Abstract
Aminoglycoside-class antibiotics bind directly to ribosomal RNA, imparting pleiotropic effects on ribosome function. Despite in-depth structural investigations of aminoglycoside–RNA oligonucleotide and aminoglycoside-ribosome interactions, mechanisms explaining the unique ribosome inhibition profiles of chemically similar aminoglycosides remain elusive. Here, using single-molecule fluorescence resonance energy transfer (smFRET) methods, we show that high-affinity aminoglycoside binding to the conserved decoding site region of the functional pre-translocation ribosome complex specifically remodels the nature of intrinsic dynamic processes within the particle. The extents of these effects, which are distinct for each member of the aminoglycoside class, strongly correlate with their inhibition of EF-G–catalyzed translocation. Neomycin, a 4,5-linked aminoglycoside, binds with lower affinity to one or more secondary binding sites, mediating distinct structural and dynamic perturbations that further enhance translocation inhibition. These new insights help explain why closely related aminoglycosides elicit pleiotropic translation activities and demonstrate the potential utility of smFRET as a tool for dissecting the mechanisms of antibiotic action.
During the elongation phase of protein synthesis, the ~2.4-MDa, two-subunit ribosome works in concert with tRNA substrates and protein factors to directionally transit the mRNA open reading frame. Central to this mechanism, aminoacyl-tRNA molecules must be selected in a codon-dependent fashion1, and both tRNA and mRNA substrates must be subsequently translocated with respect to the ribosome2. mRNA and tRNA substrates transit the 70S ribosome particle in incremental translocation steps through a ~100-Å-long, solvent-accessible channel located at the interface of large and small ribosomal subunits (50S and 30S, respectively, in bacteria). The structurally distinct aminoacyl (A), peptidyl (P) and exit (E) sites of the ribosome are rich in conserved RNA elements that stabilize tRNA and mRNA substrates in structurally characterized, ‘classical’ (C) positions through sequence- and shape-specific interactions3,4. Classically bound, the L-shaped tRNA molecules orient perpendicular to the subunit interface such that the 3′-aminoacylated CCA ends project into the 50S subunit peptidyltransferase center (PTC) while tRNA anticodons pair with mRNA codons within the small subunit decoding region5.
During synthesis, classical tRNA-ribosome interactions are remodeled through global conformational rearrangements in the pre-translocation ribosome complex that facilitate substrate motion. Conformational degrees of freedom in the particle implicated in substrate movements include ratchet-like motions at the subunit interface, as well as opening and closing cycles of the L1 stalk domain6. Such processes facilitate the formation of ‘hybrid’ (H) tRNA configurations—established intermediates in the translocation process—thus lowering the activation barrier for elongation factor-G (EF-G)-catalyzed translocation2,7–9.
Cryo-electron microscopy and smFRET investigations demonstrate that the pre-translocation ribosome complex is highly dynamic in nature6,10,11. Subunit ratcheting, L1 stalk motions and hybrid-state tRNA configurations arise through spontaneous, thermally accessible processes that may occur on distinct timescales6. Such studies have shown that tRNAs bound in the A-site and P-site achieve hybrid configurations through intrinsic conformational processes leading to either independent or coupled movements of their 3′-CCA termini out of their classical positions to adjacent binding sites in the large subunit12. Two structurally distinct hybrid-state intermediates have thus far been identified through smFRET, distinguished by relative movements between the elbow regions of A-site and P-site tRNA6,12. Movements of the P-site tRNA elbow with respect to the A-site and the ribosome, accompanied by rearrangements centered on the 3′-CCA end of A-site tRNA, are defined as hybrid state-2 (H2); coupled motions of A-site and P-site tRNA elbow regions with respect to each other and the ribosome are defined as hybrid state-1 (H1). How these distinct hybrid states specifically contribute to the mechanism of translocation remains unclear.
Consistent with hybrid tRNA configurations being accompanied by substantial conformational changes in the ribosome particle, the estimated activation barriers for achieving both H1 and H2 configurations are relatively large (about 70 kJ mol−1)12. However, single point mutations of the aminoacyl-tRNA binding sites within the PTC substantially alter transition rates into and out of classical and hybrid states12. To account for these, and many other previous demonstrations of structure-function relationships in the ribosome (reviewed in ref. 13), a metastable energy landscape model of ribosome function was recently articulated. This model puts forward a framework for understanding how even small perturbations in the ground-state energies of distinct conformational substates of the particle may contribute to the mechanism of translation regulation10,14. A key element of this model is that small-molecule effectors of the translation apparatus may generally yield detectable alterations in the structural and kinetic parameters of conformational processes in the ribosome particle at the single-molecule scale. Pre-steady state smFRET measurements of tetracycline and thiostrepton inhibition of tRNA selection have already demonstrated the potential of this approach for probing antibiotic action on the ribosome11. Steady state measurements also revealed that tRNA dynamics within the pre-translocation complex are sensitive to the nature of the nascent polypeptide, magnesium ion concentrations and saturating concentrations of the translocation inhibitor viomycin11. However, investigations providing a direct link between observed rates of motions within the pre-translocation complex and ribosome function are generally lacking.
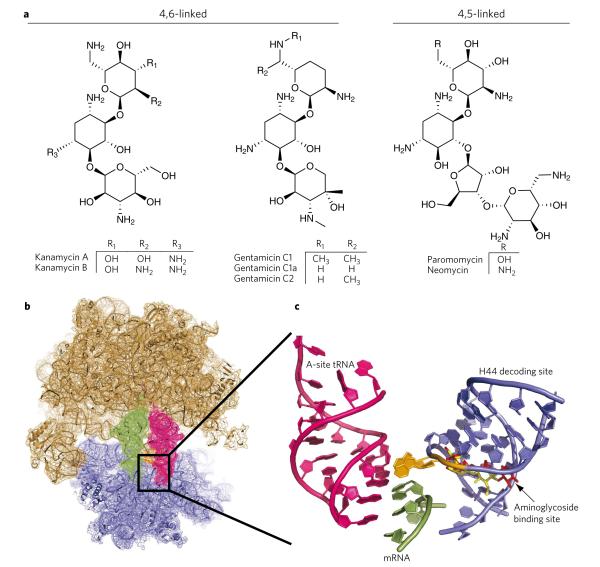
Aminoglycoside-class antibiotics are structurally related, ribosome-targeting compounds used clinically to treat infections (Fig. 1a). Members of this class exhibit pleiotropic translational effects that decrease tRNA selection accuracy15, inhibit spontaneous and EF-G–catalyzed translocation2,16–20 and affect ribosome recycling21. Aminoglycoside-induced perturbations of ribosome activities have been largely attributed to their direct binding to the ‘decoding’ site, a nearly universally conserved, asymmetric internal loop present within helix 44 (h44) of the small subunit, 16S rRNA13,22 (Fig. 1b,c). 4,5-linked and 4,6-linked aminoglycosides bind the decoding site proximal to residues A1408, A1492 and A1493 of h44 through a common binding mode mediated by structurally conserved ring I and ring II elements22 (Fig. 1c).
