Summary
Here we report the generation and analysis of genome-wide exon-level transcriptome data from 16 brain regions comprising the cerebellar cortex, mediodorsal nucleus of the thalamus, striatum, amygdala, hippocampus, and 11 areas of the neocortex. The dataset was generated from 1,340 tissue samples collected from one or both hemispheres of 57 postmortem human brains, spanning from embryonic development to late adulthood and representing males and females of multiple ethnicities. We also performed genotyping of 2.5 million SNPs and assessed copy number variations for all donors. Approximately 86% of protein-coding genes were found to be expressed using stringent criteria, and over 90% of these were differentially regulated at the whole transcript or exon level across regions and/or time. The majority of these spatiotemporal differences occurred before birth, followed by an increase in the similarity among regional transcriptomes during postnatal lifespan. Genes were organized into functionally distinct co-expression networks, and sex differences were present in gene expression and exon usage. Finally, we demonstrate how these results can be used to profile trajectories of genes associated with neurodevelopmental processes, cell types, neurotransmitter systems, autism, and schizophrenia, as well as to discover associations between SNPs and spatiotemporal gene expression. This study provides a comprehensive, publicly available dataset on the spatiotemporal human brain transcriptome and new insights into the transcriptional foundations of human neurodevelopment.
Introduction
Human neurodevelopment is a complex and precisely regulated process that unfolds over a protracted period of time1-3. Human-specific features of this process are likely to be important factors in the evolution of human specializations2,3,5,6. However, in addition to giving us remarkable cognitive and motor abilities, the formation of molecularly distinct and intricate neural circuits may have also increased our susceptibility to certain psychiatric and neurological disorders4,7-10. Furthermore, sex differences play an important role in brain development and function and are a risk factor for disorders such as autism spectrum disorders (ASD)10-14. Thus, comprehensive knowledge about the spatiotemporal dynamics of the brain transcriptome is essential for a better understanding of neurodevelopment, sexual dimorphism, and evolution, as well as our increased susceptibility to certain brain disorders.
Previous transcriptome studies of the developing human brain have included relatively small sample sizes and predominantly focused on few regions or developmental time points15-19. In this study, we explore the transcriptomes of 16 regions from developing and adult postmortem brains of clinically unremarkable donors representing males and females of multiple ethnicities.
Study design, data generation, and quality control
To investigate the spatiotemporal dynamics of the human brain transcriptome, we created a 15-period system spanning from embryonic development to late adulthood (Table 1; Supplementary Information 2.1). We sampled transient prenatal structures and immature and mature forms of 16 brain regions, including 11 NCX areas, from multiple specimens per period (Table 2; Supplementary Information 2.2; Supplementary Figs. 1-3; Supplementary Table 1). The 11 NCX areas are referred to hereafter collectively as the region NCX. We also genotyped donor's DNA using an Illumina 2.5 million SNP chip (Supplementary Fig. 4; Supplementary Table 2). Only brains from clinically unremarkable donors with no signs of large-scale genomic abnormalities were included in the study (N=57, including 39 with both hemispheres; age, 5.7 post-conceptual weeks to 82 years; sex, 31 males and 26 females; postmortem interval [PMI], 12.11±8.63 [mean±SD] hours; pH, 6.45±0.34 [mean±SD]).
Table 1. Periods of human development and adulthood as defined in this study.
| Period | Description | Age |
|---|---|---|
| 1 | Embryonic | 4≤ Age <8 Postconceptual weeks (PCW) |
| 2 | Early fetal | 8≤ Age <10 PCW |
| 3 | Early fetal | 10≤ Age <13 PCW |
| 4 | Early midfetal | 13≤ Age <16 PCW |
| 5 | Early midfetal | 16≤ Age <19 PCW |
| 6 | Late midfetal | 19≤ Age <24 PCW |
| 7 | Late fetal | 24≤ Age <38 PCW |
| 8 | Neonatal and early infancy | Birth≤ Age <6 Postnatal months (M) |
| 9 | Late infancy | 6 M≤ Age <12 M |
| 10 | Early childhood | 1≤ Age <6 Postnatal years (Y) |
| 11 | Middle and late childhood | 6≤ Age <12 Y |
| 12 | Adolescence | 12≤ Age <20 Y |
| 13 | Young adulthood | 20≤ Age <40 Y |
| 14 | Middle adulthood | 40≤ Age <60 Y |
| 15 | Late adulthood | 60 Y ≤ Age |
Table 2. Ontology and nomenclature of analyzed brain regions and NCX areas.
