Abstract
Background and Purpose
The piwi-interacting RNA (piRNA) is the most predominant RNA species in eukaryotes. The piRNA are a class of non-coding (nc) RNA that bind and degrade the RNA formed by the transposons to control the transposon -induced gene mutations. The role of piRNA after focal ischemia is not yet evaluated.
Methods
We profiled 39,727 piRNAs in the cerebral cortex of adult rats subjected to transient focal ischemia using microarrays. The RT targets of stroke-responsive piRNAs were identified with bioinformatics. To understand how piRNA are controlled, we analyzed the transcription factor (TF) binding sites in the putative promoters of 10 representative stroke-responsive piRNAs.
Results
In the ipsilateral cortex of ischemic rats, 105 piRNAs showed altered expression (54 up- and 51 down-regulated; >2.5 fold) compared to sham. Twenty five of those showed >5-fold change. A bioinformatics search showed that the transposon targets of the highly stroke-responsive piRNAs are distributed among the 20 autosomal chromosomes and there is a redundancy in the targets between the piRNAs. Furthermore, the transposon targets were observed to be highly repetitious for each piRNA across the chromosome length. Of the 159 TFs observed to have binding sites in the piRNA gene promoters, 59% belonged to 20 major families indicating that TFs control stroke-responsive piRNAs in a redundant manner.
Conclusions
The present study is the first to show that many piRNAs are expressed in adult rodent brain and several of them respond to focal ischemia.
Keywords: Non-coding RNA, Stroke, Transposons, Brain damage, Bioinformatics, Expression profiling
In eukaryotes, ∼40% of the genome is comprised of transposons which are transcribed into RNA, reverse transcribed into double-stranded DNA and inserted into new locations in the genome.1,2 As transposition mutates the protein-coding genes, a class of small non-coding (nc) RNA called PIWI-interacting RNA (piRNA; 26 to 31 nt long) selectively target and silence the RNAs formed by transposons.3 Thus, piRNA balances the fitness of the genome to maintain the genetic equilibrium. Interestingly, thousands of piRNA are known to be produced from disrupted transposons in genome regions biased towards heterochromatin.4,5
Very few studies to date evaluated the significance of ncRNA in ischemic brain damage. We and others showed that miRNA expression profiles alter extensively following focal ischemia and modulating specific miRNAs induces neuroprotection.6-10 While these studies indicate the role of ncRNA in ischemic pathophysiology, the significance of other ncRNA like piRNA is not evaluated yet. To fill this void, we profiled the expression of 39,727 piRNAs in the brains of adult rats subjected to transient middle cerebral artery occlusion (MCAO). Using bioinformatics we identified the transposon targets of representative stroke-responsive piRNAs. While piRNA control transposons, the mechanisms that control piRNA are not precisely known. A plethora of transcription factors (TFs) controls the transcription of protein-coding as well as nc genes, and many TFs are known to modulate ischemic brain damage.11-15 Hence, we analyzed the putative promoters of representative stroke-responsive piRNA genes to identify TF binding sites.
Methods
Focal ischemia
Adult, male, spontaneously hypertensive rats (SHR; 280-320g; Charles River, Wilmington, MA) used in these studies were cared for in accordance with the Guide for the Care and Use of Laboratory Animals, U.S. Department of Health and Human Services Publication number 86-23 (revised 1986). Transient MCAO was induced under isoflurane anesthesia by the intraluminal suture method as described earlier.6, 13
PiRNA microarray analysis
From each rat, the brain was sliced in a rat brain matrix to generate 1-mm sections. One section from the coordinates between +1 mm to -1 mm was quickly stained with TTC to confirm infarction. From the adjacent sections the ischemic core region was dissected from the ipsilateral cortex. Cerebral cortex from sham-operated rats served as control. Total RNA was extracted from 100 mg of each sample with RNeasy kit (Qiagen, Valencia, CA), treated with DNase, and the RNA quality and integrity were confirmed. RNA was labeled with Cy-3 and hybridized to Rat RN34 piRNA Expression Oligo microarrays (ArrayStar, Rockville, MD) that contained probes for 39,727 piRNAs selected from the NCBI database and mapped to the RN34 genome sequence using UCSC BLAST. After hybridization, the arrays were scanned with an Agilent microarray scanner. The array quality was maintained by confirming that the spot centroids were located properly at 4 corners of the array, by checking the spatial distribution of the population and nonuniformity outliers distributed across the array, by running net signal statistics to confirm the dynamic range of the signal for non-control probes, by generating histogram of signals plots to confirm the level and the shape of the signal distribution, with negative control stats (the average and SD of the net signals; mean signal minus scanner offset and the background-subtracted signals), correcting for local background inliers, and checking reproducibility statistics (%CV replicated probes). A transcript was considered detectable only if the signal intensity was higher than 3 times the maximal background signal and the spot CV (SD/signal intensity) was <0.5. The expression data files obtained by the Agilent Feature Extraction Software were imported into the GeneSpring GX software, data sets from different arrays were quantile normalized and the differentially expressed piRNAs were identified by fold-change screening with a threshold of ≥2.5 fold. Statistically significant differences between the groups were identified by the statistical measures in built in the GeneSpring based on t-test p-value method with a high stringency (fold change cutoff of >2.5 and a p-value of <0.001 to decrease the false positives.