Figure 1. Aminoglycoside-induced restructuring of the ribosome 16S decoding site.
(a) Chemical structures of kanamycin, gentamicin, paromomycin and neomycin. (b) Overview of the 70S prokaryotic ribosome structure composed of the large 50S subunit (tan) and the small 30S subunit (blue). The decoding site is boxed and contains the aminoglycoside binding portion of the 16S rRNA helix 44. (c) Enlarged view of the decoding site 16S rRNA (blue) with bound aminoglycoside antibiotics. The A1492-A1493 bases (orange) ‘turn out’ in the presence of aminoglycosides or upon cognate codon-anticodon pairing and interact with the tRNA (magenta) and mRNA (green) at the site of codon-anticodon interaction. Gentamicin (yellow) and neomycin (red) are shown bound in the decoding site. Crystal structures were constructed from Protein Data Bank entries 2J00 and 2J01 (ref. 4),with aminoglycosides mapped from 2QB9 and 2QAL (ref. 35) using PyMol software (Delano Scientific).
Aminoglycoside binding to h44 induces local rearrangements that stabilize extruded A1492 and A1493 configurations22,23 (Fig. 1c) and global rearrangements in the ribosome particle observed through biochemical means24. In extruded conformations, A1492 and A1493 can interact directly with the codon-anticodon complex to affect both tRNA selection and translocation mechanisms through ground-state stabilization20,22. Although aminoglycosides are widely recognized to affect tRNA selection by increasing miscoding rates, such effects alone are not generally associated with their bactericidal activities, as protein structure and function are generally robust to missense errors22,25,26. Instead, aminoglycoside inhibition of translocation16–20 may be a more critical determinant of drug efficacy22. Point mutations in h44, including an A1408G transition, reduce drug affinity for the decoding site and confer aminoglycoside antibiotic resistance in clinical isolates27–29. The presence of the G1408 nucleotide (Escherichia coli numbering) in human cytoplasmic ribosomes is a key specificity determinate, while aminoglycoside toxicity effects are associated with mitochondrial ribosomes bearing an adenosine residue at position 1408 (ref. 30).
Here, using smFRET, we examined members of the aminoglycoside-class antibiotics for their capacities to induce structural and kinetic alterations in conformational processes intrinsic to the pre-translocation ribosome complex. In line with the metastable energy landscape model of translation regulation, these small-molecule effectors were shown to exhibit distinct, titratable changes in the classical-hybrid tRNA exchange on single, intact ribosome particles. By correlating these activities with pre-steady state measurements of single-step translocation rates, quantitative relationships were found between aminoglycoside-induced perturbations of dynamic processes within the pre-translocation complex and the rate of translocation. These data reveal that even small changes in the activation barriers to tRNA motions on the ribosome can result in substantial translocation defects. Such observations highlight the sensitivity of smFRET experiments and the potential to leverage this tool as a means of exploring structure-function relationships of closely related antibiotics in biologically relevant contexts.
RESULTS
Kanamycin binding alters tRNA motions in the ribosome
Aminoglycoside-induced alterations in the pre-translocation ribosome complex were examined through high-spatial-resolution and high-temporal-resolution smFRET imaging of the intrinsic exchange of A-site and P-site tRNAs between classical and hybrid configurations12. Experiments were performed on pre-translocation complexes bearing tRNAfMet(Cy3-s4U8) in the P-site and fMet-Phe-tRNAPhe(Cy5-acp3U47) in the A-site using a total internal reflection–based system (Methods). Such complexes have been shown to be fully functional in both tRNA selection and translocation31. In the absence and presence of aminoglycosides, time-dependent changes in FRET reporting on the classical (C) and hybrid state exchange (H1 and H2) were recorded at 25-ms time resolution. Experiments were performed at ambient temperature (~23 °C) using buffers containing elevated magnesium ion concentration ([Mg2+]) to maximize the signal-to-noise ratio of the measurements and to ensure minimal tRNA and subunit dissociation, as well as spontaneous translocation, during the observation period (about 5 min)31. In so doing, the rates of exchange between states could be determined using automated analytical procedures, which reduce biases potentially introduced by manual data selection and increase the analytical throughput of the smFRET experiments (Methods).
As observed previously at 40-ms time resolution12, in the absence of antibiotic, A-site and P-site tRNAs dynamically exchange between C (~0.55 FRET; ~62% occupancy), H1 (~0.39 FRET; ~16% occupancy) and H2 (~0.24 FRET; ~22% occupancy) configurations (Supplementary Fig. 1). Addition of kanamycin (1 and 2), a 4,6-linked aminoglycoside, resulted in a concentration-dependent increase in classical-state occupancy (Fig. 2a), an activity previously observed for the 4,5-linked aminoglycoside paromomycin20. At saturating kanamycin concentrations32 (≥20 μM), evidence for a global dampening of transition rates was observed (Supplementary Table 1; Supplementary Fig. 2a), and classical-state occupancy increased by ~8% to ~70% total time-averaged occupancy. Corresponding reductions were observed in both H1 and H2 hybrid states, where total hybrid-state occupancy (H1 + H2) decreased by ~8% (from ~38% to ~30%). Such changes were significantly larger than day-to-day (s.d. = 3.0%, Supplementary Fig. 3) and intraday, experiment-to-experiment variability (P < 0.05, s.d. = 2.3%, Supplementary Fig. 4). Similar trends were observed with distinct pre-translocation complexes containing P-site tRNAPhe(Cy3-s4U8) and A-site NAc-Phe-Lys-tRNALys(Cy5-acp3U47) (Supplementary Fig. 5), and at lower [Mg2+] (5 mM, data not shown). As experimental variability was shown to be adequately approximated by analytical bootstrapping procedures (±1.5%) (Methods; Supplementary Fig. 6), unless otherwise stated, such methods were used for all measurements in order to estimate the errors in FRET state occupancies and calculated rates.
Figure 2. Kanamycin binding to the decoding site increases time-averaged occupancy of specific ribosome conformations.
