Abstract
In this study, we characterized early biochemical changes associated with sertraline and placebo administration and changes associated with a reduction in depressive symptoms in patients with major depressive disorder (MDD). MDD patients received sertraline or placebo in a double-blind 4-week trial; baseline, 1 week, and 4 weeks serum samples were profiled using a gas chromatography time of flight mass spectrometry metabolomics platform. Intermediates of TCA and urea cycles, fatty acids and intermediates of lipid biosynthesis, amino acids, sugars and gut-derived metabolites were changed after 1 and 4 weeks of treatment. Some of the changes were common to the sertraline- and placebo-treated groups. Changes after 4 weeks of treatment in both groups were more extensive. Pathway analysis in the sertraline group suggested an effect of drug on ABC and solute transporters, fatty acid receptors and transporters, G signaling molecules and regulation of lipid metabolism. Correlation between biochemical changes and treatment outcomes in the sertraline group suggested a strong association with changes in levels of branched chain amino acids (BCAAs), lower BCAAs levels correlated with better treatment outcomes; pathway analysis in this group revealed that methionine and tyrosine correlated with BCAAs. Lower levels of lactic acid, higher levels of TCA/urea cycle intermediates, and 3-hydroxybutanoic acid correlated with better treatment outcomes in placebo group. Results of this study indicate that biochemical changes induced by drug continue to evolve over 4 weeks of treatment and that might explain partially delayed response. Response to drug and response to placebo share common pathways but some pathways are more affected by drug treatment. BCAAs seem to be implicated in mechanisms of recovery from a depressed state following sertraline treatment.
Keywords: depression, metabotype, metabolomics, pharmacometabolomics, personalized medicine, subclassification of disease
Introduction
Major depressive disorder (MDD) is a common and potentially life-threatening psychiatric illness that is a significant cause of disability worldwide.1 Selective serotonin (5-HT) reuptake inhibitors (SSRIs) are the major class of antidepressants for the treatment of MDD. Antidepressant effects of SSRIs result from inhibition of presynaptic 5-HT transporter, followed by changes in 5-HT neurotransmission and secondary changes in other neurotransmitter systems, such as catecholamines,2, 3, 4, 5 neurotransmitter amino acids6, 7, 8, 9, 10 and acetylcholine.11 It was shown that SSRIs induce delayed transcriptional and translational changes that mediate molecular and cellular neuroplasticity.12, 13, 14 Different antidepressants, including SSRIs, following their chronic administration have been found to increase biomarkers for energy metabolism.15, 16, 17, 18
The onset of antidepressant therapeutic action typically does not occur until at least 2–4 weeks of treatment; mechanisms underlying this delayed effect of antidepressants remain largely unknown and the biochemical changes that happen over the first 4 weeks that lead to therapeutic benefit are not well defined. Response to current therapies in treating MDD varies considerably, with 40% of patients not responding and 60% not remitting after an initial trial of therapy.19 Both the delayed onset of antidepressant therapeutic action and the variability in the response prevent clinicians from predicting whether an antidepressant is going to work for a particular patient. The detailed molecular mechanisms underlying variation in treatment response in depression and the genetic and biochemical basis remain unknown although numerous studies implicate the norepinephrine and 5-HT systems.20, 21, 22, 23 The placebo effect adds complexity, as 30–40% of MDD patients respond to placebo through mechanisms that are not yet understood.24, 25 Further insights into the mechanisms of response to placebo vs response to medications are needed and an ability to identify early on who might be a responder to placebo can stream line clinical trials. Development of biomarkers that can predict early on no response should have great clinical utility.
Metabolomics enables the identification and quantification of hundreds to thousands of compounds that can report on changes in biochemical pathways and should provide novel insights into the complex metabolic mechanisms of disease and drug effects.26, 27 Metabolomics provides powerful tools for characterization of complex phenotypes involving endogenous and exogenous mechanisms and for studies of complex diseases, such as depression. We have started to apply metabolomics tools to map global plasma biochemical changes in depressive subjects in remission and in mechanisms involved in response to SSRIs. In prior studies we identified perturbations in lipid metabolism, dicarboxylic fatty acids, branched chain and other amino acids, including GABA, as well as in ketone bodies in plasma of subjects with MDD.28, 29 In a study by Ji et al.,30 we used a pharmacometabolomics-informed pharmacogenomic research strategy and identified that glycine and a single-nucleotide polymorphism (SNP) in the glycine dehydrogenase gene were associated with response to escitalopram treatment. Merging pharmacometabolomics with pharmacogenomics using ‘1000 Genomes' SNP imputation, we provided additional insights about selective serotonin reuptake inhibitor response.31 We also demonstrated that metabolic profiles prior to treatment could help distinguish responders from non-responders to sertraline and placebo.32
In this current study, a hypothesis-generating metabolomics approach was used to map global biochemical changes induced by 1 week and 4 weeks treatment with sertraline and placebo. We also correlated biochemical changes and treatment outcomes at 1 and 4 weeks as a first step towards determining which biochemical changes could be associated with the delayed response to sertraline treatment.
Methods
Study design and patients
Study participants were outpatients, 18–65 years of age, from a randomized, double-blind, flexible dosing, placebo-controlled study performed at 12 clinical sites. Patients had a primary diagnosis of non-psychotic MDD by DSM-IV criteria, with symptoms of depression present for at least 1 month prior to screening, and a total baseline score >22 on the 17-item Hamilton Rating Scale for Depression (HAMD17)30 at screening. The full exclusion criteria for patients enrolled in the study was reported before.32 Patients were randomized to treatment (oral administration) with either sertraline or placebo at a ratio of 1:1. Sertraline dosing was started at 50 mg per day at baseline (week 0), with dose increased up to 100 mg per day at week 1 and up to 150 mg per day at week 2, as seen needed by the treating clinician. The primary outcome measure was the HAMD17 score. HAMD17 scores were determined and serum samples were collected at baseline (before treatment started), 1 week and at 4 weeks after treatment; the samples were collected from fasting subjects. A complete list of the inclusion and exclusion criteria used in this study, subjects medications, have been described in our previous report.32 Briefly, no psychotropic medications were used by the participants. The study protocol was developed in accordance with the principles of the Declaration of Helsinki. All patients provided written informed consent. The study was sponsored and monitored by Pfizer (Groton, CT, USA). Each site's IRB approved and oversaw the study.