| Periods 1-2 | Periods 3 - 15 |
|---|---|
| FC: Frontal cerebral wall | OFC: Orbital prefrontal cortex |
| DFC: Dorsolateral prefrontal cortex | |
| VFC: Ventrolateral prefrontal cortex | |
| MFC: Medial prefrontal cortex | |
| M1C: Primary motor (M1) cortex | |
| PC: Parietal cerebral wall | S1C: Primary somatosensory (S1) cortex |
| IPC: Posterior inferior parietal cortex | |
| TC: Temporal cerebral wall | A1C: Primary auditory (A1) cortex |
| STC: Superior temporal cortex | |
| ITC: Inferior temporal cortex | |
| OC: Occipital cerebral wall | V1C: Primary visual (V1) cortex |
| HIP: Hippocampal anlage | HIP: Hippocampus |
| AMY: Amygdala | |
| VF: Ventral forebrain | STR: Striatum |
| MGE: Medial ganglionic eminence | |
| LGE: Lateral ganglionic eminence | |
| CGE: Caudal ganglionic eminence | |
| DIE: Diencephalon | MD: Mediodorsal nucleus of the thalamus |
| DTH: Dorsal thalamus | |
| URL: Upper (rostral) rhombic lip | CBC: Cerebellar cortex |
Transcriptome profiling was performed using total RNA extracted from a total of 1,340 dissected tissue samples (RNA integrity number [RIN], 8.83±0.93 [mean±SD]; Supplementary Tables 3 and 4). We used the Affymetrix GeneChip Human Exon 1.0 ST Array platform, which features comprehensive coverage of the human genome, with 1.4 million probe sets that assay expression across the entire transcript or individual exon, thereby providing redundancy and increasing confidence in estimates of gene-level differential expression (DEX) and differential exon usage (DEU). Descriptions of tissue sampling and quality control measures implemented throughout transcriptome data generation steps are provided in Supplementary Information 2-5 and Supplementary Figures 5-8.
Global transcriptome dynamics
Spatiotemporal gene expression
After quality control metrics were assessed, we performed quantile normalization and summarized core and unique probe sets, representing 17,565 protein-coding genes, into gene-level information. Employing stringent criteria (log2-transformed signal intensity ≥6 in at least one sample and mean DABG P<0.01 in at least one region and period) to define an “expressed” gene, we found that 15,132 (86.1%) of 17,565 genes surveyed were expressed in at least one brain region during at least one period, and 14,375 (81.8%) were expressed in at least one NCX area (Fig. 1a; Supplementary Information 6.1; Supplementary Fig. 9). To investigate the contribution of different factors to the global transcriptome dynamics, we applied multi-dimensional scaling (MDS) and principal component analysis (PCA), which revealed that regions and age (i.e., spatiotemporal dynamics) contribute more to the global differences in gene expression than other tested variables: sex, ethnicity, or inter-individual variation (Fig. 1b; Supplementary Information 6.2 and 6.3; Supplementary Figs 10 and 11).
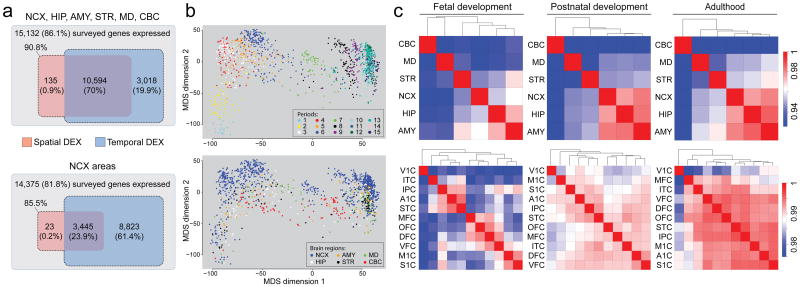
Figure 1. Global spatiotemporal dynamics of gene expression.
a, Venn diagrams representing total number of genes considered to expressed and the number of spatially and temporally DEX genes for brain regions (top) and NCX areas (bottom). b, MDS plot showing transcriptional similarity, colored by periods (top) and regions (bottom). Euclidean distance of log2-transformed signal intensity (nonmetric, stress=18.9%) was used to measure pairwise similarity. c, Heat map matrix of pairwise Spearman correlations between brain regions (top) or NCX areas (bottom) during fetal development (periods 3–7), postnatal development (periods 8–12), and adulthood (periods 13–15).
To identify genes that were spatially or temporally regulated, we employed a conservative threshold (FDR q-value<0.01 and ≥2-fold log2-transformed signal intensity difference), included PMI and RIN as technical covariates within all of our ANOVA models of differential expression, considered the influence of dissection variation, and applied a 5-fold jackknife procedure (Supplementary Information 6.4; Supplementary Figs 12 and 13). We found that 70.9% of expressed genes were spatially DEX between any two regions within at least one period, and 24.1% were spatially DEX between any two NCX areas (Fig. 1a). In contrast, 90.0% of expressed genes were temporally DEX between any two periods across regions, and 85.3% were temporally DEX between any two periods across NCX areas. Moreover, 70.0% and 23.9% of expressed genes were both spatially and temporally DEX within brain regions and within NCX areas, respectively. The bulk of spatiotemporal regulation occurred during prenatal development. For instance, 57.7% of NCX-expressed genes were temporally DEX across fetal development (periods 3-7), in contrast to 9.1% during postnatal development (periods 8-12) and a mere 0.7% during adulthood (periods 13-15). Together, these data indicate that the majority of brain-expressed protein-coding genes are temporally and, to a lesser extent, spatially regulated, and this regulation occurs predominately during development.
