Abstract
Background:
There is relatively little methylation array data available specifically for oral squamous cell carcinoma (OSCC). This study aims to compare the DNA methylome across a large cohort of tumour/normal pairs.
Methods:
DNA was extracted from 44 OSCCs and paired normal mucosa. DNA methylation analysis employed the Illumina GoldenGate high-throughput array comprising 1505 CpG loci selected from 807 epigenetically regulated genes. This data was correlated with extracapsular spread (ECS), human papilloma virus (HPV) status, recurrence and 5-year survival.
Results:
Differential methylation levels of a number of genes distinguished the tumour tissue sample from the matched normal. Putative methylation signatures for ECS and recurrence were identified. The concept of concordant methylation or CpG island methylator phenotype (CIMP) in OSCC is supported by our data, with an association between ‘CIMP-high' and worse prognosis. Epigenetic deregulation of NOTCH4 signalling in OSCC was also observed, as part of a possible methylation signature for recurrence, with parallels to recently discovered NOTCH mutations in HNSCC. Differences in methylation in HPV-driven cases were seen, but are less significant than that has been recently proposed in other series.
Conclusion:
Although OSCC seems as much an ‘epigenetic' as a genetic disease, the translational potential of cancer epigenetics has yet to be fully exploited. This data points to the application of epigenetic biomarkers and targets available to further the development of therapy in OSCC.
Keywords: oral squamous cell carcinoma, DNA methylation, microarray, prognosis
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy with a worldwide incidence of 615 000 cases per annum, of which around 300 000 are oral squamous cell carcinomas (OSCC) (Kademani, 2007). The UK incidence of OSCC has recently increased, reaching over 6000 cases per annum (CR-UK press release, 2012). It is now apparent that cancer is as much a disease of misdirected epigenetics as genetic mutations, losses and amplifications, and recent published evidence suggests that as many as 5% of known gene promoters (i.e. 1500–2000 genes) are abnormally methylated in a typical solid tumour (Schuebel et al, 2007) compared with, typically, 11 gene mutations (Sjoblom et al, 2006). Next generation sequencing in HNSCC has shown that, although common in TP53 and NOTCH pathways, somatic mutations in specific genes are relatively heterogeneous, individually occurring in <10% of cases (Agrawal et al, 2011). Detection of methylation at known target CpG sequences can be made highly sensitive, opening up the possibility of the development of minimally invasive biomarkers in body fluid surrogates (Laird, 2003; Shaw et al, 2006a). However, genome-wide methylation platforms have lagged behind other genomic methods because bisulphite conversion necessitates an inherent loss of sequence complexity resulting in lack of hybridisation specificity. Consequently, the literature has concentrated on a rather narrow cohort (Ha and Califano, 2006; Shaw, 2006) of methylated gene promoters, and the translational potential of the broader DNA methylome in HNSCC remains relatively unexplored. The concept of a CpG island methylator phenotype (CIMP) (Toyota et al, 1999), although well explored in several tumour types, has yet to be investigated in genome wide or array approach in OSCC (Shaw et al, 2007).
More recently, high-throughput, genome-wide quantitative methylation platforms have become available, allowing a more comprehensive overview of DNA methylation (Bibikova et al, 2006; Bibikova and Fan, 2009). These arrays have been subject to technical validation, showing good correlation of quantitative methylation data with single gene assays (Bibikova et al, 2006). Reassuringly, available data shows that copy number alterations have little impact on methylation data in such arrays (Houseman et al, 2009). Although some studies have concentrated on the methylation signature in HSNCC cell lines, it is now appreciated that the culture conditions introduce significant epigenetic artefacts and clinically relevant data might only be available from primary tumour tissue or xenografts (Hennessey et al, 2011). It is known that the DNA methylome differs significantly across different HNSCC subsites (Poage et al, 2011), and that there may be differences between human papillomavirus (HPV)-related and unrelated cases (Sartor et al, 2011). In an attempt to control for these known sources of variation, it is reasonable to interrogate the clinical correlates of DNA methylation in a single subsite of HNSCC. Previous methylation array studies have made useful contributions but have aggregated HNSCC sites in their analyses (Poage et al, 2010, 2011). In the case of OSCC, subsequent analyses will largely exclude the influence of HPV (Lopes et al, 2011), which has been shown to be significant in only a minority of cases, although this should be confirmed experimentally. Extracapsular spread within cervical metastases (ECS) is the single most predictive adverse feature for recurrence and survival in OSCC (Shaw et al, 2010), being far more significant than metastasis per se. Although the mechanism underlying ECS is poorly understood, there is significant support for a contributory role of the stroma (Marsh et al, 2011), perhaps through recruited cancer associated fibroblasts (Quante et al, 2011). There is also evidence for epigenetic co-ordination of these paracrine tumour/stromal interactions (Jiang et al, 2008), thus it is logical to explore the correlation between ECS and DNA methylation. In this study, we interrogate methylation profiles of a series of OSCC, comparing matched tumour and normal paired tissues. We correlate significant tumour-specific changes with clinicopathological characteristics, in particular, recurrence and survival, opening directions for future biomarker and mechanistic studies in OSCC.
Materials and methods
Clinical
Forty four OSCC patients were selected for inclusion in this study, all of whom provided informed consent under a specific institutional ethical approval (REC EC.47.01), between May 2006 and March 2007. Inclusion criteria included histologically confirmed OSCC with a treatment decision for primary surgery. At the time of surgery, 5 mm3 tumour samples were excised from resected specimens within the tumour mass, but not involving the margin. Normal samples of similar dimension were taken from the resection margin, ∼10 mm from the macroscopic tumour edge and subsequently confirmed as benign by routine histopathology. Tumour and matched normal samples were snap-frozen and stored at − 80 °C until DNA preparation. Detailed histopathological analysis was recorded for each surgical resection and the clinical outcome for each patient was recorded at 5th year follow-up. The Office of National Statistics supplied death certification details comprising the immediate cause of death plus the associated factors.
Laboratory
DNA was extracted from 2 mm3 tissue samples using DNeasy tissue kit (Qiagen, Crawley, UK). DNA concentrationand quality was measured by spectrophotometry (Nanodrop; Thermo Fisher Scientific Inc., Wilmington, DE, USA). Bisulphite treatment of each sample was undertaken using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). An aliquot of the converted DNA (corresponding to 500 ng starting gDNA) was subjected to microarray analysis (GoldenGate array-Illumina Ltd, San Diego, CA, USA), (Bibikova and Fan, 2009) examining 1505 CpG sites on 807 genes. Raw methylation data and associated clinicopathological data have been deposited in ArrayExpress (accession: E-MTAB-1298).