GC-TOF mass spectrometry
Data were collected as described in Trupp et al.33 Briefly, the study design was entered into the SetupX database.34 Samples were aliquotted and maintained at −80 °C until use, at which point 30 μl of samples were extracted and derivatized with methoxyamine and MSTFA to trimethylsilation. Fatty acid methyl esters internal standards were included in the sample. One microliter of sample was injected using a Gerstel liner exchange system onto an Agilent 6890 gas chromatograph (Santa Clara, CA, USA) interfaced to a Leco Pegasus IV time of flight mass (TOF) spectrometer. Data were processed using BinBase35 and compound spectra and retention times were matched against the Fiehn laboratory library or NIST05.36 Intensities from all identified metabolites were summed and this value was used for data normalization. These data were used for statistical analysis.
Statistical methods
Metabolic signatures of sertraline and placebo (univariate analysis)
Paired t-tests were used to determine which metabolites changed significantly from baseline to week 1 and to week 4. These analyses were conducted separately for the sertraline and placebo groups. A significant t-test indicates that a metabolite is changing differently in the two groups. To determine which metabolites changed differently between the sertraline and placebo groups, we compared 1-week change scores and 4-week change scores between the two groups using t-tests for independent groups. Metabolite values were log transformed before data analysis. Q-values37 were calculated to control for false discovery rate.
Correlation with response analysis
Pearson correlation analysis was used to examine associations of changes in metabolite levels with concurrent changes in depressive symptoms. Analyses were conducted separately for 1- and 4-week change. Correlations of the sertraline group and placebo group were compared using the two-sample Z-test after hyperbolic tangent transformation. For these tests, Q-values37 were calculated for controlling the false discovery rate.
Response-pathway oriented analysis
After testing the marginal effects of metabolites, we then aimed to identify sets of metabolites with group effects on the drug response through the following steps. First, an individually significant metabolite was chosen based on the aforementioned marginal correlation analysis; second, a small subset of metabolites of strong marginal correlations with the preselected metabolite was identified; third, a principal component regression (PCR) analysis was used to regress the drug response on several principal components. These principal components are linear combinations of the subset of metabolites and therefore represent the group effect of the subset.
Pathway enrichment and network modeling
Pathways and networks were analyzed using multiple approaches. MetaMapp38 was used to calculate metabolic networks, which were displayed using Cytoscape. MetaMapp uses KEGG reaction pairs to build a metabolic network and then adds non-mapped compounds via Tanimoto similarity.39
Results
Sample characteristics
The sertraline and placebo groups did not differ significantly in age (44±11.4 years vs 40±12.7 years, P=0.12), gender (29/43 (69%) female vs 32/46 (70%) female, P=0.83) or race (34/43 (79%) white vs 34/46 (74%) white, P=0.57). The response rate was slightly higher for sertraline than for placebo, but this difference was not statistically significant (26/43 (60%) vs 23/46 (50%)) responders for sertraline and placebo groups, respectively.
Metabolic signatures of sertraline after 1 and 4 weeks of treatment
A GC-TOF metabolomics platform was used to semiquantitatively measure 348 compounds in patient serum. Of these, 143 were known compounds, 17 compounds were identified by spectral matching to the NIST library and 188 were unknown.
One-week treatment with sertraline
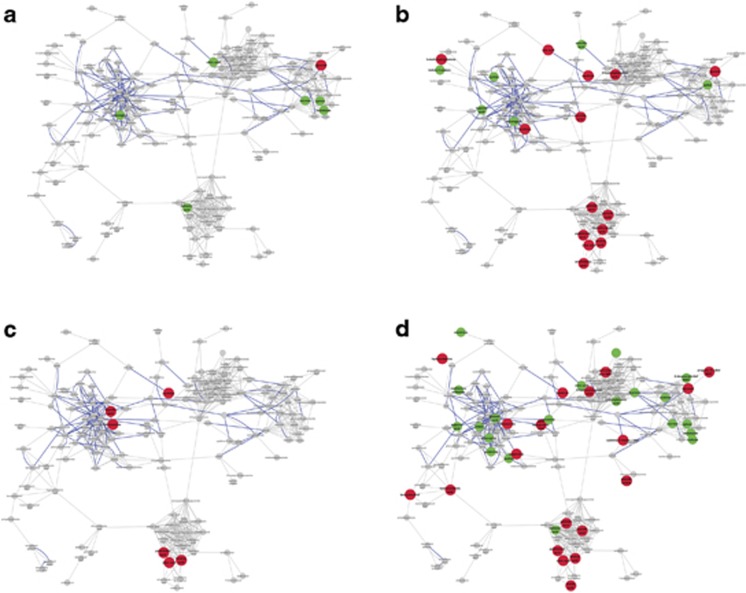
Several metabolites were changed following 1 week of treatment with sertraline; they included intermediates of TCA cycle, sugars and metabolites involved in fatty acid biosynthesis (Supplementary Table 1). All metabolites except glycerol showed an increase in concentration over this period. However, with Q-value cutoff set at 0.1 none of these changes was found to be significant. A metabolic network reconstruction highlighting the metabolites that changed after 1 week of sertraline is shown in Figure 1a.
Figure 1.
Compounds changing significantly (P<0.05) in the first (a, left top) and fourth (b, right top) week of sertraline treatment up (green) or down (red) are shown as enlarged nodes. Placebo treatment in the first week (c, left bottom) and fourth week (d, right bottom) is also shown. All compounds that were measured in the experiment are shown. Gray lines indicate network connections based upon structural similarity and bold blue lines indicate KEGG reaction pairs.
Four-week treatment with sertraline
Biochemical changes following 4-week treatment with sertraline were far more extensive than changes seen after 1 week of treatment (Table 1). Prominent effects were found on levels of fatty acids (linoleic, palmitic, arachidonic, oleic, palmitoleic and heptadecanoic acids all showed decreases) and glycerol, as well as changes in metabolites within the TCA and urea cycles (decreases in ornithine, citrulline and increase in aconitic acid). Other changes included metabolites from the purine pathway (xanthine decreased), one carbon metabolism (cysteine increased), tryptophan (5-methoxytryptamine decreased and indole-3-acetic acid increased) and a ketone body (3-hydroxybutanoic acid decreased). With Q-values cutoff controlled at 0.1, significant changes were decreased levels of linoleic, arachidonic and palmitic acids, and of ornithine and glycerol. A metabolic network reconstruction that highlights the metabolites changed after 4-week treatment with sertraline is shown in Figure 1b.