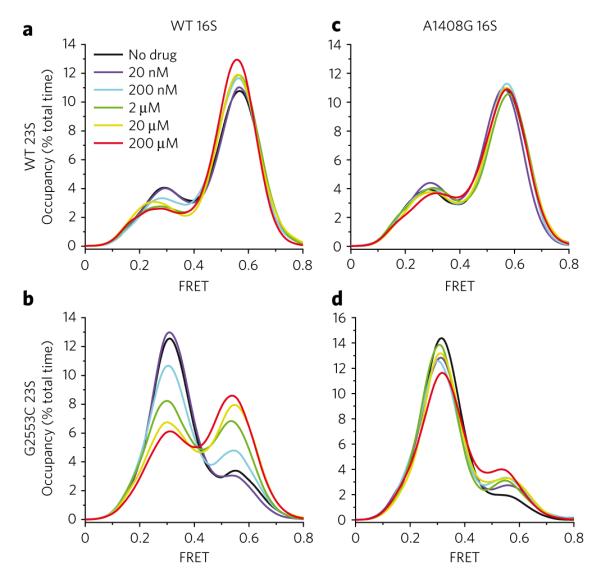
The time-averaged occupancy (Supplementary Fig. 1) in all nonzero FRET states was measured for the surface immobilized population of intact 70S E. coli ribosomes at increasing concentrations of kanamycin. The FRET values between P-site tRNAfMet(Cy3-s4U8) and A-site fMet-Phe-tRNAPhe(Cy5-acp3U47) report directly on three specific conformational intermediates in the translocation pathway: the classical state (~0.55 FRET), the hybrid 1 (~0.39 FRET) state and the hybrid 2 state (~0.24 FRET). (a–d) Histograms are shown for wild-type (a), G2553C 23S (b), A1408G 16S (c) and G2553C 23S + A1408G 16S (d) ribosomal complexes in the presence of increasing kanamycin concentrations (black to red lines). Histograms were constructed by summing FRET values over the first 50 frames of each acquired movie over all nonzero FRET states and plotting the spline-interpolated data points.
Consistent with half-maximal inhibitory concentration (IC50) values for kanamycin-induced inhibition of in vitro translation (~150 nM)33, the half-maximal effective concentration (EC50) of kanamycin-induced classical-state stabilization was estimated to be ~300 nM. By comparing the relative classical-state occupancies in the absence and presence of saturating drug concentrations, the energetic contribution of kanamycin-induced classical-state stabilization was estimated to be 1.00 ± 0.01 kJ mol−1 (Methods; Supplementary Table 2). These data are consistent with a model in which kanamycin induces a high-affinity conformation of the ribosome for A-site tRNA, preferentially stabilizing the classical state by promoting extruded conformations of residues A1492 and A1493 where interactions can occur with the minor groove of the mRNA-tRNA minihelix20,22 (Fig. 1c).
Enhancement of the kanamycin-induced signal
In addition to decoding site interactions, the classical A-site tRNA configuration (A/A) is maintained through a base-specific interaction between G2553 of the A loop, a highly conserved 23S rRNA element within the PTC, and residue C75 of the 3′-CCA terminus (reviewed in ref. 14). A G2553C mutation disrupts this native contact to specifically destabilize classical-state occupancy in favor of hybrid states8,12. Postulating that the dynamic range of the aminoglycoside-induced effects on tRNA dynamics mediated through the decoding site may be increased by shifting the free energy landscape in favor of hybrid states, we repeated kanamycin titrations on pre-translocation ribosome complexes in a G2553C-mutant background, where in the absence of kanamycin, classical-state occupancy was ~19%.
As anticipated, kanamycin-induced perturbations of the classi cal-hybrid equilibrium were substantially greater in the G2553C-mutant background than in the wild-type background. Here, at 20 μM kanamycin, classical-state occupancy increased by ~25% (versus ~8% for wild-type ribosomes) (Fig. 2b; Supplementary Fig. 6b), whereas the estimated EC50 of classical-state stabilization was unchanged (~300 nM). Thermodynamic analysis (Methods) showed that saturating kanamycin concentrations stabilized the classical state by 3.21 ± 0.31 kJ mol−1 (Supplementary Table 2), only partially compensating for the reductions observed in classical-state occupancy resulting from the single G2553-C75 base pair disruption. The differences observed in kanamycin-induced classical-state stabilization on wild-type and mutant pre-translocation complexes (1.0 kJ mol−1 versus 3.2 kJ mol−1, respectively) suggest that conformational processes in addition to the classical-hybrid exchange may also be affected by kanamycin binding and that the effect of kanamycin on these processes is enhanced in the mutant system. One plausible explanation for this observation is that both the G2553C mutation and kanamycin binding either directly or indirectly affect the subunit ratcheting equilibrium associated with hybrid states formation6,10,34. In this view, in addition to inducing local conformational changes in the decoding site that favor classical A-site tRNA binding, kanamycin may also influence the subunit ratcheting-unratcheting equilibrium in favor of the unratcheted state, lending to additional energetic stabilizing of the classical state.
Resistance mutation reduces the kanamycin signal
To test whether kanamycin-induced perturbations of the classical-hybrid tRNA equilibrium could be attributed to drug interactions localized to the decoding site, identical experiments were performed on wild-type and G2553C-mutant pre-translocation ribosome complexes also bearing an A1408G aminoglycoside resistance mutation. Ribosomes containing both A1408G and G2553C mutations were observed to occupy the classical state ~10% of the time on average (versus ~19% for the G2553C complex). Although not observed in the A1408G background alone, these data reveal that the A1408G mutation in the G2553C background reduces classical-state occupancy by ~9% (compare no drug controls in Fig. 2a,b versus Fig. 2c,d).
Consistent with kanamycin-induced perturbations in the pre-translocation complex stemming from specific interactions localized to the decoding site, kanamycin did not substantially promote classical-state occupancy on ribosomes bearing the A1408G resistance mutation (Fig. 2c; Supplementary Fig. 6c). By contrast, at the highest (200 μM) kanamycin concentrations tested, classical-state occupancy was modestly stabilized in the A1408G G2553C mutant background (~10%), although to a substantially lower extent than for the G2553C mutant ribosome (~25%) (Fig. 2b,d). These experiments showed that the principal determinants of the kanamycin-induced effects on tRNA dynamics can be attributed to high-affinity binding to the canonical h44 binding site and that the A1408G resistance mutation increases the apparent Kd to an extent consistent with previous estimates (about 10-fold to 100-fold)29. The modest classical-state stabilization observed on the A1408G G2553C mutant ribosome is in line with the increased dynamic range of this system, where even weak kanamycin interactions with h44 are evidently sufficient to yield modestly altered rates of tRNA dynamics and/or subunit ratcheting exchange. However, such effects were only reproducibly observed at 200 μM kanamycin.
Kanamycin activity observed in single ribosomes
To address whether kanamycin-induced perturbations of the classical-hybrid tRNA equilibrium were reversible, direct measurements of single G2553C mutant pre-translocation ribosomes were made during a binding-dissociation cycle (Fig. 3). Prior to kanamycin addition, individual pre-translocation complexes predominantly occupied hybrid states with rare, short-lived excursions to the classical state. Upon addition of 20 μM kanamycin, a clear switch in the dynamic mode of the ribosome was frequently observed, where classical-state occupancy became predominant and transitions to hybrid states became highly transient in nature. Upon buffer exchange, the system returned to a dynamic mode indistinguishable from the unbound state observed before kanamycin addition. These data clearly demonstrated the reversibility of the drug-ribosome interaction and supported the notion that changes in classical-state occupancy induced by kanamycin arise directly from changes in classical-hybrid equilibrium within individual ribosome complexes.
