Abstract
In biological membranes, changes in lipid composition or mechanical deformations produce defects in the geometrical arrangement of lipids, thus allowing the adsorption of certain peripheral proteins. Here, we perform molecular dynamics simulations on bilayers containing a cylindrical lipid (PC) and a conical lipid (DOG). Profiles of atomic density and lateral pressure across the bilayer show differences in the acyl chain region due to deeper partitioning of DOG compared to PC. However, such analyses are less informative for the interfacial region where peripheral proteins adsorb. To circumvent this limitation, we develop, to our knowledge, a new method of membrane surface analysis. This method allows the identification of chemical defects, where hydrocarbon chains are accessible to the solvent, and geometrical defects, i.e., voids deeper than the glycerol backbone. The size and number of both types of defects increase with the number of monounsaturated acyl chains in PC and with the introduction of DOG, although the defects do not colocalize with the conical lipid. Interestingly, the size and probability of the defects promoted by DOG resemble those induced by positive curvature, thus explaining why conical lipids and positive curvature can both drive the adsorption of peripheral proteins that use hydrophobic residues as membrane anchors.
Abbreviations: MD, molecular dynamics; DOPC, dioleoylphosphatidylcholine; DMPC, dimyristoylphosphatidylcholine, dioleoylphosphatidylserine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; DOG, dioleoylglycerol; DAG, diacylglycerol; ASA, accessible surface area
Introduction
The compact arrangement of lipids in biological membranes can be altered in several ways. Thus, membranes contain substantial amounts of lipids such as PE or DAG whose intrinsic conical shape makes them less prone to align within the bilayer than abundant cylindrical lipids such as PC (1,2). Furthermore, membranes are frequently deformed by external forces (motors, cytoskeleton, or protein coats) and adopt shapes such as tubes, discs, or vesicles, where lipid molecules are necessarily tilted (3–6). These considerations raise the question of packing defects. To what extent do the presence of conical lipids in a flat membrane or the application of an external force cause imperfections in the geometrical arrangement of lipids?
In a physiological context, the presence of lipid packing defects in membranes can be an asset. In the endoplasmic reticulum, they may facilitate the folding of nascent transmembrane proteins (7). In small transport vesicles, they create stress that makes the membrane more prone to undergo fusion (8). Finally, lipid packing defects seem essential for the membrane adsorption of various peripheral proteins (1,2,9,10). Striking examples are CTP:phosphocholine cytidylyltransferase (11–13) and ArfGAP1 (14–16) whose binding to lipid membranes is strongly favored by the presence of conical lipids and/or by positive curvature (when the protein faces a convex membrane surface).
At first glance, a lipid packing defect induced by a conical lipid in a flat membrane or by positive curvature should resemble a void in the interfacial zone (14,15,17). However, this picture is probably oversimplified. First, lipids are not stiff objects: by twisting their flexible acyl chains they could minimize unfavorable exposure to water (13). Second, lateral diffusion could attenuate the misalignment of lipid molecules: lipids with complementary shape could cluster (e.g., a cone with an inverted cone) or lipids with a specific shape could accumulate in regions of favorable geometry (e.g., a cone in a region of negative curvature). However, recent measurements as well as theoretical calculations suggest that these processes are not very efficient due to entropic cost (18–22). These various considerations emphasize the need for a better atomic description of packing defects in lipid bilayers that are under physical (bending) or chemical (presence of conical lipids) stress.
So far, only a few simulation studies with various DAG species have been reported. Recently, we showed that introducing DOG into DOPC perturbed some properties of the bilayer (e.g., thickness and order parameter) and made an embedded amphipathic helix of the ALPS (Amphipathic-Lipid-Packing-Sensor) family more flexible compared to ALPS in pure DOPC (16). Bennett et al. (23) used potentials of mean force to determine the flip-flop barrier of palmitoyloleoyl-glycerol across a pure palmitoyloleoylphosphatidylcholine (POPC) bilayer or across a model of liquid-ordered membrane. Finally, Alwarawrah et al. (24) reported simulations of dipalmitoylglycerol (DPG) within a POPC bilayer in presence or absence of cholesterol. DPG was shown to induce spaces between PC headgroups that did not localize above DPG but between the first and second nearest-neighbor PC headgroups of a DPG.
In this work, we use a combination of atomistic simulations and new, to our knowledge, geometrical tools to assess the physicochemical perturbations that are induced by the highly conical lipid DOG in a flat PC membrane (Fig. 1). We observe that although DOG increases the number and size of lipid-packing defects in the interfacial zone, the defects do not colocalize with the DOG molecules, suggesting global rather than local perturbations. Of importance, the defects induced by DOG strongly resemble those induced by positive curvature, which have been recently described by Voth and co-workers (25). This analogy explains why some peripheral proteins that use hydrophobic insertions and notably amphipathic helices of the ALPS family appear hypersensitive to both positive curvature and conical lipids (26). This latter aspect is further developed in an accompanying work (27).
Figure 1.
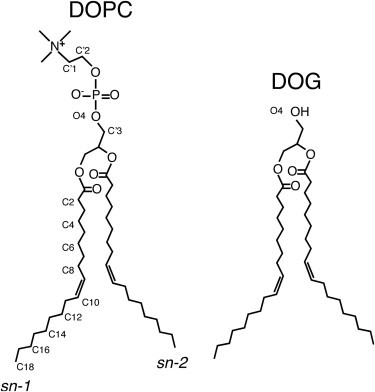
Structure of DOPC and DOG. Note that the double bond on each aliphatic tail is in a cis configuration. Each label below the phosphorous atom applies for both DOPC and DOG and for each aliphatic chain (sn-1 and sn-2).
