Abstract
Background:
To establish the contribution of eight founder alleles in three DNA damage repair genes (BRCA1, CHEK2 and NBS1) to prostate cancer in Poland, and to measure the impact of these variants on survival among patients.
Methods:
Three thousand seven hundred fifty men with prostate cancer and 3956 cancer-free controls were genotyped for three founder alleles in BRCA1 (5382insC, 4153delA, C61G), four alleles in CHEK2 (1100delC, IVS2+1G>A, del5395, I157T), and one allele in NBS1 (657del5).
Results:
The NBS1 mutation was detected in 53 of 3750 unselected cases compared with 23 of 3956 (0.6%) controls (odds ratio (OR)=2.5; P=0.0003). A CHEK2 mutation was seen in 383 (10.2%) unselected cases and in 228 (5.8%) controls (OR=1.9; P<0.0001). Mutation of BRCA1 (three mutations combined) was not associated with the risk of prostate cancer (OR=0.9; P=0.8). In a subgroup analysis, the 4153delA mutation was associated with early-onset (age ⩽60 years) prostate cancer (OR=20.3, P=0.004). The mean follow-up was 54 months. Mortality was significantly worse for carriers of a NBS1 mutation than for non-carriers (HR=1.85; P=0.008). The 5-year survival for men with an NBS1 mutation was 49%, compared with 72% for mutation-negative cases.
Conclusion:
A mutation in NBS1 predisposes to aggressive prostate cancer. These data are relevant to the prospect of adapting personalised medicine to prostate cancer prevention and treatment.
Keywords: BRCA1, NBS1, NBN, CHEK2, aggressive prostate cancer, survival
The number of known genes for which mutations clearly predispose to prostate cancer is small, and include BRCA2, BRCA1, CHEK2, NBS1 and HOXB13 (Struewing et al, 1997; Thorlacius et al, 1997; Dong et al, 2003; Edwards et al, 2003; Seppälä et al, 2003; Cybulski et al, 2004; Kirchhoff et al, 2004; Kote-Jarai et al, 2011; Ewing et al, 2012; Leongamornlert et al, 2012). Four of these, BRCA2, BRCA1, CHEK2, NBS1 (also known as Nibrin; NBN) are involved in the DNA damage response pathway (Futaki and Lui, 2001). In Poland, we have identified eight founder alleles in three DNA damage repair genes that predispose to breast cancer (Górski et al, 2005; Cybulski et al, 2011). Three founder alleles are in BRCA1 (5382insC, 4153delA, C61G), four are in CHEK2 (1100delC, IVS2+1G>A, del5395, I157T) and one variant allele (657del5) is in NBS1. To establish the contribution of eight founder alleles in three DNA damage repair genes (BRCA1, CHEK2 and NBS1) to prostate cancer in Poland, and to measure the impact of these variants on survival, we genotyped 3750 men with prostate cancer and 3956 controls.
Materials and methods
Patients
We studied men with prostate cancer who were diagnosed between 1999 and 2012 in 14 centres situated throughout Poland. This study was initiated in Szczecin in 1999 and was extended to include Białystok, Olsztyn in 2002 and Opole in 2003. Other centres began recruiting between 2005 and 2008 (Koszalin, Gdansk, Lublin, Łodź, Warszawa, Wrocław, Poznan, Rzeszów, Bydgoszcz, Zabrze). All men with prostate cancer were invited to participate. Study subjects were asked to participate at the time of diagnosis or during an outpatient visit to an oncology clinic and were unselected for age or family history. Four thousand five hundred thirty-one men were invited and of these, 3915 (86.4%) participated. All patients provided a blood sample within 6 months of diagnosis. The mean age of diagnosis was 68.8 years (range 41–96 years). A family history was taken either by the construction of a family tree or the completion of a standardised questionnaire. All first- and second-degree relatives diagnosed with prostate cancer and the ages of diagnosis were recorded. A family history of cancers in relatives was available for 3586 (92%) subjects. Four hundred sixteen men reported at least one first- or second-degree relative with prostate cancer (familial cases). In addition, information was recorded on PSA level at time of diagnosis, grade (Gleason score) and stage. The study was approved by the Ethics Committee of the Pomeranian Medical University in Szczecin, Poland.
Genotyping
DNA was isolated from 5 to 10 ml of peripheral blood. DNA was successfully isolated from 3853 (98.4%) of 3915 cases. Eight founder mutations in BRCA1, CHEK2 and NBS1 were genotyped as described previously (Cybulski et al, 2004, 2006; Górski et al, 2005). In brief, BRCA1 mutations, 4153delA and 5382insC , were detected using allele-specific oligonucleotide PCR, and C61G was detected using restriction fragment length polymorphism PCR. The CHEK2 del5395 mutation was detected by a multiplex PCR reaction. The IVS2+1G>A and I157T variants in CHEK2 were detected using restriction fragment length polymorphism PCR analysis, and the 1100delC mutation was analysed using allele-specific oligonucleotide PCR. NBS1 mutation was tested using allele-specific oligonucleotide PCR. All eight mutations were successfully analysed in 3750 of 3853 cases (97%) including 412 familial prostate cancer cases.
Controls
The control group included 3956 cancer-free men age 23–90 years (mean age, 61.2 years). The purpose of the control group was to estimate with accuracy the frequency of founder alleles of BRCA1, CHEK2 and NBS1 in the underlying Polish population. These controls were derived from four sources. The first series consisted of consisted of 603 unselected men (age range, 30–90 years; mean age, 64.2 years) selected at random from the computerised patient lists of five large family practices located in the region of Szczecin. These were invited to participate by mail and participated in 2003 and 2004. The second subgroup consisted of 1008 men from the region of Szczecin (age range, 23–87 years; mean age, 61.6 years). These men were part of a population-based study of the 1.5 million residents of West Pomerania designed to identify familial cancer clusters and were interviewed in 2007. Men with any cancer diagnosed in a first-degree relative were excluded from this control group. The third series consisted of 1301 unselected men at age above 45 (age range, 45–90 years; mean age, 61.9 years) with PSA level below 4.0 ng l−1. These men were selected randomly from a database of a population-based study of the 1.5 million residents of West Pomerania and provided blood sample between 2010 and 2012. Men with PSA above 4.0 ng l−1 and men with a positive family history of prostate cancer were excluded from this group. The fourth series included 1044 Polish men (age range, 55–66 years; mean age, 60.1 years), who participated in population colonoscopy screening programme for colorectal cancer between 2005 and 2010, and provided blood samples for DNA analysis (771 men were from Szczecin, 189 from Białystok and 84 from Łodz). The allele frequencies for all variants in our control group were not dependent on age, and the prevalence estimates of mutations in all genes were similar in younger and in older controls. The frequency of I157T in our controls (4.7%) is similar to that reported by Brennan et al (2007) in a non-overlapping series of 790 controls from Poland (5.6%). The frequency of 1100delC in our controls and in controls genotyped by Brennan et al (2007) is 0.2%. The frequency of NBS1 in our controls and in 6984 (non-overlapping) controls genotyped by Chrzanowska et al (2006) is 0.6%.