Transcriptional architecture of the human brain
To assess the transcriptional relatedness, we calculated correlation matrices of pairwise comparisons between brain regions/NCX areas (Fig. 1c) and performed unsupervised hierarchical clustering across periods 3–15, when all analyzed regions/areas can be consistently followed across time. Among regions, this analysis showed distinct and developmentally regulated clustering of NCX (combination of 11 areas), HIP and AMY, with CBC exhibiting the most distinctive transcriptional profile. At the level of NCX areas, clustering formed the following groups during fetal periods: OFC, DFC, and MFC of the prefrontal cortex; VFC and primary somatomotor cortex (S1C and M1C); and parietal-temporal perisylvian areas (IPC, A1C, and STC). V1C had the most distinctive transcriptional profile of NCX areas throughout development and adulthood. The increased correlations between NCX, HIP, AMY, and the majority of non-V1C NCX areas with age indicate that transcriptional differences are particularly pronounced during development.
Consistent with the clustering observed, CBC showed the greatest number of region-restricted or region-enriched DEX genes with 516 (4.8%) of 10,729 genes spatially DEX (Supplementary Information 6.4; Supplementary Table 5). In contrast, the number of genes highly enriched in the other regions was lower: NCX (46; 0.43%), HIP (48; 0.45%), AMY (4; 0.04%), STR (137; 1.28%), and MD (216; 2.01%). The majority of these spatially enriched genes were also temporally regulated, and some were transiently enriched during a narrow time-window, such as those featured in Supplementary Figures 14 and 15 (NCX: FLJ32063, KCNS1; HIP: CDC20B, METTL7B; AMY: TFAP2D, UTS2D; STR: C10ORF11, PTPN7; MD: CEACAM21, SLC24A5; CBC: ESRRB, ZP2). These clustering and region-enrichment results reveal that regional transcriptomes are developmentally regulated and likely reflect anatomical differences.
Spatiotemporal differential exon usage
Alternative exon usage is an important mechanism for generating transcript diversity20,21. Using the splicing ANOVA and splicing index algorithm with conservative criteria (FDR<0.01 with a minimum 2-fold splice index difference between at least two regions/areas or periods; Supplementary Information 6.5), we found that 13,647 of 15,132 (90.2%) expressed genes exhibited DEU across sampled regions (0.1%), periods (19.5%), or both (70.6%). Of 14,375 NCX genes, 88.7% exhibited DEU across sampled areas (<0.01%), periods (59.8%), or both (28.9%). The regulation of DEU also varied across time, with the vast majority of expressed genes (83.0%) exhibiting temporal DEU across fetal development, while only 0.9% and 1.4% were temporally regulated across postnatal development and adulthood, respectively.
Focusing on ANKRD32, a gene we have previously shown to express an alternative variant in the late midfetal frontal cortex17, we confirmed and extended DEU findings by revealing that while the longer isoform (ANKRD32a) was equally expressed across fetal NCX areas, the shorter isoform (ANKRD32b), comprising the last three exons, exhibited dynamic areal patterns. ANKRD32b was transiently expressed in a gradient along the anterior-posterior axis of the midfetal frontal cortex, with the highest expression in OFC and lowest in M1C. Prior to this, ANKRD32b was most highly enriched in the ITC and, to a lesser extent, in the STC. These spatiotemporal patterns disappeared after birth, when only ANKRD32a was expressed, and were not observed in the mouse NCX of equivalent periods (Supplementary Fig. 16; Supplementary Table 6). These findings illustrate the complexity of DEU in the human brain and demonstrate how specific alternative transcripts can be spatially restricted during a narrow developmental window and with inter-species differences.
Sex differences in the transcriptome
Sex-biased gene expression
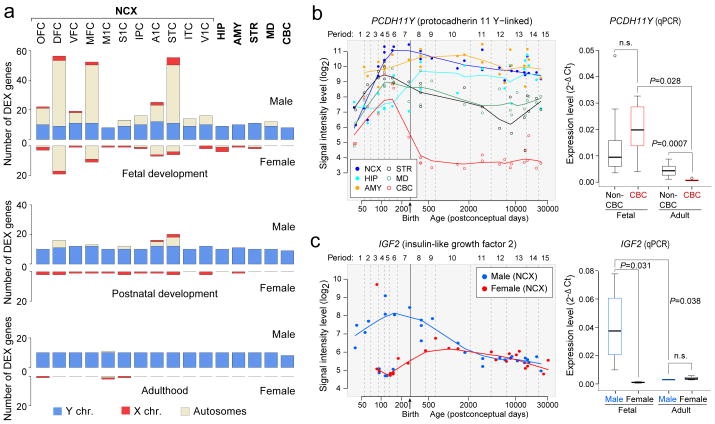
Previous studies have identified sexually dimorphic gene expression in the developing and adult human brain12-14. Analysis of our dataset using a sliding window algorithm and t-test model (FDR<0.01 with >2-fold difference in log2-transformed signal intensity; Supplementary Information 6.6), identified 159 genes, including a number of previously reported and newly uncovered genes with male- or female-bias in expression located on the Y (13), X (9), and autosomal (137) chromosomes. A large fraction (76.7%) displayed male-biased expression (Fig. 2a; Supplementary Table 7). Notable spatial differences were observed, and more genes had sex-biased expression during prenatal development than during postnatal life, with the adult brain characterized by the lowest number.