Table 1. Metabolic changes (P<0.05) after 4-week treatment with sertraline and placebo.
| Metabolite |
Sertraline |
P | Q | Metabolite |
Placebo |
P | Q | ||
|---|---|---|---|---|---|---|---|---|---|
| t-value | Percent change | t-value | Percent change | ||||||
| Linoleic acid | −3.791 | −20.471 | 0.001 | 0.036 | Elaidic acid | −5.259 | −30.801 | 5.500E−06 | 0.003 |
| Ornithine | −3.788 | −18.435 | 0.001 | 0.036 | Fructose | 4.213 | 487.726 | 1.443E−04 | 0.014 |
| Glycerol | −3.597 | −15.845 | 0.001 | 0.052 | Hippuric acid | 4.389 | 331.668 | 8.420E−05 | 0.014 |
| Arachidonic acid isomer | −3.306 | −16.947 | 0.002 | 0.088 | Linoleic acid | −4.175 | −22.794 | 1.617E−04 | 0.014 |
| Palmitic acid | −3.444 | −14.788 | 0.002 | 0.072 | Oleic acid | −4.206 | −21.281 | 1.472E−04 | 0.014 |
| Arachidonic acid | −3.123 | −15.713 | 0.004 | 0.113 | Palmitoleic acid | −4.520 | −24.734 | 5.620E−05 | 0.014 |
| Hippuric acid | 3.031 | 268.553 | 0.005 | 0.115 | Methionine sulfoxide | 3.082 | 77.910 | 0.004 | 0.113 |
| Oleic acid | −3.024 | −0.299 | 0.005 | 0.115 | Propane-1,3-diol NIST | −3.099 | −11.241 | 0.004 | 0.113 |
| Aconitic acid | 2.912 | 26.214 | 0.006 | 0.122 | α-Ketoglutaric acid | −2.997 | −11.514 | 0.005 | 0.115 |
| Cystine | 2.667 | 327.080 | 0.012 | 0.161 | Palmitic acid | −2.978 | −10.948 | 0.005 | 0.115 |
| Palmitoleic acid | −2.597 | 131.426 | 0.014 | 0.172 | Homoserine | −2.879 | −4.833 | 0.006 | 0.122 |
| 5-Methoxytryptamine | −2.530 | 425.326 | 0.016 | 0.184 | Inulobiose | 2.835 | 88.022 | 0.007 | 0.127 |
| Xanthine | −2.538 | −8.746 | 0.016 | 0.184 | Myristic acid | −2.843 | −9.520 | 0.007 | 0.127 |
| Myristic acid | −2.480 | −8.007 | 0.018 | 0.190 | Cytidine-5′-diphosphate | −2.590 | 1.726 | 0.013 | 0.172 |
| Indole-3-acetate | 2.462 | 21.503 | 0.019 | 0.191 | Glycerol | −2.535 | −7.201 | 0.015 | 0.183 |
| Heptadecanoic acid | −2.358 | −7.089 | 0.024 | 0.201 | Arabitol | 2.479 | 15.601 | 0.018 | 0.190 |
| 3-Hydroxybutanoic acid | −2.345 | 33.418 | 0.025 | 0.201 | Creatinine | 2.473 | 148.690 | 0.018 | 0.190 |
| Xylitol | 2.197 | 163.060 | 0.035 | 0.228 | Hydroxylamine | −2.481 | −8.192 | 0.018 | 0.190 |
| Hydroxycarbamate NIST | 2.152 | 298.069 | 0.039 | 0.228 | 2-Deoxytetronic acid | 2.434 | 31.055 | 0.020 | 0.193 |
| Asparagines | 2.132 | 53.085 | 0.040 | 0.228 | Histidine | 2.393 | 59.236 | 0.022 | 0.200 |
| Uric acid | −2.111 | −6.143 | 0.042 | 0.230 | Proline | 2.375 | 30.962 | 0.023 | 0.201 |
| Succinic acid | −2.067 | −5.740 | 0.046 | 0.235 | Glutamine | 2.330 | 82.385 | 0.025 | 0.201 |
| Maltose | 2.302 | 171.497 | 0.027 | 0.204 | |||||
| 3-Hydroxybutanoic acid | −2.245 | 9.976 | 0.031 | 0.213 | |||||
| Pseudo uridine | −2.158 | −5.284 | 0.037 | 0.228 | |||||
| 2-Deoxyerythritol NIST | 2.131 | 133.213 | 0.039 | 0.228 | |||||
| GABA | 2.133 | 139.734 | 0.039 | 0.228 | |||||
| Glutamic acid | 2.098 | 1264.390 | 0.042 | 0.230 | |||||
| Sucrose | 2.107 | 597.169 | 0.042 | 0.230 | |||||
| Hydrocinnamic acid | −2.089 | −6.049 | 0.043 | 0.230 | |||||
| Xanthine | −2.092 | −5.011 | 0.043 | 0.230 | |||||
| Quinic acid | 2.070 | 210.924 | 0.045 | 0.233 | |||||
| Oxoproline | −2.050 | −5.221 | 0.047 | 0.235 | |||||
| Benzylalcohol | −2.043 | 17.474 | 0.048 | 0.235 | |||||
Significant P-values are in bold.
Pathway enrichment analysis for 4-week treatment with sertraline is shown in Table 2. Several pathways involved in signaling were enriched in this analysis including transmembrane transporters of small molecules, signaling by GPCR, fatty acid receptors and transporters.
Table 2. Pathway enrichment analysis of the effect of sertraline exposure from baseline to week 4.