Statistical analysis
The prevalences of all alleles in cases and controls were compared. Odds ratios (ORs) were generated from two-by-two tables and statistical significance was assessed with the Fisher exact test or the χ2 test where appropriate. The ORs were used as estimates of relative risk. For the survival analysis, the patients were followed from the date of biopsy until death or March 2012. The vital status and the date of death were requested from the Polish Ministry of the Interior and Administration in March 2012, and were obtained in April 2012. These data were available for 3487 (93%) of 3750 men with prostate cancer.
The mean follow-up (overall, 54.4 months) was 67.5 months for BRCA1 carriers (P=0.2), 53.6 months for NBS1 carriers (P=0.9), 57.1 months for CHEK2 carriers (P=0.1), compared with 54.0 months in non-carriers. Mean follow-up was compared using t-test.
Kaplan–Meier survival curves were constructed for carriers of mutations in either of the three genes and for non-carriers. Comparison of survival curves was performed by log-rank test. For a subset of 1804 patients (including 37 NBS1 mutation carriers and 1767 non-carriers) survival data and detailed tumour characteristics were available (PSA level at diagnosis, tumour stage and Gleason score). A multivariable Cox regression analysis was performed, including age of diagnosis, year of diagnosis, PSA level at diagnosis, Gleason score and stage (T1–4) as covariates.
Results
A mutation in one of the three DNA damage repair genes was seen in 443 of 3750 (11.8%) patients with prostate cancer and in 190 of 2912 (6.5%) controls (Table 1). Strong associations were seen for both CHEK2 and NBS1. The single NBS1 mutation (657del5) was detected in 53 of 3750 unselected cases (OR=2.5; P=0.0003) and in 10 of 412 familial cases (OR=4.3; P=0.0001) compared with 23 of 3956 (0.6%) controls. A CHEK2 mutation was seen in 383 (10.2%) unselected cases of prostate cancer (OR=1.9; P<0.0001), in 59 (14.3%) familial cases (OR=2.7; P<0.0001) and in 228 (5.8%) controls. A BRCA1 mutation (three BRCA1 mutations combined) was not associated with the risk of prostate cancer (OR=0.9; P=0.8). Although not statistically significant (P=0.2), the 4153delA mutation was associated with OR of 5.3 for prostate cancer, but the other two mutations of BRCA1 were not associated with an increase in the risk of prostate cancer (OR=0.5 for the 5382insC mutation; OR=1.1 for the C61G mutation).
Table 1. Association of variant alleles in BRCA1, NBS1 and CHEK2 with prostate cancer risk.
| Controls (n=3956) No. (%) | Unselected cases (n=3750) No. (%) | OR | 95% CI | P-value | Familial cases (n=412) No. (%) | OR | 95% CI | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| Any BRCA1 mutation | 17 (0.4%) | 14 (0.4%) | 0.9 | 0.4–1.8 | 0.8 | 4 (1.0%) | 2.3 | 0.8–6.8 | 0.3 |
| 5382insC | 13 (0.3%) | 6 (0.2%) | 0.5 | 0.2–1.3 | 0.2 | 1 (0.2%) | 0.7 | 0.1–5.7 | 0.8 |
| C61G | 3 (0.08%) | 3 (0.08%) | 1.1 | 0.2–5.2 | 0.9 | 2 (0.5%) | 6.4 | 1.1–38.6 | 0.1 |
| 4153delA | 1 (0.03%) | 5 (0.13%) | 5.3 | 0.6–45.2 | 0.2 | 1 (0.2%) | 9.6 | 0.6–154 | 0.5 |
| |
|
|
|
|
|
|
|
|
|
| NBS1 mutation | |||||||||
| 657del5 | 23 (0.6%) | 53 (1.4%) | 2.5 | 1.5–4.0 | 0.0003 | 10 (2.4%) | 4.3 | 2.0–9.0 | 0.0001 |
| |
|
|
|
|
|
|
|
|
|
| Any CHEK2 mutation | 228 (5.8%) | 383 (10.2%) | 1.9 | 1.6–2.2 | <0.0001 | 59 (14.3%) | 2.7 | 2.0–3.7 | <0.0001 |
| Any CHEK2 truncating mutation | 43 (1.1%) | 84 (2.2%) | 2.1 | 1.4–3.0 | 0.0001 | 16 (3.9%) | 3.7 | 2.1–6.6 | <0.0001 |
| 1100delC | 7 (0.2%) | 21 (0.6%) | 3.2 | 1.4–7.5 | 0.009 | 4 (1.0%) | 5.5 | 1.6–19.0 | 0.01 |
| IVS2+1G>A | 21 (0.5%) | 28 (0.7%) | 1.4 | 0.8–2.5 | 0.3 | 7 (1.7%) | 3.2 | 1.4–7.7 | 0.01 |
| del5395 | 15 (0.4%) | 35 (0.9%) | 2.5 | 1.3–4.5 | 0.004 | 5 (1.2%) | 3.2 | 1.2–8.9 | 0.04 |
| |
|
|
|
|
|
|
|
|
|
| CHEK2 I157T missense mutation | 186 (4.7%) | 303 (8.1%) | 1.8 | 1.5–2.2 | <0.0001 | 43 (10.4%) | 2.4 | 1.7–3.3 | <0.0001 |
Abbreviations: CI=confidence interval; HR=hazard ratio.
Familial cases – prostate cancers in two or more first- or second-degree relatives.
One control carried two mutations (I157T and 1100delC).
Eleven cases carried two mutations (four cases had I157T and a CHEK2 truncating mutation, four carried I157T and NBS1 mutation, two carried I157T and a BRCA1 mutation, and one carried a CHEK2 truncating mutation and a BRCA1 mutation).
We also investigated the incidence of the mutations by age, dividing cases into two groups: men diagnosed at age of 60 and below, and men diagnosed at age above 60 years (Table 2). For all genes, the mutation frequencies and the ORs were higher for early-onset cases than for cases diagnosed above age of 60 years. The ORs for early-onset prostate cancer were 3.1 (P=0.003) for NBS1 mutation, 2.3 (P<0.0001) for a CHEK2 mutation and 1.9 (P=0.9) for a BRCA1 mutation. For cases diagnosed above age of 60 years the ORs were 2.4 (P=0.0009) for NBS1 mutation, 1.8 (P<0.0001) for a CHEK2 mutation and 0.7 (P=0.6) for BRCA1 mutation. Among younger cases, the OR for BRCA1 (all three variants combined) was 1.9 (P=0.9) and for the 4153delA mutation alone was 20.3 (P=0.004).