Figure 2. Sex-biased gene expression.
a, Number of sex-biased DEX genes in brain regions/NCX areas during fetal development (periods 3–7), postnatal development (periods 8–12), and adulthood (periods 13–15). b, PCDH11Y exon array signal intensity (left) and qRT-PCR validation (right) (N=5 male brains per period). c, IGF2 exon array signal intensity (left) and qRT-PCR (right) validation in NCX (N=4 per sex and period). P-values were calculated by unpaired t-test. Whiskers indicate 5th and 95th percentile.
Consistent with previous findings13,14, we found that the largest differences were attributable to Y chromosome genes, especially PCDH11Y, RPS4Y1, USP9Y, DDX3Y, NLGN4Y, UTY, EIF1AY, and ZFY, which displayed constant expression across regions and periods, with the exception of PCDH11Y down-regulation in the postnatal CBC (Fig. 2b). Interestingly, the functional homologues of these genes on the X chromosome (PCDH11X, RPS4X, USP9X, DDX3X, NLGN4X, UTX, and EIF1AX), except ZFX during fetal development, were not up-regulated in a compensatory manner in female brains (Supplementary Fig. 17).
We also found other X-linked and autosomal genes with sex-biased expression and distinct spatiotemporal patterns, including functionally uncharacterized transcripts (LOC554203, C3ORF62, FLJ35409, and DKFZP586), S100A10 (which has been linked to depression22), and IGF2 (an imprinted autosomal gene previously implicated in brain growth and cognitive function23,24) that exhibited population-level male-biased developmental expression (Fig. 2c).
Sex-biased exon usage
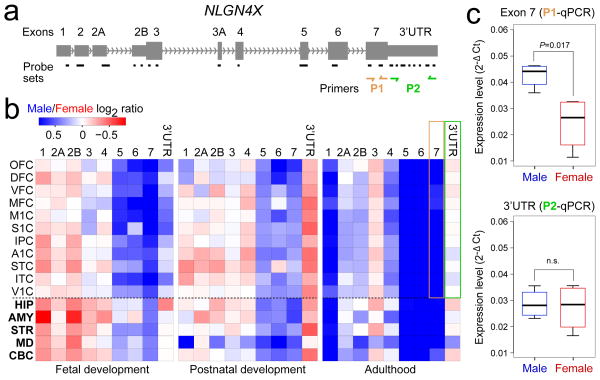
We next explored sex-biased DEU using a sliding window algorithm and splicing t-test model (FDR<0.01 and splicing index>2; Supplementary Information 6.6). We identified 155 genes (145 autosomal) that displayed sex-biased expression of probe sets encoding one or a subset of exons (Supplementary Table 8) in one or multiple regions/NCX areas. These included several members of the collagen family of genes (COL1A1, COL1A2, COL3A1, COL5A2, and COL6A3), C3, KCNH2 (a gene associated with schizophrenia25), NOTCH3 (a gene mutated in a common form of hereditary stroke disorder26), ELN (a gene located within the Williams syndrome critical region27), and NLGN4X (a X chromosome gene implicated in synapse function and associated with ASD and moderate X-linked intellectual disability11,28). Although comparably expressed in males and females at the population and gene-level (Supplementary Fig. 17), NLGN4X exhibited a significant male-bias in expression of exon 7 and, to a lesser extent, exons 1, 5 and 6 in a developmentally regulated manner (Fig. 3). Together, these findings show that developmentally and spatially regulated differences in gene- and exon-level expression exist between male and female brains.
Figure 3. Sex-biased differential exon usage.
a, Gene structure and probe set composition of NLGN4X. Depicted by yellow and green arrows are primers used for qRT-PCR validation. b, Heat map of the log2 male/female signal intensity ratio of each exon for fetal development (periods 3–7), postnatal development (periods 8–12), and adulthood (periods 13–15). Differences in expression of exon 7 (yellow frame) and 3′UTR (green frame) in adult NCX are highlighted. Note that exons 2 and 3A did not meet our expression criteria and are not represented. c, qRT-PCR validation of exon 7 and 3′UTR expression in adult NCX (N=4 per sex). P-values were calculated by unpaired t-test. Whiskers indicate 5th and 95th percentile.
