Abstract
Developing wisdom teeth are easy-accessible source of stem cells during the adulthood which could be obtained by routine orthodontic treatments. Human pulp-derived stem cells (hDPSCs) possess high proliferation potential with multi-lineage differentiation capacity compare to the ordinary source of adult stem cells1-8; therefore, hDPSCs could be the good candidates for autologous transplantation in tissue engineering and regenerative medicine. Along with these benefits, possessing the mesenchymal stem cells (MSC) features, such as immunolodulatory effect, make hDPSCs more valuable, even in the case of allograft transplantation6,9,10. Therefore, the primary step for using this source of stem cells is to select the best protocol for isolating hDPSCs from pulp tissue. In order to achieve this goal, it is crucial to investigate the effect of various isolation conditions on different cellular behaviors, such as their common surface markers & also their differentiation capacity.
Thus, here we separate human pulp tissue from impacted third molar teeth, and then used both existing protocols based on literature, for isolating hDPSCs,11-13 i.e. enzymatic dissociation of pulp tissue (DPSC-ED) or outgrowth from tissue explants (DPSC-OG). In this regards, we tried to facilitate the isolation methods by using dental diamond disk. Then, these cells characterized in terms of stromal-associated Markers (CD73, CD90, CD105 & CD44), hematopoietic/endothelial Markers (CD34, CD45 & CD11b), perivascular marker, like CD146 and also STRO-1. Afterwards, these two protocols were compared based on the differentiation potency into odontoblasts by both quantitative polymerase chain reaction (QPCR) & Alizarin Red Staining. QPCR were used for the assessment of the expression of the mineralization-related genes (alkaline phosphatase; ALP, matrix extracellular phosphoglycoprotein; MEPE & dentin sialophosphoprotein; DSPP).14
Keywords: Stem Cell Biology, Issue 69, Medicine, Developmental Biology, Cellular Biology, Bioengineering, Dental pulp tissue, Human third molar, Human dental pulp stem cells, hDPSC, Odontoblasts, Outgrown stem cells, MSC, differentiation
Introduction
Stem cells are clonogenic cells which possess two remarkable features, known as multi-potency and self-renewal15. Among all stem cells with different replicative potencies, dental stem cells as the postnatal stem cells have drawn attention in recent years because of their accessibility, plasticity, and high proliferative ability in comparison with other adult stem cells16. Characteristically, similar to the mesenchymal stem cells, dental pulp stem cells are adherent clonogenic cells which have multiple differentiation capacity into mesenchyme and/or non-mesenchyme lineages, both in vitro and in vivo.1-8,17,18 DPSCs are identified by their negative expression of hematopoietic antigens (e.g., CD45, CD34, CD14), and positive expression of CD90, CD29, CD73, CD105, CD44 and STRO-1.19,20
Easy obtained potential with minimum pain & morbidity make human DPSCs as a valuable source of MSCs compared to the ordinary sources, such as bone marrow mesenchymal stem cells21. In general, DPSCs have been isolated by either outgrowth method, i.e., migration of stem cells from pulp tissue explants (DPSC-OG)22-24, and/or by enzymatic digestion (DPSC-ED)4,6,25. Previous studies have shown that the isolation method and culture conditions can induce different populations or lineages under in vitro passages26,27. In the case of permanent teeth (pDPSCs), Huang et al revealed that enzymatic digested pDPSCs have higher proliferation potential compared to the outgrown DPSCs.26 Moreover, in the case of deciduous teeth (dDPSCs), it was demonstrated that STRO-1 & CD34 markers expressed more in dDPSC-ED in comparison with dDPSC-OG. In addition, dDPSC-ED displayed higher mineralization rate in defined osteo/odonto medium.27 Therefore, due to the outstanding potential of DPSCs in regenerative medicine, more studies will be required for better understanding of possible various populations which are derived from different isolation methods.
Here, it was attempt to introduce easy way of pulp extraction, by using one-step dental diamond disk to facilitate the process of pulp extraction. Moreover, after the isolation of human pulp-derived stem cells by applying ED or OG methods, general properties & differentiation capacity between two groups were also investigated.
Protocol
1. Prepare the Enzyme Solution and Proliferation Medium (PM)
Make Collagenase Type I Solution: Weigh out collagenase type I (12 mg/ml) and dissolve in 1 ml PBS and filter using a 0.2 μm syringe filter. Then place it 15 ml tube and keep it at -20 °C until needed.