| Pathway name | PT | OV | Metabolites | P | Q |
|---|---|---|---|---|---|
| Transmembrane transport of small molecules | R | 9 | 170 (192) | 1.21E−06 | 1.51E−03 |
| Signaling by GPCR | R | 7 | 84 (93) | 1.30E−06 | 1.51E−03 |
| Signal transduction | R | 7 | 89 (98) | 1.93E−06 | 1.51E−03 |
| Regulation of lipid metabolism by PPAR-α | R | 3 | 5 (5) | 3.60E−06 | 2.12E−03 |
| SLC-mediated transmembrane transport | R | 8 | 147 (165) | 4.85E−06 | 2.28E−03 |
| Free fatty acid receptors | R | 4 | 19 (23) | 8.34E−06 | 3.27E−03 |
| Transport of fatty acids | R | 3 | 9 (9) | 2.97E−05 | 9.96E−03 |
| GPCR ligand binding | R | 5 | 66 (75) | 9.26E−05 | 0.027 |
| Gα (q) signaling events | R | 4 | 38 (43) | 1.45E−04 | 0.038 |
| Nucleotide metabolism | W | 3 | 16 (17) | 1.91E−04 | 0.045 |
| Fatty acid, triacylglycerol and ketone body metabolism | R | 5 | 83 (89) | 2.78E−04 | 0.059 |
| Glucose homeostasis | W | 3 | 21 (21) | 4.44E−04 | 0.087 |
| Arachidonate production from DAG | R | 2 | 5 (5) | 5.32E−04 | 0.096 |
| Class A/1 (Rhodopsin-like receptors) | R | 4 | 56 (64) | 6.62E−04 | 0.111 |
Abbreviations: PCR, principal component regression; PT, source of pathway; R, Reactome; W, Wikipathways; OV, number of overlapping metabolites; P, P-value calculated for metabolites; Q, Q-value calculated for metabolites after correction for FDR.
Metabolic signatures of placebo after 1 and 4 weeks of treatment
One-week treatment
Metabolites that changed after 1-week treatment with placebo are shown in Supplementary Table 1. Changes included amino acids homoserine and aspartate, one metabolite, xanthine, from the purine pathway, and the fatty acids linoleic, oleic and palmitoleic acids. Each metabolite showed a concentration decrease. However, similarly to 1-week treatment with sertraline, none of these changes was significant with Q-value cutoff of 0.1.
Four-week treatment with placebo
Significantly more profound biochemical changes were observed after 4 weeks of treatment with placebo, compared with 1 week (Table 1). Reductions were found in levels of fatty acids (linoleic, oleic, myristic and palmitoleic acids) and glycerol. Other changes included TCA cycle metabolites (decreased α-ketoglutarate, and increased glutamine and glutamic acid), amino acids (decreased homoserine, and increased proline, GABA and glutamic acid), metabolites linked to one carbon cycle (increased creatinine and decreased homoserine) (Figure 1d). Both xanthine from the purine pathway, and the ketone body 3-hydroxybutanoic acid were decreased. With Q-values cutoff controlled at 0.1, significant changes were found for elaidic, hippuric, linoleic, oleic and palmitoleic acids and fructose.
Associations of changes in metabolites with treatment outcomes
One-week treatment with sertraline
The associations between changes in individual metabolites and treatment outcome were also examined. Correlations between changes in individual metabolites and treatment outcome as assessed using HAMD17 scores are presented in Table 3a. Decrease in 5-methoxytryptamine (5-MTPM) levels was the most significant in association with symptoms reduction following 1 week of treatment with sertraline. Symptoms reduction was also associated with decrease in levels of ribose, trehalose and cystine. Increases in the levels of arachidonic and α-ketoglutaric acids were associated with symptoms reduction after 1-week sertraline treatment.
Table 3. Correlations with treatment outcomes: correlations between biochemical changes and changes in Ham-D.
| Metabolite | Cor | P-value |
|---|---|---|
| (a) One-week sertraline | ||
| 5-Methoxytryptamine | 0.405 | 0.016 |
| Arachidonic acid isom | −0.398 | 0.018 |
| Ribose | 0.389 | 0.021 |
| Cystine | 0.370 | 0.029 |
| Trehalose | 0.358 | 0.035 |
| α-Ketoglutaric acid | −0.339 | 0.047 |
| Xylose | −0.335 | 0.049 |
| (b) One-week placebo | ||
| Phosphoric acid | 0.510 | 0.001 |
| 4-Hydroxyproline | −0.439 | 0.005 |
| Cholesterol | 0.397 | 0.011 |
| Malate | −0.323 | 0.042 |
| (c) Four-week sertraline | ||
| Valine | 0.507 | 0.002 |
| Leucine | 0.478 | 0.004 |
| Lactic acid | −0.435 | 0.009 |
| Pseudo uridine | −0.409 | 0.015 |
| Conduritol-β-epoxi | −0.406 | 0.015 |
| Isoleucine | 0.382 | 0.024 |
| Mannonic acid NIST | 0.380 | 0.024 |
| Cystine | 0.362 | 0.032 |
| Inulobiose | −0.361 | 0.033 |
| (d) Four-week placebo | ||
| Lactic acid | 0.383 | 0.015 |
| Ribose6 | −0.371 | 0.018 |
| 3-Hydroxybutanoic acid | −0.355 | 0.025 |
| Oxalic acid | 0.346 | 0.029 |
| Hydroxycarbamate NIST | 0.345 | 0.029 |
| Citrulline | −0.336 | 0.034 |
| Indole-3-acetate | 0.323 | 0.042 |
| 1-Monostearin | 0.318 | 0.046 |
Negative correlations indicate better response with increase in metabolite; positive indicates better response with decrease in metabolite.
Four-week treatment with sertraline
After 4 weeks of sertraline treatment, the most significant changes associated with symptoms reduction were decreases in the levels of branched chain amino acids (BCAAs), valine, leucine and isoleucine (Table 3c). Similarly to 1 week, at 4 weeks of sertraline treatment symptoms reduction were associated with increase in levels of cysteine. Improvement of the symptoms after 4 weeks on sertraline was also associated with increases in the levels of lactic acid and pseudouridine.
One-week treatment with placebo
After 1 week of treatment with placebo, decrease in phosphoric acid and increases in 4-hydroxyproline and malate (Table 3b) were associated with symptoms reduction. Changes in several metabolites within TCA/urea cycle also correlated with changes in depression scores.
Four-week treatment with placebo
Changes in lactic acid, sugars, oxalic acid, ketone body, indole-3-acetic acid, urea cycle intermediate cirtruline and alanine were associated with symptoms reduction at 4 weeks of placebo treatment (Table 3d). In contrast to the sertraline findings, no association between changes in levels of BCAAs was noted.
Response-pathway oriented analysis
Since valine was found to be the most significant metabolite showing correlation with response in the sertraline group, we selected a list of 26 metabolites (Supplementary Table 2) marginally correlated with valine in sertraline group including leucine, isoleucine and tyrosine. A PCR analysis with variable selection identified four principal components jointly and significantly associated with drug response. In the first principal component (Supplementary Table 3), isoleucine, leucine, methionine, valine and tyrosine possessed the largest weights. Thus, this metabolite set characterized by major amino acids explored the possible relationship between amino acids, their transport and function, and depression, suggesting that further investigation of amino acids is warranted in studying biological mechanisms of sertraline.