Co-regulated transcriptional networks
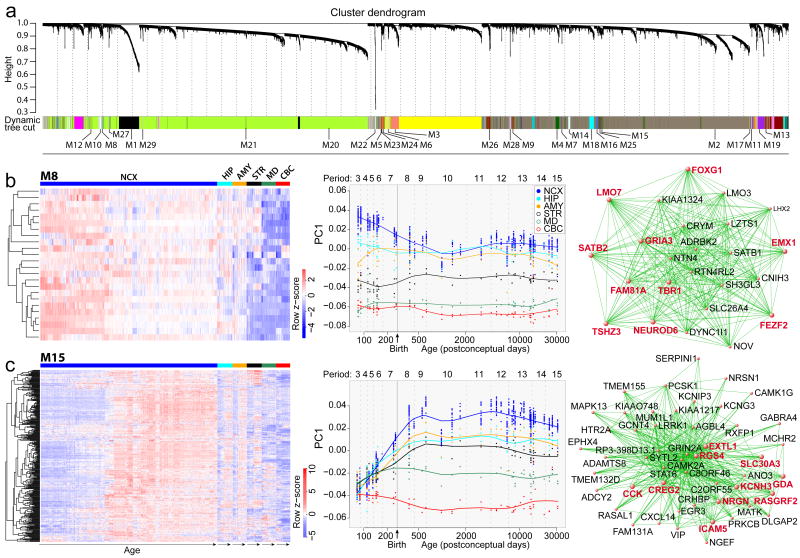
To extract additional biological information embedded in the multi-dimensional transcriptome dataset, we performed weighted gene co-expression network analysis29 to identify modules of co-regulated genes. We identified 29 modules associated with distinct spatiotemporal expression patterns and biological processes (Fig. 4a; Supplementary Information 6.7; Supplementary Table 9-11; Supplementary Figs 18-20). Among modules corresponding to specific spatiotemporal patterns, M8 consisted of 24 genes with a common developmental trend that exhibited highest levels in early fetal NCX and HIP (period 3) and then progressively declined with age until infancy (period 9) (Fig. 4b). In contrast, M15 contained 310 genes exhibiting changes in the opposite direction in the NCX, HIP, AMY, and STR (Fig. 4c). Gene ontology (GO) enrichment analysis showed that M8 genes were enriched for GO categories related to neuronal differentiation (Bonferroni-adjusted P=7.7×10-3) and transcription factors (P=5.2×10-3) (Supplementary Information 6.8; Supplementary Table 9).
Figure 4. Global co-expression networks and gene modules.
a, Dendrogram from gene co-expression network analysis of samples from period 3 to 15. Modules of co-expressed genes were assigned a color and number (M1 to M29). b, Heat map of genes in M8 (left) showing the spatiotemporal expression pattern after hierarchical clustering. The expression values for each gene are arranged in the heat map, ordered first by brain regions, then by age and last by NCX areas. Spatiotemporal pattern of M8 (middle) summarized by the first principal component (PC1) for expression of genes in the module across age. Top 50 M8 genes (right), defined by the highest intramodular connectivity, Top 10 hub genes are in red. c, Same analyses were performed for M15. Results for other modules are available in Supplemental Information.
Genes with the highest degree of connectivity within a module are termed hub genes and are expected to be functionally important within the module. M8 hub genes included transcription factors TBR1, FEZF2, FOXG1, SATB2, NEUROD6, and EMX1 (Fig. 4b), which have been functionally implicated in NCX and HIP development4,30-38. Furthermore, FOXG1 variants have been linked to Rett syndrome and intellectual disability34. Conversely, M15 GO categories included ionic channels (P=8.0×10-8) and neuroactive ligand-receptor interaction (P=4.0×10-14). Sequence variants in hub genes (Fig. 4c) have been linked to major depression (GDA)39 and to schizophrenia and affective disorders (NRGN and RGS4)7,40.
We also identified two large-scale temporally regulated modules (M20 and M2) with opposite developmental trajectories of genes co-expressed across regions, which either gradually declined (M20) or increased (M2) in expression with age (Supplementary Figs 21 and 22). M20 was enriched for GO categories related to zinc finger proteins (P=7.3×10-48) and transcription factors (P=4.8×10-50), including many ZNF and SOX family members, while M2 was enriched for categories related to membrane proteins (P=1.8×10-21) and genes involved in calcium signaling (P=8.1×10-10), synaptic transmission (P=1.6×10-6), and neuroactive ligand-receptor interaction (P=4.1×10-4), reflecting processes important in postnatal brain maturation. Their hub genes encoded transcriptional factors, modulators of chromatin state, and signal transduction proteins, all of which likely play an important role in driving the networks. Dramatic expression shifts of M20 and M2 in the opposite direction just before birth indicate that this period is associated with transcriptional changes that likely reflect environmental influences on brain development and intrinsic changes in cellular composition and functional processes.
Expression trajectories of neurodevelopment
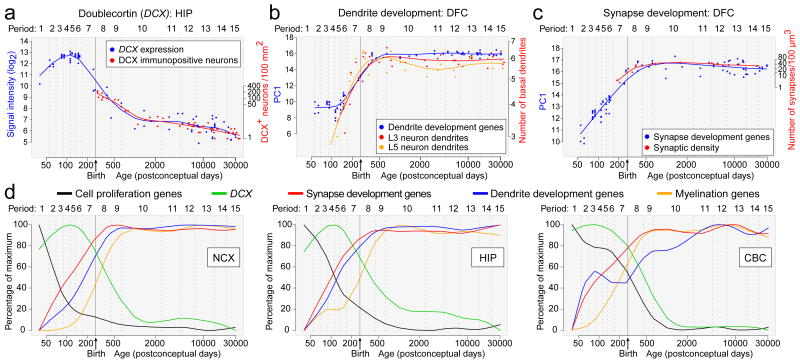
One important application for the generated dataset is to gain insight into normal and abnormal human neurodevelopment by analyzing trajectories of individual genes or groups of genes associated with a particular neurobiological category or disease. To test this strategy, we compared our expression data of DCX (a gene expressed in neuronal progenitor cells and migrating immature neurons), as well as genes associated with dendrite (MAP1A, MAPT, CAMK2A) and synapse (SYP, SYPL1, SYPL2, SYN1) development, with independently generated, non-transcriptome human datasets. The DCX expression trajectory was remarkably reminiscent of the reported changes in the density of DCX-immunopositive cells in the postnatal human hippocampus (Fig. 5a)36,40. In our transcriptome dataset, DCX expression progressively increased until early midfetal development (period 5) and then gradually declined with age until early childhood (period 10). Likewise, expression trajectories of dendrite and synapse development gene groups closely paralleled the growth of basal dendrites of DFC pyramidal neurons41 (Spearman correlation, r=0.810 for layer 3 and r=0.700 for layer 5; Fig. 5b) and DFC synaptogenesis42 (r=0.946; Fig. 5c), respectively. Steep increases in both processes occurred between late midfetal and late infancy, indicating that a considerable portion of these two processes occurs before birth and reaches a plateau around late infancy.
