Abstract
Purpose
Determine if blood mRNA expression patterns in children with newly diagnosed untreated idiopathic epilepsy are different than those in healthy controls. Determine the differential expression patterns between epilepsy subjects with generalized onset or partial onset seizures compared to healthy controls.
Methods
Whole blood was obtained from otherwise healthy pediatric subjects with newly diagnosed untreated idiopathic epilepsy along with healthy pediatric controls. mRNA was isolated and hybridized to Affymetrix HGU 133 2.0+ microarrays. Analysis was performed using Genespring. Differentially expressed gene lists resulted from comparison of i) epilepsy and control groups and ii) seizure type subgroups with controls.. Tissue expression and gene ontology analysis was performed using DAVID.
Key findings
Thirty-seven epilepsy patients and 28 controls were included. Overall, 575 genes were differentially expressed in subjects with epilepsy compared to controls. The generalized seizure subgroup versus control (GvC) gene list and the partial seizure subgroup versus control (PvC) gene list were different (p < 0.05). Tissue expression analysis identified almost half of the genes in GvC and PvC as brain based. Functional group analysis identified several biologically relevant pathways. In GvC, these included mitochondria and lymphocyte activation. In PvC, apoptosis, inflammatory defense and cell motion pathways were identified.
Significance
A unique, biologically meaningful mRNA expression pattern is detectable in whole blood of pediatric subjects with new onset and untreated epilepsy. This analysis finds many similar pathways to those identified in brain studies examining lesional intractable epilepsy. Blood mRNA expression patterns show promise as a target for biomarker development in pediatric epilepsy.
Keywords: Gene expression, mRNA, pediatric epilepsy, genomics, epilepsy
Introduction
Epilepsy is a common pediatric disease, with a lifetime prevalence of about 1% in the United States.(Russ et al., 2012) Half the people with epilepsy begin having seizures during childhood.(Wyllie E & Linehan C, 2011) The underlying pathophysiologic mechanism of epileptogenesis is not yet completely elucidated but appears to involve genome-environment interactions.(Crino, 2007) mRNA expression pattern profiling may be useful to understand these interactions since it allows for the study of genome-wide changes.(Sharp et al., 2006) Prior expression studies in epilepsy have focused on brain tissue obtained from epilepsy surgery.(Becker et al., 2002; Jamali et al., 2006; Lee et al., 2007; Xi et al., 2009) However, most children with epilepsy do not undergo epilepsy surgery, and those that do represent a unique, medically refractory population that may not be generalizable. Therefore demonstration of differential expression in blood would be important both for investigation into pathophysiology and the development of biomarkers.
The feasibility of blood biomarker development for neurological disorders has been investigated in animal models. In studying rat models of status epilepticus, stroke and trauma, Liu et al demonstrated differential genomic expression patterns for each mechanism of injury common to both brain and blood.(Liu et al., 2010) Several studies demonstrate meaningful results in human blood expression studies of neurological conditions. Unique mRNA expression patterns were measurable in patients with neurofibromatosis type 1 and tuberous sclerosis complex type 2.(Tang et al., 2004b) More recently, it was shown that blood mRNA expression patterns could be a useful biomarker for acute migraine, medication overuse headaches and menstrual-related migraine.(Hershey et al., 2012; Hershey et al., 2011; Hershey et al., 2004) In epilepsy, unique genomic expression patterns associated with chronic valproic acid administration have been demonstrated.(Tang et al., 2004a)
The current study was designed to examine the hypothesis that peripheral blood mRNA expression patterns in children with newly diagnosed untreated epilepsy were different than those in matched healthy controls. A secondary aim was to determine the differential expression patterns between subjects with generalized onset and partial onset epilepsy phenotypes compared to healthy controls.
Methods
Population
Subjects were consecutively identified through the new onset seizure clinic at Cincinnati Children’s Hospital Medical Center (CCHMC) over a 16 month period. This clinic evaluates children with a suspected first time seizure or suspected new onset epilepsy within 10 days of referral from an 8 county region of Southwestern Ohio. Subjects were eligible if they were 5.5–18 years old (inclusive newly diagnosed with idiopathic epilepsy (defined by normal development, neurologic examination and MRI with seizure and EEG characteristics consistent with the 1989 ILAE classification criteria for either syndromic or non-syndromic idiopathic epilepsy), previously untreated with anticonvulsants, had no serious chronic medical issues, were not taking any chronic central nervous system medications, and had a body mass index between the 5% and 95% (exclusive) for age. Informed consent was obtained. The first pre-drug blood samples were used for this study. The final epilepsy cohort was determined after dChip outliers (n=4) were excluded and prior to comparison to controls or seizure type analysis performed.
The patient's seizure type and epilepsy classification were classified by an epileptologist according to the 1981 and 1989 International League Against Epilepsy's Classification for Seizure, Epilepsies, and Epilepsy Syndromes.(1989) Subjects were categorized as either epilepsy with generalized onset seizures (G cohort), epilepsy with partial onset seizures (P cohort) or healthy control (C cohort) (Figure). G cohort included syndromes such as childhood absence epilepsy and juvenile myoclonic epilepsy and idiopathic generalized epilepsy. P cohort included syndromes such as benign epilepsy with centrotemporal spikes and idiopathic partial epilepsy.