Discussion
Recently, we conducted a metabolomics study to determine whether the baseline metabolic profile (that is, metabotype) of a patient with MDD would allow a prediction of patient's response to treatment with sertraline or placebo.32 In that study, utilizing an electrochemistry-based analytical platform that enables quantification of redox active compounds, we found that metabolites from phenylalanine, tryptophan, purine and tocopherol pathways enabled separation of responders and non-responders to treatment with the SSRI or placebo. The current study extends on these findings by (1) analyzing different classes of metabolites using GC-TOF mass spectrometry analytical platform; (2) identifying the biochemical changes that occur in MDD patients 1 week and 4 weeks after treatment with sertraline or placebo, and (3) identifying which changes are associated with a reduction in depressive symptoms after 1 and 4 weeks of treatment with sertraline or placebo.
Several metabolites from multiple pathways were changed in sertraline- and placebo-treated subjects. In both sertraline- and placebo-treated patients, the larger effects were observed after 4 weeks of treatment compared with 1 week. Multiple mechanisms could account for the biochemical changes observed in this study. Progression of the disease itself, as well as behavioral changes (for example, changes in diet) associated with improvement of the symptoms after treatment, could explain some of the effects such as changes in fatty acids and indices of energy metabolism. Thus, it was suggested that plasma levels of linoleic acid may serve as a possible biomarker for residual depression and anxiety in older people with previous depression;40 it was also shown earlier that increased dietary intake of oleic acid was associated with reduced risk of severe depressed mood among women while increased dietary intake of linoleic acid was associated with increased risk of severe depressed mood among men.41 It was recently found that omega-3 augmentation of SSRI citalopram treatment produced a significantly greater reduction in HAM-D scores compared with citalopram treatment alone.42 In a yeast model, sertraline has been shown to inhibit phospholipases A1 and D and activates phospholipase A2.43 It is thus possible, if the effect on phospholipases is similar in mammalian systems, that the changes in fatty acids observed in this study could be the result of sertraline inhibition and/or activation of specific phospholipases. It was also shown in primary cultures of mouse astrocytes that chronic SSRIs treatment upregulates calcium-dependent phospholipase A2 (PLA2), which releases arachidonic acid from the membrane-bound phospholipids.44, 45 Additional mechanism, involving PLA2 activation and increased release of arachidonic acid following SSRIs could involve membrane phospholipids remodeling and changes in signal transduction pathways.46, 47 The changes noted in fatty acids in the placebo group are of interest and could reflect changes in fatty acid biosynthesis or degradation as disease progresses. It can also be common to what was observed in the sertraline group.
Several studies implicated impairment of energy metabolism pathways in both pathogenesis of depression and effects of antidepressants. In an animal model of depression, levels of aconitate hydratase and pyruvate dehydrogenase were found to be regulated in opposite directions by stress and antidepressant drug treatment, thus representing molecular correlates of both vulnerability to depression and response to drug.48 Chronic administration of different antidepressants, including SSRIs, was associated with increased levels of biomarkers for energy metabolism, such as activity of brain creatine kinase, of mitochondrial respiratory chain complexes II and IV, and of citrate synthase and succinate dehydrogenase.17, 49, 50 Recently in an animal model significant changes in amino-acid metabolism related to energy metabolism via TCA cycle, and increased levels of glucose, glucose-6-phosphate and fructose-6-phosphate following chronic paroxetine treatment were found.18 Additional mechanisms related to the observed changes in energy metabolism following sertraline treatment could be related to the effects of 5-HT on glucose metabolism and glycolysis. It was demonstrated that 5-HT stimulates glucose uptake in skeletal muscle and activates skeletal muscle 6-phosphofructo-1-kinase activity in a dose-response through 5-HT2A receptors.51 Recently, it was found that 5-HT through 5-HT2A receptors controls hepatic glycolysis in mice,52 and 5-HT had stimulatory and inhibitory effects on glycogen synthesis in hepatocytes mediated by 5-HT1/2A and 5-HT2B receptors, respectively.53
We also analyzed associations between changes in individual metabolites and treatment outcome. After 1 week of sertraline treatment, the decrease in 5-methoxytryptamine and increases in arachidonic acid and α-ketoglutarate levels were the most significant in association with symptoms reduction. Association of decrease in 5-MTPM with symptoms reduction suggests potential role of methoxyindole branch of the tryptophan pathway in recovery form depressed state; currently we are investigating different branches of the pathway in the effects of SSRIs. Changes in the levels of arachidonic levels most likely reflect increase in the activity in phospholipase A2, resulting from increased 5-HT levels at earlier time point.44, 45 Association of increased levels of α-ketoglutarate with symptoms reduction is not clear; however, it may be related to the activation of energy metabolism following SSRI treatment. In support of this mechanism is the observation that improvement of symptoms at 4 weeks of treatment with sertraline was associated with decreased levels of lactic acid. The most significant changes associated with symptoms reduction after 4 weeks of treatment with sertraline were decreases in the levels of BCAAs, valine, leucine and isoleucine. In our previous study, we have demonstrated that higher levels of leucine and isoleucine were associated with depression.29 BCAAs have well-established anabolic effects on protein metabolism that involves activation of mTOR pathway. In this respect, it is particularly interesting that antidepressant effect of NMDA receptor antagonist ketamine results from activation of mTOR pathway involving BCAAs.54 In a recent study, Webhofer et al.18 found that in mice valine, leucine and isoleucine levels were increased after chronic treatment with SSRI paroxetine. Detailed mapping of effects of SSRIs, as well as of other antidepressants, on metabolism in animals and in humans should be compared at a more detailed level to enable translational research.
Of interest, there were several metabolites that closely correlated with changes in BCAAs and these included methionine and tyrosine. Methionine and the methionine cycle are closely linked to the folate cycle, to one carbon metabolism and to methylation processes that have been implicated in depression.