Make dispase Solution: Weigh out dispase (16 mg/ml) and dissolve in 1 ml PBS and filter using a 0.2 μm syringe filter. Then place it 15 ml tube and keep it at 4 °C until needed.
Make enzyme solution: Add 1 ml collagenase type I solutions (12 mg/ml) and 1 ml dispase solutions (16 mg/ml) into the 2 ml sterile PBS containing 100 mg/ml penicillin, 100 mg/ml streptomycin. Total concentration of dispase and Collagenase I in final volume should be 4 mg/ml and 3 mg/ml, receptively. Then, aliquot this volume in to four 15 ml tube, each containing 1 ml enzyme solution. Each tube could be used for one pulp digestion.
Make washing solution: Add 100 mg/ml penicillin, 100 mg/ml streptomycin into PBS.
Make basic media: Alpha modification of Eagle's medium (α-MEM) supplemented with 10% FBS & 100 units/ml penicillin, 100 mg/ml streptomycin.
Make the Proliferation Media (PM): Alpha modification of Eagle's medium (α-MEM) supplemented with 10% FBS, 100 μM L-ascorbic acid 2-phosphate, 2 mM L-glutamine, 100 units/ml penicillin, 100 mg/ml streptomycin, 0.25 mg/ml amphotericin B.
Make Odontogenic media: Alpha modification of Eagle's medium (α-MEM) supplemented with 10% FBS, 100 μM L-ascorbic acid 2-phosphate, 2 mM L-glutamine, 100 units/ml penicillin, 100 mg/ml streptomycin, 0.01 μM Dexamethasone, 5 mM β-Glycerol phosphate, 1.8 mM Monopotassium phosphate.
2. Prepare Human Dental Pulp Tissue for Dental Pulp Stem Cell Isolation
Normal human third molars were collected from adults (21-29 years of age) at the Dental Clinic of the Imam Ali under approved Institutional Review Board (IRB).
Teeth were placed into the basic medium (α-MEM supplemented with 10% FBS) and were transferred into laboratory at 4 °C.
Under the sterile condition, working within a biohazard laminar flow hood, set-up was done one 100 mm Petri dishes for each tooth to be processed. Tooth surfaces were cleaned by 70% ethanol.
While working in one of the Petri-dishes, tooth was kept by dental forceps and a scalpel blade was used to clean off debris and periodontal ligament on the roots.
Cemento-enamel junction was slowly cut by using sterilized dental disc to reveal the pulp chamber. It should be considered that cutting process must be performed slowly to reduce overheating of the dental tissue.
Pulp tissue was gently separated from the crown and root.
Pulp tissue was cut into small pieces with the aid of a scalpel blade.
3. Human Dental Pulp Isolation
Pulp tissues underwent either enzymatic dissociation of pulp tissue or direct cell outgrowth from pulp tissue explants for isolation of hDPSCs.
- Enzymatic dissociation of pulp tissue:
- Small pieces of pulp tissues were transferred into Enzyme solution for 1 hr at 37 °C. Vortex every 30 min to help breaking up tissue.
- Afterwards, large cell aggregates were removed and Single-cell suspensions were obtained by passing cells through a 70-μM cell strainer.
- Single-cell suspensions were centrifuged at 1,200 rpm for 5 min at room temperature.
- Supernatants were carefully pipetted off and pellet was re-suspended in 1 ml proliferation medium (PM). (Note: FBS in proliferation medium terminate enzymatic dissociation.)
- Single-cell suspensions of dental pulp was seeded into 25 cm2 culture flask with PM and then incubated at 37 °C in 5% CO2.
- The culture medium was changed every three days until the cell confluency was achieved.
- Direct cell outgrowth from pulp tissue explants:
- Pulp tissue was minced into 1-2-mm fragments, and each piece was placed in 25-cm2 culture flasks with PM and then incubated at 37 °C in 5% CO2. It should be considered that total volume of the PM for outgrowth of cell must support the attachment of pulp pieces for further cell outgrowth (2-3 ml per flask).
- Medium was changed after outgrowth was observed.
- The outgrown cells at confluence were sub-cultured at ratio of 1:4 (passage 1).