Healthy controls were between 5.5–18 years old (inclusive), with no history of seizures, epilepsy, chronic medical issues, medications or concomitant illnesses. Demographic information was obtained at the time of the blood draw, and no identifying information was kept for the controls. Controls were consented as part of a healthy control group in a genomic study of tic disorders and not specifically chosen as a matched cohort. Controls did not have a tic disorder or any other neurological conditions. The study was approved by the Institutional Review Board at Cincinnati Children’s Hospital Medical Center.
Blood collection, RNA isolation, and microarray hybridization and normalization
Whole blood was obtained prior to initiation of anticonvulsants. Blood sample collection, mRNA isolation, microarray hybridization and normalization were performed as described previously.(Hershey et al., 2008) Whole blood was collected into 6 Paxgene Blood RNA tubes, RNA isolated using Paxgene Blood RNA Kit and concentrated using RNeasy MinElute Cleanup Kit (Qiagen,Valencia, CA, USA <http://www.preanalytix.com/RNA-Instr.asp>). mRNA was assessed for concentration by spectrophotometry (1 mg/ml total RNA) and for quality using the ratio of 28S:18S ribosomal RNA with Agilent Bioanalyzer 2100 (<http://www.chem.agilent.com>).
Resulting RNA was hybridized to Affymetrix HGU133 2.0+ microarrays, which analyze over 47,000 probesets, (<http://www.affymetrix.com>) using the standard Affymetrix labeling protocol (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). This resulted in a single cell intensity file (CEL) for each subject. Quality of resulting unnormalized microarray data was inspected using dChip, v2005 (DNA-Chip analyzer (<http://www.dchip.org>). Microarrays with greater than 4.5% outliers were excluded. Microarrays passing quality control criteria were normalized using robust multichip average (RMA).(Hsu et al., 2007) For each chip the MAS5 algorithm was used on the Affymetrix Expression Console Program to assign present, absent or marginal calls for each probeset, resulting in a percentage present for an alpha of p=0.05.
Statistical Analysis
Experiments were performed using Genespring GX v10 (Agilent Technologies, Santa Clara, CA, USA, <www.genomics.agilent.com>). Experiment 1 examined the differential expression patterns for all epilepsy patients (G + P=E) compared to healthy controls (C). Experiment 2 examined the differential expression patterns between each seizure group. Experiment 3 compared the gene lists obtained from the GvC, PvC and EvC analyses. In each experiment the analysis produced a list of those genes which were differently expressed between study groups.
Minimum average fold change cut-offs were made at both 1.3 and 1.5 for each experiment, generating two lists of differentially expressed probesets for each experimental setup. The lists based on the 1.5 fold change were analyzed to determine if they were strict subsets of those based on the 1.3 fold change. Resulting probesets are considered to correlate with gene expression, as over two-thirds of probesets represent specific genes.(Hershey et al., 2011) An unpaired Mann-Whitney test was performed on each gene list. All genes demonstrating significantly different expression were analyzed (p<0.05, Benjamini-Hochberg False Discovery Rate, or FDR).(Benjamini Y, 1995)
Resulting gene lists were analyzed for over-representation in tissue expression, biological pathway and gene ontology using DAVID 6.7 (Database for Annotation, Visualization and Integrated Discovery, NIAID/NIH <http://david.abcc.ncifcrf.gov/>). In this analysis, a minimum EASE score of 1 was used as a cutoff for gene pathway significance. The EASE is a modified Fisher’s exact test that estimates enrichment of a given probeset compared to that expected by chance alone.
Results
Subject characteristics
The study population consisted of 65 subjects: 37 epilepsy and 28 healthy controls. The epilepsy cohort was younger (9.9 ± 3.4 years vs 11.9 ± 2.9 years, p=0.014) with slightly fewer males (54% vs.75%, p=0.12) (Table 1). The epilepsy cohort consisted of 15 subjects with generalized epilepsy and 22 subjects with partial epilepsy. Seven epilepsy subjects were taking a daily medication for a non-neurological condition (such as seasonal allergies) at the time of the blood draw. No child was on the same medication. Syndrome diagnoses include childhood absence epilepsy (n=9), juvenile myoclonic epilepsy (n=2), and benign epilepsy with centrotemporal spikes (n=3). All other participants had idiopathic generalized or idiopathic partial epilepsy. For all chips analyzed, the mean percentage of probesets called present was 40.3% (±2.8%). Results are reported for each experiment using a 1.3 minimum fold change cutoff and a 1.5 minimum fold change cutoff when a significant result was obtained using the stricter threshold. After the Mann Whitney tests the gene lists from the 1.5 fold change remained proper subsets of those resulting from the 1.3 fold change.
Table 1.
Demographics
| Epilepsy | Control | |
|---|---|---|
| # | 37 | 28 |
| Age (years) | 9.9 (±3.4) | 11.9 (±2.9)* |
| Gender (F:M) | 17:20 | 7:21 (n.s.) |
| Race | ||
| Asian | 1 | |
| Black | 4 | 2 |
| White | 32 | 26 |
p-value = 0.014 for mean difference between Epilepsy and Control groups (Student’s T-test, 2-tailed).