While there is strong evidence that MDD is influenced by inheritance,55 GWA studies for possible genetic factors that contribute to MDD risk, as well as GWA studies for genes associated with SSRI response in MDD patients, have provided somewhat disappointing results.56, 57 However, it was demonstrated that the SNP in the ABCB1 gene encoding ATP-binding cassette, subfamily B (MDR/TAP), member 1 (ABCB1) transporter protein expressed in the endothelial cells of the blood–brain barrier, is associated with severity of MDD and its response to citalopram and escitaloppram.58, 59 Recently, in GWA study Kohli et al.60 identified the SNP in a gene encoding a neuron-specific neutral amino-acid transporter, SLC6A15, as an MDD risk gene. While the exact function of the SLC6A15 SNP is not well understood, from both human and animal studies, it is known that SLC6A15 belongs to the solute carrier 6 (SLC6) gene family, which also includes the transporters for monoamines (including serotonin transporter, the primary target for SSRIs) and neurotransmitter amino acids, as well as a sodium-dependent transporter for neutral amino acids, including BCAAs.61 Earlier it was shown that in the SLC6A15 knockout mice, leucine uptake into cortical synaptosomes is significantly reduced.62 Zhou et al.63 demonstrated that SSRIs sertraline and fluoxetine have high affinity for the prokaryotic leucine transporter homolog (LeuT) of SLC6A15. Interestingly, SSRIs also bind to the homologous norepinephrine and dopamine transporters, although with much lower affinity than to their principal target serotonin transporter,64, 65 and only one or two different functional group substitutions are sufficient to convert an SSRI into a norepinephrine reuptake inhibitor with higher affinity to norepinephrine transporter.66, 67 Therefore, it is not unlikely that observed effect of sertraline on BCAAs could result from the SLC6A15 SNP in depressed patients. All these findings and the association between the improvement of depression and changes in BCAAs levels observed in our study strongly suggest potential role of BCAA in depression and in the effects of SSRI; further studies are needed to examine potential role of BCAAs in depression and the effects of different antidepressants, in both patients and animal models.
One of the limitations of this study is that despite comprehensive biochemical analysis it included relatively small number of subjects. However, this study should be viewed as hypotheses-generating to test whether metabolomics approach could provide insights into pathways implicated in SSRI and placebo response and mechanism of delayed response. Replication and validation of these findings in other clinical trials is important for future studies, however, this is outside the scope of this first report. Whether and to what extent peripheral biochemical changes reflect CNS (central nervous system) changes related to mechanisms of depression and drug effects also should be addressed in future studies. Depression is a disease that affects both brain and also the rest of the body, as is well known from the cortisol data;68, 69 therefore, it is possible that the peripheral changes reflect both direct CNS and additional CNS-linked peripheral effects and maybe direct peripheral effects also. Irrespective of the exact mechanism that leads to the changes and to response, the presence of consistent changes can be an important signature that could be the basis of further iterative understanding from both animal and human studies.
In conclusion, our data for the first time illustrate that biochemical changes following SSRI treatment are progressive and some of the pathways that are modified in the first week might lead to subsequent changes noted in the following weeks. The presence of consistent changes could provide an opportunity for developing biomarkers that can predict response or no response to antidepressant treatment.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- Prisco S, Esposito E. Differential effects of acute and chronic fluoxetine administration on the spontaneous activity of dopaminergic neurones in the ventral tegmental area. Br J Pharmacol. 1995;116:1923–1931. doi: 10.1111/j.1476-5381.1995.tb16684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bymaster FP, Zhang W, Carter PA, Shaw J, Chernet E, Phebus L, et al. Fluoxetine, but not other selective serotonin uptake inhibitors, increases norepinephrine and dopamine extracellular levels in prefrontal cortex. Psychopharmacology (Berl) 2002;160:353–361. doi: 10.1007/s00213-001-0986-x. [DOI] [PubMed] [Google Scholar]
- Dremencov E, El Mansari M, Blier P. Effects of sustained serotonin reuptake inhibition on the firing of dopamine neurons in the rat ventral tegmental area. J Psychiatry Neurosci. 2009;34:223–229. [PMC free article] [PubMed] [Google Scholar]
- Kitaichi Y, Inoue T, Nakagawa S, Boku S, Kakuta A, Izumi T, et al. Sertraline increases extracellular levels not only of serotonin, but also of dopamine in the nucleus accumbens and striatum of rats. Eur J Pharmacol. 2010;647:90–96. doi: 10.1016/j.ejphar.2010.08.026. [DOI] [PubMed] [Google Scholar]
- Golembiowska K, Dziubina A. Effect of acute and chronic administration of citalopram on glutamate and aspartate release in the rat prefrontal cortex. Polish J Pharmacol. 2000;52:441–448. [PubMed] [Google Scholar]
- Wang SJ, Su CF, Kuo YH. Fluoxetine depresses glutamate exocytosis in the rat cerebrocortical nerve terminals (synaptosomes) via inhibition of P/Q-type Ca2+ channels. Synapse. 2003;48:170–177. doi: 10.1002/syn.10200. [DOI] [PubMed] [Google Scholar]
- Bhagwagar Z, Wylezinska M, Taylor M, Jezzard P, Matthews PM, Cowen PJ. Increased brain GABA concentrations following acute administration of a selective serotonin reuptake inhibitor. Am J Psychiatry. 2004;161:368–370. doi: 10.1176/appi.ajp.161.2.368. [DOI] [PubMed] [Google Scholar]
- Marsteller DA, Barbarich-Marsteller NC, Patel VD, Dewey SL. Brain metabolic changes following 4-week citalopram infusion: increased 18FDG uptake and gamma-amino butyric acid levels. Synapse. 2007;61:877–881. doi: 10.1002/syn.20428. [DOI] [PubMed] [Google Scholar]