4. Immunophenotyping
250,000 cells/tube of both types of cells in passage 3 were incubated by PE or FITC-conjugated anti-human antibodies with their isotypes for 30 min at 4 °C in dark place. (Note: the concentration of antibodies was applied according to the manufacture protocol.)
After the incubation time, 100 μl PBS or FACS dilution solution was added to each tube and centrifuged at 1,200 rpm for 5 min in dark place.
Supernatants were removed & pellets were re-suspended in 100 μl PBS or FACS dilution in dark place.
Targeted surface markers (CD markers) were evaluated by BD FACSCalibur.
5. Induction of Differentiation of DPSCs into Mineralized Cells & Quantified Alizarin Red S Assay
DPSCs that were obtained by either enzymatic dissociation of pulp tissue (known as DPSC-ED) or direct cell outgrowth from pulp tissue explants (DPSC-OG) were sub-cultured for 3 passages.
At Passage 3, cells were seed into a 6 well plate at 5,000 cell/well.
At confluence of 60%, odontogenic medium were replaced by conventional PM. Three wells were remained as a negative control by adding growth medium.
Medium change every 3 days until day 21.
At day 21, Cells were washed with PBS and were fixed by 1 ml/well 10% paraformaldehyde for 15 min at room temperature.
Then paraformaldehyde was pipetted off, carefully and cells were washed 3 times (5-10 min for each time) with distilled H2O. (Note: Avoid disturbing the monolayer.)
After washing, 1 ml/well Alizarin red S were added for 20 min at room temperature.
Cells were washed by deionized H2O four times (5 min for each time with gentle rocking) to remove any additional Alizarin red.
1 ml/well H2O were added to prevent the cells from drying.
The plates were assumed to visual inspection and/or image acquisition.
For Alizarin Red S quantification assay, H2O were replaced by 1 ml 10% acetic acid to each well of 6-well plate & incubate for 30 min at room temperature with shaking.
Then, cell monolayer/well were scrape & both acetic acid & cells were transferred into the separate 1.5 ml micro-centrifuge tubes. (Note: each well should be transfer into a separate tube i.e. three test wells & three control wells for each ED & OG groups.)
Tubes were vortexed vigorously for 30 sec.
Heat to 85 °C for 10 min. (Note: to avoid evaporation, tubes were sealed with parafilm.)
After heating, tubes were transferred into ice for 5 min. (Note: Tubes should not be opened until become cooled.)
The slurries were centrifuged at 20,000 x g for 15 min. While centrifuging, Alizarin Red standards were made up.
Alizarin Red S standard were prepared by first diluting the dilution buffer & used for making 2-fold serial dilutions based on the table.
| High Range concentration of dye | Low Range concentration of dye | ||||||||||||
| 2 mM | 1 mM | 500 μM | 250 μM | 125 μM | 62.5 μM | 31.3 μM | 15.6 μM | 7.8 μM | 3.9 μM | 1.9 μM | 0.9 μM | 0.4 μM | Blank |
After the centrifugation, 1 ml of supernatant/tube were transferred into new separate 1.5 ml micro-centrifuge tubes.
The pH was neutralized with ~375 μl 10% ammonium hydroxide per tube. (Note: pH rage should be 4.1-4.5.)
150 μl of standard & samples (includes separate tests & controls) were added into an opaque-walled, transparent bottom 96-well plate.
The absorbance was read at 405 nm and OD values from samples were compared with standard plot for further analysis.
Representative Results
DPSCs that were obtained by enzymatic dissociation (DPSC-ED) are shown here at day 10, 15,18 (Figure 1). The colonies initiated to form on almost 3 to 5 days after the isolation.
Outgrown DPSCs (DPSC-OG) are shown in Figure 2 on day 5, 10, 13 & 18. Fibroblast-like cells began to migrate from pulp tissue into the flask by parallel ordering by almost 5 days after seeding.
DPSCs at passage 3 using both methods are shown in Figure 3. Both types of DPSCs were almost in the same size & morphology.
Immunophenotyping results in DPSC-ED and DPSC-OG (Figure 4). These results indicated the presence of mesenchymal stem cell's markers such as CD44, CD73, CD105 & CD90, and the absence of hematopoietic & endothelial markers such as CD34, CD45 & CD11b. The expressions of CD105 & CD146 (perivascular marker) were more in DPSC-OG in comparison with DPSC-ED. STRO-1 was negative in both groups. (Blue lines referred to OG group, while red lines referred to ED one, yellow lines indicated isotypes.)