Epilepsy versus Controls
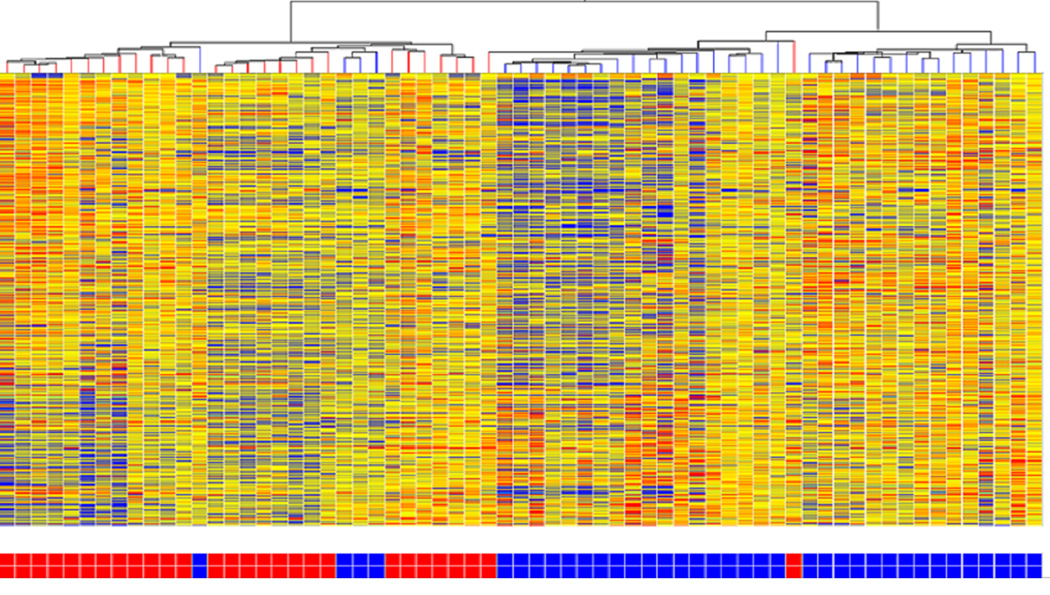
When all epilepsy patients were grouped together, 575 probesets were differentially expressed at a minimum fold change of 1.3 with 289 (50.3%) showing decreased expression. The microarray hierarchical cluster analysis revealed a distinct mRNA expression pattern in epilepsy patients compared to the controls (Fig 1).
Figure 1.
Hierarchical cluster analysis of expression patterns of all genes expressed at a significantly different level between Epilepsy (Partial and Generalized) and Controls. Epilepsy subjects are represented by blue boxes at the bottom of the figure and controls by red boxes. There is evident clustering of epilepsy subjects. Individual subjects are represented in each column. Individual probesets are represented by each row. Hierarchical cluster analysis groups subjects and probesets that are most alike together and presented as a branching pattern of subjects (top) and probesets (left). Subjects within the same branch are the most similar. For individual probesets, red indicates that the probeset is expressed at a higher level of expression than the average expression for that probeset, while blue represents a lower level of expression.
Epilepsy subgroups versus Controls
At a minimum fold change of 1.3, there were significant differences in expression between the G and C cohorts and also between the P and C cohorts; with 740 and 806 probesets identified, respectively. For the GvC analysis, 455/740 (60.8%) differentially expressed probesets had decreased expression in G compared to C. For the PvC analysis, 538/806 (66.7%) had increased expression in P compared to C. A GvP comparison yielded only 127 probeset expression differences. In G and P, most of the probesets identified were not overlapping, indicating potentially unique mRNA expression patterns.
Gene expression differences in GvC, PvC and EvC
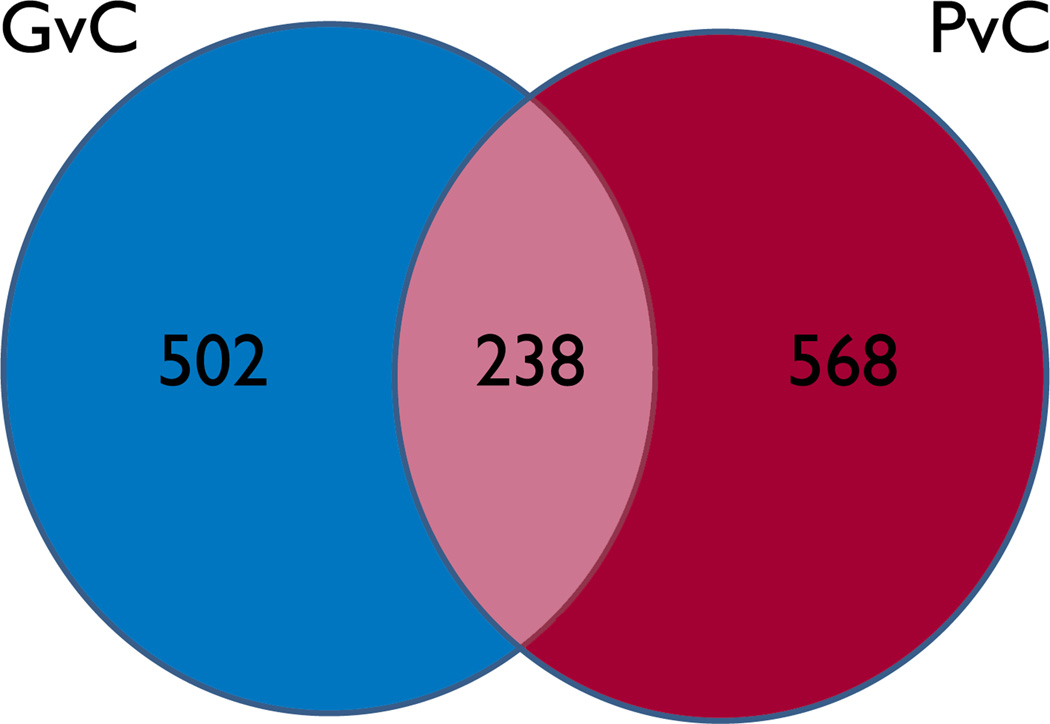
The gene lists obtained after the G versus C (GvC), P versus C (PvC) and both G and P versus C (EvC) experiments were compared for differential expression. At a minimum fold change of 1.3, 502 probesets were differentially expressed in GvC and not overlapping with PvC (Fig 2). In comparison, for PvC, 568 genes were not overlapping with GvC. There were 238 overlapping probesets (genes differentially expressed in both GvC and PvC). Only 9 genes were differentially expressed in EvC and not overlapping in either GvC or PvC. At a minimum fold change of 1.5, 63 probesets were differentially expressed in GvC and not overlapping with PvC. For PvC, 87 probesets were differentially expressed. There were only 27 differentially expressed overlapping probesets in GvC and PvC.