Figure 5. Trajectories of genes associated with neurodevelopmental processes.
a, Comparison between DCX expression in HIP and the density of DCX-immunopositive cells in the human dentate gyrus36. b, Comparison between transcriptome-based dendrite development trajectory in DFC and Golgi method-based growth of basal dendrites of layer 3 (L3) and 5 (L5) pyramidal neurons in the human DFC41. c, Comparison between transcriptome-based synapse development trajectory in DFC and density of DFC synapses calculated using electron microscopy42. For (b) and (c), PC1 for gene expression was plotted against age to represent the developmental trajectory of genes associated with dendrite or synapse development. Independent datasets were centered, scaled, and plotted on a logarithmic scale. d, PC1 value percentage of maximum PC1 value for given set of genes plotted against age to represent general trends and regional differences in several neurodevelopmental processes in NCX, HIP, and CBC.
After demonstrating the accuracy and viability of using the dataset to profile human neurodevelopment, we manually curated lists of genes associated with over 80 categories, including various neurodevelopmental processes, neural cell types, and neurotransmitter systems (Supplementary Information 6.9; Supplementary Table 12). Interesting trajectories and notable differences in their onset, rates of incline and decline, and shapes were observed within and between brain regions for in the categories, including major neurodevelopmental processes (neural cell proliferation and migration, dendrite and synapse development, and myelination; Fig. 5d), cortical GABAergic inhibitory interneurons (CALB1, CALB2, NOS1, PVALB, and VIP), and glutamate receptors (Supplementary Figs 23 and 24). As expected, two major trends were observed in neurodevelopmental trajectories: changes in expression of cell proliferation genes preceded the increase in expression of DCX, both of which decreased during perinatal development while synapse development, dendrite development, and myelination trajectories increased. Interestingly, the NCX expression trajectory for synapse development (Fig. 5c, d) did not display a drastic decline during late childhood or adolescence as previously reported for synapse density42. We also identified network modules and additional genes that are highly correlated with the categories (Supplementary Tables 13 and 14). For example, M20 and M2 were strongly correlated with neuron migration (r=0.894) and myelination (r=0.972), respectively.
In addition, our dataset allowed us to generate expression trajectories of genes commonly associated with ASD and schizophrenia. We investigated a number genes previously linked to these disorders (Supplementary Information 6.10) and observed distinct and dynamic expression patterns, especially among NCX areas, including examples for CNTNAP2, MET, NLGN4X, and NRGN shown in Supplementary Figure 25. To gain insight into potential biological functions of ASD and schizophrenia-associated genes in human neurodevelopment, we identified other genes with significantly correlated spatiotemporal expression profiles and performed GO enrichment analysis (Supplementary Tables 15 and 16). These findings reveal spatiotemporal differences associated in these expression trajectories and provide additional co-expressed genes that can be interrogated for their role in the respective processes or disorders.
Expression quantitative trait loci
Previous reports have demonstrated the connection between SNP-based genetic variation and expression quantitative trait loci (eQTLs) in the adult postmortem human brain, primarily in the cerebral cortex43-47. Our multiregional developmental dataset enabled us to search for association between SNP genotypes and spatiotemporal gene expression. We tested only for cis-eQTLs, restricting the search to SNPs within 10 kb of either transcription start site (TSS) or termination site (TES), as opposed to trans-eQTLs, which would require much larger sample sizes.
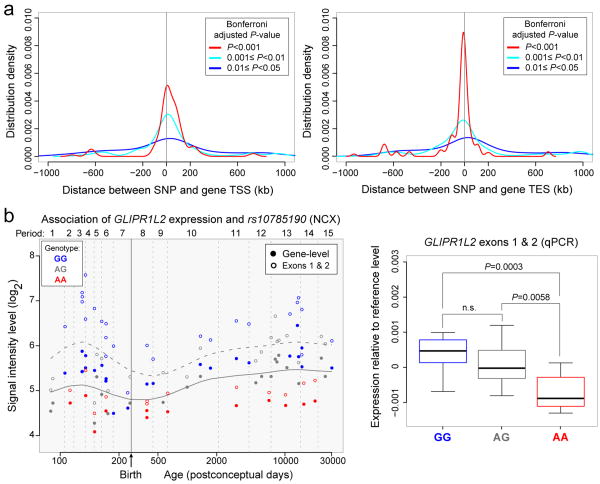
Implementing a conservative strategy (gene-wide Bonferroni correction followed by genome-wide FDR<0.1; Supplementary Information 9), we identified 39 NCX, 8 HIP, 4 AMY, 2 STR, 6 MD, and 5 CBC genes (Supplementary Table 17) with evidence of cis-eQTL, including two previously reported genes (ITGB3BP and ANKRD27)45,47. Consistent with previous studies48, associated SNPs were enriched near TSS or TES (Fig. 6a).