Alizarin Red S morphology & quantification results for evaluation of mineralized tissue formation in DPSC-ED and DPSC-OG on day 21 are shown in Figure 5. The more absorbing red color the more calcium deposition. These results indicate more calcium deposition in differentiated DPSC-ED (d) in comparison with DPSC-OG (c). (Triplicate technical replications) quantification of Alizarin Red S also indicated the significant (*) higher concentration of dye between DPSC-ED in comparison with OG groups. (Dye concentration also increased significantly (**, ***) between tests & controls of each group (ED or OG) which are indicated the calcium deposition after the differentiation process)
(Notes: A paired t-test analysis was used to determine the differences in the amount of mineralized matrix produced ED and OG groups after induction of differentiation. The results were expressed as mean ±SEM. (significance assumed for P < 0.05)
QPCR results of the expression of odontoblast and mineralized-related genes (DSPP) and (MEPE, ALP) respectively, in differentiated DPSC-ED & DPSC-OG are shown in Figure 6. These results indicated the significant up-regulation of ALP & MEPE after the differentiation of ED and OG groups. Meanwhile, ALP and MEPE expressions were significantly higher in differentiated DPSC-ED compared to the differentiated DPSC-OG. However, there is no difference in the expression of DSPP in differentiated cells between both groups (Housekeeping gene: Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHAZ)).28
(Note: The QPCR data were analyzed using the comparative CT method. A paired t-test analysis was used to determine the differences in gene expression between ED & OG groups before and after the differentiation, and also between differentiated cells of two groups. The results were expressed as mean ±SEM. (Significance assumed for P < 0.05) (Triplicate technical replications)
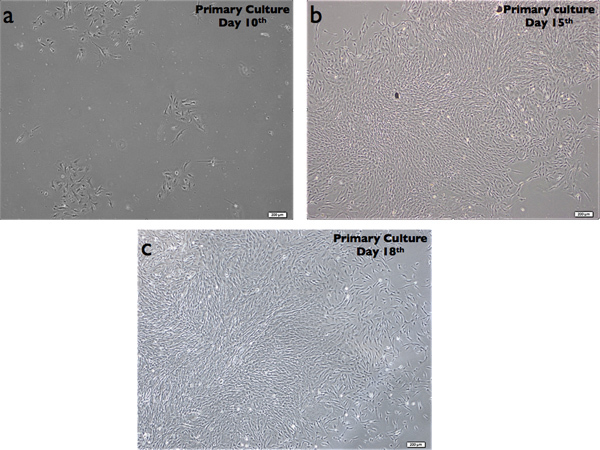 Figure 1. Enzymatic dissociated DPSC (DPSC-ED) at day 10, 15 & 18. Magnification bar =200 μm.
Figure 1. Enzymatic dissociated DPSC (DPSC-ED) at day 10, 15 & 18. Magnification bar =200 μm.
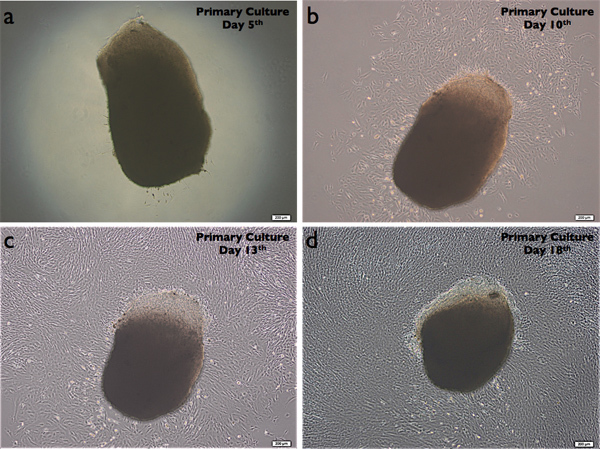 Figure 2. Outgrown DPSCs (DPSC-OG) on day 5, 10, 13 and 18. Magnification bar =200 μm.
Figure 2. Outgrown DPSCs (DPSC-OG) on day 5, 10, 13 and 18. Magnification bar =200 μm.