Figure 2.
Generalized v Control (GvC) in blue. Partial v Control (PvC) in red. Genes differently expressed in both lists shown in center. Epilepsy(G+P)vC genes not differently expressed in GvC or PvC (n=9) not shown.
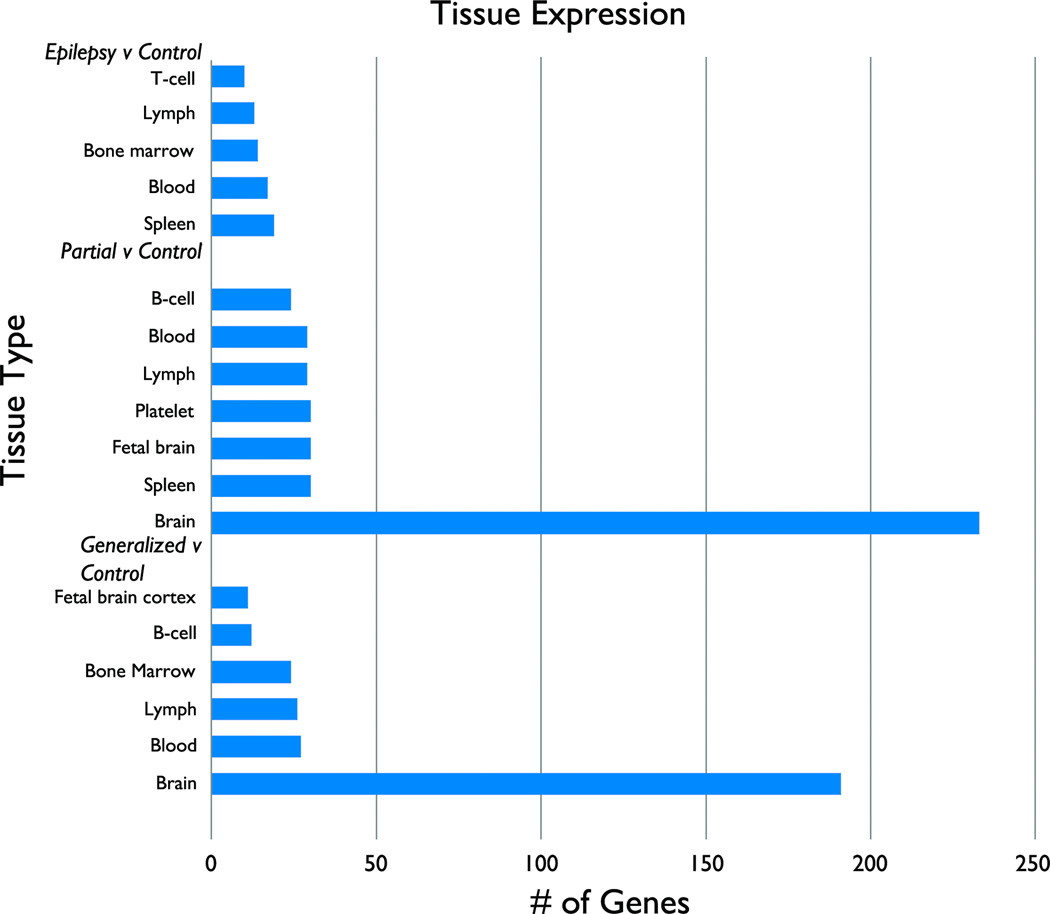
Tissue expression patterns
Using DAVID, the probesets identified in GvC, PvC and EvC were analyzed to determine which tissues were represented more than expected by chance alone. For the 502 gene list generated from GvC, two biologically relevant tissue types were represented: brain (191 genes, p=0.022) was most represented, but 27 other tissues including immunologic tissue were represented. For the 568 gene list generated from PvC, brain (233 genes, p=.0005) was again the most represented. Twenty-five other tissue types, including immunologic tissue, were represented (Fig 3).
Figure 3.
Select biologically relevant tissue expression in Epilepsy (G and P)v Control (Top); Partial v C (Middle); and Generalized v C (Bottom).
Biological pathways
DAVID was used to determine which biological pathways were over-represented in GvC, PvC and EvC. At a minimum fold change of 1.3, 22 pathways were identified in GvC, many of which were biologically relevant. These include consistently identified mitochondrial membrane and respiratory chain pathways, protein kinase, secretory and metal ion transport pathways. In PvC, 67 pathways were identified. Inflammatory and immunological “defense response” pathways, including several involved in positive regulation of lymphocyte differentiation, were identified. Additionally, apoptosis pathways were found. Further, cell motion, protein transport, secretory, protein kinase, metal ion transport (such as zinc fingers) and cell signaling pathways were consistently identified. In EvC, 22 pathways were identified. There was activation of both apoptotic and anti-apoptotic, protein kinase and phosphorylation pathways (Table 2).