Methods
System setup
Four systems composed of a fully solvated (40 water molecules per lipid) lipid bilayer were considered in this study: three pure systems composed of DMPC, POPC, or DOPC lipids, and one mixed system containing DOPC and DOG lipids at a molar ratio of 85:15 (denoted DOPC/DOG throughout this work). Note that DOG is tolerated in DOPC bilayers provided that its molar density does not exceed 15–20 mol % (28,29); beyond, its conical shape induces a transition to an inverted hexagonal phase (HII). For each composition three different patches were used: 70 lipids, 128 lipids, and 280 lipids. The DOPC/DOG mixture was obtained by replacing the head of randomly chosen DOPC molecules by a hydroxyl group. We performed the same number of DOG substitutions in each leaflet. Each bilayer was then properly equilibrated for 100 ns before a production run of 100, 200, or 700 ns (Table S1 in the Supporting Material). For pure DOPC and DOPC/DOG, additional simulations of patches of 1024 lipids were performed (5 ns of equilibration and 195 ns of production).
Force field and simulation details
The Berger force field (30) was used for all lipids. In DOPC and DOG, the cis-double bond of each acyl chain was corrected as previously suggested (31,32). For DOG, our previous model (16) was used: its parameterization is entirely based on Berger except for the charge on the hydroxyl oxygen, which is set to −0.7 e (taken from OPLS-UA (33)). In the accompanying work, a peptide is simulated with the OPLS-AA force field (34) within lipid bilayers identical to those of this work. We thus directly used the half-ε double-pairlist OPLS method (35,36), so that our lipids are directly compatible with peptides/proteins simulated with OPLS-AA. For compatibility, the TIP3P water model (37) was used. Additionally, for the patches of 280 lipids only, 120 mM of ions (Na+/Cl−) were added to be consistent with the simulations of the companion work (containing a peptide) (27).
All simulations were performed using GROMACS 4.5.3 (38) within the NPT ensemble. All systems were equilibrated with the Berendsen thermostat at 300 K (with a time constant of 0.1 ps; lipids and water coupled separately) and the Berendsen barostat at 1 bar (with a time constant of 1 ps and a compressibility of 4.5 × 10−5 bar−1) (39). Production runs were run at 300 K using the velocity-rescaling thermostat (40) (with a time constant of 0.1 ps, lipids and water coupled separately) and at 1 bar using the Parrinello-Rahman barostat (41) (with a time constant of 4 ps and a compressibility of 4.5 × 10−5 bar−1). Pressure coupling was applied semiisotropically (x and y directions coupled, z direction scaled independently from x and y). Periodic boundaries were applied in all directions. Bond lengths were constrained using the P-LINCS algorithm (38). A time step of 2 fs was used with the leapfrog integrator. Trajectories were initiated by assigning a Maxwell-Boltzmann distribution of velocities at the chosen temperature. Water molecules were kept rigid using the SETTLE algorithm (42). Lennard-Jones interactions were cutoff at 1.0 nm. The smooth particle-mesh Ewald method (43,44) was used for evaluating electrostatic interactions, with a real space cutoff of 1.0 nm, a grid of 0.12 nm, and an interpolation order of 4. The neighbor list was updated every 10 steps. Frames were saved every 10 ps (100 ps for the big patches of 1024 lipids) for subsequent analysis. All molecular graphics were generated with VMD (45).
Analysis of bilayer properties
A number of classical analyses of membrane properties were conducted, including hydrophobic thickness, lateral diffusion, bending and area compressibility modulus, lateral pressure profile, and potential of mean force of lipids across the membranes. All details on these analyses are reported in the Supporting Material.
Packing defects mapping using ASA and a new Cartesian scheme
Packing defects were defined and evaluated in three ways: i), using an approach analogous to the one used by Voth and co-workers (25), and ii), using, to our knowledge, two new methods that we introduce in this work.
The first method is based on the evaluation of the ASA of the bilayer aliphatic carbons. The ASA was evaluated on the two leaflets of 280 lipids with the program NACCESS (46) using a probe radius of 0.3 nm as done in (25). All the nonzero areas were then projected on the x-y plane and defined as defects (overlapping defects were merged into a single one).
Our new methods for evaluating packing defects consist in the following procedure. First, the plane perpendicular to the membrane normal was mapped into a grid of 0.1 nm resolution. For each grid point, we scanned the normal to the membrane plane starting from the solvent and descending up to 0.1 nm below the sn-2 carbon of the nearest glycerol. During the scanning procedure, solvent atoms were ignored and the analysis was done separately on each leaflet. At each z position of a given grid point, the presence of an eventual overlapping atom was evaluated by calculating a distance between the grid point center and the center of any atom of the bilayer. Overlap was assigned when this distance was shorter than the grid point diagonal half-length (0.07 nm) plus the van der Waals radius of the atom (see Fig. S1). Depending on the nature and depth of the first atom met during this scanning procedure (i.e., the first atom that overlaps with the grid point under consideration), we distinguished three cases: i), no defect when the atom was nonaliphatic; ii), chemical defect when the atom was an aliphatic carbon; iii), geometrical defect when the atom was an aliphatic carbon and was 0.1 nm below the sn-2 carbon of the nearest glycerol. Adjacent elementary points of similar nature were then merged, resulting in defects of various sizes. Note that geometrical defects are a subcategory of chemical defects. Further details on the two methods are given in the Supporting Material, notably the choice of van der Waals radii.