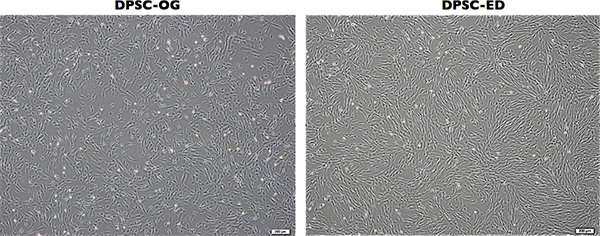 Figure 3. Outgrown DPSCs (DPSC-OG) & enzymatic digested DPSCs (DPSC-ED) Morphologies in passage 3. Magnification bar =200 μm. Click here to view larger figure.
Figure 3. Outgrown DPSCs (DPSC-OG) & enzymatic digested DPSCs (DPSC-ED) Morphologies in passage 3. Magnification bar =200 μm. Click here to view larger figure.
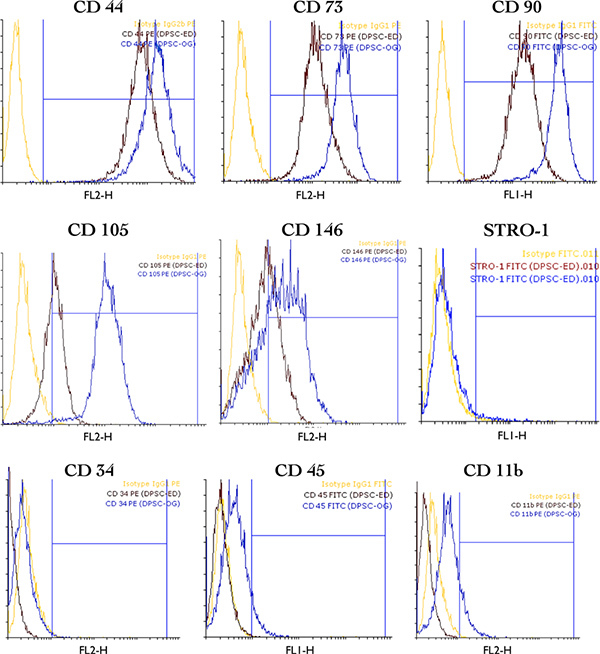 Figure 4. Overlaid Histogram of immunophenotyping results in DPSC-ED (dark red) and DPSC-OG (Blue). (Yellow lines represent isotypes). Click here to view larger figure.
Figure 4. Overlaid Histogram of immunophenotyping results in DPSC-ED (dark red) and DPSC-OG (Blue). (Yellow lines represent isotypes). Click here to view larger figure.
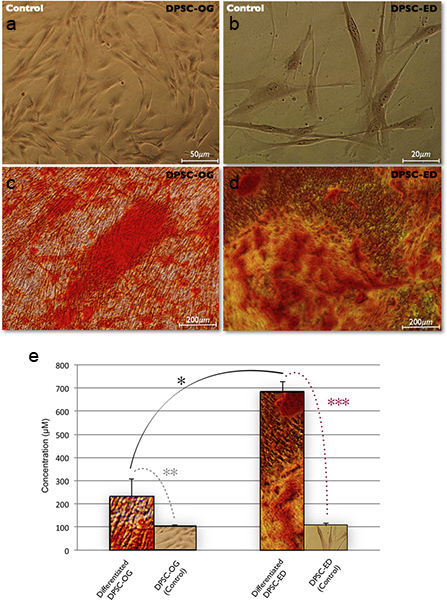 Figure 5. Alizarin Red S staining results in differentiated DPSC-OG (c), and differentiated DPSC-ED (d).(a) & (b) referred to the controls. Quantification of Alizarin Red S assay show more dye absorption in differentiated DPSC-ED compared to the differentiated OG group (e). Click here to view larger figure.
Figure 5. Alizarin Red S staining results in differentiated DPSC-OG (c), and differentiated DPSC-ED (d).(a) & (b) referred to the controls. Quantification of Alizarin Red S assay show more dye absorption in differentiated DPSC-ED compared to the differentiated OG group (e). Click here to view larger figure.