Table 2.
Select differentially expressed pathways. Pathways were selected on basis of highest relevance, highest EASE scores and recurrence on gene ontology list.
| Fold Change 1.3 | Fold Change 1.5 | |
|---|---|---|
| Generalized v Control (total # of pathways =22) | Lymphocyte activation | Lymphocyte activation |
| Mitochondrial membrane | ||
| Respiratory chain | ||
| Protein kinase | ||
| Secretory | ||
| Metal ion transport | ||
| Partial v Control (total # of pathways=67) | Lymphocyte activation | |
| Inflammatory defense | Inflammatory defense | |
| Cell motion | Cell motion | |
| Protein transport | ||
| Secretory | ||
| Protein kinase | ||
| Apoptosis | Apoptosis |
At a minimum fold change of 1.5, only 2 pathways were identified in GvC: C-type lectin and lymphocyte activation. For PvC, 5 pathways were identified. Two were apoptosis pathways. Also identified were cell motion and two inflammatory pathways.
Discussion
Differences in epilepsy
It is hypothesized that epilepsy is caused by complex interactions between the genome and the environment; few genetic disorders follow a simple Mendelian inheritance pattern.(Grill et al., 2008) This genome-environment interaction requires a variety of technologies to dissect its complexity. In our study, we demonstrate specific blood mRNA expression patterns that differentiate epilepsy (and its subgroups) from healthy controls. Based on tissue expression analysis, half of the identified genes are highly expressed in brain. This result suggests the utility of blood expression studies to detect changes associated with primarily central nervous system disorders.(Hershey et al., 2011; Jickling et al., 2010; Tian et al., 2011) The current report shows several candidate pathways differently expressed in the blood of pediatric patients with new onset epilepsy. Relevant pathways identified include mitochondrial and respiratory chain, inflammatory and defense response, cell motion, apoptosis, secretory, cell signaling, metal ion transport and protein kinase pathways.
Differences in GvC and PvC
PvC demonstrated a larger, more robust and biologically relevant gene list than GvC. Inflammatory pathways were only identified in PvC. In GvC, while there were fewer significant pathways overall, there was evidence of differential expression of nuclear mitochondrial genes not present in PvC. Differential expression of inflammatory pathways in partial epilepsy suggests there may be a larger inflammatory contribution in the promotion of epileptogenesis in these subjects. However, there may be bias in the generalized epilepsy subgroup toward nonconvulsive seizures. Of the 22 patients with generalized epilepsy, 9 had childhood absence epilepsy (CAE), and many others had a diagnosis of idiopathic generalized epilepsy which may manifest predominantly absence seizures. It is difficult to interpret the result of phenotypic variability between generalized and partial epilepsy found in our study in the context of the published literature. Previous publications have mostly studied brain in medically intractable partial epilepsy human subjects and animal models. Widescale whole genome mRNA expression studies of CAE patients have not been published. Prior work using the genetic absence epilepsy rats from Strasbourg (GAERS) model demonstrate some mRNA expression differences in more select mRNAs isolated from brain.(Lakaye et al., 2002; Powell et al., 2008)
Inflammation
Inflammation appears to be an important factor in epileptogenesis, playing a role in treatment resistance and failure to terminate seizures, resulting in status epilepticus.(Granata et al., 2011; Marchi et al., 2011; Vezzani et al., 2011) The role of inflammation is evident in several well-recognized principally immune/inflammation-mediated encephalitides.(Dalmau et al., 2008; Pardo et al., 2004; Specchio et al., 2010) However there is evidence to support the role of inflammation in other less explosive epilepsy phenotypes.
Previous mRNA expression studies in different models showed a relationship between pro-inflammatory cytokines and cell death, with the cytokines perpetuating the apoptosis pathway.(Vezzani et al., 2011) In a recent animal study by Winden et al, lesioned rats were found to have increased expression of inflammatory and cell death pathways, but subsequent analysis led the authors to conclude this was due to the effects of the lesion rather than seizures.(Winden et al., 2011) In our group of human new onset epilepsy subjects, there was dysregulation of several chemokines and pro-inflammatory factors such as tumor necrosis factor-alpha, interleukin 8, toll like receptor 8 and several chemokines including CCL2 and CXCR4. This is consistent with a recent meta-analysis of 12 large-scale expression studies of brain tissue in medically refractory patients submitted to epilepsy surgery. The authors of that study concluded that CCL2 and CXCR4 may be implicated in increased seizure susceptibility.(Mirza et al., 2011)
Mitochondria
The role of mitochondrial gene expression in epilepsy is unclear but promising. Mitochondria are uniquely vulnerable to oxidative stress, and any resulting oxidative damage may result in increased seizure susceptibility.(Waldbaum & Patel, 2010) It is increasingly recognized that several commonly used anticonvulsants can negatively affect mitochondrial function.(Berger et al., 2010) In our study, several mitochondrial genes were identified, including those involved in the respiratory chain as well as reactive oxygen species modulator 1, which has a role in formation of reactive oxygen species, and thioredoxin, which protects against oxidative stress-induced apoptosis. Ideally, in the future this information could be used to identify patients who may either 1) benefit from drugs that improve mitochondrial resistance to oxidative damage or 2) are at higher risk for no improvement or worsening with certain anticonvulsants based on mRNA expression patterns.