Bioinformatics
We analyzed the targets of the top 4 stroke-responsive piRNAs (2 upregulated and 2 downregulated) using a modified version of miRanda in piRNABank (Institute of Bioinformatics and Applied Biotechnology, Bangalore, India). Each piRNA was searched for repeat sequence targets against the transposon database of the 20 autosomal rat chromosomes (RGSC 3.4) across a representative one million base stretch (position 1 to 1 million) with a mean free energy maximum of -20.0 kcal/mol and a very stringent score threshold of 140. In addition, to obtain a chromosome-wide target distribution and to search for target redundancy, each piRNA was scanned against the entire length of a randomly selected autosomal chromosome in multiple 1 million base stretches with an interval of 20 million bases. The same threshold parameters as above were applied. We also analyzed the promoters of 10 representative stroke-responsive piRNA genes for conserved TF sites. To locate the promoters, the genomic locus for each piRNA was entered into the UCSC genome browser and scanned for the nearest promoter upstream to that particular locus (within 10kb upstream to TSS). For each piRNA, a 1 Kb sequence was analyzed for overrepresented TF binding sites using the Genomatix RegionMiner software (Genomatix Software GmbH). All TF matrices with a Z-Score of ≥2 (representing p<0.05) were considered statistically significant.
Results
Stroke-induced changes in piRNAome
In the cerebral cortex of normal rats, 9.7% (3,885 ± 296) of the 40,000 piRNAs analyzed obtained a statistically significant present call (Supplementary Fig. 1). Following 1h transient MCAO and 24h reperfusion, 105 piRNAs showed altered expression (54 up- and 51 down-regulated) by >2.5 fold compared to sham (n = 3/group). Of the 105 piRNAs altered after focal ischemia, 25 showed >5 fold change (9 up- and 16 down-regulated) and 80 showed 2.5 to 4.9 fold change (45 up- and 35 down-regulated) (Supplementary Tables 1 and 2). The top 24 stroke-responsive piRNAs are given in Table 1. For a single piRNA, piR-177729 showed the maximal increase of 36.01 fold and piR-169523 showed the maximal decrease of 13.41 fold after focal ischemia compared to sham (Table 1).
Table 1. The top 24 ischemia-responsive piRNAs.
| Upregulated | Downregulted | ||||
|---|---|---|---|---|---|
|
| |||||
| piRNA | NCBI # | Δ fold | piRNA | NCBI # | Δ fold |
| piR-177729 | DQ762407 | 36.01 | piR-169523 | DQ754201 | -13.41 |
| piR-143106 | DQ727784 | 31.26 | piR-70903 | DQ627359 | -11.07 |
| piR-173369 | DQ758047 | 12.90 | piR-88428 | DQ621316 | -9.20 |
| piR-64423 | DQ614312 | 8.78 | piR-64621 | DQ614509 | -9.18 |
| piR-176691 | DQ761369 | 6.83 | piR-182498 | DQ767176 | -8.78 |
| piR-64425 | DQ614313 | 6.23 | piR-177543 | DQ762221 | -8.67 |
| piR-71756 | DQ628212 | 5.14 | piR-170937 | DQ755615 | -8.49 |
| piR-87058 | DQ619946 | 5.06 | piR-148170 | DQ732848 | -8.24 |
| piR-62321 | DQ602209 | 4.98 | piR-167731 | DQ752409 | -8.18 |
| piR-62320 | DQ602208 | 4.43 | piR-176643 | DQ761321 | -7.76 |
| piR-88241 | DQ621129 | 4.43 | piR-82548 | DQ615436 | -6.93 |
| piR-62318 | DQ602206 | 4.29 | piR-168069 | DQ752747 | -6.19 |
The Δ fold is mean (with <20% SD in each case) in comparison to the sham group (n = 3 in each case). The complete list of piRNAs up- and down-regulated by >2.5 fold in the ischemic brain were given in Supplementary Tables 1 and 2.