As noted previously by Voth and co-workers (25), the distribution of packing defect size falls off exponentially. For all methods, the area defect distribution was plotted on a semilog scale and fitted to an exponential decay: p(A) = Be−kA, where p(A) is the probability of finding a defect area of A nm2, B is a constant, and k is the exponential decay in units of nm−2. All points below 0.05 nm2 were discarded from the fit (small defects tend to be similar whatever the composition). All probabilities strictly below 10−4 were also discarded because of their poor convergence.
Results and Discussion
Does DOG directly create voids in the interfacial zone of DOPC bilayers?
Because DOPC and DOG differ solely by their polar heads (Fig. 1), one may naively assume that replacing some DOPC molecules by DOG in a bilayer will affect the atomic density in the polar head region, although leaving the hydrophobic region unaltered. In other words, a DOPC → DOG substitution should create a void or defect corresponding to the lack of phosphocholine. However, in previous MD simulations we and other groups reported a more complex picture (16,23,24). The critical point is that DOG (here used at 15 mol %) does not occupy the same position as DOPC, but, instead, slightly sinks toward the center of the bilayer. The different partitioning of DOG and DOPC is exemplified by two observations (Fig. 2). First, carbon 2 (C2) of DOG occupies a deeper (−0.2 nm) position than the corresponding atom of DOPC (compare the red and gray atoms in Fig. 2, a and b, and the dotted profiles in Fig. 2, c and d). Second, the density profile of DOG lacks the characteristic minimum at the border between the two leaflets that is observed for DOPC, thereby indicating that the aliphatic tails of DOG partially invade the bilayer center and interdigitate (compare the red and gray dotted lines in Fig. 2, c and d).
Figure 2.
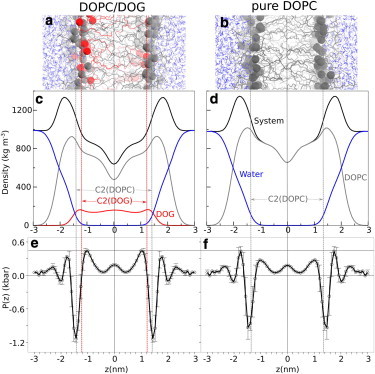
Cross-section analysis of DOPC/DOG and DOPC bilayers. The upper panels show snapshots of DOPC/DOG (85:15 mol:mol) (a) and pure DOPC (b) bilayers. All atoms are represented as wireframes except C2 atoms of DOPC and DOG, which are depicted as gray (DOPC) or red (DOG) spheres. The middle panels show density profiles for DOPC/DOG (c) and DOPC (d) bilayers with the density of the whole system in black, DOPC in gray, DOG in red, and water in blue. The vertical dotted lines coincide with the C2 carbons of DOPC (gray) and DOG (red); the vertical solid line corresponds to the center of the bilayer. The lower panels show the symmetrized lateral pressure profile (with error bars) of DOPC/DOG (e) and DOPC (f) bilayers. The vertical lines indicate the bilayer center as well as the C2 atoms of DOPC and DOG. Note that the density and pressure profiles indicate substantial changes between the two bilayers for the hydrophobic region but not for the polar region.
These observations suggest two conclusions. On the one hand, DOG should substantially affect the bulk properties of the bilayer. For example, an effect of DOG on the elastic properties of the membrane can be anticipated because the different partitioning of DOG and DOPC leads to an increase in membrane thickness (Table 1 and Fig. 2). On the other hand, how DOG affects the interfacial properties of the bilayer remains unclear. Notably, it is difficult to understand how DOG could create defects in the interfacial region while partitioning deeper than DOPC. Solving this second issue was the main motivation of this work and, as explained later, requires new analytical methods. Before presenting these new approaches, the next two sections summarize other general observations that can be extracted from a classical analysis of DOPC and DOPC/DOG simulations and which give information about the physical properties of such bilayers. Additional analysis is also presented in the Supporting Material, including evaluations of the DOPC and DOG area and volumes (Fig. S2 and Fig. S3), as well as determination of the hydration of the phosphate groups in DOPC/DOG and DOPC bilayers. As expected, DOG (Fig. 1) leads to a slightly higher hydration of the membrane polar region because oxygen O4 can act both as a hydrogen bond donor and acceptor, in contrast to oxygen O4 of DOPC, which acts only as a hydrogen bond acceptor (Fig. S4).
Table 1.
