Abstract
NCLs (neuronal ceroid lipofuscinoses) form a group of eight inherited autosomal recessive diseases characterized by the intralysosomal accumulation of autofluorescent pigments, called ceroids. Recent data suggest that the pathogenesis of NCL is associated with the appearance of fragmented mitochondria with altered functions. However, even if an impairement in the autophagic pathway has often been evoked, the molecular mechanisms leading to mitochondrial fragmentation in response to a lysosomal dysfunction are still poorly understood. In this study, we show that fibroblasts that are deficient for the TPP-1 (tripeptidyl peptidase-1), a lysosomal hydrolase encoded by the gene mutated in the LINCL (late infantile NCL, CLN2 form) also exhibit a fragmented mitochondrial network. This morphological alteration is accompanied by an increase in the expression of the protein BNIP3 (Bcl2/adenovirus E1B 19 kDa interacting protein 3) as well as a decrease in the abundance of mitofusins 1 and 2, two proteins involved in mitochondrial fusion. Using RNAi (RNA interference) and quantitative analysis of the mitochondrial morphology, we show that the inhibition of BNIP3 expression does not result in an increase in the reticulation of the mitochondrial population in LINCL cells. However, this protein seems to play a key role in cell response to mitochondrial oxidative stress as it sensitizes mitochondria to antimycin A-induced fragmentation. To our knowledge, our results bring the first evidence of a mechanism that links TPP-1 deficiency and oxidative stress-induced changes in mitochondrial morphology.
Keywords: Bcl2/adenovirus E1B 19 kDa protein interacting protein 3 (BNIP3), confocal microscopy, late infantile neuronal ceroid lipofuscinosis (LINCL), mitochondrion, mitofusins
Abbreviations: AR, aspect ratio; BNIP3, Bcl2/adenovirus E1B 19 kDa interacting protein 3; [Ca2+]mt, mitochondrial matrix Ca2+; CTL, control; Drp1, dynamin related protein 1; ER, endoplasmic reticulum; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; FF, form factor; Fis1, fission 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HADHA, hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase; HSP, heat-shock protein; IMM, inner mitochondrial membrane; KRH buffer, Krebs-Ringer Hepes buffer; LSD, lysosomal storage disease; MAM, mitochondrial associated membranes; MCFR, mean channel fluorescence ratio; MEM, minimum essential medium; MFF, mitochondrial fission factor; Mfn1, mitofusin 1; Mfn2, mitofusin 2; MLIV, mucolipidosis type IV; NCL, neuronal ceroid lipofuscinosis; LINCL, late infantile NCL; OMM, outer mitochondrial membrane; Opa1, optic atrophy factor 1; ROS, reactive oxygen species; RNAi, RNA interference; RT–qPCR, reverse transcription-quantitative PCR; siRNA, small interfering RNA; SOD, superoxide dismutase; TMRE, tetramethylrhodamine ethyl ester; TPP-1, tripeptidyl peptidase-1
INTRODUCTION
LSDs (lysosomal storage diseases) represent diverse sets of conditions resulting from an impaired uptake, sorting or digestion of the material captured by cells during endocytosis or autophagy [1]. So far, about 50 LSDs have been described. These autosomal recessive diseases can be caused by defects in soluble enzymes, in non-enzymatic lysosomal proteins or even in non-lysosomal proteins that alter lysosomal function [1]. Among these diseases, NCLs (neuronal ceroid lipofuscinoses) form a group of inherited autosomal recessive disorders known as the most common neurodegenerative diseases in children and are characterized by the intralysosomal accumulation of autofluorescent pigments called ceroids [2]. NCLs result from mutations in one of the eight CLN genes and are classified according to the onset of the disease [2]. The CLN2 gene, mutated in the LINCL (late infantile NCL), encodes TPP-1 (tripeptidyl peptidase-1), a lysosomal hydrolase, which is involved in the degradation of several proteins such as the c-subunit of ATPsynthase, neuromedin B, cholecystokinin and the pro-apoptotic protein Bid [3–7]. Like the other NCLs, LINCL is characterized by seizures, progressive mental decline, loss of vision, impairment of locomotor function and shortened lifespan [8,9]. Most of these symptoms reflect severe neurodegeneration (mostly in the hypothalamus and the cerebellum) as well as astrocyte activation and gliosis [8].
Although these monogenic diseases are rather simple in terms of causative defects, biochemical and cellular cascades of events subsequent to the original lysosomal disorder are very complex. Several common and uncommon pathogenic cascades have been described in the LSDs, such as accumulation of secondary metabolites, alteration of calcium homoeostasis, generation of oxidative stress, inflammation and modifications in lipid trafficking, autophagy and ER (endoplasmic reticulum) stress (reviewed in [10]). Several studies indicate that mitochondria are often damaged in case of LSD [11–13]. However, to our knowledge, nothing is known about the putative impact of lysosomal storage on the morphology and the functions of mitochondria in LINCL.
Mitochondria support many cellular functions, such as ATP production, regulation of calcium homoeostasis, NEFA (non-esterified fatty acid) β-oxidation, haem and cluster [Fe–S] syntheses [14]. They are very dynamic organelles that can adopt various morphologies in terms of sizes and shapes [15]. The dynamic morphology of the mitochondria ranges from an elongated network to a fragmented aspect, depending on cell conditions such as stress, cell cycle phase, energy needs and bioenergetic status of the organelle and is regulated by frequent cycles of mitochondrial fusion and fission [16]. So far, in eukaryotes, several proteins have been identified as regulating those events. Among them, Mfn1 (mitofusin 1) and Mfn2 (mitofusin 2) are two GTPases involved in the fusion of the OMM (outer mitochondrial membrane) [16]. The fusion of the IMM (inner mitochondrial membrane) is ensured by the Opa1 (optic atrophy factor 1) GTPase. Fission is mediated by the action of proteins called MFF (mitochondrial fission factor), Fis1 (fission 1) and Drp1 (dynamin-related protein 1) [16]. MFF and Fis 1 are localized in the OMM and recruit Drp1, a cytosolic GTPase that, once recruited, oligomerizes around the mitochondria and forms a constriction ring [16]. So far, mitochondrial fragmentation has been reported in several LSDs such as mucolipidosis (I, II, IV types), GM1-gangliosidosis, nephropathic cystinosis and NCL (CLN1 and CLN6 forms) [12,13,17–19]. Even if, in vitro, this fragmented morphology is not associated with a modification of mitochondrial calcium concentration or spontaneous cell death, it has been suggested that it could impair the ‘calcium buffering capacity’ of this organelle in response to a particular stimulus [18]. Indeed, Kiselyov and co-workers have shown that after cell stimulation with bradykinin, a Ca2+-mobilizing agonist, the mitochondrial calcium loading was reduced in MLIV (mucolipidosis type IV) fibroblasts when compared with control fibroblasts [19]. In contrast, a mitochondrial matrix calcium overload was observed after histamine stimulation in a model of GM1 gangliosidosis [20]. These observations reveal that the impact of a lysosomal dysfunction on the mitochondrial calcium homoeostasis could be more complex than initially proposed [20].
Furthermore, several studies suggest that the accumulation of non-degraded material in lysosomes could also modify both the expression of genes encoding mitochondrial proteins and the mitochondrial lipid content. For example, it has been shown that the expression of several subunits of the complex V (Fo/F1-ATP synthase) is increased in the cultured renal proximal tubule epithelial cells isolated from patients suffering from nephropathic cystinosis [21]. Likewise, mutations in the gene encoding the CLN6 protein would lead to an increased expression of MnSOD (manganese superoxide dismutase) (SOD2) an antioxidant enzyme located within the mitochondrial matrix [22].
Our results suggest that a TPP-1 deficiency in fibroblasts induces a fragmentation of the mitochondrial network, probably due to a reduction in the abundance of both Mfn1 and Mfn2. This accentuated fragmentation of mitochondria is not accompanied by changes either in the matrix calcium concentration or in the abundance of ROS (reactive oxygen species) whatever it is in basal or in stress-induced conditions. In contrast, an increase in the expression of BNIP3 (Bcl2/adenovirus E1B 19 kDa interacting protein 3), a pro-apoptotic BH3-only protein, described to induce mitochondrial dysfunction in several cell types [23,24], was observed in TPP-1-deficient cells. Applying an RNAi (RNA interference) approach, it appears that BNIP3 was not involved in the fragmentation of the mitochondrial network in LINCL cells in basal conditions. However, interestingly this protein seems to sensitize mitochondria of TPP-1-deficient cells to further mitochondrial fragmentation induced by antimycin A. Altogether, our results suggest that cells that display a deficiency in TPP-1 might be more sensitive to an additional stress targeting mitochondria, such as an oxidative stress induced by a respiratory chain defect.
MATERIALS AND METHODS
Cell cultures
The skin fibroblast cell lines either deficient for the TPP-1 activity (LINCL, GM09404) or control fibroblasts (CTL, GM02456) were purchased from the Coriell Institute for Medical Research. Skin fibroblasts were grown in Eagle's MEM (minimum essential medium) supplemented with 2 mM glutamine, 100 mM non-essential amino acids, 100 mM pyruvate and 10% (v/v) FBS (fetal bovine serum) (complete MEM). At confluence, skin fibroblasts were trypsinized and subcultured in 24-well, 12-well or six-well culture plates or 25 cm2 culture flasks, according to the experimental needs or 75 cm2 culture flasks (Corning) for maintaining the cells in culture.
As control, matching the nuclear background of LINCL cells, 24 h after seeding, LINCL were washed with PBS and incubated or not for 48 h with 10 nM purified recombinant TPP-1 protein (kindly provided by P. Lobel, Piscataway, NJ, U.S.A.) diluted in complete MEM. At the end of the incubation, media were removed and replaced by fresh complete MEM. All experiments were carried out 72 h later when the enzymatic activity was fully recovered (up to 150% of the enzymatic activity found in CTL cells).
TPP-1 activity assay
CTL and LINCL fibroblasts were seeded at a density of 500000 cells in 75 cm2 culture flasks. After 48 h of incubation in the presence or absence of purified recombinant TPP-1, followed by 3 days of recovery, the TPP-1 activity was assayed in cell lysates after membrane solubilization in 0.2% Triton X-100, in 50 mM acetate buffer; pH 5.0 containing 1 mM TPP-1 substrate (Ala-Ala-Phe-7-amido-4-methylcoumarin, Sigma) as previously described [25]. The reaction was stopped by the addition of 50 mM glycine buffer, pH 10.5 supplemented with 5 mM EDTA and 0.5% Triton X-100. Finally, the fluorescence was measured with a spectrofluorimeter (SLM Instruments, Inc.) (λexc, 350 nm; λem, 420 nm). Autofluorescence values were subtracted from fluorescent signals and results were normalized for protein content, determined by the Pierce 660 nm Protein Assay kit (Thermo Scientific).
Mitochondrial abundance, membrane potential and flow cytometry analyses
Mitochondrial abundance was assessed in LINCL cells loaded or not with TPP-1 enzyme. Briefly, cells were seeded in 6-well culture plates at a density of 100000 cells/well. After 48 h of incubation in the presence or absence of purified recombinant TPP-1 (10 nM), followed by 3 days of recovery, cells were stained with 100 nM MitoTracker Green FM (Molecular Probes) as previously reported [26]. Cells were analysed by flow cytometry with a FACScalibur (BD Biosciences), using the FL1-H channel. Data were processed using the BD CellQuest™ Pro software. MCFRs (mean channel fluorescence ratios) were calculated for each condition (LINCL and LINCL+TPP-1) by calculating the ratio between the Mitotraker Green mean fluorescence intensities and the mean values of autofluorescence intensities measured for their corresponding unlabelled cells, used as negative controls.