HPV testing
As some of the tumours overlapped the junction between oral cavity and oropharynx sites (in particular tonsillar pillar and soft palate), it was felt appropriate to include HPV status to clarify the underlying molecular aetiology. Tumour HPV status was determined using quantitative PCR (qPCR) analysis for HPV16 DNA presence as previously described (Schache et al, 2011). Briefly, duplicate real-time PCR reactions were conducted using primers and a FAM-MGB-labelled Taqman probe designed and optimised to amplify the E6 region of HPV16. Commercially available primers and a VIC-TAMRA-labelled probe for the housekeeping gene RNase P were included in the multiplex reaction to standardise for DNA input. The HPV16-positive cervical cancer cell line, CaSki (U.K. Health Protection Agency Culture Collections – 87020501) was used as a positive control and calibrator. The detection threshold for HPV-positive status was set at one or more copies of HPV16 E6 gene per diploid genome. A sample was only considered positive if it met the threshold in both of the duplicate runs.
Bioinformatics analysis
The array data were initially assessed for quality. One tumour sample found as an outlier in principal components analysis (PCA), together with the matching normal sample were excluded from further analysis. Initial processing of the data was conducted using the bioconductor package methylumi (Davis et al., 2012). All 84 CpG sites on the X-chromosome were excluded from data analysis to avoid gender-specific bias.
R statistical package version 2.14.1 (R, 2012) was employed to perform Wilcoxon signed-rank and Wilcoxon rank sum tests. Hierarchical clustering, PCA and survival analysis were performed using Partek Genomic Suite (Partek Inc., St Loius, MO, USA). Hypergeometric analysis of pathways enriched by the differential genes was conducted using MetaCore (GeneGo Inc, Part of Thomson Reuters, Carlsbad, CA, USA). Genes represented on the Illumina GoldenGate Cancer Panel I methylation array were used as reference in the pathway analysis. In statistical analyses, a 5% FDR cutoff was employed after multiple testing correction of P-values.
Methylation in tumour/normal paired tissue
When more than half of the samples (at least 22/43) from a group had average beta values >0.2, the CpG locus was considered methylated in that group. The Wilcoxon signed-rank test was employed to identify statistically significant differential methylation between tumour and matching normal samples using R statistical package. M-value, that is, the log2 ratio of intensities of methylated probe versus unmethylated probe, was employed for Wilcoxon rank sum or signed-rank tests, as it was recently reported that β values have severe heteroscedasticity for highly methylated or unmethylated CpG sites and M-value is a better measure for statistical analyses (Du et al, 2010). However, as β values provide more intuitive biological interpretation, differences in methylation levels were derived using average β values, representing the ratio of methylated probe intensity and overall intensity, that is, the sum of methylated and unmethylated probe intensities. An offset of 100 was added to regularise β when both probe intensities were low. Thus, for each CpG locus, differential methylation values (Δ β) were calculated by subtracting the average β values of tumour samples from the average β values of the normal samples.
Comparison was made with previously published HNSCC methylation data (Poage et al, 2010; Guerrero-Preston et al, 2011) as well as against a metaanalysis of 63 previous publications reporting mRNA expression microarray data in HNSCC (Yu et al, 2008). Limiting inclusion criteria to ⩾20 tumour/normal pairs, this analysis was restricted to four datasets published by Ye (Ye et al, 2008), Dysvik (Dysvik et al, 2006), Kuriakose (Kuriakose et al, 2004) and Ibrahim (Ibrahim et al, 2003).
Hierarchical agglomerative clustering of tumour samples was performed with Euclidean distance measure and average linkage using average β values of probes selected for differential methylation between tumour and normal samples. Tumours identified by this method as having the CpG island methylator phenotype (CIMP) were validated using Rand Index.
Tumour methylation
Methylation data from tumour samples were normalised for sequence length and GC content, and important probes were selected using LumiWCluster package, thus eliminating arbitrary detection P-value cutoffs in removing unreliable methylation values (Kuan et al, 2010). Wilcoxon rank sum test was employed in identifying statistically significant differences in methylation between samples with and without extracapsular spread (ECS); human papilloma virus (HPV)-induced tumour and HPV negative tumours; with and without lymph node metastasis.
For the generation of a methylation signature associated with recurrence, univariate Cox regression analysis was performed with methylation as predictor and freedom from recurrence (FFR) as response. Freedom from recurrence after resection was calculated from the date of surgery to the date of recurrence, with censoring of all other patients. Significant methylation markers associated with recurrence were selected by filtering on P-value <0.01. Multivariate Cox regression was also performed with additional covariates including age, ECS and HPV status. A composite marker risk score was calculated for each patient from the linear combination of the methylation values weighted by the Cox regression coefficient for each selected marker. To quantify the predictive accuracy of the composite marker, time-dependent ROC curve at 5th year was generated. Estimation of time-dependent sensitivity and specificity based on censored outcome was performed using nearest neighbours algorithm. The area under the time-dependent ROC curve (AUC) at 5th year was calculated to quantify the predictive accuracy of the methylation signature. To assess the uncertainty in the predictive accuracy of the marker, using 10 000 bootstrap replicates and the bootstrap percentile method, 95% confidence intervals (CI) of the AUC were calculated. R packages survivalROC (Heagerty and Saha, 2012) and boot (Canty, 2002) were employed for the analysis.
Results
Summary of cohort characteristics
The cohort comprised 29 males (66%) and 15 females (34%) with an age range of 29–86 years (median 60 years) at diagnosis. Clinicopathological characteristics of the cohort were similar to our larger OSCC series, 1992–2001 (Table 1), although it is clear that this cohort has higher pT and pN stage with more involved margins and involved nodes (Rogers et al, 2009). Although this bias is regrettable, it is principally related to the inclusion of fewer very small tumours (14% pT1 here, vs 25% in larger series). This reflects the understandable reticence and ethical dilemma in sacrificing the majority of very small tumour for research purposes rather than for pathological staging. Treatment of tumours was primary surgery in all cases and post-operative radiotherapy/chemoradiotherapy was given in 31 (74%) reflecting, again the rather advanced clinical stages related to the exclusion of the smallest tumours (Table 1). At the end of study, 19 (43%) of the patients had died, 11 (25%) of OSCC, 6 (14%) of other causes and 2 (5%) unknown. Amongst the 25 (57%) survivors, median follow-up data was 52 months and in all but two cases, the follow-up data was ⩾43 months. Twelve (27%) cases had histologically proven recurrence of OSCC.
Table 1. Clinicopathological characteristics of the patient samples employed in the present study compared with our previous study.
| This study ( n =44) | Rogers et al (n=489) | |
|---|---|---|
|
Gender
| ||
| Male | 29 (66) | 302 (62%) |
| Female |
15 (34) |
187 (38%) |
|
Tumour differentiation
| ||
| Poor | 3 (7) | 49 (10%) |
| Moderate | 27 (66) | 286 (60%) |
| Well |
11 (27) |
139 (29%) |
|
Deep margins
| ||
| Clear ⩾5 mm | 10 (24)** | 237 (48%) |
| Close<5 mm | 18 (44) | 170 (35%) |
| Involved |
13 (32) |
82 (17%) |
|
pT
| ||
| Tis, T1 | 6 (14)* | 134 (27%) |
| T2 | 22 (50) | 162 (33%) |
| T3 | 5 (11) | 30 (6%) |
| T4 |
11 (25) |
163 (33%) |
|
pN
| ||
| 0 | 19 (43)** | 310 (63%) |
| 1 | 6 (14) | 72 (15%) |
| 2+ |
19 (43) |
107 (22%) |
|
Perineural/perivascular invasion:
| ||
| Yes | 15 (37) | 125 (26%) |
| No |
26 (63) |
364 (74%) |
|
ECS:
| ||
| Yes | 16 (36) | 101 (21%) |
| No |
28 (64) |
388 (79%) |
|
Nodal status:
| ||
| N0 | 19 (43)* | 310 (63%) |
| N+ECS– | 9 (20) | 78 (16%) |
| N+ECS+ |
16 (36) |
101 (21%) |
|
Adjuvant radiotherapy
| ||
| Yes | 31 (74)*** | 194 (40%) |
| No |
11 (26) |
295 (60%) |
|
Recurrence
| ||
| Yes | 12 (29) | 120 (25%) |
| No | 29 (71) | 369 (75%) |
The Statistical Package for the Social Sciences (SPSS, v 18, Chicago) was used to undertake χ2 or Fisher's exact test. *P<0.05; **P<0.005; ***P<0.0001.