Apoptosis
The consistent identification of apoptosis pathways is a striking finding in children with new onset epilepsy. Dysregulation of apoptosis has been demonstrated in chronic epilepsy animal models and humans with medically refractory epilepsy. In one review of nine large-scale expression studies and humans and rodents, the most common functional category of genes identified was cell death/apoptosis.(Lukasiuk & Pitkanen, 2004) This is consistently found in epileptic brain tissue, both in mRNA expression studies and immunohistochemistry identification of apoptotic proteins.(Chamberlain & Prayson, 2008; Engel et al., 2007; Liu et al., 1999; Mielke et al., 1999; Xi et al., 2009) In our sample, subjects were seen in clinic generally several days after a seizure. The continued presence of a pro-apoptotic pattern in these patients, demonstrates the sensitivity of blood genomics for identification of biologically relevant biomarkers of central nervous system disease.
Other important pathways
Several other biologically relevant pathways were identified, including cell motion, secretory, cell signaling and metal ion transport. There were numerous genes identified involved in zinc interaction and zinc finger genes. Increased expression of zinc finger genes may be an early predictor of apoptosis.(Honkaniemi & Sharp, 1999; Winden et al., 2011)
Future directions
The current study has several limitations. Although there was a non-significant difference in gender between groups, the control cohort was two years older than the epilepsy subjects. This statistical difference is unlikely to cause the identified global difference in gene expression. A previous study identified some age-related differences in immune pathways in subjects with Tourette syndrome.(Lit et al., 2009) Secondly, differential gene expression data was generated after an arbitrary threshold cutoff (1.3 fold change) was applied. This is problematic since many important genes and pathways may not arise out of the noise. Compared to oncologic conditions, the expression pattern changes found in blood for neurological disorders are more subtle and generally lower. Sharp et al discuss the relatively lower fold change in gene expression patterns creates results that are difficult to replicate.(Sharp et al., 2006) Experience from our group indicates that the fold-change range of 1.3–1.5 is suitable for detecting differential expression patterns of neurological disorders in blood.(Hershey et al., 2012; Hershey et al., 2011) A “supervised” approach to tissue and functional pathway analysis was used. All data reaching statistical significance were analyzed, but only biologically relevant pathways were selected for closer examination and discussion. Future studies may benefit from an “unsupervised” approach, as described by Winden et al.(Winden et al., 2011)
The type of sample analyzed in an expression study impacts the data acquired. We report an analysis of whole blood, however there is a growing literature examining microRNA (non-coding RNA) in serum. Gallo et al demonstrate that most microRNAs exist within microvesicles circulating in bodily fluids, and suggest microRNAs could play an important role in biomarker development.(Gallo et al., 2012) In this regard, analyzing serum may offer an advantage to sampling individual cell types such as leukocytes, where the signal would not be present at all, or sampling whole blood, where the signal may be too dilute and lead to false negative results.
Conclusion
Differences in genomic expression can be measured in the blood of children with epilepsy; expression is already altered at the time of presentation. This study represents an early step in the important search for biomarkers of epileptogenesis and epileptogenicity.(Engel, 2011) Compared to healthy controls, there is differential expression in several important pathways including those related to inflammation, apoptosis and mitochondrial function. Only children with partial epilepsy had evidence of dysregulation of inflammatory pathways in our sample.
Acknowledgments
The authors acknowledge Don Gilbert, MD MS, for contributing data for controls.
This work was supported in part by NINDS NS044956 (TAG)
TAG receives support from the NINDS, NLM and Oxley Foundation. He has served as a paid consultant for the following companies: Supernus, Sunovion, Eisai, Upsher Smith, Questor, Lundbeck and AssureRx.
ADH has served as a paid consultant for MAP Pharma and Allergan.
KDH receives support from the NINDS and has received compensation for presentation of clinical trials results from Novartis.
Footnotes
The other authors have nothing to disclose.