Mitochondrial membrane potential was assessed in LINCL cells loaded or not with 10 nM TPP-1 enzyme. Briefly, cells were seeded in 6-well culture plates at a density of 100000 cells/well. After 48 h of incubation in the presence or absence of purified recombinant TPP-1, followed by 3 days of recovery, cells were incubated for 30 min with 25 nM TMRE (tetramethylrhodamine ethyl ester, perchlorate; Molecular Probes). Cells were then rinsed with ice-cold PBS, trypsinized using 0.25% (w/v) trypsin-EDTA (Gibco), rinsed once with PBS and resuspended in 500 μl of Hepes buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2.6H2O). Cells were analysed by flow cytometry with a FACScalibur (BD Biosciences), using the FL1-H channel. Data were processed using the BD CellQuest™ Pro software. MCFRs were calculated for each condition (LINCL and LINCL+TPP-1) by calculating the ratio between the TMRE mean fluorescence intensities and the mean values of autofluorescence intensities measured for the corresponding unlabelled cells, used as negative controls.
Western blot analyses
Western blot analyses were performed by IR fluorescence (Odyssey Scanner, Li-Cor) [27]. LINCL cells were seeded at a density of 500000 cells in 75 cm2 culture flasks. After 48 h of incubation in the presence or absence of purified recombinant TPP-1, followed by 3 days of recovery, cells were scraped in 10 ml of complete MEM and centrifuged for 5 min at 93 g. Cell pellets were washed thrice with ice-cold PBS and lysed in 20 μl of lysis buffer [7 M urea, 2 M thiourea, pH 8.5, CHAPS 4% (v/v), 60 mM DTT (dithiotreitol), 1 mM Na3VO4 (sodium orthovanadate), 10 mM PNPP (p-nitrophenyl phosphate), 10 mM 2-glycerophosphate, 5 mM sodium fluoride and Complete™]. After incubation on ice for 30 min, cell lysates were centrifuged for 10 min at 15700 g (4°C) and the supernatants were collected (clear cell lysates). Protein concentration was determined using the Pierce 660 nm Protein Assay kit (Thermo Scientific). Amounts corresponding to 15 μg of proteins were resolved on 3–8% or 12% bis-Tris gels (NuPage, Invitrogen). At the end of the migration, proteins were transferred onto PVDF membranes (Millipore). The semi-dry electrotransfer was performed during 2 h. Unspecific binding sites were then blocked by incubating the membranes for 1 h at room temperature (21°C) with 15 ml of blocking solution (Li-Cor Odyssey Infrared Imaging System blocking buffer diluted twice in PBS). Western blot analysis was performed with primary antibodies against HADHA (hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, ab54477, Abcam), β-subunit ATPsynthase (A21350, Molecular Probes), mt-HSP70 (heat-shock protein 70) (804-077-R100, Alexis Biochemical), Fis1 (no. HPA017430, Sigma), Drp1 (no. 611112, BD Transduction Laboratories), Opa1 (no. 612606, BD Transduction Laboratories), Mfn1 (no. WH0055669H4, SIGMA), Mfn2 (no. ab50838, Abcam), BNIP3 (no. ab10433, Abcam) used at a 1:1000 dilution, and incubated overnight at 4°C. For the protein detection, an IRDye-conjugated secondary antibody (Li-Cor) was used at a 1:5000 dilution. Protein loading was verified by the immunodetection of either α-tubulin or β-actin. The fluorescence intensity of the bands corresponding to the protein of interest was quantified using the Odyssey software (Li-Cor) and normalized by the fluorescence intensity of the bands corresponding to the α-tubulin or β-actin.
Gene expression and real-time RT–qPCR (reverse transcription-quantitative PCR)
LINCL fibroblasts were seeded at a density of 150000 cells per 25 cm2 culture flask. After 48 h of incubation in the presence or absence of purified recombinant TPP-1 (10 nM), followed by 3 days of recovery, total RNA was extracted using the QIAGEN RNeasy mini kit and the QIAcube machine (QIAGEN) according to the manufacturer's instructions. The concentration of RNA isolated with RNeasy kits was determined by measuring the absorbance at 260 nm in a UV-spectrophotometer. Total RNA (1μg) was reverse transcribed in mRNA using the Transcriptor first strand cDNA synthesis kit (Roche). Forward and reverse primers for BNIP3 and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were designed using the Primer Express 1.5 software (Applied Biosystems) BNIP3-F, 5′-TTTGCTGGCCATCGGATT-3′; BNIP3-R, 5′ACCAAGTCAGACTCCAGTTCTTCA-3′; GAPDH-F, 5′-ACCCACTCCTCCACCTTTGAC-3′; GAPDH-R, 5′-GTCCACCACCCTGTTGCTGTA-3′. Amplification reaction assays contained SYBR Green PCR mastermix (Roche) and primers at the optimal concentrations. Real-time PCR was performed with the Abi-Prism 7000 Sequence Detection System (Applied Biosystems) and the data were subjected to relative quantification using GAPDH as a reference gene for normalization. The ∆∆Ct method was used for each sample in order to calculate the relative fold change. Data calculation and normalization were performed according to the method of de Longueville et al. [28].
Silencing of BNIP3 expression using siRNA (small interfering RNA)
Cell transfection experiments with siRNA were performed using double-stranded RNA (SMARTpool, Dharmacon). SiGENOME non-targeting siRNA Ctl Pool 1 (D-001206-13-20) was used as a control. LINCL cells were transfected with 10 nM siRNA for BNIP3 (M-004374-04) using the DharmaFECT1 transfection reagent (Dharmacon, T-2001) according to the manufacturer's instructions. Silencing was checked 24 h post-transfection by RT of total extracted RNA, followed by real-time RT–qPCR. For the Western blot analyses, cells were lysed 24 h post-transfection in the same lysis buffer as described before.
Analysis of mitochondrial morphology
The morphology of the mitochondrial network was visualized in cells stained with 100 nM Mitotracker Green probe (Molecular Probes). CTL and LINCL fibroblasts were seeded at a density of 500000 cells in 75 cm2 culture flasks. After 24 h, LINCL cells were washed with PBS and incubated for 48 h in the presence or in the absence of purified recombinant TPP-1 protein diluted in complete MEM. At the end of the incubation, media were removed and replaced by fresh complete MEM without TPP-1. Before 48 h of staining, cells were cultured in Lab-Tek II culture chambers (VWR) at a density of 15000 cells per chamber. Cells were washed with KRH buffer (Krebs-Ringer Hepes buffer) (125 mM NaCl; 5 mM KCl; 1.3 mM CaCl2; 1.2 mM MgSO4; 25 mM Hepes; pH 7.4) and incubated for 30 min with 100 nM Mitotracker Green diluted in KRH+2% (w/v) BSA. Mitochondrial morphology was observed using confocal microscopy (λexc, 490 nm; λem, 516 nm) and analyses of mitochondrial population morphology were performed using the ImageJ software [29]. Briefly, after staining, two-dimensional micrographs of the mitochondrial networks were taken using a confocal microscope (Leica). First, the length of mitochondria or AR (aspect ratio), meaning the ratio between the bigger (major) and the smaller (minor) side for each mitochondrial fragment and the degree of reticulation or FF (form factor) defined as (Pm2)/4p Am) (where Pm is the length of the mitochondrial perimeter and Am is the area of the mitochondria), were determined using the ImageJ software. The images were processed to maximize the signal/background ratio using the automatic adjustment of brightness/contrast of the ImageJ software. After smoothing, the images were filtered using the hat 7% 7 kernel. Finally, the major, the minor, the area and the Pm of each fragment were determined using the ‘analyse particles’ function of ImageJ. Analyses were performed on at least 40 cells for each condition.
Immunofluorescence staining and confocal microscopy
LINCL fibroblasts were seeded at a density of 500000 cells in 75 cm2 culture flasks. The next day, LINCL were washed with PBS and incubated for 48 h with or without 10 nM purified recombinant TPP-1 protein diluted in complete MEM. At the end of the incubation, media were removed and replaced by fresh complete MEM without TPP-1. 24 h before the staining, cells were cultured onto glass coverslips (VWR) in 24-well culture plates (Corning). Cells were fixed for 10 min with 4% PFA (paraformaldehyde), permeabilized for 5 min with PBS/1% (v/v) Triton X-100 and then incubated overnight with antibodies against Drp1 (no. 611112, BD Transduction Laboratories) or against Fis1 (no. HPA017430, Sigma), diluted 100 times in PBS/1% (w/v) BSA. Finally, cells were incubated for 1 h at room temperature with an Alexa-labelled secondary antibody (Molecular Probes) diluted 500 times in PBS/1% (w/v) BSA and processed for confocal microscopy (Leica). Nuclei were visualized by ToPro-3 staining and confocal laser scan microscopy analyses were performed. For co-localization studies, images from a selected ROI (region of interest) were overlaid revealing the co-localized pixels. Threshold and background corrections were set based on Red-Green scattergram, generating a co-localization image (co-localized pixels in white) and pixels distribution scatter plot (co-localized yellow pixels along the diagonal). After setting background and threshold, the percentage of co-localization was calculated by Leica LAS-AF image analysis [percentage (%) of co-localization rate=co-localization area/area foreground; area foreground=area image−area background] [30].
Mitochondrial O2•− measurement
LINCL fibroblasts were seeded at a density of 20000 cells in 12-well plates. After 48 h of incubation in the presence or absence of purified recombinant TPP-1, followed by 3 days of recovery, the mitochondrial O2•− abundance was measured using 10 μM MitoSOX™ Red specific dye (Molecular Probes) according to the manufacturer's instructions. When indicated, 10 μM antimycin A was added for 15 min after cell loading with MitoSOX™. Fluorescence intensities were measured using a spectrofluorimeter (λexc, 485 nm; λem, 520 nm) and, after subtraction of the autofluorescence value from experimental values, fluorescence intensities were normalized for protein content.
Measurement of the mitochondrial matrix calcium
LINCL fibroblasts were seeded at a density of 10000 cells in 24-well plates. After 48 h of incubation in the presence or absence of purified recombinant TPP-1, followed by 3 days of recovery, the [Ca2+]mt (mitochondrial matrix Ca2+) was assessed using 2 μM X-Rhod-5F fluorescent probe according to the manufacturer's instructions. When indicated, 10 μM ionomycin, a calcium-specific ionophore known to increase the cytoplasmic calcium concentration [31], was added for 10, 20 or 30 min to cells loaded with X-Rhod-5F. Fluorescence intensities were measured using a spectrofluorimeter and fluorescence values were normalized for DNA content after subtraction of autofluorescence from experimental values, as previously described [32].
Statistical analysis
Data from at least three independent experiments are presented as means±S.D. or±S.E.M. and were analysed with the appropriate statistical test such as the Student's t test, the Mann–Whitney Rank Sum test, ANOVA-1 followed by a Dunnett's test or ANOVA 2 followed by a Holm–Sidak test, as advised by the software SigmaStat 3.1. Differences between means were only considered as statistically significant when P value was <0.05.