Promoter methylation analysis in tumour vs normal
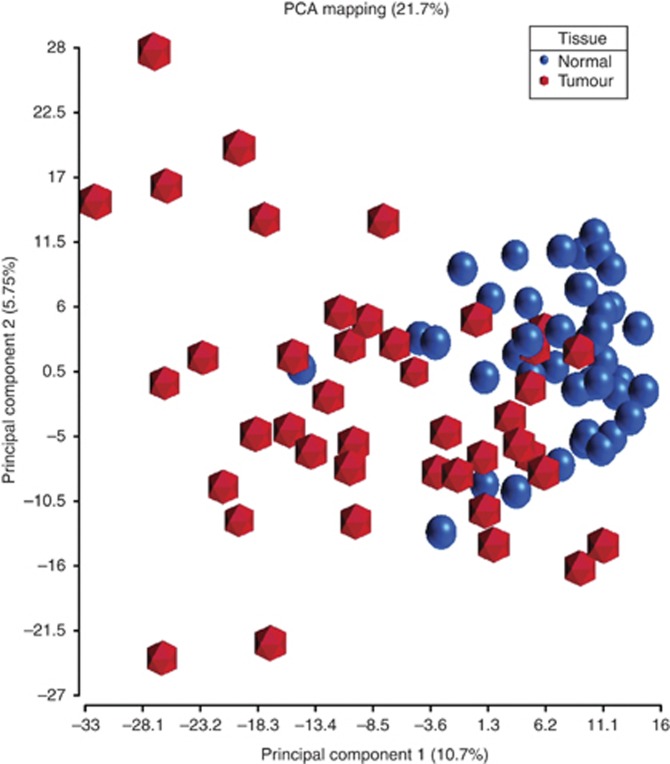
Tumour and matched normal tissues from 43 OSCC patients were analysed in this study. Unsupervised principal components analysis identified two clusters along the first principal component (accounting for 10.7% of variability in the data) that separated tumour from normal samples, with a few samples misplaced (Figure 1). There was a tighter clustering of normal samples compared with tumour, which may reflect the expected greater biological heterogeneity in the tumour samples compared with the paired oral mucosal samples. A number of markers demonstrated significantly different methylation levels between tumour and matched normal samples in Wilcoxon signed-rank test. Forty-eight probes were identified as differentially methylated when a corrected P-value (FDR) smaller than 0.05 and difference in average β values (Δβ) >+0.2 or <−0.2 were used as filtration criteria. Of these, 32 probes were hypermethylated in tumour compared with normal tissue, whereas 16 were hypomethylated in tumour (Supplementary Tables 1 and 2).
Figure 1.
Principal components analysis showing the distinction between tumour and normal samples based on methylation. Average β values from 43 tumour and matched normal samples were employed in the analysis. Separation between tumour (hexagon) and normal (sphere) samples can be visualised in the plot along the first principal component, with a few misplaced samples.
Comparison with published HNSCC methylation and mRNA expression microarray data
A comparison with two previously published HNSCC methylation array data demonstrated a number of genes differentially methylated between tumour and normal in common with our study, all showing concordant methylation status (Table 2). When the top 25 methylation markers from Poage et al (2011) were compared with our data, there were 11 common in the list. A total of 7 out of 10 hypermethylated and 4 out of 15 hypomethyalted in that study were common with our list. In comparison with Guerrero-Preston et al (2011), 8 genes were common in the differentially hypermethylated lists. A comparison of gene promoter methylation with gene expression presented in the selected microarray datasets (Ibrahim et al, 2003; Kuriakose et al, 2004; Dysvik et al, 2006; Ye et al, 2008) did not identify any genes in common with our list demonstrating greater promoter methylation in tumour. However, 5 differentially hypomethylated genes were found to be differentially expressed between tumour and normal in either all or some of the four datasets analysed, with 4 of the hypomethylated genes (SPP1, AIM2, PTHLH and IFNG) upregulated in tumour (Table 2).
Table 2. Genes showing differential methylation between tumour and normal in the present study, which are in common with other methylation or gene expression studies compared.
| Gene | Methylation status in present study | Methylation status in Poage et al | Methylation status in Guerrero-Preston et al | Expression status in Ye, Dysvik, Kuriakose or Ibrahim |
|---|---|---|---|---|
|
HOXA9
|
Hyper |
Hyper |
Hyper |
— |
|
HS3ST2
|
Hyper |
Hyper |
Hyper |
— |
|
NPY
|
Hyper |
Hyper |
Hyper |
— |
|
HTR1B
|
Hyper |
Hyper |
— |
— |
|
SOX17
|
Hyper |
Hyper |
— |
— |
|
MME
|
Hyper |
Hyper |
— |
— |
|
EPHA5
|
Hyper |
Hyper |
— |
— |
|
CALCA
|
Hyper |
— |
Hyper |
— |
|
EYA4
|
Hyper |
— |
Hyper |
— |
|
HOXA11
|
Hyper |
— |
Hyper |
— |
|
TAL1
|
Hyper |
— |
Hyper |
— |
|
WT1
|
Hyper |
— |
Hyper |
— |
|
EMR3
|
Hypo |
Hypo |
— |
— |
|
PI3
|
Hypo |
Hypo |
— |
— |
|
SPP1
|
Hypo |
Hypo |
— |
High (All) |
|
AIM2
|
Hypo |
Hypo |
— |
High (Ye) |
|
PTHLH
|
Hypo |
— |
— |
High (Ye, Kuriakose) |
|
IFNG
|
Hypo |
— |
— |
High (ibrahim) |
| CEACAM1 | Hypo | — | — | Low (Ye, Kuriakose) |
Methylation status from an Illumina GoldenGate study of 19 HNSCC tumours vs 11 non-malignant samples (Poage et al, 2010) and an Illumina HumanMethylation27 array study of 4 OSCC tissue versus 4 normal tissue (Guerrero-Preston et al, 2011) are presented. Comparison with gene expression differences in four HNSCC studies with ⩾20 tumour/ normal pairs (Ye (Ye et al, 2008), Dysvik (Dysvik et al, 2006), Kuriakose (Kuriakose et al, 2004) and Ibrahim (Ibrahim et al, 2003)) is also presented. Complete list of methylation markers significantly differential between tumour and normal in our study are presented in Supplementary Tables 1 and 2.