- Schipke CG, Heuser I, Peters O. Antidepressants act on glial cells: SSRIs and serotonin elicit astrocyte calcium signaling in the mouse prefrontal cortex. J Psychiatr Res. 2011;45:242–248. doi: 10.1016/j.jpsychires.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Mineur YS, Picciotto MR. Nicotine receptors and depression: revisiting and revising the cholinergic hypothesis. Trends Pharmacol Sci. 2010;31:580–586. doi: 10.1016/j.tips.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Lu B. Interaction between BDNF and serotonin: role in mood disorders. Neuropsychopharmacology. 2008;33:73–83. doi: 10.1038/sj.npp.1301571. [DOI] [PubMed] [Google Scholar]
- Rantamaki T, Hendolin P, Kankaanpaa A, Mijatovic J, Piepponen P, Domenici E, et al. Pharmacologically diverse antidepressants rapidly activate brain-derived neurotrophic factor receptor TrkB and induce phospholipase-Cgamma signaling pathways in mouse brain. Neuropsychopharmacology. 2007;32:2152–2162. doi: 10.1038/sj.npp.1301345. [DOI] [PubMed] [Google Scholar]
- Vidal R, Pilar-Cuellar F, dos Anjos S, Linge R, Treceno B, Vargas VI, et al. New strategies in the development of antidepressants: towards the modulation of neuroplasticity pathways. Curr Pharm Des. 2011;17:521–533. doi: 10.2174/138161211795164086. [DOI] [PubMed] [Google Scholar]
- Agostinho FR, Reus GZ, Stringari RB, Ribeiro KF, Ferraro AK, Benedet J, et al. Treatment with olanzapine, fluoxetine and olanzapine/fluoxetine alters citrate synthase activity in rat brain. Neurosci Lett. 2011;487:278–281. doi: 10.1016/j.neulet.2010.10.037. [DOI] [PubMed] [Google Scholar]
- Agostinho FR, Scaini G, Ferreira GK, Jeremias IC, Reus GZ, Rezin GT, et al. Effects of olanzapine, fluoxetine and olanzapine/fluoxetine on creatine kinase activity in rat brain. Brain Res Bull. 2009;80:337–340. doi: 10.1016/j.brainresbull.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Santos PM, Scaini G, Rezin GT, Benedet J, Rochi N, Jeremias GC, et al. Brain creatine kinase activity is increased by chronic administration of paroxetine. Brain Res Bull. 2009;80:327–330. doi: 10.1016/j.brainresbull.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Webhofer C, Gormanns P, Tolstikov V, Zieglgansberger W, Sillaber I, Holsboer F, et al. Metabolite profiling of antidepressant drug action reveals novel drug targets beyond monoamine elevation. Transl Psychiatry. 2011;1:e58. doi: 10.1038/tp.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi MH. Major depressive disorder: remission of associated symptoms. J Clin Psychiatry. 2006;67 (Suppl 6:27–32. [PubMed] [Google Scholar]
- Malhi GS, Parker GB, Greenwood J. Structural and functional models of depression: from sub-types to substrates. Acta Psychiatr Scand. 2005;111:94–105. doi: 10.1111/j.1600-0447.2004.00475.x. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Nemeroff CB. Role of serotonergic and noradrenergic systems in the pathophysiology of depression and anxiety disorders. Depress Anxiety. 2000;12 (Suppl 1:2–19. doi: 10.1002/1520-6394(2000)12:1+<2::AID-DA2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Insel TR, Wang PS. The STAR*D trial: revealing the need for better treatments. Psychiatr Serv. 2009;60:1466–1467. doi: 10.1176/ps.2009.60.11.1466. [DOI] [PubMed] [Google Scholar]
- Katz MM, Bowden CL, Frazer A. Rethinking depression and the actions of antidepressants: uncovering the links between the neural and behavioral elements. J Affect Disord. 2010;120:16–23. doi: 10.1016/j.jad.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Kirsch I. Challenging received wisdom: antidepressants and the placebo effect. McGill J Med. 2008;11:219–222. [PMC free article] [PubMed] [Google Scholar]
- Walsh BT, Seidman SN, Sysko R, Gould M. Placebo response in studies of major depression: variable, substantial, and growing. JAMA. 2002;287:1840–1847. doi: 10.1001/jama.287.14.1840. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Krishnan KR. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology. 2009;34:173–186. doi: 10.1038/npp.2008.174. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- Paige LA, Mitchell MW, Krishnan KR, Kaddurah-Daouk R, Steffens DC. A preliminary metabolomic analysis of older adults with and without depression. Int J Geriatr Psychiatry. 2007;22:418–423. doi: 10.1002/gps.1690. [DOI] [PubMed] [Google Scholar]
- Steffens DC, Wei J, Krishnan KR, Karoly ED, Mitchell MW, O'Connor CM, et al. Metabolomic differences in heart failure patients with and without major depression. J Geriatr Psychiatry Neurol. 2010;23:138–146. doi: 10.1177/0891988709358592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo R, Hebbring S, Ji Y, Zhu H, Zeng ZB, Batzler A, et al. Merging pharmacometabolomics with pharmacogenomics using ‘1000 Genomes' single-nucleotide polymorphism imputation: selective serotonin reuptake inhibitor response pharmacogenomics. Pharmacogenet Genomics. 2012;22:247–253. doi: 10.1097/FPC.0b013e32835001c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Boyle S, Matson W, Sharma S, Matson S, Zhu H, et al. Pretreatment metabotype as a predictor of response to sertraline or placebo in depressed outpatients: a proof of concept. Transl Psychiatry. 2011;32:1–7. doi: 10.1038/tp.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupp M, Zhu H, Wikoff WR, Baillie RA, Zeng ZB, Karp PD, et al. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS One. 2012;7:9. doi: 10.1371/journal.pone.0038386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz M, Fiehn O. SetupX--a public study design database for metabolomic projects. Pac Symp Biocomput. 2007. pp. 169–180. [PubMed]
- Skogerson K, Wohlgemuth G, Barupal DK, Fiehn O. The volatile compound BinBase mass spectral database. BMC Bioinformatics. 2011;12:321. doi: 10.1186/1471-2105-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Wohlgemuth G, Lee do Y, Lu Y, Palazoglu M, Shahbaz S, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA. 2009;106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadoon A, Chiu CC, McDermott L, Cunningham P, Frangou S, Chang CJ, et al. Associations of polyunsaturated fatty acids with residual depression or anxiety in older people with major depression. J Affect Disord. 2012;136:918–925. doi: 10.1016/j.jad.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Wolfe AR, Ogbonna EM, Lim S, Li Y, Zhang J. Dietary linoleic and oleic fatty acids in relation to severe depressed mood: 10 years follow-up of a national cohort. Prog Neuro-Psych Biol Psych. 2009;33:972–977. doi: 10.1016/j.pnpbp.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Gertsik L, Poland RE, Bresee C, Rapaport MH. Omega-3 fatty acid augmentation of citalopram treatment for patients with major depressive disorder. J Clin Psychopharmacol. 2012;32:61–64. doi: 10.1097/JCP.0b013e31823f3b5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey MM, Korostyshevsky D, Lee S, Perlstein EO. The antidepressant sertraline targets intracellular vesiculogenic membranes in yeast. Genetics. 2010;185:1221–1233. doi: 10.1534/genetics.110.117846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zhang S, Li M, Hertz L, Peng L. Chronic treatment of astrocytes with therapeutically relevant fluoxetine concentrations enhances cPLA2 expression secondary to 5-HT2B-induced, transactivation-mediated ERK1/2 phosphorylation. Psychopharmacology. 2009;207:1–12. doi: 10.1007/s00213-009-1631-3. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li B, Lovatt D, Xu J, Song D, Goldman SA, et al. 5-HT2B receptors are expressed on astrocytes from brain and in culture and are a chronic target for all five conventional 'serotonin-specific reuptake inhibitors'. Neuron Glia Biol. 2010;6:113–125. doi: 10.1017/S1740925X10000141. [DOI] [PubMed] [Google Scholar]