RESULTS
TPP-1 activity in LINCL cells
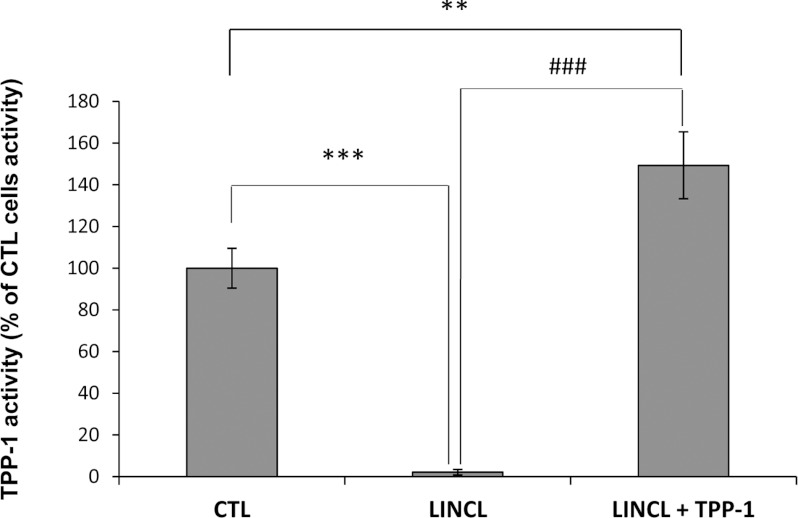
Numerous studies performed on skin fibroblasts or on other cell lines derived from patients affected by LSDs suffer from the use, as controls, of cells from healthy relatives or unrelatives. The difficulty of finding good controls might hamper the interpretation of experimental data as numerous differences (in terms of protein abundance, modifications of gene expression or changes in the activity of transcription factors, etc.) that exist between different cell lines might result from their different genetic background rather than from the effect of the lysosomal hydrolase deficiency itself. To circumvent this problem, we decided to restore the TPP-1 activity in LINCL cells, using a recombinant pro-enzyme produced by CHO (Chinese-hamster ovary) cells, as previously described [25]. As expected, when LINCL cells were incubated with 10 nM TPP-1 proenzyme for 48 h and maintained for an extra 3 days in culture, the TPP-1 activity in LINCL cells could be efficiently restored (Figure 1). Actually, the TPP-1 activity was even slightly but significantly higher than the enzymatic activity measured in healthy, but unrelated, control fibroblasts (Figure 1). While the restoration of TPP-1 activity was already observed after the 48-h incubation with the enzyme (results not shown), the extra time in culture allowed us to assess the stability of the recovery and the putative reversibility of changes induced by the enzyme deficiency.
Figure 1. TPP-1 activity in control fibroblasts, LINCL cells and LINCL loaded with recombinant TPP-1.
TPP-1 activity was measured in CTL and LINCL cells pre-incubated in the presence (LINCL+TPP-1) or in the absence (LINCL) of 10 nM recombinant TPP-1 for 48 h using the synthetic substrate Ala–Ala–Phe–AMC (AAF–AMC). Briefly, at the end of the incubation with the recombinant enzyme, cells were maintained for 72 h in culture with fresh medium without TPP-1. Cells were then homogenized in ice-cold isotonic sucrose and membranes were solubilized with Triton X-100. TPP-1 activity was assayed in 50 mM acetate buffer (pH 5.0) containing 5 mM 4-methylumbelliferyl derivative as substrate. The fluorescence of 4-methylumbelliferone released was measured in a spectrofluorimeter (λexc, 350 nm; λem, 460 nm). Results were calculated as arbitrary fluorescence units normalized for protein content (AFU/μg of proteins) and expressed in percentages of CTL cells as means±S.D. (n=3). **, ***: significantly different from CTL cells with P<0.01 and P<0.001 respectively. ###: significantly different from LINCL cells loaded with TPP-1 with P<0.001 as determined by an ANOVA-1 followed by a Dunnett's test.
TPP-1 deficiency has no impact on the mitochondrial population abundance
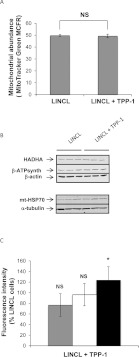
Recent data in the literature have suggested that the mitochondrial abundance was reduced in two NCL forms (CLN6 and CLN1) when compared with healthy unrelated skin fibroblasts [13]. Thus, we wanted to determine the putative effect of a TPP-1 deficiency on the mitochondrial abundance. We first assessed the mitochondrial mass by flow cytometry, in LINCL and LINCL cells pre-incubated with TPP-1 and then stained with MitoTracker Green, a fluorescent dye known to accumulate in mitochondria irrespective of the mitochondrial membrane potential [33]. Figure 2(A) shows that the mitochondrial abundance in LINCL cells is not modified upon recovery of the TPP-1 activity. Even though the use of this fluorescent probe did not allow us to detect any quantitative modification of the mitochondrial population, it is possible that modifications in the abundance of some proteins could nevertheless occur in LINCL cells. To test this hypothesis, we analysed the abundance of several mitochondrial proteins that fulfil different functions, such as oxidative phosphorylation (β-subunit of ATPsynthase), fatty acid β-oxidation (HADHA) or mitochondrial protein folding in the matrix (mt-HSP70), in LINCL and LINCL cells loaded with TPP-1. Our results show that the recovery of the TPP-1 activity has no effect either on the abundance of the β-subunit of ATPsynthase or on the abundance of HADHA (Figures 2B and 2C). However, it significantly increases the abundance of mt-HSP70, demonstrating that the abundance of some mitochondrial proteins might be reversibly changed within mitochondria of TPP-1-deficient cells (Figures 2B and 2C). We therefore decided to analyse the expression of 168 nuclear genes encoding mitochondrial proteins, using both the mitochondrial PCR array and the mitochondrial energy metabolism PCR array from SABiosciences (Qiagen) (results not shown). Among the modifications observed in TPP-1- rescued LINCL cells, only the decrease in the expression of the gene encoding the protein BNIP3 could be validated by real-time RT–qPCR (Figure 3A). Taken together, these results suggest that there are no major changes in the mitochondrial abundance and most likely no change in the biogenesis of mitochondria in cells that display a deficiency in TPP-1 activity.
Figure 2. Effects of a TPP-1 deficiency on the mitochondrial abundance.
(A) Mitochondrial population abundance was analysed in LINCL cells pre-incubated with (LINCL+TPP-1) or without (LINCL) recombinant TPP-1. Cells were then stained with 100 nM MitoTracker Green for 30 min and fluorescence was detected by flow cytometry as described in the Material and methods section. Results are expressed as MitoTracker Green Fluorescence MCFR calculated from autofluorescence signals measured for corresponding unstained cells and represent means±S.D. (n=3). NS, non-significantly different from LINCL cells as determined by a Student's t test. (B) The abundance of HADHA, β-subunit of ATPsynthase and mt-HSP70 was determined by Western blot analysis performed on 15 μg of clear cell lysates prepared from LINCL cells pre-incubated (LINCL+TPP-1) or not (LINCL) with recombinant TPP-1. Equal protein loading was controlled by the immunodetection of β-actin or α-tubulin (n=3). (C) Quantification of the abundance of β-subunit of ATP synthase (grey columns) HADHA (white columns), and mt-HSP70 (black columns) on blots presented in (B). Results are calculated as fluorescence intensity normalized for α-tubulin or β-actin and expressed in percentages of LINCL cells as means±S.D. (n=3). *: significantly different from LINCL cells with P<0.05 as determined by a Student's t test.
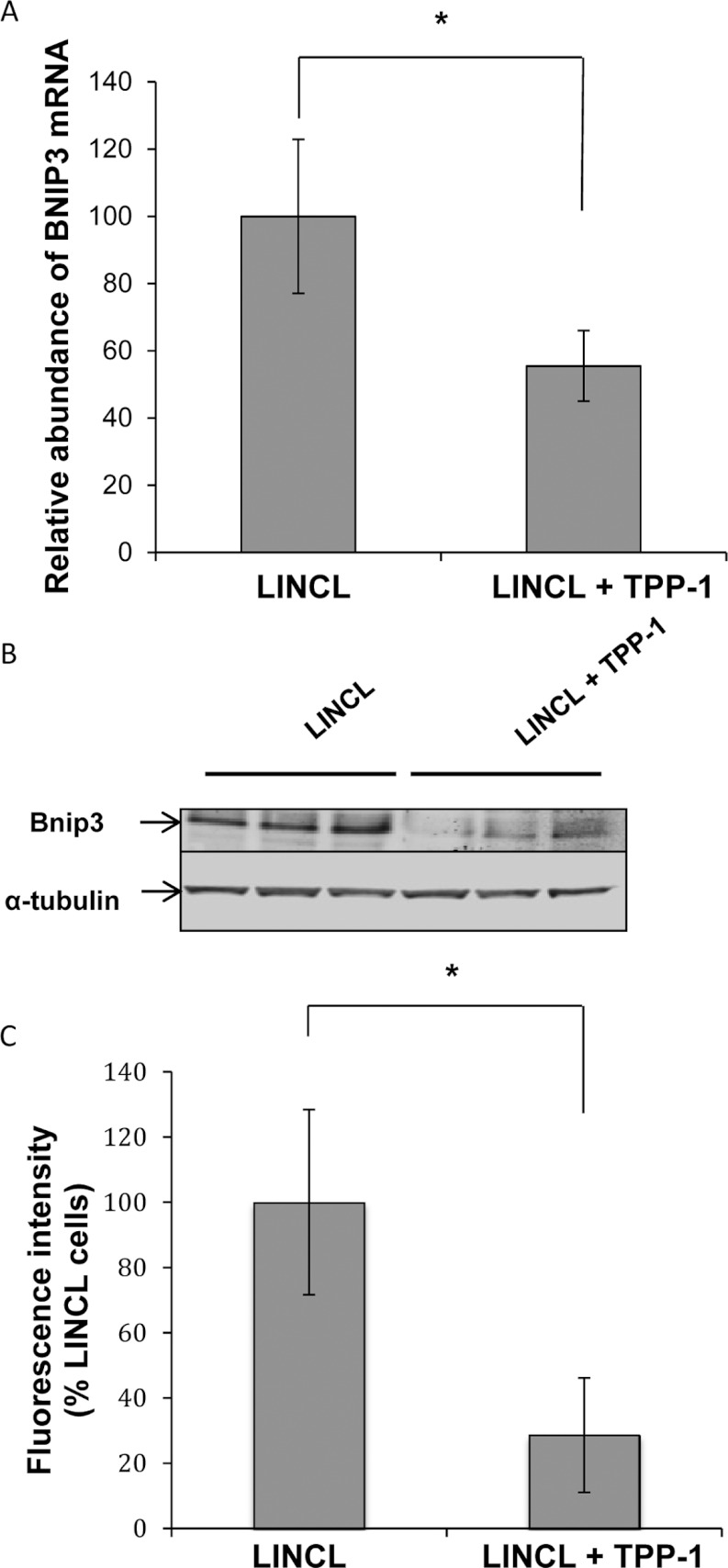
Figure 3. Effects of a TPP-1 deficiency on the expression of BNIP3.
(A) Total RNA was extracted from LINCL cells pre-incubated with (LINCL+TPP-1) or without (LINCL) TPP-1, reverse transcribed and amplified by real-time RT–qPCR in the presence of BNIP3 primers and SYBR green. GAPDH was used as reference gene for data normalization. Results are expressed in percentages of the mRNA abundance determined in LINCL cells and represent means±S.D. (n=3). *: significantly different from LINCL cells with P<0.05 as determined by a Student's t test. (B) The protein abundance of BNIP3 was determined by Western blot analysis performed on 15 μg of clear cell lysates prepared from LINCL cells pre-incubated (LINCL+TPP-1) or not (LINCL) with TPP-1 (n=3). Equal protein loading was controlled by the immunodetection of α-tubulin. (C) Quantification of the Western blot presented in (B). Fluorescence intensity of the band of interest was normalized for α-tubulin and expressed as percentages of LINCL cells. Results are means±S.D. (n=3). *: significantly different from LINCL cells with P<0.05 as determined by a Student's t test.