References
- Proposal for revised classification of epilepsies and epileptic syndromes. Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia. 1989;30:389–399. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- Becker AJ, Chen J, Paus S, Normann S, Beck H, Elger CE, Wiestler OD, Blumcke I. Transcriptional profiling in human epilepsy: expression array and single cell real-time qRT-PCR analysis reveal distinct cellular gene regulation. Neuroreport. 2002;13:1327–1333. doi: 10.1097/00001756-200207190-00023. [DOI] [PubMed] [Google Scholar]
- Benjamini YHY. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;Vol. 57(No. 1):289–300. (1995), Key: citeulike:1042553 Series B (Methodological), Vol. 57:289–300. [Google Scholar]
- Berger I, Segal I, Shmueli D, Saada A. The effect of antiepileptic drugs on mitochondrial activity: a pilot study. J Child Neurol. 2010;25:541–545. doi: 10.1177/0883073809352888. [DOI] [PubMed] [Google Scholar]
- Chamberlain WA, Prayson RA. Focal cortical dysplasia type II (malformations of cortical development) aberrantly expresses apoptotic proteins. Appl Immunohistochem Mol Morphol. 2008;16:471–476. doi: 10.1097/PAI.0b013e31815d9ac7. [DOI] [PubMed] [Google Scholar]
- Crino PB. Gene expression, genetics, and genomics in epilepsy: some answers, more questions. Epilepsia. 2007;2(48 Suppl):42–50. doi: 10.1111/j.1528-1167.2007.01066.x. [DOI] [PubMed] [Google Scholar]
- Dalmau J, Gleichman AJ, Hughes EG, Rossi JE, Peng X, Lai M, Dessain SK, Rosenfeld MR, Balice-Gordon R, Lynch DR. Anti-NMDA-receptor encephalitis: case series and analysis of the effects of antibodies. Lancet Neurol. 2008;7:1091–1098. doi: 10.1016/S1474-4422(08)70224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J., Jr Biomarkers in epilepsy: introduction. Biomark Med. 2011;5:537–544. doi: 10.2217/bmm.11.62. [DOI] [PubMed] [Google Scholar]
- Engel T, Murphy BM, Schindler CK, Henshall DC. Elevated p53 and lower MDM2 expression in hippocampus from patients with intractable temporal lobe epilepsy. Epilepsy Res. 2007;77:151–156. doi: 10.1016/j.eplepsyres.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata T, Cross H, Theodore W, Avanzini G. Immune-mediated epilepsies. Epilepsia. 2011;3(52 Suppl):5–11. doi: 10.1111/j.1528-1167.2011.03029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill MF, Losey TE, Ng YT. The Hitchhiker's guide to the child neurologist's genetic evaluation of epilepsy. Semin Pediatr Neurol. 2008;15:32–40. doi: 10.1016/j.spen.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Hershey A, Horn P, Kabbouche M, O'Brien H, Powers S. Genomic expression patterns in menstrual-related migraine in adolescents. Headache. 2012;52:68–79. doi: 10.1111/j.1526-4610.2011.02049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey AD, Burdine D, Kabbouche MA, Powers SW. Genomic expression patterns in medication overuse headaches. Cephalalgia. 2011;31:161–171. doi: 10.1177/0333102410373155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey AD, Burdine D, Liu C, Nick TG, Gilbert DL, Glauser TA. Assessing quality and normalization of microarrays: case studies using neurological genomic data. Acta Neurol Scand. 2008;118:29–41. doi: 10.1111/j.1600-0404.2007.00979.x. [DOI] [PubMed] [Google Scholar]
- Hershey AD, Tang Y, Powers SW, Kabbouche MA, Gilbert DL, Glauser TA, Sharp FR. Genomic abnormalities in patients with migraine and chronic migraine: preliminary blood gene expression suggests platelet abnormalities. Headache. 2004;44:994–1004. doi: 10.1111/j.1526-4610.2004.04193.x. [DOI] [PubMed] [Google Scholar]
- Honkaniemi J, Sharp FR. Prolonged expression of zinc finger immediate-early gene mRNAs and decreased protein synthesis following kainic acid induced seizures. Eur J Neurosci. 1999;11:10–17. doi: 10.1046/j.1460-9568.1999.00401.x. [DOI] [PubMed] [Google Scholar]
- Hsu JC, Chang J, Wang T, Steingrimsson E, Magnusson MK, Bergsteinsdottir K. Statistically designing microarrays and microarray experiments to enhance sensitivity and specificity. Brief Bioinform. 2007;8:22–31. doi: 10.1093/bib/bbl023. [DOI] [PubMed] [Google Scholar]
- Jamali S, Bartolomei F, Robaglia-Schlupp A, Massacrier A, Peragut JC, Regis J, Dufour H, Ravid R, Roll P, Pereira S, Royer B, Roeckel-Trevisiol N, Fontaine M, Guye M, Boucraut J, Chauvel P, Cau P, Szepetowski P. Large-scale expression study of human mesial temporal lobe epilepsy: evidence for dysregulation of the neurotransmission and complement systems in the entorhinal cortex. Brain. 2006;129:625–641. doi: 10.1093/brain/awl001. [DOI] [PubMed] [Google Scholar]
- Jickling GC, Xu H, Stamova B, Ander BP, Zhan X, Tian Y, Liu D, Turner RJ, Mesias M, Verro P, Khoury J, Jauch EC, Pancioli A, Broderick JP, Sharp FR. Signatures of cardioembolic and large-vessel ischemic stroke. Ann Neurol. 2010;68:681–692. doi: 10.1002/ana.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakaye B, Thomas E, Minet A, Grisar T. The genetic absence epilepsy rat from Strasbourg (GAERS), a rat model of absence epilepsy: computer modeling and differential gene expression. Epilepsia. 2002;5(43 Suppl):123–129. doi: 10.1046/j.1528-1157.43.s.5.17.x. [DOI] [PubMed] [Google Scholar]