Structural, dynamic, and elastic properties of a pure DOPC and DOPC/DOG bilayer
| Pure DOPC | DOPC/DOG | |
|---|---|---|
| Hydrophobic thickness h (nm) | 2.63 ± 0.11 | 2.89 ± 0.08 |
| DOPC diffusion coefficient D (10−8 cm2 s−1) | 8.8 ± 1.1 | 8.7 ± 0.8 |
| Compressibility KA (N m−1) | 0.331 ± 0.006 | 0.351 ± 0.027 |
| Bending modulus κ (kT) | 21.4 ± 0.6 | 14.3 ± 1.4 |
The changes in the lateral pressure profile induced by DOG in the polar head region are marginal
An alternative method to analyze the cross section of a bilayer is to compute the lateral pressure profile P(z) along its normal (13). A typical P(z) profile shows a sharp negative peak corresponding to the cohesive interfacial tension at the hydrophilic/hydrophobic interface. This peak is flanked by two positive peaks at the level of the polar headgroups (electrostatic and steric interactions, hydration repulsion) and aliphatic chains (steric interactions) (47). Thus, P(z) gives the tendency of each region to shrink or to expand and has been proposed to be a valuable parameter to assess the effect of conical lipids on the membrane surface; the prediction is that a conical lipid should reduce the pressure at the polar head region, hence facilitating protein adsorption (13).
As shown in Fig. 2, e and f, the most noticeable effect of DOG on the lateral pressure profile of a DOPC bilayer is a twofold increase of the positive peak at the level of the aliphatic tails (at ± 1.2 nm). This peak corresponds, on average, to carbons C8 and C2 of the oleoyl chain of DOPC and DOG, respectively. Surprisingly and in contrast to what has been predicted (13), this increase is insufficiently counterbalanced by a decrease in the lateral pressure in the polar head region (around ± 2.0 nm). In fact, the pressure decrease in this region is quite modest (within the error bars of our measurements) and cannot counteract the pressure increase in the aliphatic region. Instead, the overall pressure of the DOPC/DOG system is kept at zero by a further drop in the cohesive interfacial tension.
Because lateral pressure profiles and bilayer elastic properties are connected (47,48), we expected a change in rigidity and/or compressibility of the bilayer upon DOG addition. Table 1 shows the area compressibility and bending modulus of the two lipid compositions. The addition of DOG in DOPC decreases the bending modulus of the bilayer by 35% (from 21.4 to 14.3 kT) but does not significantly change its compressibility modulus. It is interesting to note that an increase in bilayer thickness is generally associated with an increase in bending rigidity (49) (e.g., 20.5 ± 2.4 kT and 29.0 ± 3.6 kT for diC18:1c9 and diC22:1, respectively). However in our case, the increase in membrane thickness does not result from an increase in acyl chain length but from the different partitioning of the two lipids in the bilayer as well as an increase of order parameter of DOPC acyl chains (16). We conclude that the different partitioning of DOG and DOPC along the membrane normal facilitate rather than impede membrane deformations.
Dynamic properties of mixed DOPC/DOG bilayers
We then evaluated the propensity of DOG and DOPC to flip-flop between leaflets. Evaluation of transbilayer movements is important in the context of defects in lipid packing: a translocating lipid should create transient perturbations in the bilayer and, conversely, a bilayer with lipid packing defects could favor flip-flop movements. However, no flip-flop of DOG and DOPC could be observed in our simulations, whose cumulative length amounted to a few microseconds. In contrast, we observed normal lateral diffusion of DOPC, which was essentially unaffected by the introduction of DOG (Table 1). Of note, Alwarawrah et al. (24) found that DPG reduces lateral diffusion of POPC within a POPC/DPG mixture, suggesting that the saturated chains of DPG but not the monounsaturated chains of DOG affect the lateral movements of PC molecules.
We determined the PMF for moving the headgroup of DOG or DOPC into the center of a pure DOPC bilayer or a DOPC/DOG bilayer. As shown in Fig. 3, DOPC and DOG retain the same energy profile in both compositions, suggesting that DOG, at 15 mol %, does not favor flip-flop movements. Note the slight shift between the curves, which is due to the difference in thickness between DOPC and DOPC/DOG bilayers, and the flatness of the energy profile in the bilayer center for DOG, which indicates that its translocation is accompanied by dehydration (i.e., as opposed to DOPC, there is no need to form a water pore to translocate DOG across the bilayer, see Fig. S5). The energy barrier for moving DOPC and DOG to the center of the bilayer was 100 and 35 kJ/mol, respectively. From these values, we estimate (23,50) the half-time of DOPC and DOG flip-flop to be ∼30 min and 100 μs, respectively; values that are well above the timescale of our simulations and in good agreement with previous MD simulations of palmitoyloleoyl-glycerol in POPC (23). We conclude from this analysis that transbilayer movements are unlikely to contribute to lipid packing defects induced by DOG.
Figure 3.
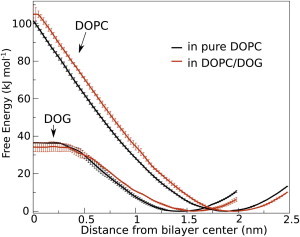
Potentials of mean force for transferring the headgroup of DOPC and DOG from water to the center of a pure DOPC or DOPC/DOG bilayer. For each PMF, the position restraint was placed on the hydroxyl or phosphate of the headgroup (for DOG and DOPC, respectively) and used to determine the x axis. The PMFs are shown with error bars.
DOG incorporation increases occurrence and size of packing defects at the hydrophilic/hydrophobic interface
The previous analyses give a time-averaged picture of the cross section of DOPC and DOPC/DOG bilayers. However, for a peripheral protein initially in the solvent, what should be more important is the instantaneous structure of the membrane surface: does it display defects at any given time and are these defects deep and large enough to facilitate protein adsorption? Therefore, we need to change our point of view by 90° and consider the structure of DOPC and DOPC/DOG membranes from the top to look at their microscopic details.