TPP-1 deficiency induces an increase in the expression of BNIP3
As the transcriptional expression of BNIP3 seemed to be down-regulated in TPP-1-rescued LINCL cells, we analysed the abundance of this protein by Western blot analysis. In accordance with the RT–qPCR results, Figures 3(B) and 3(C) show that the abundance of BNIP3 is significantly reduced in LINCL cells loaded with TPP-1 when compared with LINCL cells. BNIP3 is a member of the Bcl2 family [34], which has now also been considered as a regulator of mitochondrial morphology and function [35–37]. Indeed, the overexpression of BNIP3 can trigger mitochondrial fission by increasing the recruitment of Drp1 onto the mitochondria, but also by inhibiting Opa1 activity [36,38]. Knowing that the mitochondrial network is more fragmented in several models of lysosomal storage disorders [12,18], we next analysed the putative impact of a TPP-1 deficiency on the mitochondrial morphology in LINCL cells.
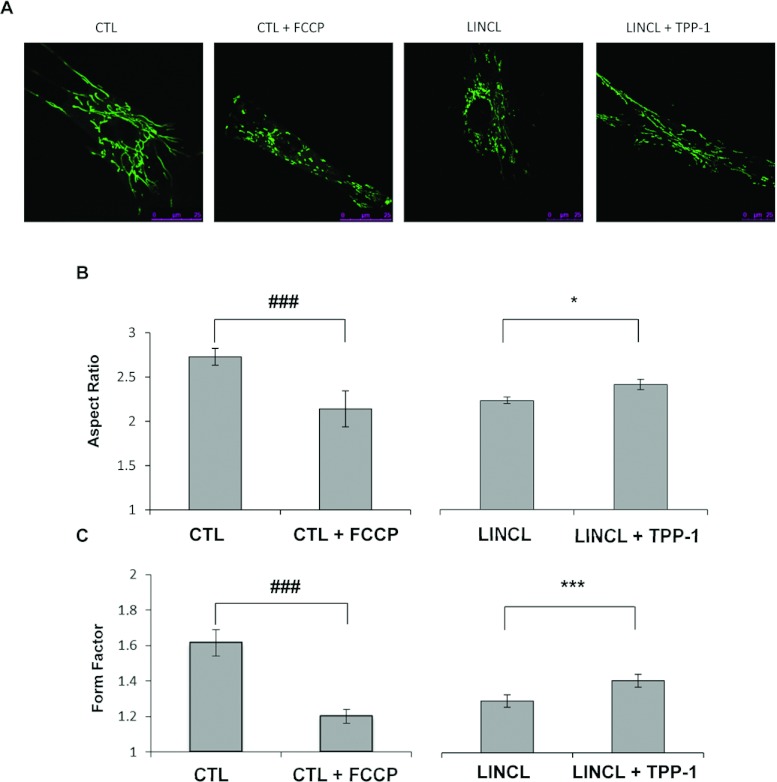
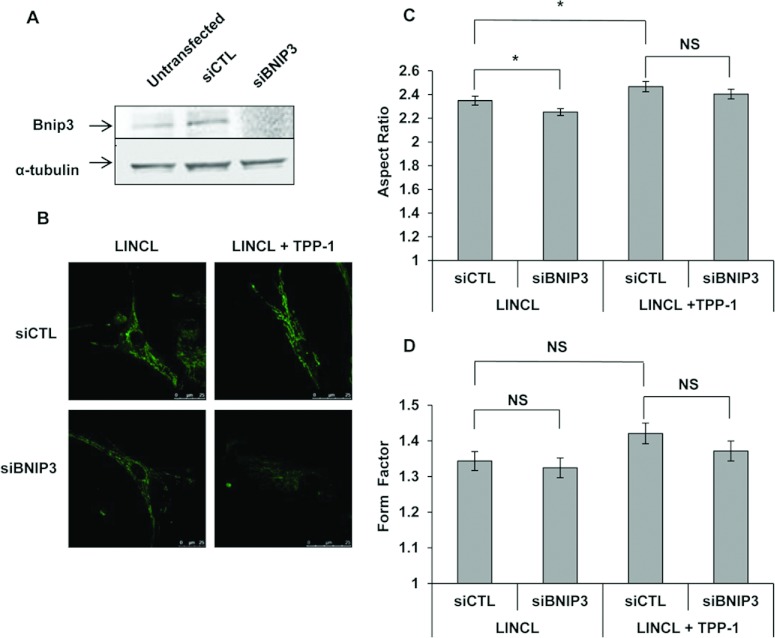
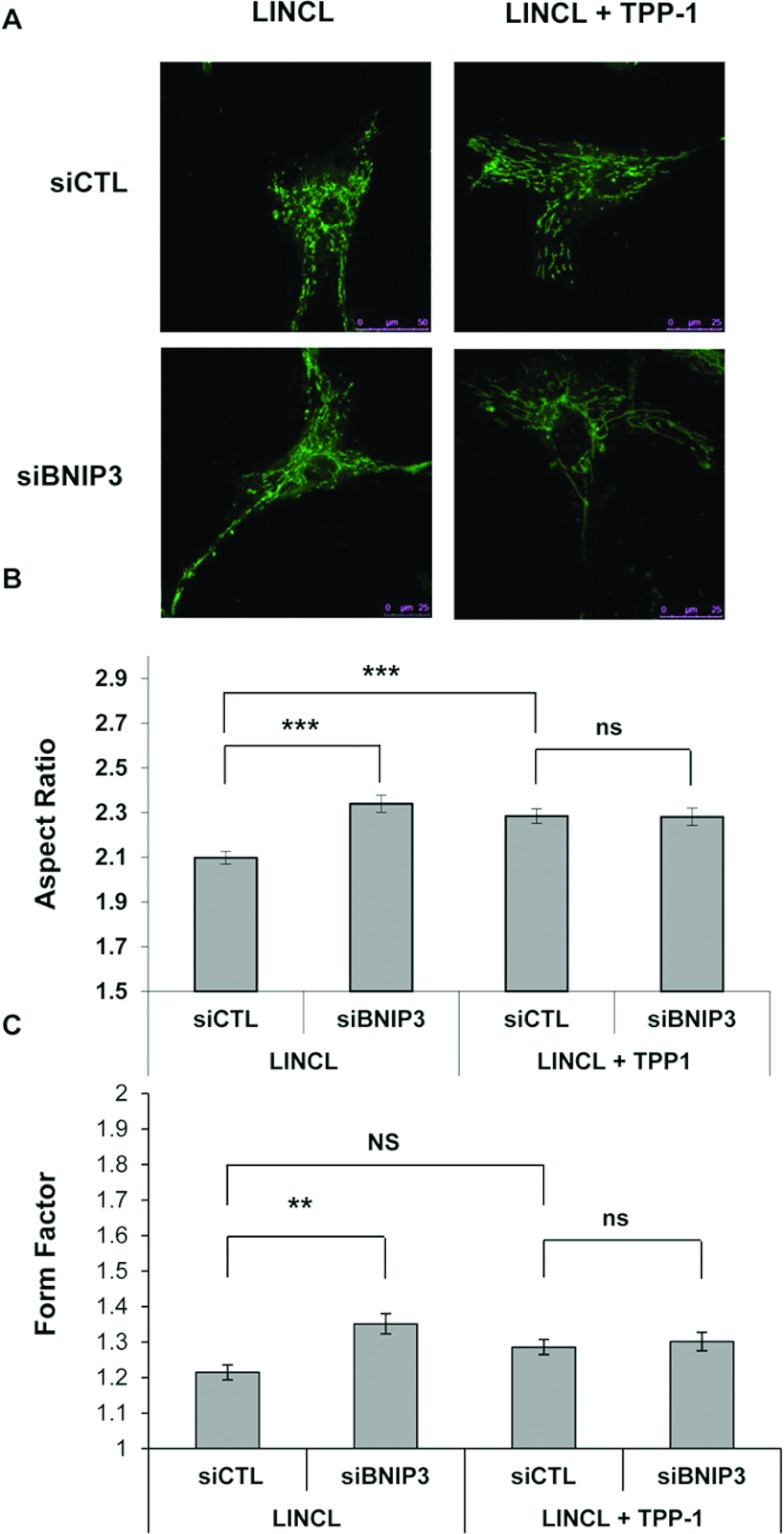
Mitochondrial network morphology was assessed in cells stained with 100 nM Mitotracker Green and visualized by fluorescence confocal microscopy. The mitochondrial network is reticulated and interconnected in healthy control cells (Figure 4A), a phenotype found in the majority of cell types [15]. A pre-incubation of these cells for 30 min with 20 μM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone), a protonophore that uncouples mitochondrial respiration from ATP production, induces a fragmentation of the mitochondrial network as previously reported [18]. These observations can be quantified on recorded micrographs using the ImageJ software. As expected, both parameters, the AR and the FF which give an indication about the length of mitochondrial segments and their degree of reticulation, respectively, are significantly reduced upon FCCP treatment (Figures 4B and 4C). More interestingly, the mitochondrial network also appears more fragmented in LINCL cells, when compared with LINCL cells that recovered a TPP-1 activity for 5 days (Figures 4A–4C). In order to determine whether or not BNIP3 is involved in the onset of the fragmented mitochondrial network observed in TPP-1-deficient cells, we analysed the morphology of the mitochondrial network in LINCL cells and LINCL cells loaded with TPP-1 after silencing the expression of BNIP3 using a RNAi approach. Western blot analysis revealed a strong decrease in the abundance of BNIP3 protein in LINCL cells 24 h post-transfection with a pool of four specific RNAs (siBNIP3) when compared with a control pool RNA (siCTL) (Figure 5A). However, the inhibition of BNIP3 expression does not reverse the mitochondrial fragmentation observed in LINCL cells (Figure 5B). Indeed, in LINCL cells transfected with siCTL, the AR is significantly higher (Figure 5C) whereas the FF tends to be higher (Figure 5D) after recovery of the TPP-1 activity, supporting the idea of a mitochondrial fragmentation in LINCL cells. However, neither the AR nor the FF are increased upon silencing of BNIP3 expression both in LINCL cells and in LINCL cells loaded with TPP-1 (Figures 5C and 5D). These results indicate that BNIP3 silencing does not reverse the mitochondrial fragmentation in LINCL cells and does not prevent the recovery of the mitochondrial network after TPP-1 loading.
Figure 4. Effects of a TPP-1 deficiency on mitochondrial morphology.
(A) Representative confocal micrographs of mitochondrial morphology of fibroblasts pre-incubated with (CTL+FCCP) or without (CTL) 20 μM FCCP for 30 min and of LINCL cells pre-incubated with (LINCL+TPP-1) or without (LINCL) TPP-1 and stained with 100 nM Mitotracker Green. (B and C) The length (or AR) and the degree of branching of mitochondria (or FF) were determined on micrographs using the ImageJ software as described in the Material and methods section. Results are expressed in either AR (B) or FF ratio (C) and represent means±S.E.M. (n≥ 59 from four independent experiments). ###: significantly different from CTL cells with P<0.001; *, ***: statistically different from LINCL cells as determined by a Mann–Whitney Rank Sum test with P<0.05 and P<0.001, respectively.
Figure 5. Effects of BNIP3 silencing on the mitochondrial morphology of LINCL and LINCL cells that recovered TPP-1 activity.
(A) Efficiency of siRNA-mediated knock down of BNIP3 in LINCL cells. The abundance of BNIP3 was determined by Western blotting analysis performed on 15 μg of clear cell lysates prepared from untransfected LINCL cells and cells transfected either with a pool of four specific siRNAs targeting BNIP3 (siBNIP3) or with a control pool of siRNAs (siCTL). Equal protein loading was controlled by the immunodetection of α-tubulin. (B) Representative confocal micrographs of mitochondrial morphology of LINCL and LINCL cells with restored TPP-1 activity, transfected either with a control pool of siRNAs (siCTL) or with a pool of four specific siRNAs targeting BNIP3 (siBNIP3) and then stained with 100 nM Mitotracker Green. The length (AR) and the degree of branching of mitochondria (FF) were determined on micrographs using the ImageJ software, as described in the Material and methods section. Results are expressed in either AR (C) or FF ratio (D) as means±S.E.M. (n≥50 from three independent experiments). *: significantly different from LINCL cells transfected with the siCTL with P<0.05, as determined by an ANOVA-2 followed by a Holm–Sidak test and by a Mann–Whitney Rank Sum test for the AR and the FF, respectively. NS, not significantly different from LINCL cells transfected with the siCTL.