Figure 3. Changes in population time-averaged state occupancy are caused by drug-induced changes in single ribosomes.
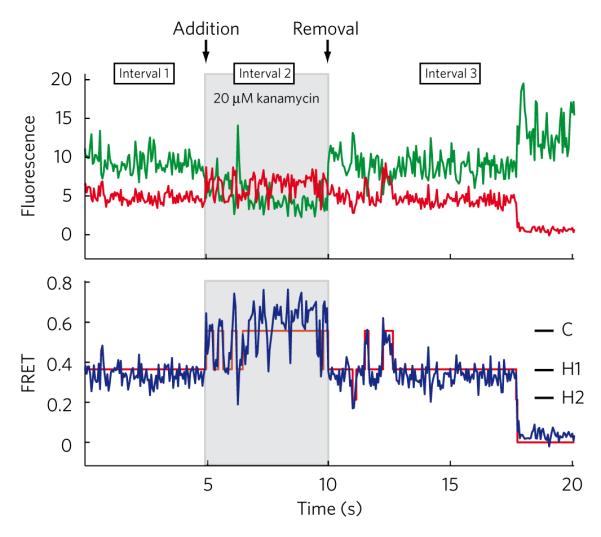
Single-molecule fluorescence traces were collected for surface-immobilized ribosomes containing the G2553C mutation over three 5-s intervals: (i) in the absence of drug, (ii) following a 2-min incubation in 20 μM kanamycin and (iii) following removal of kanamycin with drug-free buffer for 2 min. Shown are donor (green line) and acceptor (red line) fluorescence traces (top panel), along with calculated FRET trajectory (blue) and idealization (red line).
Most aminoglycosides stabilize the classical state
To test whether classical-state stabilization was an activity common to all 4,5- and 4,6-linked aminoglycosides, representative compounds from both classes (4,5-linked—paromomycin (3) and neomycin (4); 4,6-linked—gentamicin (5, 6 and 7)) were examined at saturating concentrations (20 μM) on both wild-type and G2553C-mutant pre-translocation complexes. As observed for kanamycin, gentamicin and paromomycin increased classical-state occupancy on the wild-type pre-translocation ribosome complex by ~5% and ~2%, respectively (Fig. 4a). These relatively subtle effects were again amplified by the G2553C mutation, where gentamicin and paromomycin were observed to increase classical-state occupancy by ~14% and ~12%, respectively (Fig. 4b). Both experiments revealed that classical-state stabilization was an activity common to both 4,5- and 4,6-linked aminoglycoside classes, where energetic distinctions observed on the G2553C mutant ribosome again suggested such compounds may also influence the subunit ratcheting equilibrium associated with hybrid states formation. Here, differences observed in the extent of classical-state stabilization for the various aminoglycosides may reflect subtle distinctions in either their binding energies and/or their structure-specific activities.
Figure 4. Alterations in tRNA dynamics are a common effect of aminoglycoside binding to the ribosome.
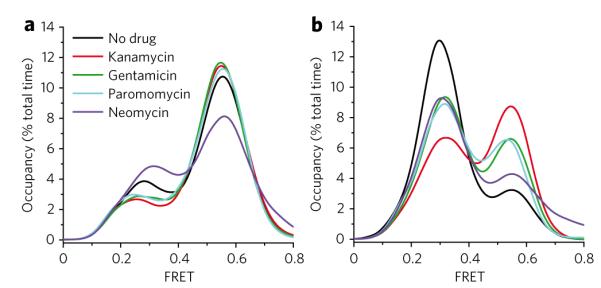
(a,b) Population time-averaged state occupancy was measured for wild-type ribosomes (a) and ribosomes containing the large subunit G2553C mutation (b) in the presence of 20 μM aminoglycosides representing both the 4,6-linked (kanamycin, gentamicin) and the 4,5-linked (paromomycin, neomycin) structural families.
Under identical conditions, neomycin, which differs from paromomycin by an –OH to –NH2 substitution at the 6′ position of ring I, decreased classical-state occupancy on the wild-type pre-translocation ribosome complex by ~10% (Fig. 4a). In contrast, classical-state stabilization was observed in the G2553C background, consistent with the other aminoglycosides (Fig. 4b). These observations suggested either that neomycin exhibits activities distinct from the other aminoglycosides through interactions with the canonical decoding site or that neomycin binds to one or more secondary binding sites within the pre-translocation complex.
Neomycin acts at multiple sites within the ribosome
Earlier equilibrium dialysis experiments17 and recent crystallographic studies35 provide evidence for one neomycin binding site in each of the two ribosomal subunits. The recent crystallographic data revealed that neomycin binds to both the h44 decoding site and the major groove of H69 of the large subunit 23S rRNA. High-affinity h44 binding and lower-affinity H69 binding were inferred, where low neomycin concentrations are expected to saturate the canonical h44 binding site, while higher neomycin concentrations may be necessary to additionally saturate the H69 site. Based on the present observations, it may be speculated that high-affinity neomycin binding stabilizes classical tRNA positions while binding at the H69 site may ultimately lead to a net stabilization of hybrid states. To test this hypothesis, neomycin titration experiments were performed on wild-type and A1408G-bearing pre-translocation complexes. Supporting the existence of two distinct neomycin binding sites within the 70S particle that differ substantially in apparent affinity, on the wild-type system low concentrations of neomycin were observed to modestly increase classical-state occupancy (~6% at 100 nM), while higher neomycin concentrations reduced classical-state occupancy in favor of hybrid states (~10% at 20 μM) (Fig. 5a).
Figure 5. Neomycin alters tRNA dynamics on the ribosome in a bimodal fashion.
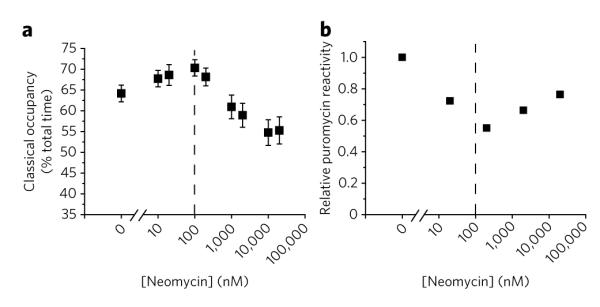
(a) smFRET traces obtained in the presence of increasing neomycin concentrations were idealized to a four-state hidden Markov model (Methods). Time-averaged classical-state occupancy is shown versus neomycin concentration along with standard errors calculated from 1,000 bootstrap samples. (b) Puromycin was delivered to ribosomal complexes containing tRNAfMet in the P-site and Cy3-DSP-Met-Phe-tRNAPhe in the A-site in the presence of increasing concentrations of neomycin. Fluorescence intensity decays with time as fluorescently labeled peptide is released from the surface (Methods; Supplementary Fig. 7). Decreased rates of puromycin reactivity indicate decreased hybrid-state occupancy. Mean relative rates of puromycin reactivity (n = 2, s.d. = 0.05) calculated from the total normalized change in fluorescence over a 10-min period are shown versus neomycin concentration.
