport of butyrate forms the basis for the tumor suppressive function of the transporter. Life Sci. 2006;78:2419–25. doi: 10.1016/j.lfs.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 11.Thangaraju M, Gopal E, Martin PM, Ananth S, Smith SB, Prasad PD, et al. SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res. 2006;66:11560–4. doi: 10.1158/0008-5472.CAN-06-1950. [DOI] [PubMed] [Google Scholar]
- 12.Ganapathy V, Thangaraju M, Gopal E, Martin PM, Itagaki S, Miyauchi S, et al. Sodium-coupled monocarboxylate transporters in normal tissues and in cancer. AAPS J. 2008;10:193–9. doi: 10.1208/s12248-008-9022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thangaraju M, Karunakaran SK, Itagaki S, Gopal E, Elangovan S, Prasad PD, et al. Transport by SLC5A8 with subsequent inhibition of histone deacetylase 1 (HDAC1) and HDAC3 underlies the antitumor activity of 3-bromopyruvate. Cancer. 2009;115:4655–66. doi: 10.1002/cncr.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. 2010;285:27601–8. doi: 10.1074/jbc.M110.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Kasturi L, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci U S A. 2003;100:8412–7. doi: 10.1073/pnas.1430846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueno M, Toyota M, Akino K, Suzuki H, Kusano M, Satoh A, et al. Aberrant methylation and histone deacetylation associated with silencing of SLC5A8 in gastric cancer. Tumour Biol. 2004;25:134–40. doi: 10.1159/000079145. [DOI] [PubMed] [Google Scholar]
- 17.Ganapathy V, Gopal E, Miyauchi S, Prasad PD. Biological functions of SLC5A8, a candidate tumour suppressor. Biochem Soc Trans. 2005;33:237–40. doi: 10.1042/BST0330237. [DOI] [PubMed] [Google Scholar]
- 18.Hong C, Maunakea A, Jun P, Bollen AW, Hodgson JG, Goldenberg DD, et al. Shared epigenetic mechanisms in human and mouse gliomas inactivate expression of the growth suppressor SLC5A8. Cancer Res. 2005;65:3617–23. doi: 10.1158/0008-5472.CAN-05-0048. [DOI] [PubMed] [Google Scholar]
- 19.Porra V, Ferraro-Peyret C, Durand C, Selmi-Ruby S, Giroud H, Berger-Dutrieux N, et al. Silencing of the tumor suppressor gene SLC5A8 is associated with BRAF mutations in classical papillary thyroid carcinomas. J Clin Endocrinol Metab. 2005;90:3028–35. doi: 10.1210/jc.2004-1394. [DOI] [PubMed] [Google Scholar]
- 20.Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer. 2006;119:2322–9. doi: 10.1002/ijc.22110. [DOI] [PubMed] [Google Scholar]
- 21.Paroder V, Spencer SR, Paroder M, Arango D, Schwartz S, Jr, Mariadason JM, et al. Na(+)/monocarboxylate transport (SMCT) protein expression correlates with survival in colon cancer: molecular characterization of SMCT. Proc Natl Acad Sci U S A. 2006;103:7270–5. doi: 10.1073/pnas.0602365103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JY, Zheng W, Kim D, Cheng JQ, Kumar N, Ahmad N, et al. Candidate tumor suppressor gene SLC5A8 is frequently down-regulated by promoter hypermethylation in prostate tumor. Cancer Detect Prev. 2007;31:359–65. doi: 10.1016/j.cdp.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Bennett KL, Karpenko M, Lin MT, Claus R, Arab K, Dyckhoff G, et al. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008;68:4494–9. doi: 10.1158/0008-5472.CAN-07-6509. [DOI] [PubMed] [Google Scholar]
- 24.Park JY, Helm JF, Zheng W, Ly QP, Hodul PJ, Centeno BA, et al. Silencing of the candidate tumor suppressor gene solute carrier family 5 member 8 (SLC5A8) in human pancreatic cancer. Pancreas. 2008;36:e32–9. doi: 10.1097/MPA.0b013e3181630ffe. [DOI] [PubMed] [Google Scholar]
- 25.Whitman SP, Hackanson B, Liyanarachchi S, Liu S, Rush LJ, Maharry K, et al. DNA hypermethylation and epigenetic silencing of the tumor suppressor gene, SLC5A8, in acute myeloid leukemia with the MLL partial tandem duplication. Blood. 2008;112:2013–6. doi: 10.1182/blood-2008-01-128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr CA, Dunne R, Hines BM, Zucker M, Cosgrove L, Ruszkiewicz A, et al. Measuring the combinatorial expression of solute transporters and metalloproteinases transcripts in colorectal cancer. BMC Res Notes. 2009;2:164. doi: 10.1186/1756-0500-2-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin HY, Park HY, Radlein S, Mahajan NP, Sellers TA, Zachariah B, et al. Protein expressions and genetic variations of SLC5A8 in prostate cancer risk and aggressiveness. Urology. 2011;78:971, e1–9. doi: 10.1016/j.urology.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, Brena RM, Gruidl M, Zhou J, Huang T, Plass C, et al. CpG island hypermethylation profiling of lung cancer using restriction landmark genomic scanning (RLGS) analysis. Cancer Biomark. 2005;1:193–200. doi: 10.3233/cbm-2005-12-307. [DOI] [PubMed] [Google Scholar]