To do so, we use three methods (Fig. 4). The first approach is similar to that introduced by Voth and co-workers (25) and is based on the evaluation of the ASA of the bilayer aliphatic carbons using a spherical probe of 0.3 nm in radius (approximately the size of a bulky residue) (see Fig. 4 b). The second and third approaches are based on a Cartesian scheme (see Fig. S1). First, the bilayer surface is mapped into a grid (x, y) whose pixels have a surface area of 0.01 nm2. For each pixel, we scan the normal to the membrane plane (z) from the solvent toward the center of the bilayer. We define two types of lipid packing defects: i), chemical defects, when the first atom that is met along the normal is an aliphatic carbon (Fig. 4 c); and ii), geometrical defects, when the first atom that is met is an aliphatic carbon and it is 0.1 nm below the sn-2 carbon of the nearest glycerol (Fig. 4 d). Thus, geometrical defects are a subcategory of chemical defects; the former correspond to voids in the bilayer, whereas the latter correspond to regions where aliphatic chains are exposed to the solvent (see schemes in Fig. 4).
Figure 4.
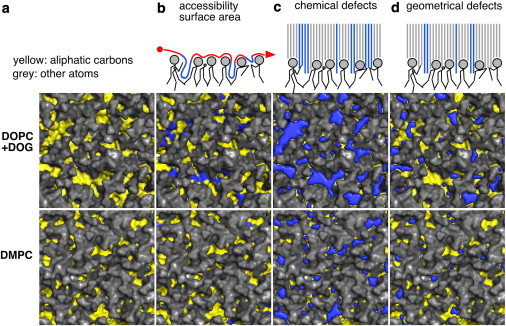
Detection of lipid packing defects. The three methods to identify packing defects in the interfacial zone of model membranes are schematized in the top row. The other panels show snapshots of the surface of DOPC/DOG and DMPC bilayers with polar heads in gray and aliphatic carbon atoms in yellow. For each composition, the columns are arranged as follows: (a) the pure bilayer; (b) the bilayer with the packing defects (blue) computed with the ASA method; (c and d) the bilayer with the chemical (c) or geometrical (d) defects (blue) computed with our new Cartesian scheme. The ASA method uses of a probe of 0.3 nm in radius and maps the accessible aliphatic carbon atoms. The Cartesian schemes probe the bilayer vertically by squares of 0.01 nm2 and look for all accessible aliphatic carbon atoms (chemical defects) or only those that are below the glycerol level (geometrical defects). See text for further details.
The different panels of Fig. 4 show top views of DOPC/DOG and DMPC bilayers with defects as revealed by the ASA method (Fig. 4 b) and by our Cartesian scheme for both chemical (Fig. 4 c) and geometrical (Fig. 4 d) defects. These various representations immediately suggest that, whatever the method used, the lipid packing defects are more abundant and larger in DOPC/DOG than in DMPC bilayers, two extreme cases in terms of lipid packing.
Similar analyses were performed on thousands of snapshots of DMPC, POPC, DOPC, and DOPC/DOG bilayers, leading to the statistical analysis shown in Fig. 5. For all lipid compositions, the distribution of defects follows roughly a single exponential function above a threshold value of 0.05 nm2 (below this value, all bilayers display the same distribution). As noted previously, all distributions suggest a second regime (which does not follow a pure exponential decay) for very large defects (25). Of importance, the exponential decay constant decreases when saturated acyl chains are replaced by monounsaturated ones and when DOG was introduced in the bilayer; the probability of occurrence of large defects follows the order: DMPC < POPC < DOPC < DOPC/DOG. The three methods agree remarkably well on this trend, but they identify defects of different sizes. Chemical defects essentially correspond to all regions where aliphatic atoms are visible from the membrane surface, regardless of their depth (compare Fig. 4, a and c). Geometrical defects are smaller because of their more restrictive definition (Fig. 4 d). Finally, the ASA method detects defects of medium size (Fig. 4 b) but the results depend on the choice of the probe radius (here 0.3 nm like in (25)); changing this value results in different defect areas. The main advantage of the ASA approach is that it is not sensitive to the orientation of the defects because the probe does not follow a preferential direction, whereas our Cartesian-based approach detects a vertical defect more readily than an oblique one because the membrane is scanned along its normal. The Cartesian-based approach combines two advantages: a fine resolution (0.1 nm) without the need to tune a probe size and the possibility to identify defects not only by their chemistry, but also by their depth. In the accompanying work (27), we show that geometrical defects, which closely resemble voids, are crucial for the partitioning of peptides that use hydrophobic insertions.
Figure 5.
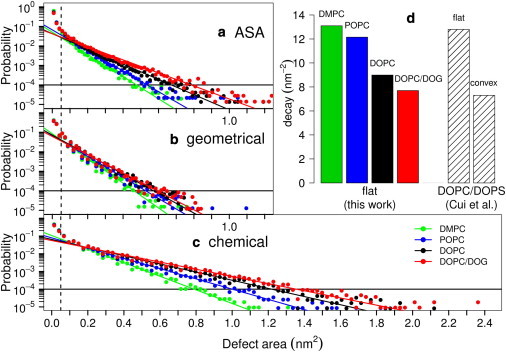
Size distribution of packing defects in model membrane systems. The various plots show the probability of finding a defect of defined size as obtained by the ASA method (a; like in (25)) or by the Cartesian schemes for geometrical (b) or chemical (c) defects. The color code is as follows: green, DMPC; blue, POPC; black, DOPC; red, DOPC/DOG. Each solid line is a fit to an exponential distribution for defects larger than 0.05 nm2 and with probability ≥10−4. The fits give the following decay constants (in units of nm−2) for DMPC, POPC, DOPC, and DOPC/DOG, respectively: (a) 13.1, 12.1, 9.0, and 7.7; (b) 14.6, 12.4, 11.5, and 10.8; (c) 8.8, 6.7, 5.3, and 4.7. The decay constants obtained in panel (a) are plotted in panel (d) and compared to those obtained by Cui et al. (25) on a DOPC/DOPS patch either in a flat or in a convex conformation.