Table 2. Association of variant alleles in BRCA1, NBS1 and CHEK2 with prostate cancer risk, by age.
| Controls (n=3956) No. (%) | Cases diagnosed at age ⩽60 years (n=619) No. (%) | OR | 95% CI | P-value | Cases diagnosed at age >60 years (n=3131) No. (%) | OR | 95% CI | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| Any BRCA1 mutation | 17 (0.4%) | 5 (0.8%) | 1.9 | 0.7–5.1 | 0.9 | 9 (0.3%) | 0.7 | 0.3–1.5 | 0.6 |
| 5382insC | 13 (0.3%) | 1 (0.2%) | 0.5 | 0.1–3.8 | 0.8 | 5 (0.2%) | 0.5 | 0.2–1.4 | 0.2 |
| C61G | 3 (0.08%) | 1 (0.2%) | 2.1 | 0.2–20.5 | 1.0 | 2 (0.06%) | 0.8 | 0.1–5.0 | 0.8 |
| 4153delA | 1 (0.03%) | 3 (0.5%) | 20.3 | 2.0–185.6 | 0.004 | 2 (0.06%) | 2.5 | 0.2–27.9 | 0.8 |
| |
|
|
|
|
|
|
|
|
|
| NBS1 mutation | |||||||||
| 657del5 | 23 (0.6%) | 11 (1.8%) | 3.1 | 1.5–6.4 | 0.003 | 43 (1.4%) | 2.4 | 1.4–4.0 | 0.0009 |
| |
|
|
|
|
|
|
|
|
|
| Any CHEK2 mutation | 228 (5.8%) | 77 (12.4%) | 2.3 | 1.8–3.1 | <0.0001 | 306 (9.8%) | 1.8 | 1.5–2.1 | <0.0001 |
| |
|
|
|
|
|
|
|
|
|
| Any CHEK2 truncating mutation | 43 (1.1%) | 16 (2.6%) | 2.4 | 1.4–4.3 | 0.004 | 68 (2.2%) | 2.0 | 1.4–3.0 | 0.0004 |
| 1100delC | 7 (0.2%) | 4 (0.6%) | 3.7 | 1.1–12.6 | 0.08 | 17 (0.5%) | 3.1 | 1.2–7.4 | 0.02 |
| IVS2+1G>A | 21 (0.5%) | 4 (0.6%) | 1.2 | 0.4–3.6 | 0.9 | 24 (0.8%) | 1.4 | 0.8–2.6 | 0.3 |
| del5395 | 15 (0.4%) | 8 (1.3%) | 3.4 | 1.5–8.2 | 0.007 | 27 (0.9%) | 2.3 | 1.2–4.3 | 0.01 |
| |
|
|
|
|
|
|
|
|
|
| CHEK2 I157T missense mutation | 186 (4.7%) | 62 (10.0%) | 2.3 | 1.7–3.0 | <0.0001 | 241 (7.7%) | 1.7 | 1.4–2.1 | <0.0001 |
Abbreviations: CI=confidence interval; HR=hazard ratio.
ORs and P-values are calculated with respect to controls as reference group.
The characteristics of the prostate cancer cases in the 443 carriers and 3307 non-carriers are presented in Table 3. There were few significant differences between subgroups; men with a CHEK2 mutation were diagnosed with prostate cancer on average 1.4 years younger than non-carriers (67.5 vs 68.9; P=0.003). Prostate cancers of advanced stage (T4) were more common in carriers of a NBS1 mutation than in non-carriers (19.5% vs 7.7% P=0.01).
Table 3. Clinical characteristics of prostate cancers in carries of variant alleles in BRCA1, NBS1, CHEK2 and in non-carriers.
| BRCA1 mutation carriers (n=14) | P-value | NBS1mutation carriers (n=53) | P-value | CHEK2 mutation carriers (n=383) | P-value | Mutation-negative cases (n=3307) | |
|---|---|---|---|---|---|---|---|
|
Age of diagnosis | |||||||
| Mean | 68.3 | 0.8 | 67.3 | 0.2 | 67.5 | 0.003 | 68.9 |
| |
|
|
|
|
|
|
|
|
PSA level at diagnosis | |||||||
| Median | 14.5 | 0.6 | 10.7 | 0.8 | 10.9 | 0.3 | 11.2 |
| ⩽4.0 | 10.0% (1/10) | 0.9 | 2.8% (1/36) | 0.9 | 5.3% (13/247) | 0.7 | 4.5% (112/2474) |
| 4.1–10 | 30.0% (3/10) | 0.7 | 47.2% (17/36) | 0.5 | 40.5% (100/247) | 1.0 | 40.4% (999/2474) |
| 10.1–20.0 | 20.0% (2/10) | 1.0 | 25.0% (9/36) | 0.8 | 26.3% (65/247) | 0.7 | 25.0% (619/2474) |
| >20.0 | 40.0% (4/10) | 0.7 | 25.0% (9/36) | 0.6 | 27.9% (69/247) | 0.5 | 30.1% (744/2474) |
| |
|
|
|
|
|
|
|
|
Gleason score | |||||||
| <7 | 36.4% (4/11) | 0.5 | 43.6% (20/46) | 0.3 | 55.2% (142/257) | 0.3 | 51.9% (1341/2584) |
| 7 | 45.4% (5/11) | 0.4 | 28.2% (13/46) | 1.0 | 26.1% (67/257) | 0.4 | 29.0% (749/2584) |
| >7 | 18.2% (2/11) | 0.8 | 28.2% (13/46) | 0.1 | 18.7% (48/257) | 0.9 | 19.1% (494/2584) |
| |
|
|
|
|
|
|
|
|
Stage | |||||||
| T3 | 30% (3/10) | 0.4 | 14.6% (6/41) | 0.8 | 18.6% (40/215) | 1.0 | 17.8% (362/2029) |
| T4 | 20% (2/10) | 0.2 | 19.5% (8/41) | 0.01 | 6.0% (13/215) | 1.0 | 7.7% (156/2029) |
| |
|
|
|
|
|
|
|
|
Family history of prostate cancer | |||||||
| Positive | 30.8% (4/13) | 0.07 | 20.0% (10/50) | 0.07 | 16.9% (59/332) | 0.001 | 11.0% (341/3095) |
P-values for mutation carriers are calculated with respect to non-carriers as reference group.
In addition, tumours of Gleason grade 8–10 were more frequent in men with a NBS1 mutation than in non-carriers (28.4% vs 19.1%), but this difference was not statistically significant (P=0.1).