Figure 6. Association between SNPs and gene expression.
a, b, SNP distribution around TSS (a) and TES (b) of the associated genes based on several Wald test P-value cut offs after gene-wide Bonferroni correction. c, GLIPR1L2 expression association with rs10785190 genotype, a SNP located in exon 1. The horizontal solid and dashed LOWESS curves are the developmental trend of gene and exon 1 and 2 expression, respectively. d, qRT-PCR validation of exon 1 and 2 expression in NCX for each genotype (N=14 GG, 14 AG, 8 AA) plotted relative to the local regression smooth curve to facilitate comparison across developmental periods. P-values were calculated by unpaired t-test. Whiskers indicate 5th and 95th percentile.
An example of a significant association in NCX, MD, and CBC is rs10785190 and GLIPR1L2, a member of the glioma pathogenesis-related 1 family of genes49. The expression differences were observed at the level of the whole transcript and exons 1 and 2, the only exons we observed to be expressed at appreciable levels in the NCX (Fig. 6b). The NCX likely had more cis-eQTLs than other regions due to its smaller variation in gene expression resulting from the averaged expression of 11 areas. Many eQTLs identified as significant in NCX also have similar associations in other regions, though they were not statistically significant after the conservative genome-wide correction (Supplementary Table 17). Thus, we have identified polymorphic regulators of transcription in different regions across development, potentially providing insights into inter-individual differences and genetic control of the brain transcriptome.
Discussion
We have described the generation and analysis of the transcriptome and corresponding genotyping dataset covering multiple regions of the developing and adult brain. This dataset (available at www.humanbraintranscriptome.org), along with an accompanying study50, provides opportunities for a variety of further investigations and comparisons with other transcriptome-related datasets of both healthy and diseased states.
Our analysis revealed several important aspects of the human brain transcriptome and expands current knowledge of the transcriptional events in human neurodevelopment. Consistent with many previous studies in other species, we found that gene expression and exon usage have complex, dynamically regulated patterns across time and space and are far more prominent than differences between sexes, ethnicities, or individuals despite their underlying genomic differences.
We confirmed and expanded on previous findings of sexual dimorphism in gene expression and exon usage, including several disease-related genes. These findings offer possible transcriptional mechanisms underlying sex differences in the incidence, prevalence, and severity of many disorders. We also demonstrated how the dataset can be used to profile trajectories of genes associated with specific neurobiological categories or disorders, many of which would not likely be evident in the transcriptomes of commonly studied model systems. Coupled with analysis of co-expressed genes in the dataset, these provide information about when and where these genes are expressed in the brain, which can help infer their function. Our data can enhance genome-wide association and linkage studies by narrowing the focus to the candidate genes that are specifically expressed during development or restricted to a specific region known to be preferentially affected.
We show associations between specific SNPs and expression levels in different regions of the developing human brain, indicating that genetic variation contributes to inter-individual transcriptome variability across regions and development. While the current number of specimens in our dataset restricted our power to detect many of the possible eQTLs, we have identified a set of cis-eQTLs, including many not previously reported, that may provide insights into expression-regulatory elements operating in the brain.
Although these findings highlight the complexity of gene expression and exon usage in the human brain, there are several potential limitations in our data that warrant discussion. Foremost, we used stringent criteria in order to faithfully characterize general transcriptional patterns and minimize false positives, rather than to capture all the changes that may occur. Also, we analyzed dissected tissue samples that contain multiple cell types, thus diluting the genetic contribution and dynamic range of genes expressed in a more cell type specific manner. Current limitations prevent us from using cell type specific approaches in systematically analyzing the spatiotemporal transcriptome. Furthermore, the number of brains and regions analyzed so far is not sufficient to fully investigate the whole magnitude of transcriptional changes or the full range of eQTLs. Application of sequencing technology will allow even more in-depth analysis of the transcriptome and discovery of novel and low-expressing transcripts and their associated spatiotemporal patterns. Finally, while specific patterns of expression are often linked to equally specialized biological processes, it is important to keep in mind that the relationship between mRNA and protein levels is not always linear or translated into apparent phenotypic differences. As these concerns are addressed in the future with the addition of samples to our dataset and the generation of new datasets from human and nonhuman brains throughout development, it will be possible to uncover further insights into the transcriptional foundations of human brain development and evolution.
Methods Summary
The Supplementary Information 3-9 provides a full description of tissue acquisition and processing, data generation, validation, and analyses.