Targets of stroke-responsive piRNAs
The major function of the piRNAs is to silence the RNAs formed by transposons to negate their mutagenic effects. The transposons are highly repetitious in nature and several piRNAs can target a single transposon as well as a piRNA can target multiple transposons. We conducted a bioinformatics prediction of the targets of 2 piRNAs upregulated (piR-177729 and piR-143106) and 2 piRNAs down-regulated (piR-169523 and piR-70903) in the ischemic brain. For each piRNA, targets were searched in twenty 1 million base stretches (1 to 1 million bases of each of the 20 autosomal chromosomes). The number of targets of the 4 piRNAs ranged from 6 to 31 with an average of 11.08 per million bases (Table 2). Surprisingly all the transposon targets of these 4 piRNAs identified in the present study exclusively belonged to the class retrotransposons (RTs). There is a high-level of redundancy in the RTs targeted by the 4 piRNAs. The 4 piRNAs targeted 51 classes of transposons spread among the 20 chromosomes, of which 26 classes were conserved among at least 3 of the 4 piRNAs analyzed (Table 3). Furthermore, 11 of those RTs were repeated from 4 to 15 times in each of the chromosomes for each piRNA (Table 3). The redundancy of target RTs was also confirmed by the observation that out of the 80 individual RTs targeted by piR-177729, 61 are repeats of 12 classes (76%) (Table 3). A similar 67% to 70% redundancy was also observed for piR-143106 (122 out of 181 are repeats of 12 RT classes), piR-169523 (151 out of 227 are repeats of 14 classes) and piR-70903 (160 out of 227 are repeats of 15 classes) (Table 3). When RT targets were searched in the entire length of representative chromosomes (in 1 million base streches at an interval of 20 million bases in each case) for the 4 piRNAs tested, we observed an average of 12.2 RTs/million bases (Table 4). Consistent with the RT redundancy observed across chromosomes, RT targets are also highly repetitious within in each chromosome for all the 4 piRNAs (data not shown). For piR-169523, 74 out of 116 RTs (64%) are 3 to 5 repeats of 17 RT classes (Table 5).
Table 2. The number of target RTs of 4 stroke-responsive piRNAs distributed in bases 1 to 1 million of the 20 autosomoal chromosomes.
| Chromosome | piR-177729 | piR-143106 | piR-169523 | piR-70903 |
|---|---|---|---|---|
|
| ||||
| # of targets | ||||
| 1 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 13 | 0 |
| 3 | 6 | 14 | 14 | 20 |
| 4 | 13 | 0 | 14 | 21 |
| 5 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 |
| 7 | 9 | 17 | 23 | 20 |
| 8 | 0 | 9 | 0 | 20 |
| 9 | 0 | 24 | 24 | 20 |
| 10 | 15 | 11 | 13 | 14 |
| 11 | 0 | 18 | 25 | 22 |
| 12 | 12 | 18 | 18 | 19 |
| 13 | 0 | 12 | 0 | 0 |
| 14 | 0 | 0 | 0 | 20 |
| 15 | 7 | 28 | 31 | 15 |
| 16 | 17 | 9 | 21 | 13 |
| 17 | 14 | 14 | 22 | 16 |
| 18 | 0 | 17 | 21 | 17 |
| 19 | 16 | 13 | 19 | 20 |
| 20 | 0 | 18 | 20 | 20 |
Table 3. RT targets of 4 piRNAs repeated in the 20 autosomal chromosomes.