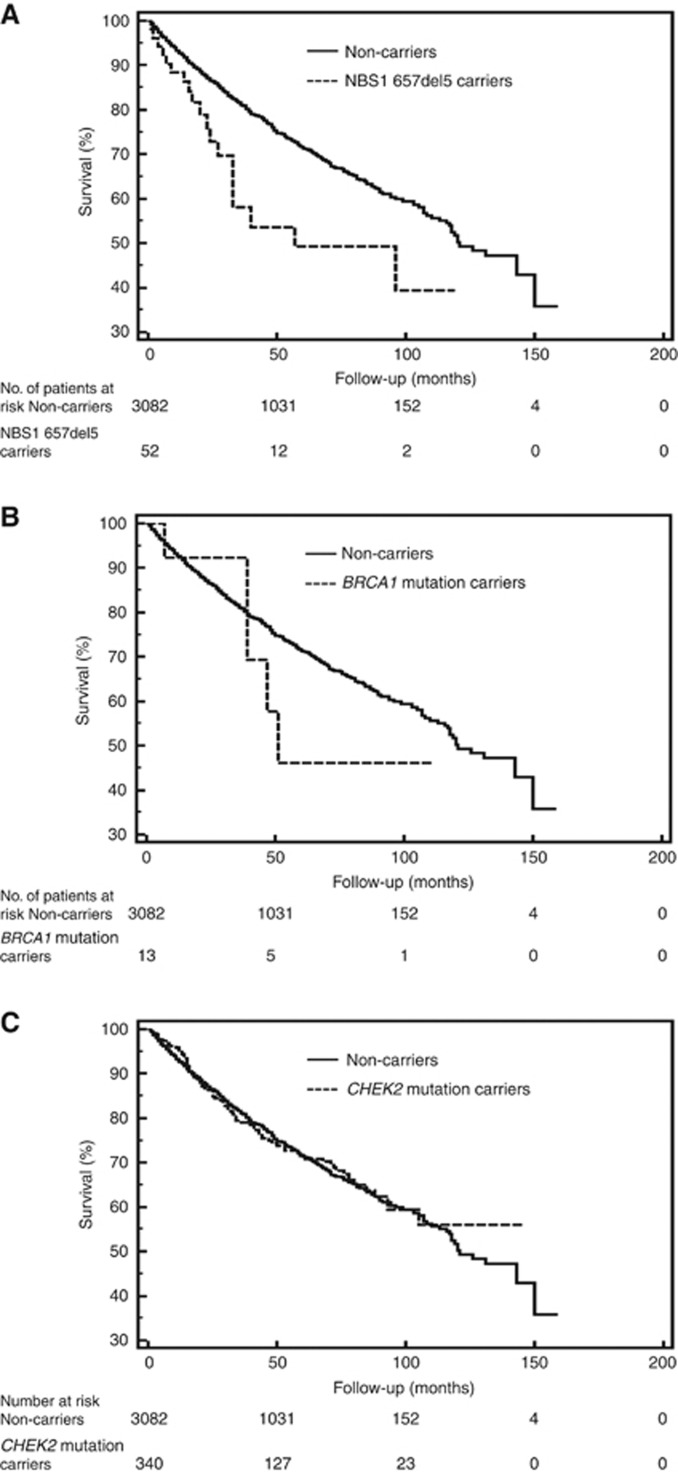
Data on survival was available for 3487 men with prostate cancer (Table 4). There were five deaths (38.5%) recorded in 13 carriers of a BRCA1 mutation, 19 deaths (36.5%) in 52 carriers of a NBS1 mutation, 87 deaths (25.6%) in 340 men with a CHEK2 mutation and 755 deaths (24.5%) in 3082 non-carriers. Kaplan–Meier survival curves for mutation carriers and non-carriers are shown in Figure 1. The mortality experience was significantly worse for carriers of a NBS1 mutation, compared with non-carriers (HR=1.85; P=0.008). The poor relative survival for carriers of an NBS1 mutation was particularly apparent in the first 5 years after diagnosis (HR=2.08; P=0.002). The 5-year survival for carriers of a NBS1 mutation was 49%, compared with 72% for non-carrier controls. After adjusting for age, year of diagnosis, PSA, stage and grade, the HR for mortality associated with a NBS1 mutation was 1.86 (95% CI, 1.05–3.32; P=0.04). Of the 52 carriers of a NBS1 mutation, 19 (36.5%) have died, on average 24.3 months after diagnosis. The characteristics of the patients and the corresponding tumours for the 19 fatal cases among NBS1 mutation carriers is presented in Table 5.
Table 4. Survival of men with a mutation in BRCA1, NBS1 and CHEK2 and in non-carriers.
| BRCA1 mutation (n=13) | NBS1 mutation (n=52) | CHEK2 mutation (n=340) | Mutation-negative cases (n=3082) | |
|---|---|---|---|---|
| Mean follow-up (months) |
67.5 |
53.6 |
57.1 |
54.0 |
| Proportion of deceased (%) |
38.5 |
36.5 |
25.6 |
24.5 |
| Median survival (months) |
51 |
57 |
122 |
121 |
| 5-Year survival (%) |
46 |
49 |
71 |
72 |
| 10-Year survival (%) |
46 |
39 |
56 |
52 |
| HR |
1.48 |
1.85 |
0.99 |
1.0a |
| 95% CI |
0.51–4.30 |
1.17–2.91 |
0.80–1.24 |
— |
| P-value | 0.38 | 0.008 | 0.95 | — |
Abbreviations: CI=confidence interval; HR=hazard ratio.
HR, 95% CI and P-values are calculated by log-rank test.
Reference value.
Figure 1.
Kaplan–Meier curves of prostate cancer patients with: (A) mutation in NSB1 (n=52); (B) mutation in BRCA1 (n=13); (C) mutation in CHEK2 (n=340), compared with prostate cancer patients with no mutation (non-carriers; n=3082).
Table 5. The characteristics of the patients and the corresponding tumours for the 19 fatal cases with the 657del5 mutation in NBS1.
| NBS1 mutation-positive lethal prostate cancer cases (n=19) | ||
|---|---|---|
| Age (years) |
Mean age (range) |
69.1 (52–86) |
| |
|
|
| Age group |
⩽60 |
21.1% (4/19) |
| 61–70 | 31.6% (6/19) | |
| >70 | 47.3% (9/19) | |
| PSA | Median (range) | 20.5 (6–190) |
| ⩽4.0 | 0.0% (0/12) | |
| 4.1–10.0 | 33.3% (4/12) | |
| 10.1–20.0 | 16.7% (2/12) | |
| |
>20.0 |
50.0% (6/12) |
| |
|
|
| Gleason score |
<7 |
40.0% (6/15) |
| 7 | 26.7% (4/15) | |
| >7 | 33.3% (5/15) | |
| |
|
|
| Stage |
T3 |
23.1% (3/13) |
| T4 | 38.4% (5/13) | |
| |
|
|
| Family history of prostate cancer | Positive | 11.8% (2/17) |
Abbreviation: PSA=prostate-specific antigen.
The survival experience of carriers of a BRCA1 mutation was also relatively poor, but this subgroup was small (n=13 BRCA1 mutation carriers) and the difference was not statistically significant (HR=1.48; P=0.38). Survival in men with a CHEK2 mutation was similar to that of non-carriers (HR=0.99 and P=0.95).