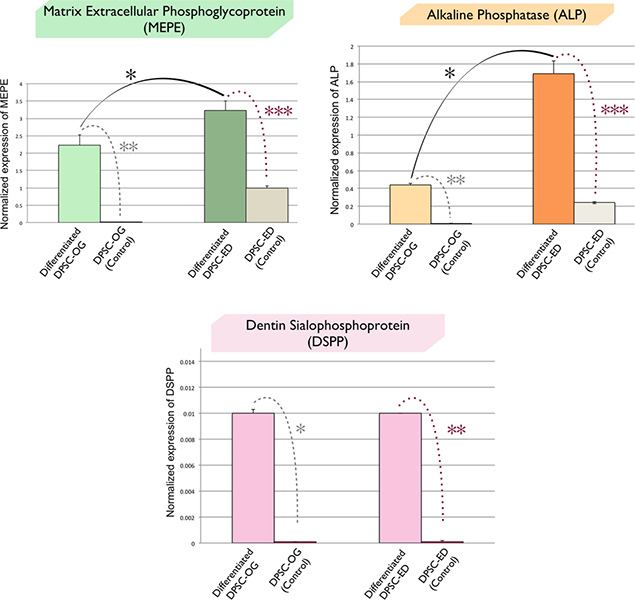 Figure 6. QPCR results of the expression of odontoblast and mineralized-related genes (DSPP) and (MEPE, ALP) respectively, in differentiated DPSC-ED & DPSC-OG. The expression of MEPE, ALP, & DSPP in differentiated cells compare to the control confirmed the differentiation of both groups into odontoblast (a-c). These results indicated the significant up-regulation of ALP & MEPE after the differentiation of ED and OG groups (a & b). Meanwhile, ALP and MEPE expressions were significantly higher in differentiated DPSC-ED compared to the differentiated DPSC-OG (a & b). However, there is no difference in the expression of DSPP in differentiated cells of both groups (c) (Housekeeping gene: Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein; YWHAZ). Click here to view larger figure.
Figure 6. QPCR results of the expression of odontoblast and mineralized-related genes (DSPP) and (MEPE, ALP) respectively, in differentiated DPSC-ED & DPSC-OG. The expression of MEPE, ALP, & DSPP in differentiated cells compare to the control confirmed the differentiation of both groups into odontoblast (a-c). These results indicated the significant up-regulation of ALP & MEPE after the differentiation of ED and OG groups (a & b). Meanwhile, ALP and MEPE expressions were significantly higher in differentiated DPSC-ED compared to the differentiated DPSC-OG (a & b). However, there is no difference in the expression of DSPP in differentiated cells of both groups (c) (Housekeeping gene: Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein; YWHAZ). Click here to view larger figure.
Discussion
This protocol describes the process of isolation and characterization of hDPSCs from dental pulp using two methods, enzymatic dissociation and direct outgrowth of stem cells from pulp tissue explants. In addition, in vitro differentiation of these cells into odontoblasts, was assessed by quantitative Alizarin Red S assay & QPCR.
Existing protocols for isolating pulp tissue from human tooth had been used various instruments such as pliers (bone forceps)9, extirpation needle29, Gracey curette30, dental fissure burs4 , etc. Those methods are time-consuming, while considering low level of overheating of pulp by adding water continuously. However, in current technique, dental diamond disc is used for the isolation of pulp tissue, which is a rapid way to cut tooth with minimum contamination in one-step process, and minimizes the overheating of pulp during a cutting process. Moreover, hDPSCs are finally differentiated into odontoblasts that have been never compared in the case of human permanent teeth by discussed aim. Comparative assessment of differentiated hDPSCs (ED & OG groups) by quantitative Alizarin Red S assay indicated the higher mineralization potential of hDPSC-ED in comparison with OG group. These results also confirmed by QPCR. DSPP, ALP and MEPE as the mineralization markers up-regulated after the odontoblast differentiation in both groups. On the other hand, the expressions of MEPE & ALP were higher in differentiated hDPSC-ED compared to the control. In agreement with previous study about deciduous teeth27, stem cells derived from permanent teeth by using ED or OG isolation methods also exhibit similar differentiation behaviors under the same conditions. Moreover, for better comparative evaluation, in this study, both hDPSC-ED & hDPSC-OG were isolated from the same individuals.