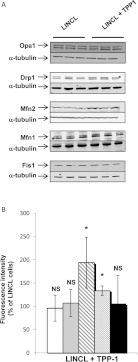
TPP-1 deficiency induces a decrease in the abundance of Mfn1 and Mfn2 proteins
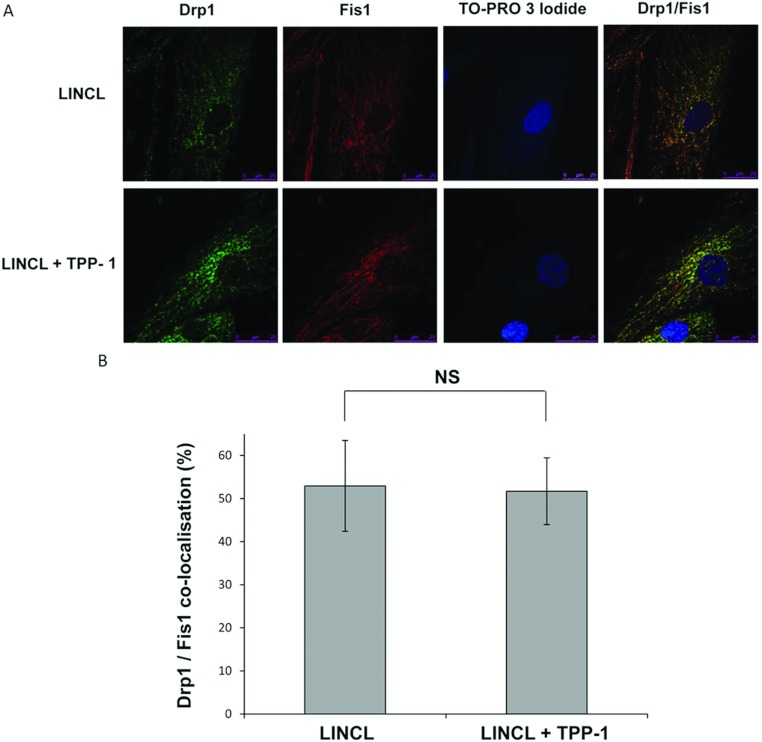
To get information about putative mechanisms leading to mitochondria fragmentation in LINCL cells, we next quantified the abundance of both Mfn1 and Mfn2 (regulating fusion events of OMM), Opa1 (regulating fusion events of the IMM), Fis1 and Drp1 (controlling fission events), the major proteins regulating the morphology of the mitochondrial reticulum [16]. The Western blot analysis revealed that the abundance of the proteins Fis1, Drp1, and Opa1 is comparable in LINCL and LINCL cells loaded with TPP-1 (Figure 6). In contrast, the recovery of the TPP-1 activity in LINCL cells significantly increases the total abundance of Mfn1 and Mfn2 (Figure 6). As mentioned before, Drp1 is a cytosolic GTPase that is recruited onto mitochondria upon activation [39]. Given its mechanism of action, we wondered whether Drp1 could be differentially targeted to mitochondria of LINCL cells, even if the total abundance of this protein is unchanged in these cells. To test this hypothesis, we analysed the putative co-localization between Drp1 and the mitochondrial protein Fis1 (Figure 7A), a protein known to participate in the mitochondrial recruitment of this GTPase [16]. However, the percentage of co-localization between these two proteins is not modified in LINCL cells when compared with LINCL cells loaded with TPP-1 (Figure 7B) suggesting that Drp1 is not differentially recruited onto the mitochondria of LINCL cells. It is thus unlikely that these proteins regulating fission events might contribute to the observed fragmentation of the mitochondrial network in LINCL cells. Altogether, our findings suggest that the more fragmented mitochondrial network observed in LINCL cells would more likely result from a deficit of mitochondrial fusion than from an enhanced fission of mitochondria.
Figure 6. Effects of a TPP-1 deficiency on the abundance of major regulators of mitochondrial dynamics.
(A) The abundance of Opa1, Drp1, Mfn2, Mfn1 and Fis1 was determined by fluorescence Western blot analysis on 15 μg of clear cell lysates prepared from LINCL cells pre-incubated with (LINCL+TPP-1) or without (LINCL) TPP-1 (n=3). Equal protein loading was controlled by the immunodetection of α-tubulin. Asterisks denote non-specific and uncharacterized bands. (B) Quantification of the abundance of Opa1 (white columns), Drp1 (grey columns), Mfn2 (hatched lines), Mfn1 (dots) and Fis1 (black columns) on the blots shown in (A). Results are calculated as fluorescence intensity normalized for α-tubulin and expressed as a percentage of LINCL cells as means±S.D. (n=3). *: significantly different from LINCL cells with P<0.05, as determined by a Student's t test.
Figure 7. Cellular distribution of Drp1/Fis1 and colocalization of these proteins as visualized by immunofluorescence confocal microscopy.
(A) Representative micrographs of Drp1 and/or Fis1 immunostaining taken with a confocal microscope. LINCL cells were cultured on coverslips and were incubated with (LINCL+TPP-1) or without (LINCL) TPP-1 before being fixed, permeabilized and incubated with antibodies raised against Fis1-(red) or Drp1-(green). In merged micrographs, the appearance of the yellow fluorescence signals reflects the colocalization between both proteins. Nuclei were stained with TO-PRO-3 iodide (blue fluorescence). (B) The co-localization between Drp1 and Fis1 was quantified using the co-localization module of Leica LAS AF software. The results are expressed as the percentages of co-localization (means±S.D.; n=20). NS, non-significantly different from LINCL cells, as determined by a Student's t test.
Mitochondrial membrane potential, abundance of O2•− and mitochondrial matrix calcium concentration
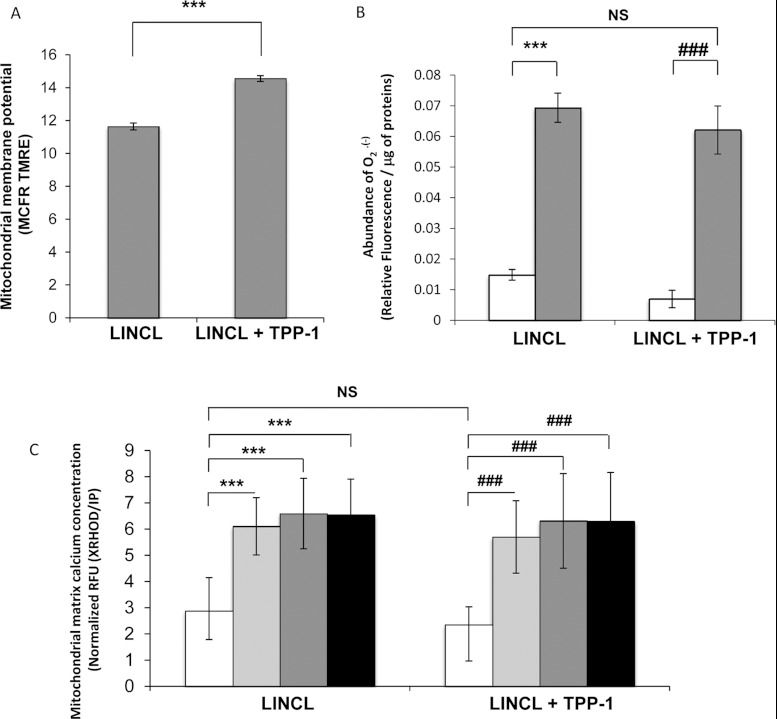
The fragmentation of mitochondria is often, but not always [40], associated with a decrease in the mitochondrial membrane potential. It is also known that mitofusins (Mfn1 and Mfn2) can be targeted for degradation in response to IMM depolarization [41]. Therefore we analysed the mitochondrial membrane potential in cells loaded with 25 nM TMRE (low concentration in non-quenching conditions) using flow cytometry. As shown in Figure 8(A), the mitochondrial membrane potential is significantly increased in LINCL cells that recovered a TPP-1 activity. This result suggests that the mitochondrial membrane potential is lower in LINCL cells, a finding in agreement with a more fragmented mitochondria phenotype in these cells. It should also be emphasized that the change in the TMRE fluorescent signal for TPP-1-loaded cells is unlikely to result from a difference in mitochondrial mass between LINCL and LINCL cells loaded with TPP-1 as we failed to detect any significant difference in the abundance of the organelle between these cells with the Mitotracker Green staining (Figure 2A).
Figure 8. Effects of a TPP-1 deficiency on the mitochondrial membrane potential, mitochondrial matrix calcium concentration and mitochondrial O2•− content.
(A) The mitochondrial membrane potential was analysed in LINCL cells pre-incubated with (LINCL+TPP-1) or without (LINCL) recombinant TPP-1, using the specific mitochondrial membrane potential probe TMRE. Cells were stained or not (to allow autofluorescence determination from cells without dye) with 25 nM TMRE for 30 min and fluorescence was analysed by flow cytometry. Results are expressed as TMRE fluorescence MCFR as means±S.D. (n=3), ***: significantly different from LINCL cells with P<0.001 as determined by a Student's t test. (B) The mitochondrial O2•− content was assessed in LINCL cells pre-incubated with (LINCL+TPP-1) or without (LINCL) TPP-1 using the specific mitochondrial MitoSOX™ Red dye. Cells were stained (white and grey columns) or not (to allow autofluorescence determination from cells without dye) with 10 μM MitoSOX™ Red specific dye for 10 min and fluorescence intensities were measured using a spectrofluorimeter (λexc, 485 nm; λem, 520 nm). As a positive control, cells were incubated for 15 min with 10 μM antimycin A (grey columns) while untreated cells were maintained for 15 min without molecule (white columns). Results were normalized for protein content and expressed in relative fluorescence arbitrary unit/μg of proteins as means±S.D. (n=3). ***, ###: significantly different from untreated LINCL and LINCL+TPP-1 cells, respectively, with P<0.001 as determined by an ANOVA-2 followed by a Holm-Sidak's t test. NS, non-significantly different from LINCL cells. (C) The mitochondrial calcium concentration in LINCL cells pre-incubated with (LINCL+TPP1) or without (LINCL) recombinant TPP-1 was determined using X-Rhod-5F, a specific mitochondrial matrix calcium probe. Cells were stained or not (to allow autofluorescence determination from cells without dye) with 2 μM X-Rhod-5F fluorescent probe for 30 min and fluorescence intensities were measured in a spectrofluorimeter (λexc, 580 nm; λem, 612 nm). To test mitochondrial calcium buffering capacity, cells were incubated with 10 μM ionomycin for 10 min (grey columns), 20 min (dark grey columns) or 30 min (black columns) or left untreated (white columns). Results are expressed as Relative Fluorescence Units (RFU) normalized for DNA content determined by propidium iodide (IP) staining as means±S.D. (n=9). ***, Significantly different from LINCL cells with P<0.001, ###: significantly different from LINCL+TPP-1 cells as determined by a Mann–Whitney Rank Sum test. NS, non-significantly different from LINCL cells.