The bimodal nature of neomycin-induced alterations in the pre-translocation complex was confirmed using a previously established, fluorescence-based, puromycin-release assay (Methods). In the absence of A-site tRNA, puromycin (8) reacts rapidly when fluorescently labeled peptidyl-tRNA occupies the classical P-site12,31 (Supplementary Fig. 7). However, in the pre-translocation complex, puromycin reactivity is known to be substantially reduced due to the transfer of the fluorescently labeled nascent chain to the newly incorporated aa-tRNA. Residual reactivity of the pre-translocation complex with puromycin reflects the intrinsically dynamic nature of the system, where A-site peptidyl-tRNA reacts with puromycin when the hybrid state is occupied8,12,31,36. Here, in a manner that paralleled the bimodal nature observed through smFRET observations (Fig. 5a), neomycin decreased puromycin reactivity in the pre-translocation complex at concentrations up to ~200 nM, while promoting a recovery in puromycin reactivity at concentrations >200 nM (~25% increase at 10 μM) (Fig. 5b; Supplementary Fig. 7).
These data supported the existence of at least two distinct neomycin binding sites with separable activities: a high-affinity binding site (~100 nM EC50) that stabilizes classical tRNA configurations and a low-affinity binding site (>1 μM EC50) that stabilizes A/P hybrid-state tRNA configurations. Consistent with this notion, at low neomycin concentrations (<200 nM), specific reductions in transition rates out of the classical state were observed, while at higher neomycin concentrations (>200 nM), substantial reductions were observed in the rates into and out of both classical and hybrid states (Supplementary Fig. 2b). In the A1408G resistance background, low-concentration neomycin effects were substantially abrogated, while high-concentration neomycin effects were unchanged (Supplementary Fig. 2b). Thus, neomycin binding to the high-affinity site stabilizes classical tRNA configurations through interactions with the canonical h44 decoding region. Low-affinity neomycin binding to one or more secondary sites appeared to operate, to a substantial extent, independently of the decoding region to stabilize hybrid tRNA configurations. While these data provide evidence for at least one additional neomycin binding site within the pre-translocation complex that is consistent with the previously proposed H69 binding site35, additional experiments will be required to pinpoint the specific sites responsible for hybrid states stabilization and the molecular basis of classical-hybrid state exchange rate dampening when these secondary sites are occupied.
Correlating tRNA dynamics with translocation inhibition
To test the relationship between aminoglycoside-induced changes in the classical-hybrid equilibrium and translocation inhibition, pre-steady state measurements of EF-G–catalyzed translocation were performed under identical experimental conditions in the absence and presence of specific aminoglycoside concentrations19 (Supplementary Fig. 8). In these experiments a strong correlation was found between the extent of classical-state occupancy increase and translocation inhibition (Pearson correlation coefficient >0.91, P < 0.02) and between the rate exiting the classical state and translocation rate (Fig. 6a,b; Supplementary Table 3). Low neomycin concentrations, where increased classical-state occupancy was observed (100 nM), fit this trend. In line with previous reports20, and the notion that hybrid states formation lies along the translocation reaction coordinate2,7,9, these data suggest that ground-state stabilization of classical tRNA positions by aminoglycoside-class antibiotics is an important and general mechanism for translocation inhibition, where ~1.5-fold to 3-fold reductions in the rates tRNAs exit the classical state result in ~3-fold to 6-fold reductions in translocation rates.
Figure 6. Stabilization of the classical state is strongly correlated with inhibition of translocation by decoding site–binding aminoglycosides.
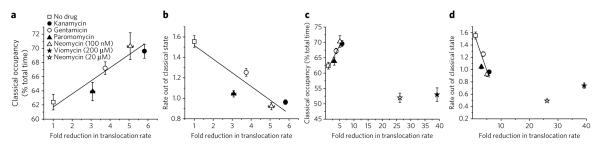
(a) Correlation of time-averaged classical-state occupancy versus the fold-reduction of the single-step translocation rate of wild-type ribosomes in the presence of drug (20 μM, unless otherwise noted) revealed a strong correlation with classical-state occupancy. (b) A strong correlation is also observed between translocation rates and the rate constant of transitioning from the classical to hybrid states, as calculated using hidden Markov modeling (Methods). (c,d) Translocation inhibition for 20 μM neomycin (c) and 200 μM viomycin (d) is shown on the expanded axis and was similar to that observed for the no EF-G control. Error bars on translocation rates represent the s.d. of triplicate experiments. Error bars on state occupancy and transition rates were calculated in each experiment from 1,000 bootstrap samples of smFRET traces.
At higher neomycin concentrations (≥ 20 μM), where transition rates out of hybrid and classical states were both substantially reduced (Supplementary Fig. 2b), a net stabilization of hybrid states was observed in conjunction with much greater translocation inhibition (Fig. 6c,d). These results were grossly similar, in both extent and mechanism, to the inhibition observed in the presence of high concentrations (200 μM) of the well-established translocation inhibitor viomycin (9), a cyclic peptide thought to bind proximal to the subunit interface near H69 and h44 (reviewed in ref. 2)37. Bulk and smFRET studies have both shown that viomycin stabilizes ratcheted ribosome conformations34,38, but these studies conflict regarding its effects on P-site tRNA. Bulk studies suggest that viomycin stabilizes the P/E hybrid state, while smFRET measurements suggest that viomycin stabilizes classical tRNA configurations39,40.
Like neomycin, viomycin exhibited a bimodal titration behavior in its effects on tRNA dynamics. At lower viomycin concentrations (< 2 μM), hybrid states were stabilized, while higher concentrations substantially restored classical-state occupancy (Supplementary Fig. 9). Contrary to previous reports40, at high concentrations (>100 μM), viomycin yielded a net stabilization of hybrid states (an increase of ~12% total occupancy). However, consistent with earlier work2,39,40 and on par with the effects observed at 20 μM neomycin where multiple molecules may be bound to the pre-translocation complex, ~3-fold to 5-fold reductions were observed in the rates exiting both hybrid and classical states.