| RT | piR-177729 | piR-143106 | piR-169523 | piR-70903 |
|---|---|---|---|---|
| L1_Rn | 8 | 14 | 14 | 15 |
| L1_Rat1 | 7 | 13 | 14 | 15 |
| Lx | 4 | 12 | 14 | 13 |
| L1_Rn2 | 3 | 12 | 12 | 12 |
| L1_Rat4 | 6 | 12 | 9 | 9 |
| L1_Rat2 | 1 | 11 | 13 | 11 |
| Lx8 | 6 | 10 | 8 | 9 |
| L1_Rat3 | 4 | 9 | 9 | 8 |
| Lx9 | 2 | 8 | 7 | 10 |
| Lx6 | 4 | 7 | 11 | 8 |
| Lx2 | 1 | 7 | 9 | 9 |
| L1_Mur1 | 1 | 7 | 6 | 5 |
| Lx5 | 5 | 6 | 10 | 10 |
| Lx7 | 4 | 6 | 7 | 8 |
| Lx3B | 1 | 6 | 5 | 7 |
| Lx2B | 5 | 5 | 9 | 8 |
| L1_Mur2 | 3 | 5 | 8 | 4 |
| L1VL4a | 4 | 5 | 7 | 9 |
| Lx3A | 1 | 4 | 7 | 10 |
| Lx4B | 2 | 4 | 7 | 8 |
| Lx4A | -- | 4 | 6 | 4 |
| RNHAL1 | 1 | 3 | 11 | 9 |
| L2 | 4 | 3 | 7 | 8 |
| L1VL4 | -- | 3 | 5 | 11 |
| L1M2 | 1 | 3 | 5 | 3 |
| Lx3C | 2 | 2 | 7 | 4 |
RT, Retrotransposon. All targets presented belong to the RT class long interspersed repetitive elements (LINEs). The numbers indicate the cumulative total repeats of a RT present in 1 to 1 million bases of all 20 chromosome. Targets conserved among at least 3 out of the 4 piRNAs are presented.
Table 4. The number of target RTs of 4 stroke-responsive piRNAs distributed across the length of representative chromosomes.
| piR-177729 | piR-143106 | piR-169523 | piR-70903 | ||
|---|---|---|---|---|---|
|
| |||||
| Position in CHR | # of targets/million bases | ||||
|
| |||||
| Start base | End base | CHR 10 | CHR 3 | CHR 7 | CHR 11 |
| 1 | 1,000,000 | 15 | 14 | 23 | 24 |
| 21,000,000 | 22,000,000 | 8 | 0 | 21 | 19 |
| 42,000,000 | 43,000,000 | 13 | 18 | 0 | 21 |
| 63,000,000 | 64,000,000 | 6 | 0 | 20 | 21 |
| 84,000,000 | 85,000,000 | 7 | 0 | 0 | 17 |
| 105,000,000 | 106,000,000 | 6 | 5 | 24 | -- |
| 126,000,000 | 127,000,000 | -- | 0 | 28 | -- |
| 147,000,000 | 148,000,000 | -- | 12 | -- | -- |
| 168,000,000 | 169,000,000 | -- | 7 | -- | -- |
|
| |||||
| Average | 9.16 | 6.22 | 16.57 | 20.4 | |
The chromosome lengths (in million bases) obtained from the NCBI database are 106, 169, 127 and 87 for CHR 10, CHR 3, CHR 7 and CHR 11, respectively.
Table 5. RT targets of piR-169523 repeated in the length of CHR 7.
| RT | 1 to 1 Mil | 21 Mil to 22 Mil | 62 Mil to 63 Mil | 105 Mil to 106 Mil | 126 Mil to 127 Mil | # of repeats |
|---|---|---|---|---|---|---|
| LX | P | P | P | P | P | 5 |
| LX5 | P | P | P | P | P | 5 |
| LX6 | P | P | P | P | P | 5 |
| LX8 | P | P | P | P | P | 5 |
| LX9 | P | P | P | P | P | 5 |
| L1_Rat1 | P | P | P | P | P | 5 |
| L1_Rn | P | P | P | P | P | 5 |
| L1_Rn2 | P | P | P | P | P | 5 |
| L1_Mur1 | P | P | P | P | 4 | |
| L1_Rat2 | P | P | P | P | 4 | |
| L1_Rat4 | P | P | P | P | 4 | |
| L2 | P | P | P | P | 4 | |
| LX4B | P | P | P | P | 4 | |
| LX2B | P | P | P | P | 4 | |
| LX3B | P | P | P | P | 4 | |
| LX4A | P | P | P | 3 | ||
| L1_Mur2 | P | P | P | 3 |
The length of Chromosome 7 is 127 million bases. Mil, million, P, Present. No targets were observed in stretches 42 to 43 million and 84 to 85 million.