Interestingly, in models of MLIV or nephropathic cystinosis, mitochondrial fragmentation and aberrations have also been associated with a decrease in the mitochondrial calcium buffering capacity and/or with an increased production of ROS [18,19]. To assess the putative effect of the morphological changes on these parameters, we first quantified the mitochondrial abundance of superoxide anion radicals O2•− in LINCL and LINCL cells loaded with TPP-1 cells using the MitoSOX™ fluorescent dye [42]. We found that the O2•− abundance, though slightly lower in LINCL cells that recovered TPP-1 activity, was not significantly different from fluorescent signals measured in LINCL cells (Figure 8B). As a positive control, we also incubated cells for 15 min with 10 μM antimycin A, an inhibitor of the complex III of the respiratory chain, known to induce ROS production at this concentration [43]. In these conditions, as expected, the O2•− production is increased (Figure 8B). However, the increase is comparable for both LINCL cells and LINCL cells that recovered TPP-1 activity. These results suggest that, in terms of ROS production, both LINCL and LINCL cells loaded with TPP-1 for 5 days respond similarly to an inducer of mitochondrial oxidative stress.
Then, using X-Rhod-5F, a specific fluorescent probe allowing the measurement of mitochondrial matrix calcium concentration [44], we determined the putative effects of a TPP-1 deficiency on the [Ca2+]mt concentration homoeostasis (Figure 8C). We observed that the basal [Ca2+]mt was not modified by the TPP-1 deficiency. In order to assess the buffering capacity of mitochondria, LINCL and LINCL fibroblasts that recovered TPP-1 activity were loaded with X-Rhod-5F and incubated for 10, 20 and 30 min with 10 μM ionomycin, a calcium-specific ionophore known to induce changes in the cytosolic calcium concentration [31]. After a challenge with ionomycin, a strong and significant increase in the [Ca2+]mt was observed. However, this increase was comparable in both LINCL and LINCL cells loaded with TPP-1, suggesting that the calcium buffering capacity is not altered in TPP-1-deficient fibroblasts (Figure 8C).
BNIP3 sensitizes LINCL cells to antimycin A-induced mitochondrial fragmentation
The final question we addressed in this study was about the role of BNIP3 overexpression in LINCL cells. Several studies have shown that, when ectopically overexpressed, BNIP3 is detected in the mitochondria as a single monomer loosely attached to the OMM [45]. Although the molecular mechanisms involved in the activation of BNIP3 are not completely elucidated, it has been reported that it requires an increase in cytosolic pH, in cytosolic calcium concentration or generation of an oxidative stress [46]. These modifications would trigger BNIP3 embedding into the OMM, its dimerization and, in fine, mitochondrial dysfunction. It is therefore possible that even if this protein is overexpressed in LINCL fibroblasts, it might be kept in an inactive state requiring appropriate stimuli to become biologically active. This hypothesis could explain why this protein does not seem to participate in the fragmentation of mitochondria in LINCL cells. In order to test this hypothesis, we analysed the mitochondrial morphology in cells exposed to high concentrations of antimycin A. LINCL and LINCL cells loaded with TPP-1 cells were first incubated with different concentrations of antimycin A to determine the most appropriate concentration to induce changes in mitochondrial morphology. We noticed that a 1 h incubation with 25 μM antimycin A caused a mitochondrial fragmentation significantly higher in LINCL cells when compared with LINCL cells loaded with TPP-1 (results not shown). Then, we repeated the same experiment on fibroblasts in which BNIP3 expression was silenced with siRNA (Figure 9A). The observations of the mitochondrial morphology in cells pre-incubated with antimycin A for 1 h clearly revealed a more important fragmentation of the mitochondrial network in LINCL cells incubated with a control siRNA when compared with pre-incubated LINCL cells in which the expression of BNIP3 was silenced, as demonstrated by the significant increase in both the AR and the FF parameters (Figures 9A and B). In contrast, in LINCL cells that recovered TPP-1 for 5 days, BNIP3 silencing has no impact on either the AR or the FF after addition of antimycin A. This could be linked to the lower expression of BNIP3 in these cells (Figure 3). These results suggest that although BNIP3 does not seem to be involved in the fragmentation of the mitochondrial network in LINCL cells in basal conditions, it could contribute to enhancing this process during mitochondrial oxidative stress conditions.
Figure 9. Effects of BNIP3 silencing on antimycin A-induced mitochondrial fragmentation in LINCL cells.
(A) Representative confocal micrographs of mitochondrial morphology in LINCL and LINCL cells that recovered TPP-1 activity. At the end of the incubations, cells were transfected for 24 h with either a pool of four specific siRNAs targeting BNIP3 (siBNIP3) or with a control pool of siRNAs (siCTL), incubated with 25 μM antimycin A for 1 h and eventually loaded with 100 nM Mitotracker Green before analysis. The length (AR) and the degree of branching of mitochondria (FF) were determined using the ImageJ software, as described in the Material and methods section. Results are expressed in either AR (B) or FF ratio (C) as means±S.E.M. (n≥40 from three independent experiments). **,***: significantly different from LINCL cells transfected with the siCTL with P<0.01 or 0.001, respectively, as determined by a Mann–Whitney Rank Sum test. NS, non-significantly different. NS, non-significantly different from LINCL cells pre-incubated with TPP-1 and transfected with the siCTL.
DISCUSSION
Several studies show that the mitochondrial population exhibits morphological and functional alterations in response to a lysosomal dysfunction and, therefore, suggest that mitochondrial impairment could be a common characteristic in the pathogenesis of LSDs [12,13,17–19]. These monogenic diseases, notably the NCLs, are often associated with neurodegeneration [7]. Interestingly, modifications of the mitochondrial morphology leading to alterations of mitochondrial functions, such as ATP production, have been associated with the pathogenesis of several neurodegenerative disorders caused by the intracellular accumulation of proteins [47]. For example, in AD (Alzheimer's disease), the NO (nitric oxide) produced in response to β-amyloid protein accumulation was shown to trigger mitochondrial fission, synaptic loss and neuronal damage through the activation of Drp1 by S-nitrosylation [48]. Likewise, in Huntington's disease, the accumulating mutant Huntingtin protein also increased the activity of this GTPase, leading to an excessive mitochondrial fragmentation and, in fine, to synaptic degeneration [49]. Moreover, pharmacological molecules that stimulate mitochondrial biogenesis were also reported to have beneficial effects on the neuronal survival and atrophy, suggesting an essential role of mitochondrial dysfunction in the pathogenesis of these diseases [50,51]. So far, while modifications in the dynamics of mitochondria have been reported in several LSDs, the molecular mechanisms that have been proposed were mainly related to a defect in the selective degradation of damaged mitochondria by the autophagic process (also called mitophagy) [18,52,53]. To our knowledge, the expression of regulators of mitochondrial fusion and fission events has never been studied in the context of these pathologies.
The aim of this study was to analyse the putative impact of a TPP-1 deficiency on the mitochondrial population in fibroblasts. To allow a more reliable interpretation of the results, the control fibroblastic cells were constituted of TPP-1-deficient cells in which TPP-1 activity was restored using a protocol previously described [25]. This kind of control is essential to avoid drawing conclusions from differences that could be merely due to differences in the genetic backgrounds between fibroblast cell lines used for controls and lysosomal deficiency. However, one cannot completely exclude that some modifications produced by the enzymatic deficiency persist even after enzyme recovery because some alterations might require higher dose and/or longer incubation time to be reversed or could be irreversible. In addition, the substrates of the deficient enzyme that accumulate upon enzymatic deficiency might undergo a chemical modification such as oxidation, preventing their degradation when the enzymatic activity is recovered [54].
We first analysed the effect of a TPP-1 deficiency on the abundance and the morphology of the mitochondrial population. Our results show that even though the abundance of mitochondria, determined by the Mitotracker Green staining followed by FACS analysis, is not modified in LINCL fibroblasts, these cells clearly exhibit a reversible fragmentation of the mitochondrial network. These findings support the conclusions of Kiselyov and co-workers [18]. The mitochondrial fragmentation seems to be a common feature in the LSDs since it has been described in many cell types that display various and unrelated LSDs [12,13,17–19]. As mentioned above, a first hypothesis to explain changes in morphology in cells affected by LSDs is that the impairment of the autophagic process induces the accumulation of undegraded and ‘old’ fragmented mitochondria. This hypothesis relies on the observation that the mitochondrial fragmentation in MLIV fibroblasts (deficient for the mucolipin 1) can be reproduced in healthy cells incubated with 3-MA (3-methyl adenine), an inhibitor of the class III phosphatidylinositol 3-kinase activity known to inhibit autophagy [18]. However, it has also been suggested that stimulation, rather than inhibition, of the autophagic process could lead to the fragmentation of the mitochondrial network observed in MLII and MLIII skin fibroblasts. Indeed, Sakai and co-workers showed that both the mitochondrial morphology and the mitochondrial membrane potential were restored following the inhibition of autophagosome formation with 3-MA [53]. However, our preliminary results do not support any induction or impairment of this process either in the TPP-1-deficient cells or in the brain from 120-days-old CLN2 −/− mice, since we failed to observe any conversion of LC3-I into LC3-II, a frequently used marker of macroautophagy (results not shown). Furthermore, a deficient autophagic mechanism is expected to provoke a global accumulation of mitochondria. In our experimental conditions, a global accumulation of mitochondria was not observed in LINCL cells as determined by the abundance analysis of mitochondrial population stained with the Mitotracker Green fluorescent probe (Figure 2). Another hypothesis proposed by Simonati and co-workers to explain the fragmentation of mitochondria in LSDs, is that lysosomal biogenesis and proliferation observed in response to the accumulation of undegraded substrate could induce a mechanical stress on the mitochondrial membrane, causing a decrease in the mitochondrial membrane potential and, in fine, mitochondrial fragmentation [13].
The major findings of our study lie in the mechanisms by which the mitochondrial fragmentation could occur in LINCL cells. Indeed, we found that this phenomenon is associated with a decrease in the abundance of Mfn1 and Mfn2, whereas the abundance of Drp1 and Fis1 (two proteins involved in mitochondria fission) is unchanged, suggesting that mitochondrial fragmentation in cells that display a TPP-1 deficiency could result from a deficit of fusion events. It is noteworthy that a differential recruitment of Drp1 to Fis1 in LINCL cells is also unlikely as no modifications in the co-localization of these two proteins could be observed between LINCL cells and cells that recovered TPP-1 activity.
Interestingly, in our research of proteins that are differentially expressed in response to a TPP-1 deficiency, we identified BNIP3, a BH3-only protein Bcl2 member whose expression has been described to induce mitochondrial dysfunction characterized by mitochondrial fragmentation, mitochondrial membrane potential depolarization and opening of the mitochondrial transition pore that can, in fine, lead to cell death [23,24]. As a first attempt to confirm the overexpression of BNIP3 in a more relevant model, we also analysed the expression of the gene encoding this protein in the brain of TPP-1-deficient mice. This experiment did not allow us to show any increase in BNIP3 expression (results not shown). It should be noted that the neurodegeneration observed in the brain of the TPP-1-deficient mice has been described to affect particular brain regions such as the cerebellum (in which the Purkinje cells are the most affected cells [9]). Immunochemistry studies of the BNIP3 expression in specific areas of the brain of CLN2 −/− mice over the time course of the disease development would be necessary to determine whether this protein could be involved in the neurodegeneration observed during the LINCL disease or if the differential expression is restricted to fibroblasts.