Low concentrations of viomycin (<2 μM) reduced both the rate and the extent of EF-G–catalyzed translocation (Supplementary Fig. 8). At 100 nM viomycin, where hybrid states were stabilized by ~10%, a ~1.5-fold reduction in translocation rate was observed along with a ~2-fold reduction in the translocated fraction. At 2 μM viomycin, where hybrid states were stabilized by ~20%, the rate and extent of translocation inhibition were comparable to the no EF-G control. Thus, while the aminoglycosides examined and viomycin have grossly similar effects in their reduction of tRNA dynamics within the pre-translocation complex (~1.2-fold to 4-fold), in their primary binding modes these antibiotics likely inhibit translocation through distinct mechanisms. Here, two speculative conclusions can be made. First, the nearly complete inhibition of translocation observed at both high viomycin and neomycin concentrations, where previous data17,35,41 and our titration experiments argue multiple drug molecules may be bound, suggests that primary and secondary binding site occupancy may operate additively with respect to tRNA dynamics but synergistically on the extent of translocation inhibition. Second, the binding of antibiotics outside of the h44 decoding site, which have the net effect of stabilizing hybrid-state tRNA configurations, may exhibit more pronounced translocation inhibition.
DISCUSSION
Control of translation is central to the regulation of gene expression in the cell. While potentially achieved through numerous molecular mechanisms, the metastable energy landscape view of protein synthesis10,14 suggests that perturbations in ground-state energies of distinct substates within the ribosome complex, and/or changes in energy barriers separating conformational transitions, may be critical determinants of ribosome regulation. A prediction of this model is that translation rates may be substantially altered at the systems level by even modest changes in dynamic conformational events within the pre-translocation complex.
Here, we demonstrate through smFRET measurements that the rates of intrinsic tRNA motions, and their global positioning, within the pre-translocation complex provide sensitive reporters of 4,5- and 4,6-class aminoglycoside antibiotic action (Fig. 7). In a concentration-dependent fashion, consistent with the IC50 values observed for translation inhibition33, members of both aminoglycoside classes were observed to stabilize A-site and P-site tRNA in classical configurations relative to hybrid states through direct interactions with the canonical h44 decoding site. Consistent with the metastable energy landscape model of translation, the binding of a single ~750-Da small molecule to the ~2.4-MDa ribosome, which induced relatively small increases in classical-state stability (about 1–4 kJ mol−1, corresponding to ~1.5-fold to ~3-fold reductions in the rates exiting classical states), led to the inhibition of translocation by as much as 6-fold (Fig. 6). While translocation rates increased substantially at higher temperature (37 °C) and lower [Mg2+], the aminoglycoside-induced translocation inhibition observed was shown to be largely independent of these parameters (Supplementary Fig. 10). The correspondence between the titration behaviors observed and the IC50 values of the antibiotics examined suggests that even modest reductions in translocation rates, perhaps accompanied by enhanced frame shifting, “hopping,” and “drop-off”26,42, may be sufficient to elicit their bactericidal activities. Energetically, such effects are on the same order as a single base pairing interaction12, substantially smaller than those estimated through aminoglycoside-RNA oligonucleotide measurements43. It is presently unclear why others have previously measured a 160-fold inhibition of translocation by paromomycin20, whereas a ~4-fold to 6-fold inhibition was observed here. Such differences may stem from the inclusion of physiological polyamines in our experiments and/or distinctions stemming from the bacterial origins of our ribosome complexes (BL21 versus MRE600) or the procedures for their preparation. Future experiments will be necessary to thoroughly examine these questions.
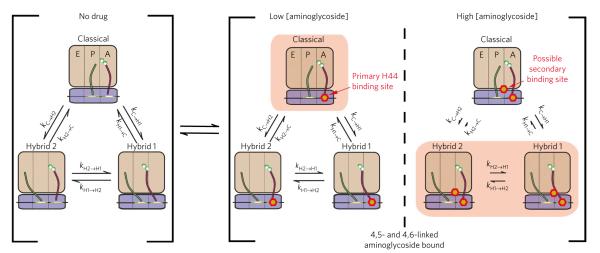
Figure 7. Kinetic scheme of aminoglycoside-induced changes in tRNA dynamics.
This scheme describes the changes in the rates of transitions between the classical state, the hybrid 1 state and the hybrid 2 state following aminoglycoside binding to the ribosome complex. 50S (tan) and 30S (blue) subunits are shown, as well as the A-site tRNA (red) and the P-site tRNA (green). At low concentrations, aminoglycosides bind with high affinity to the primary h44 binding site. High concentrations of aminoglycosides can lead to binding at a low-affinity second binding site. Arrow lengths represent the rate of transition relative to the drug-free system, and the predominantly stabilized state is highlighted (pink box).
Aminoglycoside-induced classical-state stabilization generally supports a stochastic gating mechanism44, wherein the binding of both 4,5- and 4,6-linked aminoglycosides to the h44 decoding site favors an extra-helical conformation of residues A1492-A1493 to stabilize A-site tRNA in its classical configuration relative to the hybrid states (Fig. 7, center panel). The observation that aminoglycoside binding at the A-site decoding region tends to increase the stability of P-site tRNA in its classical position suggests either that the decoding site is restructured in the classical-to-hybrid state transition or that it is structurally and functionally linked to the P-site. Such an effect may reflect a kinetic linkage between the classical-hybrid tRNA exchange and the subunit ratcheting-unratcheting equilibrium anticipated to restructure the A-site and the P-site. Evidence of allostery in the ribosome has been previously demonstrated with regard to (i) reciprocal relationships between the affinities of A-site and P-site tRNA45, (ii) functional linkages between the A-site and E-site tRNA binding sites46, and (iii) functional consequences of codon-anticodon mismatches in the P-site and E-site on both tRNA selection and release mechanisms at the A-site47.
While the primary binding site and principal mode of action of the aminoglycosides localizes to the canonical h44 decoding region, binding at secondary sites observed at elevated neomycin concentrations (>2 μM) yielded net increases in hybrid-state occupancy and significantly greater reductions in translocation rates not unlike the effects observed for viomycin. Such observations are consistent with secondary neomycin binding sites inducing distinct conformational events in the ribosome similar to those associated with viomycin binding. Consistent with this notion, early studies showed that 10 μM neomycin stabilized EF-G binding to the ribosome48 and that EF-G binds preferentially to the ratcheted ribosome complex49,50. The proposed existence of two neomycin binding sites34 (h44 and H69) proximal to the putative sites of viomycin interaction37, as well as points around which tRNAs and ribosomal subunits pivot during motion10,35, is compatible with this notion (Supplementary Fig. 11). The present data support a model in which h44 aminoglycoside binding may preferentially stabilize classical tRNA configurations and unratcheted ribosome conformations, while H69 aminoglycoside binding may preferentially stabilize both hybrid P/E tRNA and ratcheted subunit configurations34 (Fig. 7, right panel). Here, the binding of multiple drug molecules to the ribosome may additively or synergistically impact the energy landscape governing tRNA motions as well as the rate of translocation.
While evidence for secondary binding sites were not observed here for the other aminoglycosides examined, structural evidence suggests that paromomycin may also bind both the canonical h44 and H69 sites (V. Ramakrishnan, MRC Laboratory of Molecular Biology, personal communication). Here, a lack of evidence for secondary binding sites may be attributed to: (i) affinity differences between Thermus thermophilus and E. coli ribosome complexes yet to be examined, (ii) lower affinity interactions of the other aminoglycosides with both primary and secondary binding sites, or (iii) competing h44 and H69 effects that mask observable changes in tRNA position. Here, a more detailed examination of additional parameters such as the global dampening of tRNA and/or other ribosome dynamics may provide a better measure of higher order drug binding events.