Expressions of MSC markers and the absence of heamatopietic/endothelial markers identify both types of hDPSCs as mesenchymal stem cells. On the other hand, CD 105 /CD 146 were higher in DPSC-OG compared to the ED groups. Despite the existing evidence according to the presence of STRO-1 in DPSCs31, our results showed no expression of STRO-1 in both groups. However, the significant diversity in surface marker expression in different studies might be due to the either cultivation conditions such as passage number, composition of media, etc. or interindividual variations between different donors. The absence of STRO-1 in both groups may indicate that the expression of this marker might not directly affect odontoblast differentiation potential (not shown in video).
Different hDPSCs isolation condition might bring different differentiation capacity. These differences might be due to the existence of different populations in the pulp tissue. Thus, selecting the isolation methods might be useful for targeted-tissue specific repair & regeneration for future clinical approaches. Our results indicated the higher mineralization capacity of enzymatic digested DPSC compared to the outgrown ones. However, additional studies, such as the assessment of the formation of tubular structure in vivo & in vitro, are required to determine which of these two procedures is applicable for functional dentin/pulp tissue regeneration therapies.
Disclosures
No conflicts of interest declared.
Acknowledgments
We gratefully acknowledge Dr. Leila Shakeri & Dr. Aref Dournaei for their critical discus and Mr. Mohammad Reza Khadem Sharif for his technical supports.
References
- Volponi AA, Pang Y, Sharpe PT. Stem cell-based biological tooth repair and regeneration. Trends Cell Biol. 2010;20:715–722. doi: 10.1016/j.tcb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosrat IV, Widenfalk J, Olson L, Nosrat CA. Dental pulp cells produce neurotrophic factors, interact with trigeminal neurons in vitro, and rescue motoneurons after spinal cord injury. Dev. Biol. 2001;238:120–132. doi: 10.1006/dbio.2001.0400. [DOI] [PubMed] [Google Scholar]
- Gandia C, et al. Human dental pulp stem cells improve left ventricular function, induce angiogenesis, and reduce infarct size in rats with acute myocardial infarction. Stem Cells. 2008;26:638–645. doi: 10.1634/stemcells.2007-0484. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano A, d'Aquino R, Laino G, Papaccio G. Dental pulp stem cells: a promising tool for bone regeneration. Stem Cell Rev. 2008;4:21–26. doi: 10.1007/s12015-008-9013-5. [DOI] [PubMed] [Google Scholar]
- Kerkis I, et al. Early transplantation of human immature dental pulp stem cells from baby teeth to golden retriever muscular dystrophy (GRMD) dogs: Local or systemic. J. Transl. Med. 2008;6:35. doi: 10.1186/1479-5876-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyekwelu O, Seppala M, Zoupa M, Cobourne MT. Tooth development: 2. Regenerating teeth in the laboratory. Dent. Update. 2007;34:20–29. doi: 10.12968/denu.2007.34.1.20. [DOI] [PubMed] [Google Scholar]
- Cordeiro MM, et al. Dental pulp tissue engineering with stem cells from exfoliated deciduous teeth. J. Endod. 2008;34(08):962–969. doi: 10.1016/j.joen.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Pierdomenico L, et al. Multipotent mesenchymal stem cells with immunosuppressive activity can be easily isolated from dental pulp. Transplantation. 2005;80:836–842. doi: 10.1097/01.tp.0000173794.72151.88. [DOI] [PubMed] [Google Scholar]
- de Mendonca Costa A, et al. Reconstruction of large cranial defects in nonimmunosuppressed experimental design with human dental pulp stem cells. J. Craniofac. Surg. 2008;19:204–210. doi: 10.1097/scs.0b013e31815c8a54. [DOI] [PubMed] [Google Scholar]
- Tirino V, et al. Methods for the identification, characterization and banking of human DPSCs: current strategies and perspectives. Stem Cell Rev. 2011;7:608–615. doi: 10.1007/s12015-011-9235-9. [DOI] [PubMed] [Google Scholar]
- Tirino V, Paino F, De Rosa A, Papaccio G. Identification, isolation, characterization, and banking of human dental pulp stem cells. Methods Mol. Biol. 2012;879:443–463. doi: 10.1007/978-1-61779-815-3_26. [DOI] [PubMed] [Google Scholar]