Knowing the multiple role of BNIP3 in the regulation of mitochondrial morphology, we tested the putative involvement of this protein in the mitochondrial fragmentation observed in LINCL cells. Using an RNAi approach, we show that BNIP3 is not involved in the mitochondrial fragmentation found in TPP-1-deficient cells (Figure 4), indirectly lending support to a role for Mfn1 and Mfn2 proteins in the change of mitochondrial morphology in response to TPP-1 deficiency. The relative abundance of the mRNA encoding these proteins is not modified in LINCL cells (results not shown). Therefore we can hypothesize that the reduced abundance of both Mfn1 and Mfn2 in LINCL cells could result from post-translational modifications reducing their half-life time. Interestingly, it has been recently shown that mitochondrial depolarization induced by the CCCP (carbonyl cyanide m-chlorophenylhydrazone; a protonophore) leads to the recruitment of the E3 ubiquitin ligase PARKIN onto the mitochondria. The ubiquitinylated mitofusin proteins would next be targeted to proteosomal degradation [41], a process that could explain why we observe a difference in the abundance of mitofusin proteins in LINCL cells without any change in the abundance of their transcripts. Even though Drp1 is most probably not involved in the mitochondrial fragmentation observed in LINCL cells, one cannot completely rule out the possibility that the activity of this protein can be specifically modified at the OMM. Indeed, McBride and co-workers recently showed that SNEP5-mediated Drp1 deSUMOylation on the mitochondrial membrane enhances its anchoring into functional oligomers. This leads to an increase in overall recycling rates of this protein, facilitating mitochondria fission without any modification of the mitochondrial abundance of Drp1 [55].
Mitochondrial fragmentation observed in LINCL cells is associated with a decrease in the mitochondrial membrane potential (Figure 8A). Nevertheless, neither the basal mitochondrial matrix calcium concentration nor the calcium buffering capacity (Figure 8B) and the mitochondrial ROS abundance (Figure 8C) seem to be changed in cells affected by the enzymatic defect. Our results could therefore suggest that contrary to the hypothesis initially proposed by Kiselyov and co-workers [18], a defect in calcium handling by the mitochondria is not a general feature of LSDs. However, in that study, the authors impaired calcium homoeostasis using bradykinin, which induces Ca2+ release from the ER [56] whereas, in our study, we used ionomycin, a Ca2+-specific ionophore that has been isolated from the bacterium Streptomyces conglobatus [57]. Perhaps, this could explain why in our model, the mitochondrial fragmentation in LINCL cells is not associated with a decrease in the mitochondrial calcium buffering capacity. Indeed, it is well known that mitochondrial Ca2+ uptake from the ER requires close and physical interactions between both organelles through contact sites or MAMs (mitochondrial associated membranes) [58]. The lack of Ca2+ uptake by mitochondria in response to bradykinin or other calcium agonists, such as histamine, suggests that the interactions between the mitochondria and the ER could be altered in cells with LSDs [59]. Recently, d’Azzo and co-workers [20] showed, in a β-galactosidase-deficient mouse model, that the accumulation of GM1-ganglioside inside the MAMs induces the clustering of the IP3 receptor-1 (inositol trisphosphate receptor-1), VDAC (voltage-dependent anion channel) and Grp75, thereby leading to the formation of Ca2+ mega pores between the two organelles. The authors showed that the treatment of β-galactosidase-deficient cells with histamine results in a Ca2+ overload in mitochondria, leading in fine to apoptosis [20]. Interestingly, Mfn2 is another protein essential for the tethering between the ER and the mitochondria and for the efficiency of mitochondrial Ca2+ buffering in response to stimuli that generate inositol-1,4,5-trisphosphate [60]. The reduced abundance of that protein in LINCL cells therefore suggests that the interactions between both organelles might be less frequent in LINCL cells.
The last question we wanted to address in this study was to determine the biological role of the overexpressed BNIP3 in LINCL cells. Knowing that BNIP3 can be expressed at a high level without being functionally active [34], we decided to artificially increase mitochondrial ROS production using antimycin A to trigger its activation. Our results clearly indicate that, although BNIP3 does not seem to be involved in the mitochondrial fragmentation observed in a basal state in LINCL cells, it sensitizes LINCL cells to antimycin A-induced mitochondrial fragmentation, probably because it is activated in the presence of ROS. Supporting this interpretation, activation of BNIP3 in response to an oxidative stress has been described in other experimental models, such as cardiac myocytes exposed to ischaemia and reperfusion or C6 glioma cells treated with H2O2 [23,61]. In C6 glioma cells exposed to H2O2, it has been shown that BNIP3 is over-expressed and forms oligomers that in turn, mediate the suppression of the mTOR (mammalian target of rapamycin) pathway and the activation of autophagy, leading to cell death [61]. Knowing that the involvement of apoptosis in cell death in NCL diseases is still a matter of debate, the idea that BNIP3 could mediate and enhance mitochondrial dysfunction in cells from patients suffering from LINCL in response to external (oxidative) stress conditions is interesting and their potential role in the neurodegeneration observed in NCLs should be tested in the future. Interestingly, a recent study has revealed that TPP-1-deficient cells exhibit a defect in TNFα (tumour necrosis factor α)-induced Bid cleavage, cytochrome c release and caspase 9 and caspase 3 activation [6]. These authors also mentioned that this resistance is no longer observed when cells are treated with ceramide or staurosporine, two apoptosis inducers acting through mitochondrial dysfunction, suggesting that this pathway could be involved in the induced neurodegeneration of cells affected by a TPP-1 deficiency [6]. The fact that BNIP3-mediated cell death was shown to be independent of caspase 3 and cytochrome c release in some cell types [24,62], argues in favour of a putative role of BNIP3 in the cell death observed in LINCL cells exposed to an external stress. As proposed by Zhao and co-workers, the BNIP3-mediated cell death would mainly involve the nuclear translocation of endonuclease G, an apoptotic DNase when released from mitochondria [63].
Eventually, several questions remain to be addressed at the end of this study such as the identification of the transcription factor(s) and cell signalling leading to BNIP3 overexpression in LINCL cells. So far, the expression of the gene-encoding BNIP3 is known to be regulated by the activity of several transcription factors. Among them, HIF-1 (hypoxia-inducible factor-1) [64], FOXO3 (forkhead box O3) [65], E2F1 [66] and more recently, ATF3 (activating transcription factor 3) [67], have been described as transcriptional activators, unlike p53 [68] and NF-κB (nuclear factor κB) [69] that would act as repressors. The molecular mechanisms leading to the increased expression of BNIP3 and the study of the role of this protein in the sensitivity of LINCL cells to induced apoptosis should be determined in the future to get a more precise view of the role of this protein in LSDs.
AUTHOR CONTRIBUTION
All the authors contributed to the study design and data interpretation and analysis. Guillaume Van Beersel performed the experiments related to the analysis of the mitochondrial network morphology (both basal state and in response to antimicin A) and the statistical analysis, as well as the paper. Other experiments related to calcium and ROS production were performed by Stephane Demine and Eliane Tihon. Isabelle Hamer helped to draft the paper. Thierry Arnould and Michel Jadot conceived the study, participated in its design and data interpretation and also revised the paper.
ACKNOWLEDGEMENTS
We thank Peter Lobel (Center for Advanced Biotechnology and Medicine, Piscataway, NJ, U.S.A.) for the gift of the recombinant TPP-1 pro-enzyme and of samples from brain CLN2−/− mice.
FUNDING
This work was supported by the Action de Recherche Concertée-Fédération Wallonie-Bruxelles [grant number ARC#326]; and the Fonds National de la Recherche Scientifique (FSR-FNRS) [grant number 2.4650.06]. G.V.B. is a Research Fellow of the FSR-FNRS.
References
- 1.Walkley S. U., Vanier M. T. Secondary lipid accumulation in lysosomal disease. Biochim. Biophys. Acta. 2009;1793:726–736. doi: 10.1016/j.bbamcr.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Getty A. L., Pearce D. A. Interactions of the proteins of neuronal ceroid lipofuscinosis: clues to function. Cell Mol. Life Sci. 2011;68:453–474. doi: 10.1007/s00018-010-0468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohar I., Sleat D. E., Jadot M., Lobel P. Biochemical characterization of a lysosomal protease deficient in classical late infantile neuronal ceroid lipofuscinosis (LINCL) and development of an enzyme-based assay for diagnosis and exclusion of LINCL in human specimens and animal models. J. Neurochem. 1999;73:700–711. doi: 10.1046/j.1471-4159.1999.0730700.x. [DOI] [PubMed] [Google Scholar]
- 4.Bernardini F., Warburton M. J. Lysosomal degradation of cholecystokinin-(29–33)-amide in mouse brain is dependent on tripeptidyl peptidase-I: implications for the degradation and storage of peptides in classical late-infantile neuronal ceroid lipofuscinosis. Biochem. J. 2002;366:521–529. doi: 10.1042/BJ20020467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezaki J., Tanida I., Kanehagi N., Kominami E. A lysosomal proteinase, the late infantile neuronal ceroid lipofuscinosis gene (CLN2) product, is essential for degradation of a hydrophobic protein, the subunit c of ATP synthase. J. Neurochem. 1999;72:2573–2582. doi: 10.1046/j.1471-4159.1999.0722573.x. [DOI] [PubMed] [Google Scholar]
- 6.Autefage H., Albinet V., Garcia V., Berges H., Nicolau M. L., Therville N., Altie M. F., Caillaud C., Levade T., Andrieu-Abadie N. Lysosomal serine protease CLN2 regulates tumor necrosis factor-alpha-mediated apoptosis in a Bid-dependent manner. J. Biol. Chem. 2009;284:11507–11516. doi: 10.1074/jbc.M807151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sleat D. E., Donnelly R. J., Lackland H., Liu C. G., Sohar I., Pullarkat R. K., Lobel P. Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science. 1997;277:1802–1805. doi: 10.1126/science.277.5333.1802. [DOI] [PubMed] [Google Scholar]
- 8.Worgall S., Kekatpure M. V., Heier L., Ballon D., Dyke J. P., Shungu D., Mao X., Kosofsky B., Kaplitt M. G., Souweidane M. M., et al. Neurological deterioration in late infantile neuronal ceroid lipofuscinosis. Neurology. 2007;69:521–535. doi: 10.1212/01.wnl.0000267885.47092.40. [DOI] [PubMed] [Google Scholar]
- 9.Sleat D. E., Wiseman J. A., El-Banna M., Kim K. H., Mao Q., Price S., Macauley S. L., Sidman R. L., Shen M. M., Zhao Q., et al. A mouse model of classical late-infantile neuronal ceroid lipofuscinosis based on targeted disruption of the CLN2 gene results in a loss of tripeptidyl-peptidase I activity and progressive neurodegeneration. J. Neurosci. 2004;24:9117–9126. doi: 10.1523/JNEUROSCI.2729-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitner E. B., Platt F. M., Futerman A. H. Common and uncommon pathogenic cascades in lysosomal storage diseases. J. Biol. Chem. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei H., Zhang Z., Saha A., Peng S., Chandra G., Quezado Z., Mukherjee A. B. Disruption of adaptive energy metabolism and elevated ribosomal p-S6K1 levels contribute to INCL pathogenesis: partial rescue by resveratrol. Hum. Mol Genet. 2011;20:1111–1121. doi: 10.1093/hmg/ddq555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takamura A., Higaki K., Kajimaki K., Otsuka S., Ninomiya H., Matsuda J., Ohno K., Suzuki Y., Nanba E. Enhanced autophagy and mitochondrial aberrations in murine G(M1)gangliosidosis. Biochem. Biophys. Res. Commun. 2008;367:616–622. doi: 10.1016/j.bbrc.2007.12.187. [DOI] [PubMed] [Google Scholar]