Packing defects induced by DOG incorporation or by positive curvature are similar
Because incorporating conical lipids into a flat membrane or imposing positive curvature represent two alternative ways to promote the binding of some peripheral proteins (9,14,15), we next wanted to compare the packing defects associated with these two different conditions. As shown in Fig. 5 d, the change in defect size distribution when going from a flat DMPC to a flat DOPC/DOG membrane (this study) closely matches the change in defect size distribution observed by Voth and co-workers (25) when going from a flat DOPC/DOPS to a convex (radius = 20 nm) DOPC/DOPS membrane. This comparison suggests that the structure of the interfacial zone of a convex membrane strongly resembles that of a flat membrane containing conical lipids. In line with this hypothesis, the two types of bilayers share other features. In bilayers containing conical lipids or displaying positive curvature, the lipid polar heads are more disordered and solvent accessible, whereas the acyl chains are more ordered (16,51). This analogy has, however, some limitations. Notably, bending a membrane is generally associated with a decrease in membrane thickness (51), whereas incorporating a conical lipid induces membrane thickening (Fig. 2, Table 1). Thus, a membrane containing conical lipids is a good mimetic of a curved membrane for the interfacial zone, not for the central region.
It seems obvious that packing defects induced by membrane curvature should be evenly distributed on the bilayer surface. In contrast, DOG-induced defects may colocalize with the conical lipids themselves. To assess this second possibility, we evaluated the overlap between packing defects and the acyl chains of the DOG or DOPC lipids (Table 2). For this, we first determined the relative surfaces occupied by DOG and DOPC acyl chains. For each surface, we then determined the fraction of area that is occupied by lipid packing defects. Strikingly and as shown in Table 2, this fraction was not significantly higher in the DOG area than in the DOPC area. Therefore, geometrical and chemical packing defects are evenly distributed on a flat DOPC/DOG bilayer and do not colocalize with DOG although DOG favors the formation of defect. We conclude that incorporation of DOG lipids into a flat DOPC bilayer mimics the effect of inducing a positive curvature because the spatial localization of the conical lipids inside the bilayer rapidly becomes uncorrelated with packing defects (packing defects are highly dynamic objects and have short life times around 10–100 ps), which are thus ubiquitous and randomly distributed on the bilayer surface.
Table 2.
Spatial distribution of packing defects in a DOPC/DOG bilayer
| Nature of packing defects | DOG (%)a | DOPC (%)b |
|---|---|---|
| Geometrical | 1.4 ± 0.5 | 1.7 ± 0.4 |
| Chemical | 5.4 ± 1.3 | 5.4 ± 0.7 |
After determination of the relative surfaces occupied by DOG and by DOPC acyl chains, we determined the percentage of each surface that corresponds to chemical or geometrical packing defects. In this analysis, we retain only defects larger than 0.1 nm2. The obtained values show that packing defects are not enriched in the surface occupied by DOG compared to DOPC (the distributions overlap, see Figure S6). A snapshot illustrating this idea is shown in Figure S7.
Fraction of area occupied by packing defects within the total area of DOG lipids.
Fraction of area occupied by packing defects within the total area of DOPC lipids.
Conclusions
The concept of lipid packing defects has been instrumental to explain some observations on peripheral proteins. In brief, it is assumed that voids in the membrane interfacial zone, which are induced either by positive curvature or by conical lipids, act as binding sites for exposed hydrophobic residues of the protein (1,9,13,52). However, this picture remains elusive at the atomic level. First, the flexibility and movements of lipid molecules could attenuate the geometrical defects. Second, the analogy between defects induced by positive curvature and defects induced by conical lipids is deceptively simple. Imposition of a positive curvature necessarily affects all lipid molecules (like in a fan), whereas introduction of conical lipids may have a more local effect.
Here we show that, as far as the surface structure of the membrane is concerned, there is a striking similarity between the defects induced by conical lipids and by positive curvature. This similarity arises from the fact that conical lipids do not induce defects at their exact location but instead impose a stress over the entire membrane surface that leads to defects of various sizes, randomly distributed over the surface.
In a physiological context, this finding is important because it suggests that large lipid packing defects may be restricted to membranes of the early secretory pathway. Indeed, not only do these organelles exhibit regions of high curvatures (such as the tubules of the endoplasmic reticulum or the transport vesicles of the Golgi apparatus), but also their bulk lipid composition is favorable for the formation of large defects (unlike the bulk composition of late secretory membranes) (9). Studies in yeast suggest that several lipid metabolic pathways that desaturate the acyl chains or modulate the sizes of the polar head groups maintain a high ratio between conical and nonconical lipids in these organelles as compared to the plasma membrane (53–55).