Pathway analysis
Hypergeometric analysis of pathways and processes altered due to aberrant promoter methylation in tumour identified the role of VDR in transcriptional regulation of genes involved in osteoporosis (P=0.03) as a significant canonical pathway along with glutathione metabolism (P=0.04). On a process network level, a network representing response to hypoxia and oxidative stress (P=0.03) was significantly enriched. However, since the pathway and network analysis was conducted on a limited set of genes selected, interpretation of the results needs caution.
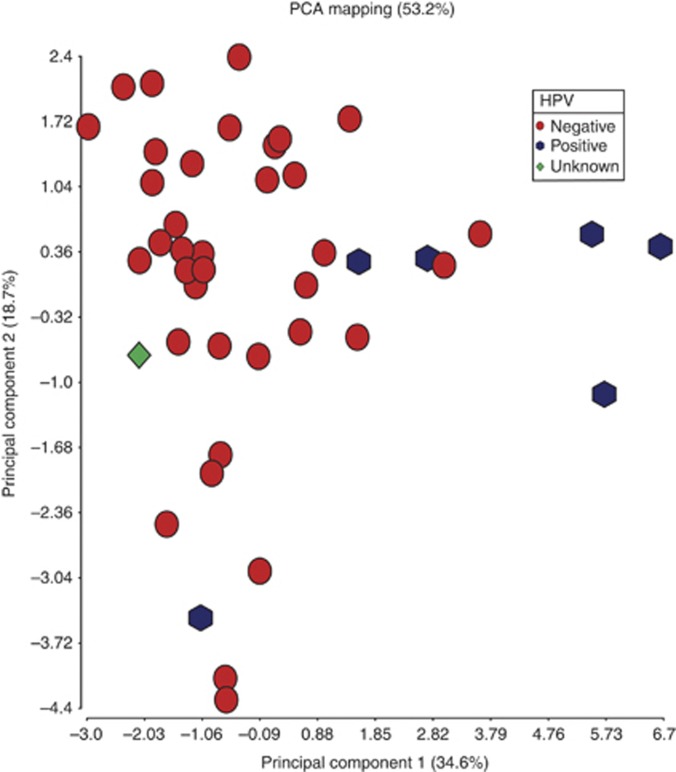
HPV status
HPV-associated HNSCC has distinct demographic, biology and prognosis (Schache et al, 2011; Shaw and Robinson, 2011), and there is growing evidence that the DNA methylation regulatory pathways are targets for E6 and E7 oncogenes (Burgers et al, 2007). Accordingly, differences in promoter methylation between HPV(+) and HPV(−) tumours were sought. Genome-wide methylation differences between these two classes were previously noted in squamous cell carcinoma cell lines (Sartor et al, 2011). In our study, PCA and unsupervised hierarchical clustering were capable of distinguishing the two classes based on methylation data in tumour samples, though to a lesser extent (Figure 2), although this may be slightly underpowered as only six samples were HPV-16 DNA positive (13%). However, Wilcoxon rank sum test identified nine probes with higher methylation levels in the HPV(+) and five probes with higher methylation levels in HPV(−) tumours (P<0.05, Δβ >|0.2|). Genes methylated at a higher level in HPV(+) were ALOX12, CCNA1, CTSL1, DAPK1, F2R, FLT1, CADM1, PDX1 and SEZ6L whereas FLJ20712, HIC1, MT1A, OSM and PDGFB were methylated more in HPV(−) samples. It is of note that CCNA1 and CTSL1 were also identified in the previous cell line study as highly methylated and with lower expression in HPV(+) compared with HPV(−) (Sartor et al, 2011).
Figure 2.
Principal components analysis showing HPV(+) (hexagon) and HPV(−) (sphere) tumour samples. Average β values from 43 tumour samples were employed in the analysis. One sample with unknown HPV status is also displayed.
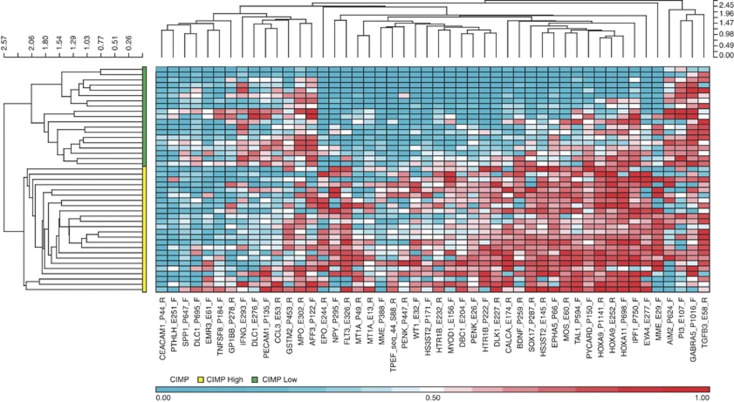
CpG island methylator phenotype (CIMP)
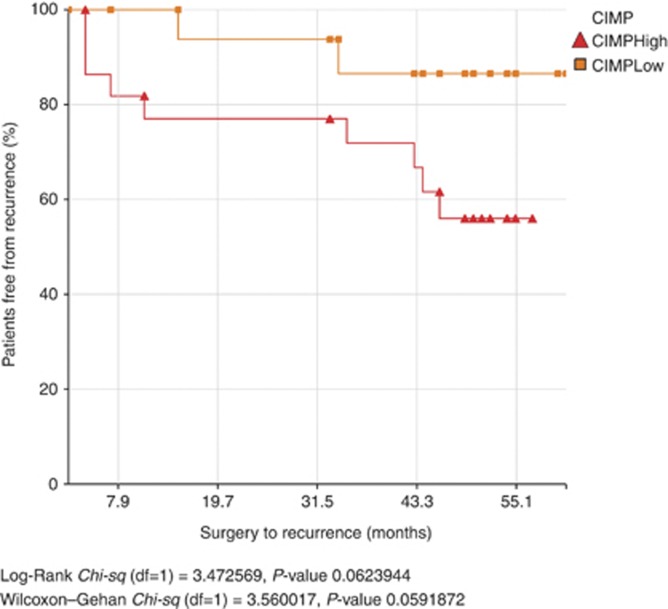
CIMP denotes the concordant tumour-specific DNA methylation observed in subgroup of tumours, and is associated with distinct clinicopathological characteristics. When the 48 differentially methylated promoters in this cohort were clustered based on average β values using hierarchical agglomerative clustering, two visibly distinct clusters were identified. One cluster comprising 19 tumour samples had mostly low β values (median=0.3) and was designated as ‘CIMP-low'. The other cluster with 24 tumour samples had mostly high β values (median=0.56) and was designated as ‘CIMP-high' (Figure 3). A PCA plot of average β values from the 48 markers demonstrated a visible distinction between CIMP-high and CIMP-low groups of tumour samples along the first principal component (Supplementary Figure 1). External cluster validation also showed good agreement between clustering and the CIMP classes (Rand Index: 0.75). A Kaplan–Meier curve using freedom from recurrence (FFR) as endpoint demonstrated a statistically non-significant difference between the CIMP groups (Figure 4), with a trend towards worse prognosis in patients belonging to the CIMP-high category (Log-rank P=0.06). This is in contrast to our previous observation based on only 10 selected gene promoters (Shaw et al, 2007). However, CIMP was not an independent factor determining survival (P=0.3) when a multivariate Cox proportional hazards regression was conducted with other covariates such as ECS and HPV status.