Discussion
The most noteworthy observation is the remarkably poor short-term survival of men with prostate cancer and NBS1 mutation. We have confirmed our earlier work that describes NBS1 as a prostate cancer susceptibility gene (Cybulski et al, 2004), and we have extended our findings by documenting the aggressive nature of the associated tumours. The NBS1 657del5 founder allele is present in ∼1 in 170 individuals in Poland and is associated with a three-fold increased risk of prostate cancer. Cancers in carriers of the NBS1 657del5 founder mutation are typically aggressive; ∼30% were of Gleason grade 8 or above and approximately one-half of the patients with this allele died within 5 years of diagnosis. Compared with men with no mutation, the relative survival rate at 5 years was only 48%. The aggressive behaviour of these cancers was not entirely attributable to the grade, after adjustment for age, grade, stage and PSA, the NBS1-associated cancers had a relatively poor survival (HR=1.86; 95% CI, 1.05–3.32; P=0.04). In Poland, ∼1.4% of prostate cancers are attributable to a mutation of NBS1 and 5.5% are due to CHEK2 mutations. However, in terms of prognosis, the cancers in carriers of CHEK2 mutations are not distinguishable from cancers in the population at large. To our knowledge, this is the first study to describe the clinical characteristics and survival of men with prostate cancer and a mutation in NBS1 and CHEK2.
There is no organised prostate screening programme in Poland and the majority of the patients in this study presented because of symptoms or because of an abnormal digital rectal examination. Our results are of interest in considering whether or not prostate cancer screening is warranted in Poland, and if so, if a screening programme should be universal or personalised (i.e., targeted to those at high risk). Personalised screening might incorporate two phases – the first phase would screen men for the five susceptibility alleles described here. Men with a mutation in one of the two genes would then be a candidate for PSA-based prostate cancer screening. In particularly men with an NBS1 mutation might be screened aggressively, perhaps including a random biopsy. In our study, the earliest age of onset among men with a NBS1 mutation was 50 years and among men with a CHEK2 mutation was 45 years. However, the principal limitation of this personalised model is that only 12% of new cases of prostate cancer in Poland occur in men with one of these mutations and therefore the potential to reduce the overall cancer burden is limited. Also, the benefit or prostate screening on reducing mortality in average risk men using the conventional PSA test has not been proven and has not been evaluated in men with predisposing mutations (Djulbegovic et al, 2010, 2012; Schröder et al, 2012). Only one study called IMPACT (Identification of Men with a genetic predisposition to ProstAte Cancer: Targeted screening in BRCA1/2 mutation carriers and controls; http://www.impact-study.co.uk) investigated prostate cancer screening targeted at men with a known genetic predisposition to the disease. Preliminary analysis of the data from the IMPACT study supports the rationale for continued PSA screening in such men (Mitra et al, 2011). The IMPACT study only referred to germline mutations in BRCA1 and BRCA2, and it is not know if men with CHEK2 and NBS1 mutations should be thus screened, but are good candidates for study.
NBS1, also known as Nibrin (NBN), is the gene for Nijmegen breakage syndrome (NBS), a rare autosomal recessive disorder that is characterised by immunodeficiency, chromosomal instability and sensitivity to ionising radiation (Varon et al, 1998). The 657del5 mutation is responsible for 90% of all reported cases of NBS to date (Varon et al, 2000). On the basis of the geographic distribution of reported clinical cases of Nijmegen syndrome, the distribution of 657del5 allele of NBS1 is not worldwide, and this allele is most common in Slavic populations of Eastern Europe. Other truncating mutations of NBS1 (698del4 of English origin, 835del4 of Italian origin, 842insT of Mexican origin, 1142delC of Canadian origin, and Q326X of Dutch origin) were detected in 10% of NBS patients (Varon et al, 1998), but their geographic extend and their role in prostate cancer susceptibility has not been established. Only one previous study explored the association between NBS1 657del5 and prostate cancer risk. In that study, the 657del5 allele was seen in 7 (0.23%) of 3037 men with prostate cancer and in none of 990 unaffected controls in the United States (Hebbring et al, 2006). The clinical characteristics of the mutation-positive cases is not described.
The NBS1 founder allele is predicted to result in a truncated protein of 219 of 754 amino acids (p26) (Maser et al, 2001). However, the 657del5 allele also creates an aberrant translation initiation site, which generates a partially functional variant of the NBS1 protein (p70). Null mutations in MRE11 and RAD50, which encode binding partners of NBS1, are lethal in vertebrates, and mouse Nbs1-null mutants are inviable (Maser et al, 2001). Therefore, it is likely that truly null mutations of NBS1 (in homozygous state) do not cause Nijmegen syndrome, but only those mutations that encode partially active Nibrin (such as the 657del5 mutation) are pathogenic for the syndrome. However, null mutations of NBS1 in heterozygous state are not lethal, and may well predispose to cancer. It is therefore possible that cancer-associated mutations of NBS1 are different from NBS-related mutations. NBS1 needs to resequenced in cancer patients to describe full spectrum of cancer predisposing mutations in different ethnic groups (regardless of geographic distribution of NBS syndrome). Of note, recently, 94 unrelated familial prostate cancer cases from the United States were screened by next-generation sequencing (whole exome-sequencing). A novel truncating mutation of NBN, 2117 C>G mutation that results in a premature stop at codon 706 (S706X) was detected in one family, and the mutation partially cosegregated with prostate cancer in this family (Zuhlke et al, 2012).
The situation of NBS1 carriers is similar to that of men with prostate cancer with a mutation in BRCA2 (in Poland, there are no known founder mutations in BRCA2 and the gene was not part of the current analysis). Men with prostate cancer and a BRCA2 mutation have a poor prognosis despite conventional therapy (prostatectomy, hormonal therapy, radiation therapy), and most BRCA2 carriers with prostate cancer will succumb to their disease (Sigurdsson et al, 1997; Narod et al, 2008; Edwards et al, 2010; Thorne et al, 2011). Both NBS1 and BRCA2 genes act in DNA damage repair signalling pathway, and mutations in the two genes (in homozygous state) cause inherited recessive clinical syndromes, such as NBS (NBS1 mutation), Fanconi anaemia (BRCA2 mutation), which are characterised by spontaneous chromosomal instability, immunodeficiency and a predisposition to cancer (Digweed, 1993; Varon et al, 1998; Howlett et al, 2002). It will be of interest to determine whether mutations of other genes for the chromosomal instability syndromes (BLM gene for Bloom syndrome, FA genes for Fanconi anaemia, ATM gene for ataxia telangiectasia) also predispose to aggressive prostate cancer.
It is also important to determine whether therapy beyond the conventional therapy is valuable for men with prostate cancer and a NBS1 mutation (or a BRCA2 mutation). Similarly to BRCA2, NBS1 appears to act as a classical tumour-suppressor gene (Willems et al, 2008). Biallelic NBS1 inactivation was observed in most tumours in 657del5 carriers and the cancers that develop in the prostates of carriers are functionally homozygous for the mutation (Cybulski et al, 2004). The product of the NBS1 gene is an integral component of the Mre11/Rad50/NBS1 nuclease complex, which has a role in the initial processing of double-strand DNA breaks before repair by homologous recombination (Petrini, 1999; Lee and Paull, 2004, 2005). If double-strand DNA breaks are not recognised, then the repair is impaired. Therefore, men with prostate cancer and NBS1 mutation (or a BRCA2 mutation) might benefit from treatment with cisplatin and PARP1 inhibitors.