- 29.Costello JF, Hong C, Plass C, Smiraglia DJ. Restriction landmark genomic scanning: analysis of CpG islands in genomes by 2D gel electrophoresis. Methods Mol Biol. 2009;507:131–48. doi: 10.1007/978-1-59745-522-0_11. [DOI] [PubMed] [Google Scholar]
- 30.Okuizumi H, Takamiya T, Okazaki Y, Hayashizaki Y. Restriction landmark genome scanning. Methods Mol Biol. 2011;791:101–12. doi: 10.1007/978-1-61779-316-5_8. [DOI] [PubMed] [Google Scholar]
- 31.Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–7. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 32.Okino ST, Pookot D, Li LC, Zhao H, Urakami S, Shiina H, et al. Epigenetic inactivation of the dioxin-responsive cytochrome P4501A1 gene in human prostate cancer. Cancer Res. 2006;66:7420–8. doi: 10.1158/0008-5472.CAN-06-0504. [DOI] [PubMed] [Google Scholar]
- 33.Plass C, Weichenhan D, Catanese J, Costello JF, Yu F, Yu L, et al. An arrayed human not I-EcoRV boundary library as a tool for RLGS spot analysis. DNA Res. 1997;4:253–5. doi: 10.1093/dnares/4.3.253. [DOI] [PubMed] [Google Scholar]
- 34.Smiraglia DJ, Fruhwald MC, Costello JF, McCormick SP, Dai Z, Peltomaki P, et al. A new tool for the rapid cloning of amplified and hypermethylated human DNA sequences from restriction landmark genome scanning gels. Genomics. 1999;58:254–62. doi: 10.1006/geno.1999.5840. [DOI] [PubMed] [Google Scholar]
- 35.Smiraglia DJ, Kazhiyur-Mannar R, Oakes CC, Wu YZ, Liang P, Ansari T, et al. Restriction landmark genomic scanning (RLGS) spot identification by second generation virtual RLGS in multiple genomes with multiple enzyme combinations. BMC Genomics. 2007;8:446. doi: 10.1186/1471-2164-8-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng X, Wang F, Zhang L, Zhang WM. Loss of heterozygosity combined with promoter hypermethylation, the main mechanism of human MutL Homolog (hMLH1) gene inactivation in non-small cell lung cancer in a Chinese population. Tumori. 2009;95:488–94. doi: 10.1177/030089160909500414. [DOI] [PubMed] [Google Scholar]
- 37.Ogiwara H, Kohno T, Nakanishi H, Nagayama K, Sato M, Yokota J. Unbalanced translocation, a major chromosome alteration causing loss of heterozygosity in human lung cancer. Oncogene. 2008;27:4788–97. doi: 10.1038/onc.2008.113. [DOI] [PubMed] [Google Scholar]
- 38.Tseng RC, Chang JW, Hsien FJ, Chang YH, Hsiao CF, Chen JT, et al. Genomewide loss of heterozygosity and its clinical associations in non small cell lung cancer. Int J Cancer. 2005;117:241–7. doi: 10.1002/ijc.21178. [DOI] [PubMed] [Google Scholar]
- 39.Ho WL, Chang JW, Tseng RC, Chen JT, Chen CY, Jou YS, et al. Loss of heterozygosity at loci of candidate tumor suppressor genes in microdissected primary non-small cell lung cancer. Cancer Detect Prev. 2002;26:343–9. doi: 10.1016/s0361-090x(02)00088-0. [DOI] [PubMed] [Google Scholar]
- 40.Brim H, Kumar K, Nazarian J, Hathout Y, Jafarian A, Lee E, et al. SLC5A8 gene, a transporter of butyrate: a gut flora metabolite, is frequently methylated in African American colon adenomas. PLoS One. 2011;6:e20216. doi: 10.1371/journal.pone.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schagdarsurengin U, Gimm O, Dralle H, Hoang-Vu C, Dammann R. CpG island methylation of tumor-related promoters occurs preferentially in undifferentiated carcinoma. Thyroid. 2006;16:633–42. doi: 10.1089/thy.2006.16.633. [DOI] [PubMed] [Google Scholar]
- 42.Babu E, Ramachandran S, CoothanKandaswamy V, Elangovan S, Prasad PD, Ganapathy V, et al. Role of SLC5A8, a plasma membrane transporter and a tumor suppressor, in the antitumor activity of dichloroacetate. Oncogene. 2011;30:4026–37. doi: 10.1038/onc.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.