- Lee LH, Shui G, Farooqui AA, Wenk MR, Tan CH, Ong WY. Lipidomic analyses of the mouse brain after antidepressant treatment: evidence for endogenous release of long-chain fatty acids. Int J Neuropsychopharmacol. 2009;12:953–964. doi: 10.1017/S146114570900995X. [DOI] [PubMed] [Google Scholar]
- Rapoport SI. In vivo fatty acid incorporation into brain phosholipids in relation to plasma availability, signal transduction and membrane remodeling. J Mol Neurosci. 2001;16:243–261. doi: 10.1385/JMN:16:2-3:243. [DOI] [PubMed] [Google Scholar]
- Mallei A, Giambelli R, Gass P, Racagni G, Mathe AA, Vollmayr B, et al. Synaptoproteomics of learned helpless rats involve energy metabolism and cellular remodeling pathways in depressive-like behavior and antidepressant response. Neuropharmacology. 2011;60:1243–1253. doi: 10.1016/j.neuropharm.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Reus GZ, Stringari RB, Rezin GT, Fraga DB, Daufenbach JF, Scaini G, et al. Administration of memantine and imipramine alters mitochondrial respiratory chain and creatine kinase activities in rat brain. J Neural Transm. 2012;119:481–491. doi: 10.1007/s00702-011-0718-2. [DOI] [PubMed] [Google Scholar]
- Scaini G, Santos PM, Benedet J, Rochi N, Gomes LM, Borges LS, et al. Evaluation of Krebs cycle enzymes in the brain of rats after chronic administration of antidepressants. Brain Res Bull. 2010;82:224–227. doi: 10.1016/j.brainresbull.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Coelho WS, Costa KC, Sola-Penna M. Serotonin stimulates mouse skeletal muscle 6-phosphofructo-1-kinase through tyrosine-phosphorylation of the enzyme altering its intracellular localization. Mol Genet Metab. 2007;92:364–370. doi: 10.1016/j.ymgme.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Coelho WS, Da Silva D, Marinho-Carvalho MM, Sola-Penna M. Serotonin modulates hepatic 6-phosphofructo-1-kinase in an insulin synergistic manner. Int J Biochem Cell Biol. 2012;44:150–157. doi: 10.1016/j.biocel.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Tudhope SJ, Wang CC, Petrie JL, Potts L, Malcomson F, Kieswich J, et al. A novel mechanism for regulating hepatic glycogen synthesis involving serotonin and cyclin-dependent kinase-5. Diabetes. 2012;61:49–60. doi: 10.2337/db11-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Dorzab J, Winokur G, Cadoret R. Depressive disease. Evidence favoring polygenic inheritance based on an analysis of ancestral cases. Arch Gen Psychiatry. 1972;27:320–327. doi: 10.1001/archpsyc.1972.01750270030004. [DOI] [PubMed] [Google Scholar]
- Psychiatric GCCC, Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, et al. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lee MS, Lee JH, Kim SW, Kang RH, Choi MJ, et al. MRP1 polymorphisms associated with citalopram response in patients with major depression. J Clin Psychopharmacol. 2010;30:116–125. doi: 10.1097/JCP.0b013e3181d2ef42. [DOI] [PubMed] [Google Scholar]
- Lin KM, Chiu YF, Tsai IJ, Chen CH, Shen WW, Liu SC, et al. ABCB1 gene polymorphisms are associated with the severity of major depressive disorder and its response to escitalopram treatment. Pharmacogenet Genomics. 2011;21:163–170. doi: 10.1097/FPC.0b013e32833db216. [DOI] [PubMed] [Google Scholar]
- Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70:252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer S. The SLC6 orphans are forming a family of amino acid transporters. Neurochem Int. 2006;48:559–567. doi: 10.1016/j.neuint.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Drgonova J, Liu QR, Hall FS, Krieger RM, Uhl GR. Deletion of v7-3 (SLC6A15) transporter allows assessment of its roles in synaptosomal proline uptake, leucine uptake and behaviors. Brain Res. 2007;1183:10–20. doi: 10.1016/j.brainres.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Law CJ, Reith ME, Wang DN. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nat Struct Mol Biol. 2009;16:652–657. doi: 10.1038/nsmb.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshleman AJ, Carmolli M, Cumbay M, Martens CR, Neve KA, Janowsky A. Characteristics of drug interactions with recombinant biogenic amine transporters expressed in the same cell type. J Pharmacol Exp Ther. 1999;289:877–885. [PubMed] [Google Scholar]
- Tatsumi M, Groshan K, Blakely RD, Richelson E. Pharmacological profile of antidepressants and related compounds at human monoamine transporters. Eur J Pharmacol. 1997;340:249–258. doi: 10.1016/s0014-2999(97)01393-9. [DOI] [PubMed] [Google Scholar]
- Eildal JN, Andersen J, Kristensen AS, Jorgensen AM, Bang-Andersen B, Jorgensen M, et al. From the selective serotonin transporter inhibitor citalopram to the selective norepinephrine transporter inhibitor talopram: synthesis and structure-activity relationship studies. J Med Chem. 2008;51:3045–3048. doi: 10.1021/jm701602g. [DOI] [PubMed] [Google Scholar]
- Wong DT, Bymaster FP. Development of antidepressant drugs. Fluoxetine (Prozac) and other selective serotonin uptake inhibitors. Adv Exp Med Biol. 1995;363:77–95. [PubMed] [Google Scholar]
- Herbert J. Cortisol and depression: three questions for psychiatry. Psychol Med. 2012;8:1–21. doi: 10.1017/S0033291712000955. [DOI] [PubMed] [Google Scholar]
- Vreeburg SA, Hoogendijk WJ, van Pelt J, Derijk RH, Verhagen JC, van Dyck R, et al. Major depressive disorder and hypothalamic-pituitary-adrenal axis activity: results from a large cohort study. Arch Gen Psychiatry. 2009;66:617–626. doi: 10.1001/archgenpsychiatry.2009.50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.