We saw little effect of a BRCA1 mutation (all three mutations combined) on prostate cancer risk (OR=0.9; P=0.8), whereas several previous studies suggested an effect (Ford et al, 1994; Struewing et al, 1997; Warner et al, 1999; Thompson and Easton, 2002a; Giusti et al, 2003; Leongamornlert et al, 2012). However, we observed excess of BRCA1 mutations in men with early-onset prostate cancer (⩽60 years), but this did not achieve statistical significance (OR=1.9; P=0.9) possibly because of small numbers. Our data are in line with the results of the Breast Cancer Linkage Consortium, which reported an increase in prostate cancer risk in carriers of a BRCA1 mutation aged <65 years with a RR of 1.82 (95% CI 1.01–3.29, P=0.05), but no risk increase was seen for men ⩾65 years (Thompson and Easton, 2002a). It is also interesting that in our study, only the 4153delA mutation was associated with increased risk of unselected prostate cancer (OR=5.3, P=0.2) and of early-onset (⩽60 years) prostate cancer (OR=20.3; P=0.004), but the other two BRCA1 mutations (5382insC and C61G) were not. This is a subgroup analysis and may be due to chance, but it is also possible that there is a genotype–phenotype effect in the BRCA1 gene, wherein only some mutations (such as 4153delA) are pathogenic for prostate cancer. Genotype–phenotype correlations have been suggested for breast and ovarian cancer risk in BRCA1 carriers (Gayther et al, 1995; Holt et al, 1996; Neuhausen et al, 1996; Thompson and Easton, 2002b; Rennert et al, 2005; Gronwald et al, 2006). It is also possible that the risk of prostate cancer may vary by the type and/or location of the BRCA1 mutation, but further studies are needed.
In conclusion, our results provide compelling evidence that a founder mutation in NBN predisposes to aggressive prostate cancers. The data presented here raise important questions about the prospect of adapting personalised medicine to prostate cancer prevention.
Acknowledgments
This study was funded by resources for the science of the Polish Ministry of Science and Higher Education as requested research grant PBZ-MNiSW-05/I/2007/02. We thank Daria Zanoza, Piotr Chłosta and Jarosław Jaskulski for their support in this study.
Appendix
Other members of the Polish Hereditary Prostate Cancer Consortium
B Masojć1, T van de Wetering1, P Serrano-Fernández1, M Puszyński2, M Soczawa2, M Słojewski2, S Archimowicz3, M Kordowski3, M Życzkowski11, W Domagała12, M Chosia12, A Borówka16, J Bagińska18, K Krajka18, M Szwiec19, M Stawicka20, O Haus21, H Janiszewska21, A Stembalska22 and MM Sąsiadek22
1International Hereditary Cancer Centre, Department of Genetics and Pathology, Pomeranian Medical University, Szczecin, Poland
2Clinic of Urology, Pomeranian Medical University, Szczecin, Poland
3Division of Urology, Maria Skłodowska-Curie Hospital, Szczecin, Poland
11Department of Urology, Medical University of Silesia, Zabrze, Poland
12Department of Urology, Ministry of Internal Affairs and Administration Hospital, Łódź
16Centre for Research on Womens Health, Toronto Ontario, Canada
18Clinic of Urology, Medical University, Gdansk, Poland
19Regional Oncology Hospital, Olsztyn, Poland
20NZOZ Centre for Medical Genetics, Poznan, Poland
21Department of Clinical Genetics, Nicolaus Copernicus University, Bydgoszcz, Poland
22Department of Genetics, Wroclaw Medical University, Wrocław, Poland
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Brennan P, McKay J, Moore L, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chow WH, Rothman N, Chabrier A, Gaborieau V, Odefrey F, Southey M, Hashibe M, Hall J, Boffetta P, Peto J, Peto R, Hung RJ. Uncommon CHEK2 missense variant and reduced risk of tobacco-related cancers: Case control study. Hum Mol Genet. 2007;16:1794–1801. doi: 10.1093/hmg/ddm127. [DOI] [PubMed] [Google Scholar]
- Chrzanowska KH, Piekutowska-Abramczuk D, Popowska E, Gładkowska-Dura M, Małdyk J, Syczewska M, Krajewska-Walasek M, Goryluk-Kozakiewicz B, Bubała H, Gadomski A, Gaworczyk A, Kazanowska B, Kołtan A, Kuźmicz M, Luszawska-Kutrzeba T, Maciejka-Kapuścińska L, Stolarska M, Stefańska K, Sznurkowska K, Wakulińska A, Wieczorek M, Szczepański T, Kowalczyk J. Carrier frequency of mutation 657del5 in the NBS1 gene in a population of Polish pediatric patients with sporadic lymphoid malignancies. Int J Cancer. 2006;118:1269–1274. doi: 10.1002/ijc.21439. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Górski B, Debniak T, Gliniewicz B, Mierzejewski M, Masojć B, Jakubowska A, Matyjasik J, Złowocka E, Sikorski A, Narod SA, Lubiński J. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64:1215–1219. doi: 10.1158/0008-5472.can-03-2502. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Wokołorczyk D, Huzarski T, Byrski T, Gronwald J, Górski B, Debniak T, Masojć B, Jakubowska A, Gliniewicz B, Sikorski A, Stawicka M, Godlewski D, Kwias Z, Antczak A, Krajka K, Lauer W, Sosnowski M, Sikorska-Radek P, Bar K, Klijer R, Zdrojowy R, Małkiewicz B, Borkowski A, Borkowski T, Szwiec M, Narod SA, Lubiński J. A large germline deletion in the Chek2 kinase gene is associated with an increased risk of prostate cancer. J Med Genet. 2006;43:863–866. doi: 10.1136/jmg.2006.044974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Wokołorczyk D, Jakubowska A, Huzarski T, Byrski T, Gronwald J, Masojć B, Debniak T, Górski B, Blecharz P, Narod SA, Lubiński J. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol. 2011;29:3747–3752. doi: 10.1200/JCO.2010.34.0778. [DOI] [PubMed] [Google Scholar]
- Digweed M. Human genetic instability syndromes: single gene defects with increased risk of cancer. Toxicol Lett. 1993;67:259–281. doi: 10.1016/0378-4274(93)90061-2. [DOI] [PubMed] [Google Scholar]