The sensitivity and experimental throughput of the single-molecule methods described here enabled titrations that revealed functional differences between structurally related aminoglycosides when bound to the intact 70S particle. These studies expand on a growing body of evidence11 demonstrating that smFRET can be used to directly image drug activities via perturbations of the ribosome energy landscape. It remains to be determined whether the activities of all aminoglycosides can be differentiated using this method and whether the activities of other antibiotic classes can be observed by similar means. Here, distinct labeling sites within the ribosome, in combination with site-directed mutagenesis strategies similar to those used here, may be necessary. Additional enhancements in the detection of drug activity may also be possible by screening distinct sample compositions and experimental conditions. Because activity assays of this nature require only minimal amounts of material, smFRET methods may serve as useful screening technologies for exploring more detailed and exhaustive titration experiments than are possible using bulk techniques. This is particularly relevant when considering the difficulties associated with preparing pure, mutant ribosome complexes necessary for studying resistance mutations. The development of new fluorophore labeling methods, improved instrumentation and analytical tools that enhance signal-to-noise ratios and total experimental throughput may be essential to such pursuits. Platform technologies enabling high-throughput experiments may ultimately provide a general approach for exploring a diverse range of ligand-biomolecule interactions and their relationship to biological and therapeutic regulation.
METHODS
Reagents
Kanamycin (EMD), paromomycin (Sigma), neomycin (Sigma), gentamicin (Sigma), puromycin (Sigma) and viomycin (United States Pharmacopeia) were used at the purity stated by the commercial suppliers (≥ 97%). All experiments were performed in Tris-polymix buffer (pH 7.5) containing 50 mM Tris Acetate, pH 7.5, 15 mM Mg(OAc)2, 100 mM KCl, 5 mM NH4OAc, 0.5 mM CaCl2, 0.1 mM EDTA, 5 mM putrescine and 1 mM spermidine. These conditions provide a balance of maximal in vitro translation performance while minimizing the rates of tRNA and subunit dissociation, as well as spontaneous translocation events that otherwise compromise the smFRET measurement31.
Bulk, pre-steady state measurements
Puromycin reactivity experiments and measurements of bulk single-step translocation are described in the Supplementary Methods.
Aminoacylation and fluorescent labeling of tRNA
tRNAfMet and tRNAPhe from E. coli strain MRE600 were obtained from commercial sources (Sigma). Aminoacylation, formylation and fluorescent labeling of tRNA were performed as previously described31. Cy3 and Cy5 dyes (GE Healthcare) were site-specifically attached through either maleimide or N-hydroxysuccinimide chemistry to tRNAfMet(s4U8), tRNAPhe(acp3U47), tRNAPhe(s4U8) or tRNALys(acp3U47) at naturally occurring modified base residues located near the elbow region of the tRNA body. tRNAPhe and tRNALys were charged using recombinant phenylalanyl tRNA synthetase (PheRS) and lysyl tRNA synthetase (LysRS) prepared as previously described12,31. Dye-labeled tRNAs prepared in this manner are fully competent in tRNA selection, translocation and peptide bond formation31.
Preparation of ribosome complexes
50S-MS2 tagged and 30S-MS2 tagged ribosomal subunits were isolated from E. coli strain BL21(DE3) as previously described51. The G2553C and A1408G point mutations were made using the Quick change site-directed mutagenesis kit (Stratagene). Initiation complexes were prepared using purified 50S and 30S subunits (1 μM each) initiated in vitro on 5′-biotinylated gene 32-derived mRNA (5′-biotin-CAA CCU AAA ACU UAC ACA CCC UUA GAG GGA CAA UCG AUG UUC AAA GUC UUC AAA GUC AUC) (Dharmacon) in the presence of IF-1 (2 μM), IF-2 (2 μM), IF-3 (2 μM), 2 mM GTP and either fMet-tRNAfMet(Cy3-s4U8) or NAc-Phe-tRNAPhe(Cy3-s4U8) (2 μM) in Tris-polymix buffer31.
Single-molecule fluorescence experiments and data processing
smFRET data were acquired using a prism-based total internal reflection (TIR) microscope as previously described12. All experiments were performed in the presence of an oxygen scavenging environment (1 unit μl−1 glucose oxidase, 8 units μl−1 catalase, 0.1% v/v glucose) containing triplet-state quenching compounds (1 mM Trolox, 1 mM cyclooctatetraene, 1 mM nitrobenzyl alcohol).
Ribosome complexes (1 nM) programmed with biotinylated mRNA were surface immobilized following brief incubation within PEG-passivated, strepatividin-coated quartz microfluidic devices12. Pre-translocation complexes were prepared by incubating surface-immobilized initiation complexes with either the ternary complex of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U47) or EF-Tu(GTP)Lys-tRNALys (Cy5-acp3U47) (50 nM) for 2 min in Tris-polymix buffer (pH 7.5, see above), as previously described12. The Cy3 fluorophore linked to tRNAfMet was excited by total internal reflection of a single frequency light source (Ventus 532 nM, Laser Quanta). Photons emitted from Cy3 and Cy5 were collected using a 1.2 NA 60× water-immersion objective (Nikon), where optical treatments were used to spatially separate Cy3 and Cy5 frequencies onto a cooled, back-thinned CCD (Cascade 512B, Photometrics). Fluorescence data were acquired using MetaMorph software (Universal Imaging Corporation) at a rate of 40 frames per second (25 ms integration). FRET trajectories were calculated from fluorescence trajectories using the formula FRET = ICy5/(ICy3 + ICy5), where ICy3 and ICy5 represent the Cy3 and Cy5 fluorescence intensities, respectively.
Fluorescence and FRET traces were selected for analysis using freely available smFRET automated analysis software (data not shown) implemented in Matlab (The MathWorks) using the following criteria: a single catastrophic photobleaching event, at least 8:1 signal-to-background noise ratio (SNR), less than 4 donor fluorophore blinking events, a correlation coefficient between donor/acceptor < 0.5, at least one data point with FRET ≥ 0.39, and a lifetime of at least 15 frames (375 ms) in any FRET state ≥ 0.15. Each smFRET experimental population contains n > 500 selected traces. Each FRET trajectory was idealized to a hidden Markov model using the segmental k-means algorithm52. As previously described12, a four state model was used to determine the lifetimes of classical (C), hybrid state-1 (H1) and hybrid state-2 (H2). Standard deviations for each FRET state in the model were set to 0.061, the value observed for dye-labeled DNA oligonucleotide samples (data not shown). A three-state model was used to determine the lifetimes of classical and the combined hybrid states (H1 + H2) for experiments where distinctions between H1 and H2 were unnecessary (Supplementary Table 2) or where the kinetic parameters of H1 and H2 could be sensitive to idealization errors (Supplementary Fig. 2). Time-averaged occupancies in each FRET state were calculated from the total dwell times in each state divided by the total dwell time in all nonzero FRET states. Kinetic parameters for conformational transitions were estimated from the dwell times observed in each state before transition using the maximum interval likelihood algorithm implemented in QuB52.