Figure 3.
Hierarchical clustering heatmap and dendrogram of tumour samples based on methylation markers differential between tumour and normal tissues. Average β values from the selected 48 probes were used for hierarchical agglomerative clustering using Euclidean distance and average linkage. CpG island methylator phenotype (CIMP) groups are shown by the two clusters on the dendrogram left of the heatmap, the bottom cluster designated as ‘CIMP-high' and the top as ‘CIMP-low'.
Figure 4.
Kaplan–Meier plot of freedom from recurrence (FFR) showing a trend towards worse prognosis by patients in the CIMP-high group. FFR in CIMP-high and CIMP-low groups were not significantly different in log-rank test (P=0.06).
Methylation signature for extracapsular spread (ECS)
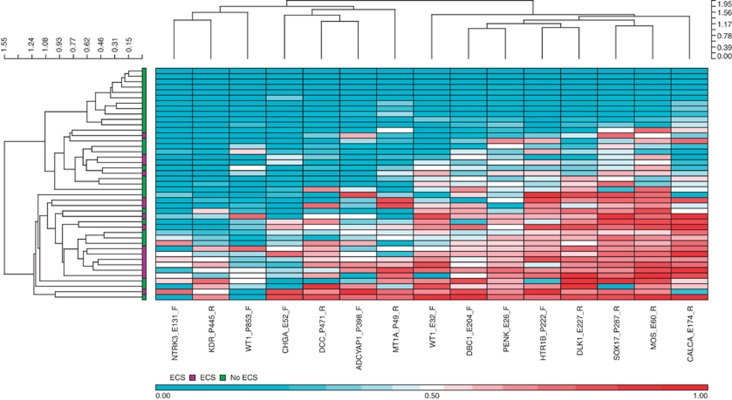
Wilcoxon rank sum test identified 15 probes differentially methylated in patients with extracapsular spread when compared with patients without ECS (P<0.05, Delta-beta >|0.2|); Table 3).Two distinct clusters were observed in hierarchical clustering of the samples based on the markers selected (Figure 5), one containing samples from patients without ECS exclusively and the other a mix of both ECS and non-ECS samples. Genes differentially methylated between ECS and non-ECS patient samples included DCC, CALCA and WT1.
Table 3. Methylation markers differential between patients with and without extracapsular spread (ECS).
| Gene | Loci of methylation | Strand | Δβ | P -value (FDR) |
|---|---|---|---|---|
|
ADCYAP1
|
P398 |
F |
0.28 |
0.008 |
|
CALCA
|
E174 |
R |
0.33 |
0.016 |
|
CHGA
|
E52 |
F |
0.24 |
0.025 |
|
DBC1
|
E204 |
F |
0.21 |
0.021 |
|
DCC
|
P471 |
R |
0.34 |
0.008 |
|
DLK1
|
E227 |
R |
0.34 |
0.023 |
|
HTR1B
|
P222 |
F |
0.31 |
0.001 |
|
KDR
|
P445 |
R |
0.29 |
0.042 |
|
MOS
|
E60 |
R |
0.31 |
0.005 |
|
MT1A
|
P49 |
R |
0.30 |
0.003 |
|
NTRK3
|
E131 |
F |
0.21 |
0.039 |
|
PENK
|
E26 |
F |
0.21 |
0.011 |
|
SOX17
|
P287 |
R |
0.29 |
0.003 |
|
WT1
|
E32 |
F |
0.38 |
0.010 |
| WT1 | P853 | F | 0.24 | 0.004 |
P-values from Wilcoxon rank sum test were corrected for multiple testing (FDR) and differences in average beta values (Δβ) between the two groups are presented alongwith the details of methylation probes. Only differentially hypermethylated probes in patients with ECS passed the filtration criteria (Δβ >0.2 and FDR<0.05)
Figure 5.
Hierarchical clustering heatmap and dendrogram of tumour samples based on methylation markers differential between patients with and without extracapsular spread. Average β values from the selected 15 probes were used for hierarchical agglomerative clustering using Euclidean distance and average linkage.
Methylation signature of recurrence
Cox proportional hazards regression with freedom from recurrence (FFR) identified a 17-gene signature associated with recurrence (P<0.01; Table 4). A multivariate Cox regression including covariates age, ECS and HPV status also found the signature genes to be statistically significant (P<0.05) except MT1A (0.068) and ATP10A (0.057). When a risk score generated from linear combination of the marker values weighted with regression coefficients was assessed using time-dependent ROC analysis with nearest neighbours method for censored outcome data, a good concordance of the risk grouping was observed with recurrence (AUC of time-dependent ROC at 5th year: 95.7%, 95%CI: 88.07–99.92). With a P-value cutoff of 0.01, the multivariate Cox regression analysis identified a 5-gene signature comprising MLLT6_P957_F, MSH2_P1008_F, NOTCH4_E4_F, PCGF4_P92_R and TFPI2_E141_F. This signature also was capable and demonstrated good performance (AUC of time-dependent ROC at 5th year: 92.2%, 95%CI: 81.4–99.3). Out of the five genes, three were the same as identified from univariate analysis: MLLT6, MSH2 and NOTCH4. Predictive accuracy of the 3-gene model was also high, though the uncertainty associated with the prediction was also high (AUC of time-dependent ROC at 5th year: 92.8%, 95%CI: 80.3–99.3).
Table 4. Methylation marker signature of recurrence (FFR) derived from univariate Cox regression.