- Djulbegovic M, Beyth RJ, Neuberger MM, Stoffs TL, Vieweg J, Djulbegovic B, Dahm P. Screening for prostate cancer: systematic review and meta-analysis of randomised controlled trials. BMJ. 2010;341:4543–4543. doi: 10.1136/bmj.c4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djulbegovic M, Neuberger MM, Dahm P. Prostate-cancer mortality after PSA screening. N Engl J Med. 2012;366:2228–2229. doi: 10.1056/NEJMc1204298. [DOI] [PubMed] [Google Scholar]
- Dong X, Wang L, Taniguchi K, Wang X, Cunningham JM, McDonnell SK, Qian C, Marks AF, Slager SL, Peterson BJ, Smith DI, Cheville JC, Blute ML, Jacobsen SJ, Schaid DJ, Tindall DJ, Thibodeau SN, Liu W. Mutations in CHEK2 associated with prostate cancer risk. Am J Hum Genet. 2003;72:270–280. doi: 10.1086/346094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SM, Evans DG, Hope Q, Norman AR, Barbachano Y, Bullock S, Kote-Jarai Z, Meitz J, Falconer A, Osin P, Fisher C, Guy M, Jhavar SG, Hall AL, O'Brien LT, Gehr-Swain BN, Wilkinson RA, Forrest MS, Dearnaley DP, Ardern-Jones AT, Page EC, Easton DF, Eeles RA, UK Genetic Prostate Cancer Study Collaborators and BAUS Section of Oncology Prostate cancer in BRCA2 germline mutation carriers is associated with poorer prognosis. Br J Cancer. 2010;103:918–924. doi: 10.1038/sj.bjc.6605822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SM, Kote-Jarai Z, Meitz J, Hamoudi R, Hope Q, Osin P, Jackson R, Southgate C, Singh R, Falconer A, Dearnaley DP, Ardern-Jones A, Murkin A, Dowe A, Kelly J, Williams S, Oram R, Stevens M, Teare DM, Ponder BA, Gayther SA, Easton DF, Eeles RA, Cancer Research UK/Bristish Prostate Group UK Familial Prostate Cancer Study Collaborators; British Association of Urological Surgeons Section of Oncology Two percent of men with early-onset prostate cancer harbor germline mutations in the BRCA2 gene. Am J Hum Genet. 2003;72:1–12. doi: 10.1086/345310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, Gielzak M, Partin AW, Shanmugam V, Izatt T, Sinari S, Craig DW, Zheng SL, Walsh PC, Montie JE, Xu J, Carpten JD, Isaacs WB, Cooney KA. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE. Risks of cancer in BRCA1-mutation carriers. Lancet. 1994;343:692–695. doi: 10.1016/s0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- Futaki M, Lui JM. Chromosome breakage syndromes and the BRCA1 genome surveillance complex. Trends Mol Med. 2001;7:560–565. doi: 10.1016/s1471-4914(01)02178-5. [DOI] [PubMed] [Google Scholar]
- Gayther SA, Warren W, Mazoyer S, Russell PA, Harrington PA, Chiano M, Seal S, Hamoudi R, van Rensburg EJ, Dunning AM, Love R, Evans G, Easton D, Clayton D, Stratton MR, Ponder BA. Germline mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nat Genet. 1995;11:428–433. doi: 10.1038/ng1295-428. [DOI] [PubMed] [Google Scholar]
- Giusti RM, Rutter JL, Duray PH, Freedman LS, Konichezky M, Fisher-Fischbein J, Greene MH, Maslansky B, Fischbein A, Gruber SB, Rennert G, Ronchetti RD, Hewitt SM, Struewing JP, Iscovich J. A twofold increase in BRCA mutation related prostate cancer among Ashkenazi Israelis is not associated with distinctive histopathology. J Med Genet. 2003;40:787–792. doi: 10.1136/jmg.40.10.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski B, Cybulski C, Huzarski T, Byrski T, Gronwald J, Jakubowska A, Stawicka M, Gozdecka-Grodecka S, Szwiec M, Urbański K, Mituś J, Marczyk E, Dziuba J, Wandzel P, Surdyka D, Haus O, Janiszewska H, Debniak T, Tołoczko-Grabarek A, Medrek K, Masojć B, Mierzejewski M, Kowalska E, Narod SA, Lubiński J. Breast cancer predisposing alleles in Poland. Breast Cancer Res Treat. 2005;92:19–24. doi: 10.1007/s10549-005-1409-1. [DOI] [PubMed] [Google Scholar]
- Gronwald J, Huzarski T, Byrski B, Medrek K, Menkiszak J, Monteiro AN, Sun P, Lubinski J, Narod SA. Cancer risks in first degree relatives of BRCA1 mutation carriers: effects of mutation and proband disease status. J Med Genet. 2006;43:424–428. doi: 10.1136/jmg.2005.036921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbring SJ, Fredriksson H, White KA, Maier C, Ewing C, McDonnell SK, Jacobsen SJ, Cerhan J, Schaid DJ, Ikonen T, Autio V, Tammela TL, Herkommer K, Paiss T, Vogel W, Gielzak M, Sauvageot J, Schleutker J, Cooney KA, Isaacs W, Thibodeau SN. Role of the Nijmegen breakage syndrome 1 gene in familial and sporadic prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:935–938. doi: 10.1158/1055-9965.EPI-05-0910. [DOI] [PubMed] [Google Scholar]
- Holt JT, Thompson ME, Szabo C, Robinson-Benion C, Arteaga CL, King MC, Jensen RA. Growth retardation and tumour inhibition by BRCA1. Nat Genet. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D'Andrea AD. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Kirchhoff T, Kauff ND, Mitra N, Nafa K, Huang H, Palmer C, Gulati T, Wadsworth E, Donat S, Robson ME, Ellis NA, Offit K. BRCA mutations and risk of prostate cancer in Ashkenazi Jews. Clin Cancer Res. 2004;10:2918–2921. doi: 10.1158/1078-0432.ccr-03-0604. [DOI] [PubMed] [Google Scholar]
- Kote-Jarai Z, Leongamornlert D, Saunders E, Tymrakiewicz M, Castro E, Mahmud N, Guy M, Edwards S, O'Brien L, Sawyer E, Hall A, Wilkinson R, Dadaev T, Goh C, Easton D, UKGPCS Collaborators. Goldgar D, Eeles R. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer. 2011;105:1230–1234. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science (New York, NY) 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science (New York, NY) 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Leongamornlert D, Mahmud N, Tymrakiewicz M, Saunders E, Dadaev T, Castro E, Goh C, Govindasami K, Guy M, O'Brien L, Sawyer E, Hall A, Wilkinson R, Easton D, UKGPCS Collaborators. Goldgar D, Eeles R, Kote-Jarai Z. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer. 2012;106:1697–1701. doi: 10.1038/bjc.