- 13.Pezzini F., Gismondi F., Tessa A., Tonin P., Carrozzo R., Mole S. E., Santorelli F. M., Simonati A. Involvement of the mitochondrial compartment in human NCL fibroblasts. Biochem. Biophys. Res. Commun. 2011;416:159–164. doi: 10.1016/j.bbrc.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Michel S., Wanet A., De Pauw A., Rommelaere G., Arnould T., Renard P. Crosstalk between mitochondrial (dys)function and mitochondrial abundance. J. Cell. Physiol. 2012;227:2297–2310. doi: 10.1002/jcp.23021. [DOI] [PubMed] [Google Scholar]
- 15.Kuznetsov A. V., Hermann M., Saks V., Hengster P., Margreiter R. The cell-type specificity of mitochondrial dynamics. Int. J. Biochem. Cell Biol. 2009;41:1928–1939. doi: 10.1016/j.biocel.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Palmer C. S., Osellame L. D., Stojanovski D., Ryan M. T. The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell. Signal. 2011;23:1534–1545. doi: 10.1016/j.cellsig.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Raben N., Wong A., Ralston E., Myerowitz R. Autophagy and mitochondria in Pompe disease: nothing is so new as what has long been forgotten. Am. J. Med. Genet. C, Semin. Med. Genet. 2012;160:13–21. doi: 10.1002/ajmg.c.31317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jennings J. J., Jr, Zhu J. H., Rbaibi Y., Luo X., Chu C. T., Kiselyov K. Mitochondrial aberrations in mucolipidosis Type IV. J. Biol. Chem. 2006;281:39041–39050. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 19.Sansanwal P., Sarwal M. M. Abnormal mitochondrial autophagy in nephropathic cystinosis. Autophagy. 2010;6:971–973. doi: 10.4161/auto.6.7.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sano R., Annunziata I., Patterson A., Moshiach S., Gomero E., Opferman J., Forte M., d’Azzo A. GM1-ganglioside accumulation at the mitochondria-associated ER membranes links ER stress to Ca(2+)-dependent mitochondrial apoptosis. Mol. Cell. 2009;36:500–511. doi: 10.1016/j.molcel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sansanwal P., Li L., Hsieh S. C., Sarwal M. M. Insights into novel cellular injury mechanisms by gene expression profiling in nephropathic cystinosis. J. Inherit. Metab. Dis. 2010;33:775–786. doi: 10.1007/s10545-010-9203-6. [DOI] [PubMed] [Google Scholar]
- 22.Heine C., Tyynela J., Cooper J. D., Palmer D. N., Elleder M., Kohlschutter A., Braulke T. Enhanced expression of manganese-dependent superoxide dismutase in human and sheep CLN6 tissues. Biochem. J. 2003;376:369–376. doi: 10.1042/BJ20030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubli D. A., Ycaza J. E., Gustafsson A. B. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem. J. 2007;405:407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vande Velde C., Cizeau J., Dubik D., Alimonti J., Brown T., Israels S., Hakem R., Greenberg A. H. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 2000;20:5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L., Lobel P. Production and characterization of recombinant human CLN2 protein for enzyme-replacement therapy in late infantile neuronal ceroid lipofuscinosis. Biochem. J. 2001;357:49–55. doi: 10.1042/0264-6021:3570049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson-Fritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews S. T., Plaisance E. P., Kim T. Imaging systems for westerns: chemiluminescence vs. infrared detection. Methods Mol. Biol. 2009;536:499–513. doi: 10.1007/978-1-59745-542-8_51. [DOI] [PubMed] [Google Scholar]
- 28.de Longueville F., Atienzar F. A., Marcq L., Dufrane S., Evrard S., Wouters L., Leroux F., Bertholet V., Gerin B., Whomsley R., et al. Use of a low-density microarray for studying gene expression patterns induced by hepatotoxicants on primary cultures of rat hepatocytes. Toxicol. Sci. 2003;75:378–392. doi: 10.1093/toxsci/kfg196. [DOI] [PubMed] [Google Scholar]
- 29.De Vos K. J., Sheetz M. P. Visualization and quantification of mitochondrial dynamics in living animal cells. Methods Cell Biol. 2007;80:627–682. doi: 10.1016/S0091-679X(06)80030-0. [DOI] [PubMed] [Google Scholar]
- 30.Zinchuk V., Zinchuk O., Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem. Cytochem. 2007;40:101–111. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elzi D. J., Bjornsen A. J., MacKenzie T., Wyman T. H., Silliman C. C. Ionomycin causes activation of p38 and p42/44 mitogen-activated protein kinases in human neutrophils. Am. J. Physiol. Cell Physiol. 2001;281:C350–360. doi: 10.1152/ajpcell.2001.281.1.C350. [DOI] [PubMed] [Google Scholar]
- 32.Vanneste J., van den Bosch de Aguilar P. Mitochondrial alterations in the spinal ganglion neurons in ageing rats. Acta Neuropathol. 1981;54:83–87. doi: 10.1007/BF00691335. [DOI] [PubMed] [Google Scholar]
- 33.Pendergrass W., Wolf N., Poot M. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A. 2004;61:162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 34.Chinnadurai G., Vijayalingam S., Gibson S. B. BNIP3 subfamily BH3-only proteins: mitochondrial stress sensors in normal and pathological functions. Oncogene. 2008;27(Suppl. 1):S114–127. doi: 10.1038/onc.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burton T. R., Henson E. S., Baijal P., Eisenstat D. D., Gibson S. B. The pro-cell death Bcl-2 family member, BNIP3, is localized to the nucleus of human glial cells: implications for glioblastoma multiforme tumor cell survival under hypoxia. Int. J. Cancer. 2006;118:1660–1669. doi: 10.1002/ijc.21547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y., Lee H. Y., Hanna R. A., Gustafsson A. B. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1924–1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rikka S., Quinsay M. N., Thomas R. L., Kubli D. A., Zhang X., Murphy A. N., Gustafsson A. B. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landes T., Emorine L. J., Courilleau D., Rojo M., Belenguer P., Arnaune-Pelloquin L. The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep. 2010;11:459–465. doi: 10.1038/embor.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura K., Nemani V. M., Azarbal F., Skibinski G., Levy J. M., Egami K., Munishkina L., Zhang J., Gardner B., Wakabayashi J., et al. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. J. Biol. Chem. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D. F., Karbowski M., Youle R. J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay P., Rajesh M., Yoshihiro K., Hasko G., Pacher P. Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 2007;358:203–208. doi: 10.1016/j.bbrc.2007.04.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dikalov S. I., Li W., Doughan A. K., Blanco R. R., Zafari A. M. Mitochondrial reactive oxygen species and calcium uptake regulate activation of phagocytic NADPH oxidase. Am. J. Physiol. Regul. Integr. Compar. Physiol. 2012;302:R1134–1142. doi: 10.1152/ajpregu.00842.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paredes R. M., Etzler J. C., Watts L. T., Zheng W., Lechleiter J. D. Chemical calcium indicators. Methods. 2008;46:143–151. doi: 10.1016/j.ymeth.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubli D. A., Quinsay M. N., Huang C., Lee Y., Gustafsson A. B. Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am. J. Physiol. Heart Circ. Physiol. 2008;295:H2025–2031. doi: 10.1152/ajpheart.00552.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graham R. M., Thompson J. W., Wei J., Bishopric N. H., Webster K. A. Regulation of Bnip3 death pathways by calcium, phosphorylation, and hypoxia-reoxygenation. Antioxid. Redox Signal. 2007;9:1309–1315. doi: 10.1089/ars.2007.1726. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Su B., Siedlak S. L., Moreira P. I., Fujioka H., Wang Y., Casadesus G., Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho D. H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., Lipton S. A. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy P. H., Shirendeb U. P. Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington's disease. Biochim. Biophys. Acta. 2012;1822:101–110. doi: 10.1016/j.bbadis.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng B., Wang X., Su B., Lee H. G., Casadesus G., Perry G., Zhu X. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer's disease. J. Neurochem. 2012;120:419–429. doi: 10.1111/j.1471-4159.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johri A., Calingasan N. Y., Hennessey T. M., Sharma A., Yang L., Wille E., Chandra A., Beal M. F. Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington's disease. Hum. Mol. Genet. 2012;21:1124–1137. doi: 10.1093/hmg/ddr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Pablo-Latorre R., Saide A., Polishhuck E. V., Nusco E., Fraldi A., Ballabio A. Impaired parkin-mediated mitochondrial targeting to autophagosomes differentially contributes to tissue pathology in lysosomal storage diseases. Hum. Mol. Genet. 2012;21:1770–1781. doi: 10.1093/hmg/ddr610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Otomo T., Higaki K., Nanba E., Ozono K., Sakai N. Inhibition of autophagosome formation restores mitochondrial function in mucolipidosis II and III skin fibroblasts. Mol. Genet. Metab. 2009;98:393–399. doi: 10.1016/j.ymgme.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Seehafer S. S., Pearce D. A. You say lipofuscin, we say ceroid: defining autofluorescent storage material. Neurobiol. Aging. 2006;27:576–588. doi: 10.1016/j.neurobiolaging.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Zunino R., Braschi E., Xu L., McBride H. M. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J. Biol. Chem. 2009;284:17783–17795. doi: 10.1074/jbc.M901902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farmer S. G., Burch R. M. Biochemical and molecular pharmacology of finin receptors. Annu. Rev. Pharmacol. Toxicol. 1992;32:511–536. doi: 10.1146/annurev.pa.32.040192.002455. [DOI] [PubMed] [Google Scholar]
- 57.Liu W. C., Slusarchyk D. S., Astle G., Trejo W. H., Brown W. E., Meyers E. Ionomycin, a new polyether antibiotic. J. Antibiot. 1978;31:815–819. doi: 10.7164/antibiotics.31.815. [DOI] [PubMed] [Google Scholar]
- 58.Giorgi C., De Stefani D., Bononi A., Rizzuto R., Pinton P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int. J. Biochem. Cell Biol. 2009;41:1817–1827. doi: 10.1016/j.biocel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizzuto R., Duchen M. R., Pozzan T. Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci. STKE. 2004;2004:re1. doi: 10.1126/stke.2152004re1. [DOI] [PubMed] [Google Scholar]
- 60.de Brito O. M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 61.Byun Y. J., Kim S. K., Kim Y. M., Chae G. T., Jeong S. W., Lee S. B. Hydrogen peroxide induces autophagic cell death in C6 glioma cells via BNIP3-mediated suppression of the mTOR pathway. Neurosci. Lett. 2009;461:131–135. doi: 10.1016/j.neulet.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J., Ye J., Altafaj A., Cardona M., Bahi N., Llovera M., Canas X., Cook S. A., Comella J. X., Sanchis D. EndoG links Bnip3-induced mitochondrial damage and caspase-independent DNA fragmentation in ischemic cardiomyocytes. PLoS ONE. 2011;6:e17998. doi: 10.1371/journal.pone.0017998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao S. T., Chen M., Li S. J., Zhang M. H., Li B. X., Das M., Bean J. C., Kong J. M., Zhu X. H., Gao T. M. Mitochondrial BNIP3 upregulation precedes endonuclease G translocation in hippocampal neuronal death following oxygen-glucose deprivation. BMC Neurosci. 2009;10:113. doi: 10.1186/1471-2202-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sowter H. M., Ratcliffe P. J., Watson P., Greenberg A. H., Harris A. L. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- 65.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Yurkova N., Shaw J., Blackie K., Weidman D., Jayas R., Flynn B., Kirshenbaum L. A. The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ. Res. 2008;102:472–479. doi: 10.1161/CIRCRESAHA.107.164731. [DOI] [PubMed] [Google Scholar]
- 67.Zmuda E. J., Viapiano M., Grey S. T., Hadley G., Garcia-Ocana A., Hai T. Deficiency of Atf3, an adaptive-response gene, protects islets and ameliorates inflammation in a syngeneic mouse transplantation model. Diabetologia. 2010;53:1438–1450. doi: 10.1007/s00125-010-1696-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng X., Liu X., Zhang W., Xiao W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J. 2011;30:3397–3415. doi: 10.1038/emboj.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaw J., Zhang T., Rzeszutek M., Yurkova N., Baetz D., Davie J. R., Kirshenbaum L. A. Transcriptional silencing of the death gene BNIP3 by cooperative action of NF-κB and histone deacetylase 1 in ventricular myocytes. Circ. Res. 2006;99:1347–1354. doi: 10.1161/01.RES.0000251744.06138.50. [DOI] [PubMed] [Google Scholar]