TF binding sites in the piRNA gene promoters
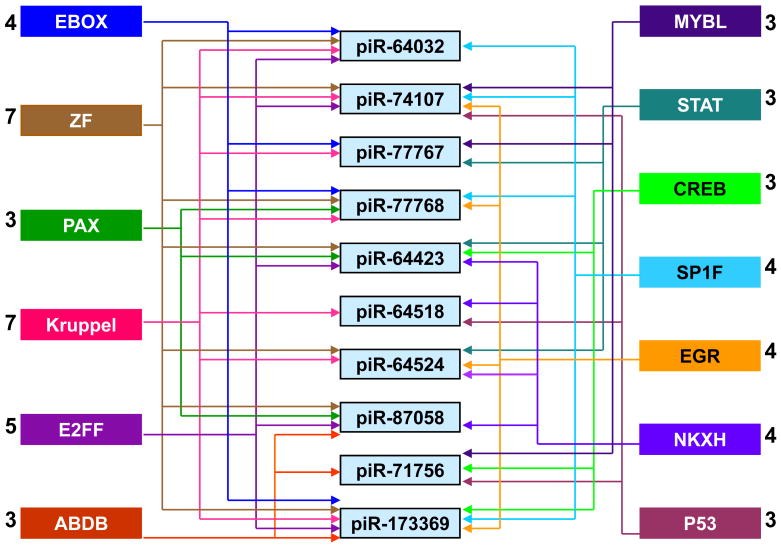
In the promoters of 10 stroke-responsive piRNA genes, we observed binding sites for 159 TFs (Supplementary Table 3) and 93 (59%) of them belonged to 20 families of TFs (Supplementary Table 4). Of these 13 TF families targeted the 10 piRNA genes in a redundant manner (Fig. 1). The zinc finger family and the Kruppel family targeted 7 of the 10 piRNA gene promoters and the E2FF family targeted 5 of the 10 piRNAs (Fig. 1). The promoter of piR-173369 gene showed the maximum of 46 TF binding sites (Supplementary Table 5).
Fig. 1.
There is a redundancy in the TF families that have binding sites in piRNA gene promoters. The number next to each TF family indicates the number of piRNA gene promoters it targets.
Discussion
In brief, the present study shows that focal ischemia significantly influences piRNA expression in rat brain. The down-stream transposon targets of the stroke-responsive piRNAs are highly repetitious and distributed throughout the genome. Furthermore, the promoters of the stroke-responsive piRNA genes contain several TF binding sites and there is a redundancy in the TF families that target these piRNAs.
Protein-coding genes represent <2% of the eukaryotic genome as ∼98% of the transcriptional output is nc RNA that include miRNA, piRNA, small interfering RNA and long non-coding RNA.16 As ncRNAs are the master controllers of the transcription and translation that decides the organ-specific and cell-specific protein repertoire any disruption in their function could lead to severe compromises in cellular homeostasis.17,18 Recent studies demonstrated that stroke profoundly alters miRNAome and modulating specific miRNAs induce neuroprotection.11-15 The present study shows that stroke also influences other ncRNAs like piRNA.
Bioinformatics analysis showed that the targets of the stroke-responsive piRNAs are highly repetitious. This redundancy of function is similar to that is known to exist for miRNAs and their target mRNAs. Bioinformatics analysis further showed that the TF families that might control the stroke-responsive piRNAs are also redundant. For example zinc finger family of TFs was observed to have binding sites in the gene promoters of 7 out of 10 piRNAs evaluated.