A second important implication of the analogy between the packing defects induced by conical lipids and by membrane curvature concerns MD simulations. Performing all-atom simulations on curved membranes requires high computational resources and poses issues such as establishing simple periodic conditions. In contrast, simulations on small flat membrane patches are more amenable to simulations. In the case of sensors of membrane curvature that use hydrophobic insertions, performing simulations on flat membranes containing conical lipids should help to dissect the mechanism of membrane adsorption. In the accompanying work (27), we use this approach to study how ALPS motifs sense packing defects in membranes.
Acknowledgments
We thank Luca Monticelli for having provided the topology of DOPC as well as the coordinates of a DOPC bilayer, Samuli Ollila for useful discussions on the lateral pressure profile, and Alenka Copic for comments on the manuscript. This work was granted access to the HPC resources of CINES under the allocation c2011-076720 and c2012-076720 made by GENCI (Grand Equipement National de Calcul Intensif).
This work was supported by the Swiss National Science Foundation grant PBELP3_141118 (to S.V.), by the French Government grant MENRT (to L.V.), and by an ERC advanced grant 268888 (to B.A.). DPT is an Alberta Innovates Health Solutions (AIHS) Scientist and Alberta Innovates Technology Futures Strategic Chair in (Bio)Molecular Simulation. D.W.B. was supported by studentships from AIHS and the Natural Sciences and Engineering Research Council (NSERC; Canada). Work in DPT’s group is supported by NSERC. Some calculations were carried out on Compute Canada facilities.
Supporting Material
References
- 1.Epand R.M. Lipid polymorphism and protein-lipid interactions. Biochim. Biophys. Acta. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farsad K., De Camilli P. Mechanisms of membrane deformation. Curr. Opin. Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 4.McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 5.Baumgart T., Capraro B.R., Das S.L. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu. Rev. Phys. Chem. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonny B. Membrane deformation by protein coats. Curr. Opin. Cell Biol. 2006;18:386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Bretscher M.S., Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 8.Chernomordik L.V., Kozlov M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 9.Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 10.Madsen K.L., Bhatia V.K., Stamou D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 2010;584:1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 11.Attard G.S., Templer R.H., Jackowski S. Modulation of CTP:phosphocholine cytidylyltransferase by membrane curvature elastic stress. Proc. Natl. Acad. Sci. USA. 2000;97:9032–9036. doi: 10.1073/pnas.160260697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies S.M., Epand R.M., Cornell R.B. Regulation of CTP: phosphocholine cytidylyltransferase activity by the physical properties of lipid membranes: an important role for stored curvature strain energy. Biochemistry. 2001;40:10522–10531. doi: 10.1021/bi010904c. [DOI] [PubMed] [Google Scholar]
- 13.van den Brink-van der Laan E., Killian J.A., de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Antonny B., Huber I., Cassel D. Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J. Biol. Chem. 1997;272:30848–30851. doi: 10.1074/jbc.272.49.30848. [DOI] [PubMed] [Google Scholar]
- 15.Bigay J., Gounon P., Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 16.González-Rubio P., Gautier R., Fuchs P.F. Amphipathic-Lipid-Packing-Sensor interactions with lipids assessed by atomistic molecular dynamics. Biochim. Biophys. Acta. 2011;1808:2119–2127. doi: 10.1016/j.bbamem.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 17.Hatzakis N.S., Bhatia V.K., Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 18.Cooke I.R., Deserno M. Coupling between lipid shape and membrane curvature. Biophys. J. 2006;91:487–495. doi: 10.1529/biophysj.105.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derganc J. Curvature-driven lateral segregation of membrane constituents in Golgi cisternae. Phys. Biol. 2007;4:317–324. doi: 10.1088/1478-3975/4/4/008. [DOI] [PubMed] [Google Scholar]
- 20.Kamal M.M., Mills D., Howard J. Measurement of the membrane curvature preference of phospholipids reveals only weak coupling between lipid shape and leaflet curvature. Proc. Natl. Acad. Sci. USA. 2009;106:22245–22250. doi: 10.1073/pnas.0907354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorre B., Callan-Jones A., Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. USA. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian A., Baumgart T. Sorting of lipids and proteins in membrane curvature gradients. Biophys. J. 2009;96:2676–2688. doi: 10.1016/j.bpj.2008.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett W.F.D., Tieleman D.P. Molecular simulation of rapid translocation of cholesterol, diacylglycerol, and ceramide in model raft and nonraft membranes. J. Lipid Res. 2012;53:421–429. doi: 10.1194/jlr.M022491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alwarawrah M., Dai J., Huang J. Modification of Lipid Bilayer Structure by Diacylglycerol: A Comparative Study of Diacylglycerol and Cholesterol. J. Chem. Theory Comput. 2012;8:749–758. doi: 10.1021/ct200790q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui H., Lyman E., Voth G.A. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys. J. 2011;100:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drin G., Casella J.F., Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 27.Vanni S., Vamparys L., Antonny B. Amphipatic Lipid Packing Sensor Motifs: Probing Bilayer Defects with Hydrophobic Residues. Biophys. J. 2013;104:575–584. doi: 10.1016/j.bpj.2012.11.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S., Rand R.P. Modification by diacylglycerol of the structure and interaction of various phospholipid bilayer membranes. Biochemistry. 1986;25:2882–2889. doi: 10.1021/bi00358a022. [DOI] [PubMed] [Google Scholar]