Supplementary Material
Acknowledgments
We thank A. Belanger, V. Imamovic, R. Johnson, P. Larton, S. Lindsay, B. Poulos, J. Rajan, D. Rimm, and R. Zielke for assistance with tissue acquisition, D. Singh for technical assistance, I. Kostovic and Z. Petanjek for Golgi measurements, P. Levitt for suggesting the ITC, and D. Karolchik and A. Zweig for help in creating tracks for the UCSC Genome browser. We also thank A. Beckel-Mitchener, M. Freund, M. Gerstein, D. Geschwind, T. Insel, M. Judas, J. Knowles, E. Lein, P. Levitt, N. Parikshak and members of the Sestan laboratory for discussion and criticism. Tissue was obtained from several sources including the Human Fetal Tissue Repository at the Albert Einstein College of Medicine, the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, the Laboratory of Developmental Biology at the University of Washington (supported by HD000836 grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development), and the Joint MRC/Wellcome Trust Human Developmental Biology Resource (http://hdbr.org) at the IHG, Newcastle Upon Tyne (UK funding awards G0700089 and GR082557). Support for pre-doctoral fellowships was provided by the China Scholarship Council (Y.Z.), the Portuguese Foundation for Science and Technology (A.M.M.S.), the Samsung Scholarship Foundation (Y.S.), Fellowship of the German Academic Exchange Service – DAAD (S.M.), and the NIDA grant DA026119 (T.G.). This work was supported by grants from the US National Institutes of Health (MH081896, MH089929, NS051869), Kavli Foundation, NARSAD, and the James S. McDonnell Foundation Scholar Award (N.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Accession code. National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO accession number GSE25219) and database of Genotypes and Phenotypes (dbGAP; accession number pending).
Author Contributions: H.J.K., Y.I.K., A.M.M.S., M.P., K.A.M., G.S., Y.S., M.B.J., Z.K., S.M., S.F., S.U. and N.S. performed and analyzed the experiments. F.C., Y.Z., X.X., M.L., T.G., and M.R. analyzed the data. D.R.W., S.M., T.H.H., M.R., J.E.K. and N.S. examined and interpreted the results. Z.K., A.M.M.S., M.P., G.S., S.N.L, S.L, A.V., T.H.H., A.H., J.E.K., and N.S. participated in tissue procurement and examination. N.S. conceived and designed the study and wrote the manuscript, which all authors commented and edited.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Kostovic I, Judas M. Prolonged coexistence of transient and permanent circuitry elements in the developing cerebral cortex of fetuses and preterm infants. Dev Med Child Neurol. 2006;48:388–393. doi: 10.1017/S0012162206000831. [DOI] [PubMed] [Google Scholar]
- 2.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubenstein JL. Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry. 2010 doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill RS, Walsh CA. Molecular insights into human brain evolution. Nature. 2005;437:64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- 5.Preuss T, Cáceres M, Oldham M, Geschwind D. Human brain evolution: insights from microarrays. Nat Rev Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- 6.Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 7.Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 8.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 9.Insel T. Rethinking schizophrenia. Nature. 2010;468:187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- 10.State M. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vawter M, et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weickert C, et al. Transcriptome analysis of male-female differences in prefrontal cortical development. Mol Psychiatry. 2009;14:558–561. doi: 10.1038/mp.2009.5. [DOI] [PubMed] [Google Scholar]
- 14.Reinius B, Jazin E. Prenatal sex differences in the human brain. Mol Psychiatry. 2009;14:987, 988–989. doi: 10.1038/mp.2009.79. [DOI] [PubMed] [Google Scholar]
- 15.Abrahams B, et al. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc Natl Acad Sci U S A. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun T, Walsh C. Molecular approaches to brain asymmetry and handedness. Nat Rev Neurosci. 2006;7:655–662. doi: 10.1038/nrn1930. [DOI] [PubMed] [Google Scholar]
- 17.Johnson M, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somel M, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ip B, et al. Investigating gradients of gene expression involved in early human cortical development. J Anat. 2010;217:300–311. doi: 10.1111/j.1469-7580.2010.01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Svenningsson P, et al. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- 23.Chen DY, et al. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehtinen MK, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huffaker SJ, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med. 2009;15:509–518. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joutel A, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 27.Ewart AK, et al. Hemizygosity At The elastin locus in a developmental disorder, Williams-syndrome. Nature Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 28.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 30.Hevner R, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 31.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 32.Chen B, Schaevitz L, McConnell S. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Rasin M, Kwan K, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariani F, et al. FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet. 2008;83:89–93. doi: 10.1016/j.ajhg.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwan K, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoth R, et al. Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PLoS One. 2010;5:e8809. doi: 10.1371/journal.pone.0008809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han W, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna WL, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perroud N, et al. Genome-wide association study of increasing suicidal ideation during antidepressant treatment in the GENDEP project. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.70. doi:tpj201070 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Stefansson H, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petanjek Z, Judas M, Kostović I, Uylings H. Lifespan alterations of basal dendritic trees of pyramidal neurons in the human prefrontal cortex: a layer-specific pattern. Cereb Cortex. 2008;18:915–929. doi: 10.1093/cercor/bhm124. [DOI] [PubMed] [Google Scholar]
- 42.Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 43.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinzen EL, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biology. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C, et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Molecular Psychiatry. 2010;15:779–784. doi: 10.1038/mp.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibbs JR, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 48.Webster JA, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren C, Ren CH, Li L, Goltsov AA, Thompson TC. Identification and characterization of RTVP1/GLIPR1-like genes, a novel p53 target gene cluster. Genomics. 2006;88:163–172. doi: 10.1016/j.ygeno.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 50.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex across the lifespan. Nature. 2011 doi: 10.1038/nature10524. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.