The role played by piRNA in maintaining the normal cellular homeostasis is enormous. It is estimated that eukaryotic genome contains >2.5 million interspersed repeat elements and most of them are highly active transposable RTs. As piRNAs silence the transposable elements, any disruption in the piRNAome can be disastrous to the cell. The miRNAs are the most studied ncRNAs and recent studies showed that cerebral miRNAs respond quickly to focal ischemia and the effect of ischemia is more extensive on miRNAome than observed previously for mRNAome (>40% miRNAs versus <5% mRNAs alter after focal ischemia).6,7,11,19 Furthermore, modulating individual miRNAs was reported to have profound effects on post-ischemic brain damage.9,10,15 In rodents, the number of piRNAs identified so far exceeds the number of known miRNAs by several fold and thus the network of permutations and combinations of piRNAs and their targets could be much higher than the miRNAs and their targets. As per bioinformatics predictions, a piRNA can target multiple transposons that contain the consensus binding sites throughout the genome and a transposon can contain multiple binding sites for several piRNAs. As the transposon-induced mutagenesis is a major disabler of protein coding genes, this redundancy might be the nature's adaptation for a proper physiological balance at the genomic level. Our present studies on piRNAs together with the previous studies on miRNAs indicate that ncRNAs need to be factored-in to understand the molecular mechanisms of ischemic brain damage.
To our knowledge this is the first study on cerebral piRNAome in experimental stroke. The pathophysiological significance of piRNAs in stroke-induced brain damage is not known presently. While the present studies indicate that stroke alters piRNAome, we do not have any evidence to show that these changes mediate ischemic brain damage. However, we predict that stroke might also influence transposons and altered piRNAome is a response to counter the increased transposon activity to control mutagenesis in the ischemic brain. On the other hand, altered piRNA levels might lead to a disruption of normal transposon network leading to pathophysiologic changes after stroke. Future functional studies with knock-in/knockout technologies might clarify these issues as well as might show if altered piRNAome and/or altered transposon activity contribute to ischemic brain damage. Our studies are first to indicate that stroke alters piRNAome and designing drugs to manipulate piRNA and perhaps other ncRNA might be essential to counter the stroke-induced mortality and morbidity.
Supplementary Material
Acknowledgments
Sources of funding: These studies were partially funded by NIH grant NS061071 and American Heart Grant MSN124341.
Footnotes
Disclosures: None
References
- 1.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gogvadze E, Buzdin A. Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci. 2009;66:3727–42. doi: 10.1007/s00018-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halic M, Moazed D. Transposon silencing by piRNAs. Cell. 2009;138:1058–60. doi: 10.1016/j.cell.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–87. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–66. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 8.Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, Jiang X, Wang Y, Chen YE. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010;30:6398–408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, Chen YE. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapadia R, Tureyen K, Bowen KK, Kalluri H, Johnson PF, Vemuganti R. Decreased brain damage and curtailed inflammation in transcription factor CCAAT/enhancer binding protein beta knockout mice following transient focal cerebral ischemia. J Neurochem. 2006;98:1718–31. doi: 10.1111/j.1471-4159.2006.04056.x. [DOI] [PubMed] [Google Scholar]
- 12.Tureyen K, Brooks N, Bowen K, Svaren J, Vemuganti R. Transcription factor early growth response-1 induction mediates inflammatory gene expression and brain damage following transient focal ischemia. J Neurochem. 2008;105:1313–24. doi: 10.1111/j.1471-4159.2008.05233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tureyen K, Kapadia R, Bowen KK, Satriotomo I, Liang J, Feinstein DL, Vemuganti R. Peroxisome proliferator-activated receptor-gamma agonists induce neuroprotection following transient focal ischemia in normotensive, normoglycemic as well as hypertensive and type-2 diabetic rodents. J Neurochem. 2007;101:41–56. doi: 10.1111/j.1471-4159.2006.04376.x. [DOI] [PubMed] [Google Scholar]
- 14.Nichols TC. NF-kappaB and reperfusion injury. Drug News Perspect. 2004;17:99–104. doi: 10.1358/dnp.2004.17.2.829042. [DOI] [PubMed] [Google Scholar]
- 15.Alexander M, Forster C, Sugimoto K, Clark HB, Vogel S, Ross ME, Ladecola C. Interferon regulatory factor-1 immunoreactivity in neurons and inflammatory cells following ischemic stroke in rodents and humans. Acta Neuropathol. 2003;105:420–424. doi: 10.1007/s00401-002-0658-x. [DOI] [PubMed] [Google Scholar]
- 16.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–38. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- 18.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 19.Lu XC, Williams AJ, Yao C, Berti R, Hartings JA, Whipple R, Vahey MT, Polavarapu RG, Woller KL, Tortella FC, Dave JR. Microarray analysis of acute and delayed gene expression profile in rats after focal ischemic brain injury and reperfusion. J Neurosci Res. 2004;77:843–57. doi: 10.1002/jnr.20218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.