- 29.Orädd G., Lindblom G., Ljusberg-Wahren H. Phase diagram of soybean phosphatidylcholine-diacylglycerol-water studied by x-ray diffraction and 31P- and pulsed field gradient 1H-NMR: evidence for reversed micelles in the cubic phase. Biophys. J. 1995;68:1856–1863. doi: 10.1016/S0006-3495(95)80362-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger O., Edholm O., Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachar M., Brunelle P., Rauk A. Molecular dynamics simulation of a polyunsaturated lipid bilayer susceptible to lipid peroxidation. J. Phys. Chem. B. 2004;108:7170–7179. [Google Scholar]
- 32.Martinez-Seara H., Róg T., Vattulainen I. Influence of cis double-bond parametrization on lipid membrane properties: how seemingly insignificant details in force-field change even qualitative trends. J. Chem. Phys. 2008;129:105103. doi: 10.1063/1.2976443. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen W.L. Optimized intermolecular potential functions for liquid alcohols. J. Chem. Phys. 1986;90:1276–1284. [Google Scholar]
- 34.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 35.Chakrabarti N., Neale C., Pomès R. An iris-like mechanism of pore dilation in the CorA magnesium transport system. Biophys. J. 2010;98:784–792. doi: 10.1016/j.bpj.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tieleman D.P., MacCallum J.L., Monticelli L. Membrane protein simulations with a united-atom lipid and all-atom protein model: lipid-protein interactions, side chain transfer free energies and model proteins. J. Phys. Condens. Matter. 2006;18:S1221–S1234. doi: 10.1088/0953-8984/18/28/S07. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 38.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 39.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 40.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101–014107. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 41.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 42.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 43.Darden T., York D., Pedersen L. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 44.Essmann U., Perera L., Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 45.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Grap. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 46.Hubbard S.J., Thornton J.M. Department of Biochemistry and Molecular Biology, University College London; London, UK: 1993. NACCESS. [Google Scholar]
- 47.Ollila S., Hyvönen M.T., Vattulainen I. Polyunsaturation in lipid membranes: dynamic properties and lateral pressure profiles. J. Phys. Chem. B. 2007;111:3139–3150. doi: 10.1021/jp065424f. [DOI] [PubMed] [Google Scholar]
- 48.Ollila O.H.S., Risselada H.J., Marrink S.J. 3D pressure field in lipid membranes and membrane-protein complexes. Phys. Rev. Lett. 2009;102:078101. doi: 10.1103/PhysRevLett.102.078101. [DOI] [PubMed] [Google Scholar]
- 49.Rawicz W., Olbrich K.C., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett W.F.D., MacCallum J.L., Tieleman D.P. Thermodynamic analysis of the effect of cholesterol on dipalmitoylphosphatidylcholine lipid membranes. J. Am. Chem. Soc. 2009;131:1972–1978. doi: 10.1021/ja808541r. [DOI] [PubMed] [Google Scholar]
- 51.Risselada H.J., Marrink S.J. Curvature effects on lipid packing and dynamics in liposomes revealed by coarse grained molecular dynamics simulations. Phys. Chem. Chem. Phys. 2009;11:2056–2067. doi: 10.1039/b818782g. [DOI] [PubMed] [Google Scholar]
- 52.Bhatia V.K., Hatzakis N.S., Stamou D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin. Cell Dev. Biol. 2010;21:381–390. doi: 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Boumann H.A., Gubbens J., de Kroon A.I. Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol. Biol. Cell. 2006;17:1006–1017. doi: 10.1091/mbc.E05-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mousley C.J., Tyeryar K.R., Bankaitis V.A. The Sec14-superfamily and the regulatory interface between phospholipid metabolism and membrane trafficking. Biochim. Biophys. Acta. 2007;1771:727–736. doi: 10.1016/j.bbalip.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneiter R., Brügger B., Kohlwein S.D. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edholm O., Nagle J.F. Areas of molecules in membranes consisting of mixtures. Biophys. J. 2005;89:1827–1832. doi: 10.1529/biophysj.105.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wohlert J., Edholm O. Dynamics in atomistic simulations of phospholipid membranes: Nuclear magnetic resonance relaxation rates and lateral diffusion. J. Chem. Phys. 2006;125:204703. doi: 10.1063/1.2393240. [DOI] [PubMed] [Google Scholar]
- 58.Kumar S., Rosenberg J.M., Kollman P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 59.Ollila, O. H. S. 2010. Lateral pressure in lipid membranes and its role in function of membrane proteins. PhD thesis. Tampere University of Technology, Finland.
- 60.Waheed Q., Edholm O. Undulation contributions to the area compressibility in lipid bilayer simulations. Biophys. J. 2009;97:2754–2760. doi: 10.1016/j.bpj.2009.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 62.Brandt E.G., Braun A.R., Edholm O. Interpretation of fluctuation spectra in lipid bilayer simulations. Biophys. J. 2011;100:2104–2111. doi: 10.1016/j.bpj.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindahl E., Edholm O. Mesoscopic undulations and thickness fluctuations in lipid bilayers from molecular dynamics simulations. Biophys. J. 2000;79:426–433. doi: 10.1016/S0006-3495(00)76304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schrodinger, L. 2010. The PyMOL Molecular Graphics System, Version 1.3r1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