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- Mitra AV, Bancroft EK, Barbachano Y, Page EC, Foster CS, Jameson C, Mitchell G, Lindeman GJ, Stapleton A, Suthers G, Evans DG, Cruger D, Blanco I, Mercer C, Kirk J, Maehle L, Hodgson S, Walker L, Izatt L, Douglas F, Tucker K, Dorkins H, Clowes V, Male A, Donaldson A, Brewer C, Doherty R, Bulman B, Osther PJ, Salinas M, Eccles D, Axcrona K, Jobson I, Newcombe B, Cybulski C, Rubinstein WS, Buys S, Townshend S, Friedman E, Domchek S, Ramon Y, Cajal T, Spigelman A, Teo SH, Nicolai N, Aaronson N, Ardern-Jones A, Bangma C, Dearnaley D, Eyfjord J, Falconer A, Grönberg H, Hamdy F, Johannsson O, Khoo V, Kote-Jarai Z, Lilja H, Lubinski J, Melia J, Moynihan C, Peock S, Rennert G, Schröder F, Sibley P, Suri M, Wilson P, Bignon YJ, Strom S, Tischkowitz M, Liljegren A, Ilencikova D, Abele A, Kyriacou K, van Asperen C, Kiemeney L, IMPACT Study Collaborators. Easton DF, Eeles RA. Targeted prostate cancer screening in men with mutations in BRCA1 and BRCA2 detects aggressive prostate cancer: preliminary analysis of the results of the IMPACT study. BJU Int. 2011;107:28–39. doi: 10.1111/j.1464-410X.2010.09648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod SA, Neuhausen S, Vichodez G, Armel S, Lynch HT, Ghadirian P, Cummings S, Olopade O, Stoppa-Lyonnet D, Couch F, Wagner T, Warner E, Foulkes WD, Saal H, Weitzel J, Tulman A, Poll A, Nam R, Sun P, Hereditary Breast Cancer Study Group. Danquah J, Domchek S, Tung N, Ainsworth P, Horsman D, Kim-Sing C, Maugard C, Eisen A, Daly M, McKinnon W, Wood M, Isaacs C, Gilchrist D, Karlan B, Nedelcu R, Meschino W, Garber J, Pasini B, Manoukian S, Bellati C. Rapid progression of prostate cancer in men with a BRCA2 mutation. Br J Cancer. 2008;99:371–374. doi: 10.1038/sj.bjc.6604453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhausen SL, Mazoyer S, Friedman L, Stratton M, Offit K, Caligo A, Tomlinson G, Cannon-Albright L, Bishop T, Kelsell D, Solomon E, Weber B, Couch F, Struewing J, Tonin P, Durocher F, Narod S, Skolnick MH, Lenoir G, Serova O, Ponder B, Stoppa-Lyonnet D, Easton D, King MC, Goldgar DE. Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am J Hum Genet. 1996;58:271–280. [PMC free article] [PubMed] [Google Scholar]
- Petrini JH. The mammalian Mre11-Rad50-nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am J Hum Genet. 1999;64:1264–1269. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennert G, Dishon S, Rennert HS, Fares F. Differences in the characteristics of families with BRCA1 and BRCA2 mutations in Israel. Eur J Cancer Prev. 2005;14:357–361. doi: 10.1097/00008469-200508000-00008. [DOI] [PubMed] [Google Scholar]
- Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Páez A, Määttänen L, Bangma CH, Aus G, Carlsson S, Villers A, Rebillard X, van der Kwast T, Kujala PM, Blijenberg BG, Stenman UH, Huber A, Taari K, Hakama M, Moss SM, de Koning HJ, Auvinen A, ERSPC Investigators Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä EH, Ikonen T, Mononen N, Autio V, Rökman A, Matikainen MP, Tammela TL, Schleutker J. CHEK2 variants associate with hereditary prostate cancer. Br J Cancer. 2003;89:1966–1970. doi: 10.1038/sj.bjc.6601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Thorlacius S, Tomasson J, Tryggvadottir L, Benediktsdottir K, Eyfjörd JE, Jonsson E. BRCA2 mutation in Icelandic prostate cancer patients. J Mol Med. 1997;75:758–761. doi: 10.1007/s001090050162. [DOI] [PubMed] [Google Scholar]
- Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, Timmerman MM, Brody LC, Tucker MA. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- Thompson D, Easton DF, Breast Cancer Linkage Consortium Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002a;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- Thompson D, Easton D, Breast Cancer Linkage Consortium Variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomarkers Prev. 2002b;11:329–336. [PubMed] [Google Scholar]
- Thorlacius S, Sigurdsson S, Bjarnadottir H, Olafsdottir G, Jonasson JG, Tryggvadottir L, Tulinius H, Eyfjörd JE. Study of a single BRCA2 mutation with high carrier frequency in a small population. Am J Hum Genet. 1997;60:1079–1084. [PMC free article] [PubMed] [Google Scholar]
- Thorne H, Willems AJ, Niedermayr E, Hoh IM, Li J, Clouston D, Mitchell G, Fox S, Hopper JL, Kathleen Cunningham Consortium for Research in Familial Breast Cancer Consortium. Bolton D. Decreased prostate cancer-specific survival of men with BRCA2 mutations from multiple breast cancer families. Cancer Prev Res (Phila) 2011;4:1002–1010. doi: 10.1158/1940-6207.CAPR-10-0397. [DOI] [PubMed] [Google Scholar]
- Warner E, Foulkes W, Goodwin P, Meschino W, Blondal J, Paterson C, Ozcelik H, Goss P, Allingham-Hawkins D, Hamel N, Di Prospero L, Contiga V, Serruya C, Klein M, Moslehi R, Honeyford J, Liede A, Glendon G, Brunet JS, Narod S. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst. 1999;91:1241–1247. doi: 10.1093/jnci/91.14.1241. [DOI] [PubMed] [Google Scholar]
- Willems AJ, Dawson SJ, Samaratunga H, De Luca A, Antill YC, Hopper JL, Thorne HJ, kConFab Investigators Loss of heterozygosity at the BRCA2 locus detected by multiplex ligation-dependent probe amplification is common in prostate cancers from men with a germline BRCA2 mutation. Clin Cancer Res. 2008;14:2953–2961. doi: 10.1158/1078-0432.CCR-07-5237. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanová E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Varon R, Seemanova E, Chrzanowska K, Hnateyko O, Piekutowska-Abramczuk D, Krajewska-Walasek M, Sykut-Cegielska J, Sperling K, Reis A. Clinical ascertainment of Nijmegen breakage syndrome (NBS) and prevalence of the major mutation, 657del5, in three Slav populations. Eur J Hum Genet. 2000;8:900–902. doi: 10.1038/sj.ejhg.5200554. [DOI] [PubMed] [Google Scholar]
- Zuhlke KA, Johnson AM, Okoth LA, Stoffel EM, Robbins CM, Tembe WA, Salinas CA, Zheng SL, Xu J, Carpten JD, Lange EM, Isaacs WB, Cooney KA.2012Identification of a novel NBN truncating mutation in a family with hereditary prostate cancer Fam Cancer; e-pub ahead of print 5 August 2012 [DOI] [PMC free article] [PubMed]