| |
Univariate
|
Multivariate
|
||||||
|---|---|---|---|---|---|---|---|---|
| Probe | HR | LowCI | UpCI | P -value | HR | LowCI | UpCI | P -value |
|
NOTCH4_E4_F
|
2.45 |
1.44 |
4.17 |
0.001 |
2.48 |
1.28 |
4.79 |
0.007 |
| IRAK3_P13_F |
1.41 |
1.15 |
1.74 |
0.001 |
1.31 |
1.05 |
1.64 |
0.016 |
|
MLLT6_P957_F
|
3.56 |
1.58 |
8.04 |
0.002 |
3.50 |
1.47 |
8.34 |
0.005 |
| MT1A_P600_F |
2.04 |
1.27 |
3.30 |
0.003 |
1.70 |
0.96 |
3.00 |
0.068 |
| CD9_P585_R |
3.27 |
1.45 |
7.37 |
0.004 |
3.14 |
1.23 |
8.02 |
0.017 |
|
MSH2_P1008_F
|
7.66 |
1.84 |
31.94 |
0.005 |
9.46 |
1.78 |
50.38 |
0.008 |
| ASCL1_E24_F |
1.45 |
1.12 |
1.89 |
0.006 |
1.48 |
1.05 |
2.11 |
0.027 |
| SLIT2_E111_R |
1.54 |
1.13 |
2.11 |
0.006 |
1.64 |
1.08 |
2.49 |
0.020 |
| TBX1_P885_R |
1.52 |
1.13 |
2.06 |
0.006 |
1.40 |
1.04 |
1.89 |
0.026 |
| ESR2_E66_F |
1.41 |
1.10 |
1.81 |
0.006 |
1.29 |
1.00 |
1.67 |
0.046 |
| MYH11_P22_F |
1.51 |
1.12 |
2.03 |
0.006 |
1.43 |
1.04 |
1.97 |
0.029 |
| TNFRSF10C_P7_F |
1.72 |
1.16 |
2.55 |
0.007 |
1.62 |
1.08 |
2.43 |
0.020 |
| POMC_P400_R |
1.42 |
1.10 |
1.84 |
0.008 |
1.38 |
1.03 |
1.83 |
0.029 |
| NES_P239_R |
1.75 |
1.16 |
2.65 |
0.008 |
1.53 |
1.04 |
2.23 |
0.029 |
| GJB2_P791_R |
1.71 |
1.15 |
2.55 |
0.008 |
1.65 |
1.10 |
2.47 |
0.016 |
| ATP10A_P147_F |
2.01 |
1.19 |
3.39 |
0.009 |
1.68 |
0.98 |
2.87 |
0.057 |
| FRZB_E186_R | 1.45 | 1.09 | 1.91 | 0.009 | 1.33 | 1.02 | 1.74 | 0.036 |
Multivariate Cox regression was also performed with additional co-variates including age, ECS and HPV status. Hazard ratios (HR), lower (LowCI) and upper (UpCI) confidence intervals as well as P-values for the selected probes from univariate and multivariate Cox regression are displayed. Probes displayed in bold had P<0.01 in multivariate Cox regression as well.
Discussion
In this study, we explored the epigenetic landscape of series of oral squamous cell carcinomas (OSCC) using an array approach. Significant difference in methylation levels of a number of genes distinguished tumour tissue samples from their matched normal, demonstrating epigenetic modifications acquired in the development of tumours. It was observed that a number of these differentially methylated genes in tumours were also identified in other recent investigations in HNSCC (Poage et al, 2010; Guerrero-Preston et al, 2011), with three genes in common with both these studies: HOXA9, HS3ST2 and NPY. In addition, EYA4 and WT1 were also among the list of genes selected for methylation in oral cancer tissue and pre-operative saliva in a study using similar array as in the present study (Viet and Schmidt, 2008). It is notable that all these common genes had consistently concordant methylation status, even though the studies differed in terms of heterogeneity of tumours, experimental design or methylation array platform employed. Methylation of one such gene, HOXA9 that codes for a homeobox protein, has been suggested as a biomarker for early detection of OSCC (Guerrero-Preston et al, 2011). In addition to HNSCC, methylation-associated silencing of the HS3ST2 gene has been observed in human breast, colon, lung and pancreatic cancers (Miyamoto et al, 2003) as well as in haematological neoplasms (Martin-Subero et al, 2009). Interestingly, there was no correlation between those gene promoter methylation targets and commonly downregulated genes as noted in the publicly available datasets. This may be explained by the multiplicity of molecular mechanisms regulating gene expression, only one of which is DNA methylation. Similarly, a metaanalysis of gene expression data from OSCC samples with matched normal samples, identified commonality with our list of 16 genes differentially hypomethylated between tumour and normal tissues. Among the genes showing lower levels of methylation and higher levels of expression in tumour was SPP1, which codes for osteopontin. This secreted glycoprotein has an important role in determining the oncogenic potential of many cancers and its increased expression is reported to correlate with tumour progression and metastasis (Senger et al, 1983; Brown et al, 1994; Sodek et al, 2000; Weber, 2001).
The identified putative methylation signatures for ECS and recurrence in this study require meticulous confirmation in an independent cohort, however, they may offer the promise of translation into diagnostic and prognostic biomarkers in tumour and surrogate tissue. In particular, epigenetic deregulation of NOTCH signalling in OSCC has been demonstrated as part of a methylation signature for recurrence. This complements the recent discovery of common NOTCH1, NOTCH2 and NOTCH3 mutations in HNSCC (Agrawal et al, 2011) using next generation sequencing. The NOTCH family of genes code for evolutionarily conserved intercellular signalling pathways which regulating cell–cell interactions. Further functional exploration of this pathway in HNSCC and identification of druggable targets are self-evidently a priority in translation of these findings to the clinic. Genes differentially methylated between ECS and non-ECS patient samples included DCC, CALCA and WT1. Interestingly the role of DCC as an epigenetically regulated tumour suppressor gene in HNSCC has already been documented in detail (Carvalho et al, 2006; Guerrero-preston et al, 2011). WT1 promoter methylation has subsequently been proven in our own group to be a clinically valuable biomarker in lung cancer screening (Nikolaidis et al, 2012) and has also been identified similarly as a biomarker for cervical cancer (Lai et al, 2008). CALCA promoter methylation has recently been established as an independent adverse prognostic factor in leukaemia (Ismail et al, 2011).
Tissue processing with macromolecule extraction was performed on whole biopsy tissue rather than on microdissected tissue and the composition of both the epithelial and the stromal compartments was unknown. Epithelial or stromal specific methylation patterns have been observed previously in both breast and prostate and so, a more compartment-specific approach may provide additional insights than a whole-tissue approach (Rodriguez-Canales et al, 2007; Qiu et al, 2008). The findings in particular of NOTCH involvement in this mixed cell population of undefined neoplastic cellularity could have greater significance if the epithelial component alone was utilised. The relevance of the epigenome from mixed whole tissue may however be more likely to reflect in vivo tumour phenotypes as these are reliant upon epithelial and mesenchymal interactions.
One of the aims of this study was to explore the occurrence of tumour subgroups with concordant methylation levels at multiple loci, known as CpG island methylator phenotype (CIMP) (Toyota et al, 1999). A wide range of genetic and environmental factors may lead to CIMP, including the increased expression and abnormal targeting of DNA methyltransferases (DNMTs) (Etoh et al, 2004; Kim et al, 2006; Teodoridis et al, 2008). We observed distinct clustering of tumour samples into CIMP-high and CIMP-low groups with genes differentially methylated between tumour and normal tissues. We have previously investigated the association of CIMP with allelic imbalance, pathology and clinical behaviour of OSCC (Shaw et al, 2007). In that study, CIMP was shown to correlate with a marked inflammatory response and less aggressive tumour biology (Shaw et al, 2007). However, in the present study, we observed a trend for CIMP-high tumours to be associated with poor prognosis, though CIMP was not an independent factor in predicting prognosis. This difference in observation may stem from the difference in technology and markers used, as in the previous study we had used only 10 genes involved in a variety of cellular processes. Although the current observation of CIMP-high tumours presenting poor survival, requires external validation with other HNSCC or OSCC series, it is in line with many other studies in various cancers (Maruyama et al, 2001; Wei et al, 2002; Brock et al, 2003; Roman-Gomez et al, 2004; Abe et al, 2005).