From these values, the relative changes in free energy between states (ΔΔG) were determined according to the equation ΔΔG = −RTln(keq), where R is the gas constant, T is temperature in Kelvin and keq is the ratio of state populations at equilibrium.
Experimental and analytical error analysis
Experimental error was measured by performing a limited subset of smFRET experiments in triplicate, and an analytical bootstrapping method was used to approximate these errors in the remaining data (see Supplementary Methods).
Accession codes
Protein Data Bank: Ribosome structures used in this work were deposited as part of previous studies under entries 2J00 and 2J01 (ref. 4) and 2QB9 and 2QAL (ref. 35).
Supplementary Material
Acknowledgments
The authors thank J. Munro for comments and suggestions during the preparation of this manuscript and for the generation of ribosome structural diagrams. We thank all members of the laboratory for their constructive comments during preparation of this manuscript, as well as R. Green (Johns Hopkins School of Medicine, Howard Hughes Medical Institute) for providing MS2-tagged rRNA constructs. This work was supported by the US National Institute of General Medical Sciences (5R01GM079238-03), New York State Foundation for Science, Technology and Innovation, and a US National Science Foundation CAREER award. M.B.F. is a trainee in the Weill Cornell/Rockefeller University/Sloan-Kettering Tri-Institutional MD-PhD Program supported by US National Institutes of Health Medical Scientist Training Program grant GM07739. D.S.T. is supported by the Tri-Institutional Training Program in Computational Biology and Medicine.
Footnotes
Author contributions M.B.F. and S.C.B. designed the research. M.B.F. performed the smFRET experiments. D.S.T. implemented the automated analysis software, and D.S.T. and M.B.F. performed the data analysis. R.B.A. prepared the dye-labeled tRNA and ribosome complexes and reagents for the smFRET experiments. M.B.F., D.S.T. and S.C.B. prepared the manuscript.
Additional information Supplementary information and chemical compound information is available online at http://www.nature.com/naturechemicalbiology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Rodnina MV, Gromadski KB, Kothe U, Wieden HJ. Recognition and selection of tRNA in translation. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 2.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem. Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 4.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 5.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 angstrom resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 6.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard S. Navigating the ribosome’s metastable energy landscape. Trends Biochem. Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 8.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc. Natl. Acad. Sci. USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu. Rev. Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- 12.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green R, Noller HF. Ribosomes and translation. Annu. Rev. Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 14.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies J, Davis BD. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics: the effect of drug concentration. J. Biol. Chem. 1968;243:3312–3316. [PubMed] [Google Scholar]
- 16.Cabanas MJ, Vazquez D, Modolell J. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem. Biophys. Res. Commun. 1978;83:991–997. doi: 10.1016/0006-291x(78)91493-6. [DOI] [PubMed] [Google Scholar]
- 17.Misumi M, Nishimura T, Komai T, Tanaka N. Interaction of kanamycin and related antibiotics with the large subunit of ribosomes and the inhibition of translocation. Biochem. Biophys. Res. Commun. 1978;84:358–365. doi: 10.1016/0006-291x(78)90178-x. [DOI] [PubMed] [Google Scholar]
- 18.Fredrick K, Noller H. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 19.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 20.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 21.Hirokawa G, et al. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puglisi JD, et al. Aminoglycoside antibiotics and decoding. In: Garrett RA, et al., editors. The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. American Society for Microbiology Press; Washington, DC: 2000. pp. 419–429. [Google Scholar]
- 23.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 24.Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J. Mol. Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- 25.Fast R, Eberhard TH, Ruusala T, Kurland CG. Does streptomycin cause an error catastrophe? Biochimie. 1987;69:131–136. doi: 10.1016/0300-9084(87)90245-8. [DOI] [PubMed] [Google Scholar]
- 26.Kurland CG. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 27.Sander P, Prammananan T, Böttger EC. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 1996;22:841–848. doi: 10.1046/j.1365-2958.1996.01532.x. [DOI] [PubMed] [Google Scholar]
- 28.Prammananan T, et al. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 1998;177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 29.Recht MI, Douthwaite S, Puglisi JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu DH, Litovchick A, Rando RR. Stereospecificity of aminoglycoside-ribosomal interactions. Biochemistry. 2002;41:10499–10509. doi: 10.1021/bi026086l. [DOI] [PubMed] [Google Scholar]
- 33.Sucheck SJ, et al. Design of bifunctional antibiotics that target bacterial rRNA and inhibit resistance-causing enzymes. J. Am. Chem. Soc. 2000;122:5230–5231. [Google Scholar]
- 34.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 35.Borovinskaya MA, et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 36.Semenkov Y, Shapkina T, Makhno V, Kirillov S. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Lett. 1992;296:207–210. doi: 10.1016/0014-5793(92)80380-y. [DOI] [PubMed] [Google Scholar]
- 37.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 38.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan D, Kirillov S, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys. J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada T, Teshima T, Shiba T, Nierhaus KH. The translocation inhibitor tuberactinomycin binds to nucleic acids and blocks the in vitro assembly of 50S subunits. Nucleic Acids Res. 1980;8:5767–5777. doi: 10.1093/nar/8.23.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell. 2007;99:475–487. doi: 10.1042/BC20070037. [DOI] [PubMed] [Google Scholar]
- 43.Kaul M, Pilch DS. Thermodynamics of aminoglycoside-rRNA recognition: the binding of neomycin-class aminoglycosides of the A site of 16S rRNA. Biochemistry. 2002;41:7695–7706. doi: 10.1021/bi020130f. [DOI] [PubMed] [Google Scholar]
- 44.Vaiana AC, Sanbonmatsu KY. Stochastic gating and drug-ribosome interactions. J. Mol. Biol. 2008;386:648–661. doi: 10.1016/j.jmb.2008.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karimi R, Ehrenberg M. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 1996;15:1149–1154. [PMC free article] [PubMed] [Google Scholar]
- 46.Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 47.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campuzano S, Vazquez D, Modolell J. Dissociation of guanosine nucleotide-elongation factor G-ribosome complexes. Biochemistry. 1979;18:1570–1574. doi: 10.1021/bi00575a029. [DOI] [PubMed] [Google Scholar]
- 49.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 50.Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 51.Youngman EM, Green R. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Qin F. Principles of single-channel kinetic analysis. Methods Mol. Biol. 2007;403:253–286. doi: 10.1007/978-1-59745-529-9_17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.