- Eslaminejad MB, Vahabi S, Shariati M, Nazarian H. In vitro Growth and Characterization of Stem Cells from Human Dental Pulp of Deciduous Versus Permanent Teeth. J. Dent. (Tehran) 2010;7:185–195. [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ling J, Wu L, Liu L, Xiao Y. Expression of mineralization markers in dental pulp cells. J. Endod. 2007;33:703–708. doi: 10.1016/j.joen.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Nombela-Arrieta C, Ritz J, Silberstein LE. The elusive nature and function of mesenchymal stem cells. Nat. Rev. Mol Cell Biol. 2011;12:126–131. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J. Dent. Res. 2009;88:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano A, et al. Scaffold's surface geometry significantly affects human stem cell bone tissue engineering. J. Cell Physiol. 2008;214:166–172. doi: 10.1002/jcp.21175. [DOI] [PubMed] [Google Scholar]
- d'Aquino R, et al. Human dental pulp stem cells: from biology to clinical applications. J. Exp. Zool. B. Mol. Dev. Evol. 2009;312:408–415. doi: 10.1002/jez.b.21263. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Feki A, Papaccio G, Caton J. Dental pulp stem cells, niches, and notch signaling in tooth injury. Adv. Dent. Res. 2011;23:275–279. doi: 10.1177/0022034511405386. [DOI] [PubMed] [Google Scholar]
- Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J. Bone Miner. Res. 2003;18:696–704. doi: 10.1359/jbmr.2003.18.4.696. [DOI] [PubMed] [Google Scholar]
- Tomic S, et al. Immunomodulatory properties of mesenchymal stem cells derived from dental pulp and dental follicle are susceptible to activation by toll-like receptor agonists. Stem Cells Dev. 2011;20:695–708. doi: 10.1089/scd.2010.0145. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, et al. Mineralized nodule formation by cultures of human dental pulp-derived fibroblasts. Arch. Oral Biol. 1992;37:1045–1055. doi: 10.1016/0003-9969(92)90037-9. [DOI] [PubMed] [Google Scholar]
- About I, et al. Human dentin production in vitro. Exp. Cell Res. 2000;258:33–41. doi: 10.1006/excr.2000.4909. [DOI] [PubMed] [Google Scholar]
- Couble ML, et al. Odontoblast differentiation of human dental pulp cells in explant cultures. Calcif. Tissue Int. 2000;66:129–138. doi: 10.1007/pl00005833. [DOI] [PubMed] [Google Scholar]
- Onishi T, Kinoshita S, Shintani S, Sobue S, Ooshima T. Stimulation of proliferation and differentiation of dog dental pulp cells in serum-free culture medium by insulin-like growth factor. Arch. Oral Biol. 1999;44(99):361–371. doi: 10.1016/s0003-9969(99)00007-2. [DOI] [PubMed] [Google Scholar]
- Huang GT, Sonoyama W, Chen J, Park SH. In vitro characterization of human dental pulp cells: various isolation methods and culturing environments. Cell Tissue Res. 2006;324:225–236. doi: 10.1007/s00441-005-0117-9. [DOI] [PubMed] [Google Scholar]
- Bakopoulou A, et al. Assessment of the impact of two different isolation methods on the osteo/odontogenic differentiation potential of human dental stem cells derived from deciduous teeth. Calcif. Tissue Int. 2011;88:130–141. doi: 10.1007/s00223-010-9438-0. [DOI] [PubMed] [Google Scholar]
- Curtis KM, et al. EF1alpha and RPL13a represent normalization genes suitable for RT-qPCR analysis of bone marrow derived mesenchymal stem cells. BMC Mol. Biol. 2010;11:61. doi: 10.1186/1471-2199-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek J, et al. Dental pulp stem cells and their characterization. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2009;153:31–35. doi: 10.5507/bp.2009.005. [DOI] [PubMed] [Google Scholar]
- d'Aquino R, et al. Human postnatal dental pulp cells co-differentiate into osteoblasts and endotheliocytes: a pivotal synergy leading to adult bone tissue formation. Cell Death Differ. 2007;14:1162–1171. doi: 10.1038/sj.cdd.4402121. [DOI] [PubMed] [Google Scholar]
- Atari M, et al. Isolation of pluripotent stem cells from human third molar dental pulp. Histol. Histopathol. 2011;26:1057–1070. doi: 10.14670/HH-26.1057. [DOI] [PubMed] [Google Scholar]