Although the epigenetic signature associated with HPV-driven HNSCC has been recently highlighted (Richards et al, 2009; Sartor et al, 2011), our data offers a somewhat less convincing distinction in the DNA methylome between HPV(+) and HPV(−) cases than these reports may imply. However, it has to be noted that we had only very few samples from HPV(+) patients. We have recently documented that many HPV (+) cases in our clinical population are also smokers (Schache et al, 2011), and although this seems to have little impact on prognostic benefit, this may add genotoxic stress and blur the lines drawn between smoking-related and HPV(+) cases. Notably, the CCNA1 promoter methylation seen in HPV(+) cases reported by Sartor (Sartor et al, 2011), is confirmed in our data, whereas epigenetic downregulation of this gene has been shown to be previously common in OSCC by pyrosequencing methylation assay (Shaw et al, 2006b, 2008). The differences in DNA methylome and underlying perturbations of epigenetic regulatory cellular machinery await further confirmation, elucidation and possible translational application.
We present methylation array data for a significant cohort of OSCC cases, incorporating detailed pathology and long-term clinical outcomes in keeping with our previous approach (Rogers et al, 2009). The benefits of using carefully curated snap-frozen tissue from paired tumour and normal tissues are self-evident in undertaking this work. Although comprehensive validation has not been attempted using single gene methylation assays, the reliability of this array in similar settings has been previously demonstrated (Bibikova et al, 2006; Bibikova and Fan, 2009). Additionally, comparison with expression array datasets (Yu et al, 2008) and the limited methylation array data available in OSCC (Viet and Schmidt, 2008; Poage et al, 2010; Guerrero-Preston et al, 2011) has been carried out, with confirmation of a number of targets. Although the spectrum of OSCC presented here is skewed towards larger tumours where tissue procurement is safely achieved, in other respects, this cohort represents a consecutive series treated uniformly according to written and audited protocols. The tissue procurement is associated with an academic tertiary centre responsible for treatment of all OSCC cases from a discrete geographical population of 2.5 million.
It is evident that epigenetic regulation features as a major mechanism in the carcinogenesis of OSCC and that several pathways appear to be specifically epigenetically deregulated in the cases showing greatest biological aggression that often prove refractory to aggressive conventional therapies. The potential to target epigenetics with novel agents seems a rational approach. Additionally, the translational capacity of epigenetic biomarker panels may be clinically applicable in screening, diagnosis, prediction of response and prognostication.
Acknowledgments
PVJ is supported by Cancer Research UK.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abe M, Ohira M, Kaneda A, Yagi Y, Yamamoto S, Kitano Y, Takato T, Nakagawara A, Ushijima T. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65 (3:828–834. [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333 (6046:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, Goldmann T, Seifart C, Jiang W, Barker DL, Chee MS, Floros J, Fan JB. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16 (3:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MV, Gou M, Akiyama Y, Muller A, Wu TT, Montgomery E, Deasel M, Germonpre P, Rubinson L, Heitmiller RF, Yang SC, Forastiere AA, Baylin SB, Herman JG. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin Cancer Res. 2003;9 (8:2912–2919. [PubMed] [Google Scholar]
- Brown LF, Papadopoulos-Sergiou A, Berse B, Manseau EJ, Tognazzi K, Perruzzi CA, Dvorak HF, Senger DR. Osteopontin expression and distribution in human carcinomas. Am J Pathol. 1994;145 (3:610–623. [PMC free article] [PubMed] [Google Scholar]
- Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26 (11:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AL, Chuang A, Jiang WW, Lee J, Begum S, Poeta L, Zhao M, Jeronimo C, Henrique R, Nayak CS, Park HL, Brait MR, Liu C, Zhou S, Koch W, Fazio VM, Ratovitski E, Trink B, Westra W, Sidransky D, Moon CS, Califano JA. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66 (19:9401–9407. doi: 10.1158/0008-5472.CAN-06-1073. [DOI] [PubMed] [Google Scholar]
- Canty A.2002Resampling methods in R: the boot package R News 2(32–7. . http://home.jesus.ox.ac.uk/∼clifford/CIS/AngeloCanty.boot.pdf [Google Scholar]
- Davis S, Bilke S, Triche T, Bootwalla M.2012Methylumi: Handle Illumina methylation dataR package version 2.0.13
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dysvik B, Vasstrand EN, Lovlie R, Elgindi OA, Kross KW, Aarstad HJ, Johannessen AC, Jonassen I, Ibrahim SO. Gene expression profiles of head and neck carcinomas from Sudanese and Norwegian patients reveal common biological pathways regardless of race and lifestyle. Clin Cancer Res. 2006;12 (4:1109–1120. doi: 10.1158/1078-0432.CCR-05-0115. [DOI] [PubMed] [Google Scholar]
- Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, Kitano S, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164 (2:689–699. doi: 10.1016/S0002-9440(10)63156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Preston R, Soudry E, Acero J, Orera M, Moreno-Lopez L, Macia-Colon G, Jaffe A, Berdasco M, Ili-Gangas C, Brebi-Mieville P, Fu Y, Engstrom C, Irizarry RA, Esteller M, Westra W, Koch W, Califano J, Sidransky D. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res. 2011;4 (7:1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha PK, Califano JA. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7 (1:77–82. doi: 10.1016/S1470-2045(05)70540-4. [DOI] [PubMed] [Google Scholar]
- Heagerty PJ, Saha P.2012SurvivalROC: Time-dependent ROC curve estimation from censored survival dataR package v. 1.0.0
- Hennessey PT, Ochs MF, Mydlarz WW, Hsueh W, Cope L, Yu W, Califano JA. Promoter methylation in head and neck squamous cell carcinoma cell lines is significantly different than methylation in primary tumors and xenografts. PLoS One. 2011;6 (5:e20584. doi: 10.1371/journal.pone.0020584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Christensen BC, Karagas MR, Wrensch MR, Nelson HH, Wiemels JL, Zheng S, Wiencke JK, Kelsey KT, Marsit CJ. Copy number variation has little impact on bead-array-based measures of DNA methylation. Bioinformatics. 2009;25 (16:1999–2005. doi: 10.1093/bioinformatics/btp364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim SO, Aarsaether N, Holsve MK, Kross KW, Heimdal JH, Aarstad JH, Liavaag PG, Elgindi OA, Johannessen AC, Lillehaug JR, Vasstrand EN. Gene expression profile in oral squamous cell carcinomas and matching normal oral mucosal tissues from black Africans and white Caucasians: the case of the Sudan vs. Norway. Oral Oncol. 2003;39 (1:37–48. doi: 10.1016/s1368-8375(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Ismail EA, El-Mogy MI, Mohamed DS, El-Farrash RA. Methylation pattern of calcitonin (CALCA) gene in pediatric acute leukemia. J Pediatr Hematol Oncol. 2011;33 (7:534–542. doi: 10.1097/MPH.0b013e3181f46bc4. [DOI] [PubMed] [Google Scholar]