- Lee TS, Mane S, Eid T, Zhao H, Lin A, Guan Z, Kim JH, Schweitzer J, King-Stevens D, Weber P, Spencer SS, Spencer DD, de Lanerolle NC. Gene expression in temporal lobe epilepsy is consistent with increased release of glutamate by astrocytes. Mol Med. 2007;13:1–13. doi: 10.2119/2006-00079.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lit L, Enstrom A, Sharp FR, Gilbert DL. Age-related gene expression in Tourette syndrome. J Psychiatr Res. 2009;43:319–330. doi: 10.1016/j.jpsychires.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Rong Y, Baudry M, Schreiber SS. Status epilepticus induces p53 sequence-specific DNA binding in mature rat brain. Brain Res Mol Brain Res. 1999;63:248–253. doi: 10.1016/s0169-328x(98)00285-x. [DOI] [PubMed] [Google Scholar]
- Lukasiuk K, Pitkanen A. Large-scale analysis of gene expression in epilepsy research: is synthesis already possible? Neurochem Res. 2004;29:1169–1178. doi: 10.1023/b:nere.0000023604.91584.6c. [DOI] [PubMed] [Google Scholar]
- Marchi N, Granata T, Freri E, Ciusani E, Ragona F, Puvenna V, Teng Q, Alexopolous A, Janigro D. Efficacy of anti-inflammatory therapy in a model of acute seizures and in a population of pediatric drug resistant epileptics. PLoS One. 2011;6:e18200. doi: 10.1371/journal.pone.0018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke K, Brecht S, Dorst A, Herdegen T. Activity and expression of JNK1, p38 and ERK kinases, c-Jun N-terminal phosphorylation, and c-jun promoter binding in the adult rat brain following kainate-induced seizures. Neuroscience. 1999;91:471–483. doi: 10.1016/s0306-4522(98)00667-8. [DOI] [PubMed] [Google Scholar]
- Mirza N, Vasieva O, Marson AG, Pirmohamed M. Exploring the genomic basis of pharmacoresistance in epilepsy: an integrative analysis of large-scale gene expression profiling studies on brain tissue from epilepsy surgery. Hum Mol Genet. 2011;20:4381–4394. doi: 10.1093/hmg/ddr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo CA, Vining EP, Guo L, Skolasky RL, Carson BS, Freeman JM. The pathology of Rasmussen syndrome: stages of cortical involvement and neuropathological studies in 45 hemispherectomies. Epilepsia. 2004;45:516–526. doi: 10.1111/j.0013-9580.2004.33103.x. [DOI] [PubMed] [Google Scholar]
- Powell KL, Kyi M, Reid CA, Paradiso L, D'Abaco GM, Kaye AH, Foote SJ, O'Brien TJ. Genetic absence epilepsy rats from Strasbourg have increased corticothalamic expression of stargazin. Neurobiol Dis. 2008;31:261–265. doi: 10.1016/j.nbd.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Russ SA, Larson K, Halfon N. A national profile of childhood epilepsy and seizure disorder. Pediatrics. 2012;129:256–264. doi: 10.1542/peds.2010-1371. [DOI] [PubMed] [Google Scholar]
- Sharp FR, Lit L, Xu H, Apperson M, Walker W, Wong B, Gilbert DL, Hershey A, Glauser TA. Genomics of brain and blood: progress and pitfalls. Epilepsia. 2006;47:1603–1607. doi: 10.1111/j.1528-1167.2006.00809.x. [DOI] [PubMed] [Google Scholar]
- Specchio N, Fusco L, Claps D, Vigevano F. Epileptic encephalopathy in children possibly related to immune-mediated pathogenesis. Brain Dev. 2010;32:51–56. doi: 10.1016/j.braindev.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Tang Y, Glauser TA, Gilbert DL, Hershey AD, Privitera MD, Ficker DM, Szaflarski JP, Sharp FR. Valproic acid blood genomic expression patterns in children with epilepsy - a pilot study. Acta Neurol Scand. 2004a;109:159–168. doi: 10.1046/j.1600-0404.2003.00253.x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Schapiro MB, Franz DN, Patterson BJ, Hickey FJ, Schorry EK, Hopkin RJ, Wylie M, Narayan T, Glauser TA, Gilbert DL, Hershey AD, Sharp FR. Blood expression profiles for tuberous sclerosis complex 2, neurofibromatosis type 1, and Down's syndrome. Ann Neurol. 2004b;56:808–814. doi: 10.1002/ana.20291. [DOI] [PubMed] [Google Scholar]
- Tian Y, Apperson ML, Ander BP, Liu D, Stomova BS, Jickling GC, Enriquez R, Agius MA, Sharp FR. Differences in exon expression and alternatively spliced genes in blood of multiple sclerosis compared to healthy control subjects. J Neuroimmunol. 2011;230:124–129. doi: 10.1016/j.jneuroim.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Vezzani A, Aronica E, Mazarati A, Pittman QJ. Epilepsy and brain inflammation. Exp Neurol. 2011 doi: 10.1016/j.expneurol.2011.09.033. [DOI] [PubMed] [Google Scholar]
- Waldbaum S, Patel M. Mitochondria, oxidative stress, and temporal lobe epilepsy. Epilepsy Res. 2010;88:23–45. doi: 10.1016/j.eplepsyres.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden KD, Karsten SL, Bragin A, Kudo LC, Gehman L, Ruidera J, Geschwind DH, Engel J., Jr A systems level, functional genomics analysis of chronic epilepsy. PLoS One. 2011;6:e20763. doi: 10.1371/journal.pone.0020763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie ECG, Gidal BE, Goodkin HP, Linehan CBA. Epidemiologic aspects of epilepsy. In: Wyllie E, Cascino GD, Gidal BE, Goodkin HP, editors. Wyllie's Treatment of Epilepsy: Principles and Practice. Lippincott: Williams & Wilkins; 2011. pp. 2–10. [Google Scholar]
- Xi ZQ, Xiao F, Yuan J, Wang XF, Wang L, Quan FY, Liu GW. Gene expression analysis on anterior temporal neocortex of patients with intractable epilepsy. Synapse. 2009;63:1017–1028. doi: 10.1002/syn.20681. [DOI] [PubMed] [Google Scholar]