- Jiang L, Gonda TA, Gamble MV, Salas M, Seshan V, Tu S, Twaddell WS, Hegyi P, Lazar G, Steele I, Varro A, Wang TC, Tycko B. Global hypomethylation of genomic DNA in cancer-associated myofibroblasts. Cancer Res. 2008;68 (23:9900–9908. doi: 10.1158/0008-5472.CAN-08-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kademani D. Oral cancer. Mayo Clin Proc. 2007;82 (7:878–887. doi: 10.4065/82.7.878. [DOI] [PubMed] [Google Scholar]
- Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, Kim DH. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer. 2006;107 (5:1042–1049. doi: 10.1002/cncr.22087. [DOI] [PubMed] [Google Scholar]
- Kuan PF, Wang S, Zhou X, Chu H. A statistical framework for Illumina DNA methylation arrays. Bioinformatics. 2010;26 (22:2849–2855. doi: 10.1093/bioinformatics/btq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose MA, Chen WT, He ZM, Sikora AG, Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, Elango EM, Delacure MD, Chen FA. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61 (11:1372–1383. doi: 10.1007/s00018-004-4069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, Liu J, Chan MW, Chu TY, Sun CA, Chang CC, Yu MH. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer. 2008;123 (1:161–167. doi: 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3 (4:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Lopes V, Murray P, Williams H, Woodman C, Watkinson J, Robinson M. Squamous cell carcinoma of the oral cavity rarely harbours oncogenic human papillomavirus. Oral Oncol. 2011;47 (8:698–701. doi: 10.1016/j.oraloncology.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Marsh D, Suchak K, Moutasim KA, Vallath S, Hopper C, Jerjes W, Upile T, Kalavrezos N, Violette SM, Weinreb PH, Chester KA, Chana JS, Marshall JF, Hart IR, Hackshaw AK, Piper K, Thomas GJ. Stromal features are predictive of disease mortality in oral cancer patients. J Pathol. 2011;223 (4:470–481. doi: 10.1002/path.2830. [DOI] [PubMed] [Google Scholar]
- Martin-Subero JI, Ammerpohl O, Bibikova M, Wickham-Garcia E, Agirre X, Alvarez S, Bruggemann M, Bug S, Calasanz MJ, Deckert M, Dreyling M, Du MQ, Durig J, Dyer MJS, Fan J, Gesk S, Hansmann M, Harder L, Hartmann S, Klapper W, Kuppers R, Montesinos M, Nagel I, Pott C, Haferlach C, Cigudosa JC, Siebert R. A comprehensive microarray-based DNA methylation study of 367 hematological neoplasms. PLoS ONE. 2009;4 (9:e6986. doi: 10.1371/journal.pone.0006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zochbauer-Muller S, Farinas AJ, Vakar-Lopez F, Minna JD, Sagalowsky A, Czerniak B, Gazdar AF. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61 (24:8659–8663. [PubMed] [Google Scholar]
- Miyamoto K, Asada K, Fukutomi T, Okochi E, Yagi Y, Hasegawa T, Asahara T, Sugimura T, Ushijima T. Methylation-associated silencing of heparan sulfate D-glucosaminyl 3-O-sulfotransferase-2 (3-OST-2) in human breast, colon, lung and pancreatic cancers. Oncogene. 2003;22 (2:274–280. doi: 10.1038/sj.onc.1206146. [DOI] [PubMed] [Google Scholar]
- Nikolaidis G, Raji OY, Markopoulou S, Gosney JR, Bryan J, Warburton C, Walshaw M, Sheard J, Field JK, Liloglou T. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012;77 (22:5692–5701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poage GM, Christensen BC, Houseman EA, McClean MD, Wiencke JK, Posner MR, Clark JR, Nelson HH, Marsit CJ, Kelsey KT. Genetic and epigenetic somatic alterations in head and neck squamous cell carcinomas are globally coordinated but not locally targeted. PLoS One. 2010;5 (3:e9651. doi: 10.1371/journal.pone.0009651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poage GM, Houseman EA, Christensen BC, Butler RA, Avissar-Whiting M, McClean MD, Waterboer T, Pawlita M, Marsit CJ, Kelsey KT. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clin Cancer Res. 2011;17 (11:3579–3589. doi: 10.1158/1078-0432.CCR-11-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, Trivett M, Thompson ER, Ramakrishna M, Gorringe KL, Polyak K, Haviv I, Campbell IG. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40 (5:650–655. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, Friedman R, Varro A, Tycko B, Wang TC. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19 (2:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press Release CRUK 2012Oral cancer cases past 6,000 a year http://www.cancerresearchuk.org/cancer-info/news/archive/pressrelease/2012-03-16-hpv-push-oral-cancer-cases-past-6000-a-year
- R Development Core Team . R: A Language and Environment for Statistical Computing. Austria R Foundation for Statistical Computing: Vienna; 2012. [Google Scholar]
- Richards KL, Zhang B, Baggerly KA, Colella S, Lang JC, Schuller DE, Krahe R. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS One. 2009;4 (3:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Canales J, Hanson JC, Tangrea MA, Erickson HS, Albert PS, Wallis BS, Richardson AM, Pinto PA, Linehan WM, Gillespie JW, Merino MJ, Libutti SK, Woodson KG, Emmert-Buck MR, Chuaqui RF. Identification of a unique epigenetic sub-microenvironment in prostate cancer. J Pathol. 2007;211 (4:410–419. doi: 10.1002/path.2133. [DOI] [PubMed] [Google Scholar]
- Rogers SN, Brown JS, Woolgar JA, Lowe D, Magennis P, Shaw RJ, Sutton D, Errington D, Vaughan D. Survival following primary surgery for oral cancer. Oral Oncol. 2009;45 (3:201–211. doi: 10.1016/j.oraloncology.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J, Jimenez-Velasco A, Castillejo JA, Agirre X, Barrios M, Navarro G, Molina FJ, Calasanz MJ, Prosper F, Heiniger A, Torres A. Promoter hypermethylation of cancer-related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood. 2004;104 (8:2492–2498. doi: 10.1182/blood-2004-03-0954. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Dolinoy DC, Jones TR, Colacino JA, Prince ME, Carey TE, Rozek LS. Genome-wide methylation and expression differences in HPV(+) and HPV(−) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6 (6:777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schache AG, Liloglou T, Risk JM, Filia A, Jones TM, Sheard J, Woolgar JA, Helliwell TR, Triantafyllou A, Robinson M, Sloan P, Harvey-Woodworth C, Sisson D, Shaw RJ. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res. 2011;17 (19:6262–6271. doi: 10.1158/1078-0432.CCR-11-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, van Engeland M, Ting AH, Jair K, Yu W, Toyota M, Imai K, Ahuja N, Herman JG, Baylin SB. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3 (9:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger DR, Asch BB, Smith BD, Perruzzi CA, Dvorak HF. A secreted phosphoprotein marker for neoplastic transformation of both epithelial and fibroblastic cells. Nature. 1983;302 (5910:714–715. doi: 10.1038/302714a0. [DOI] [PubMed] [Google Scholar]
- Shaw R. The epigenetics of oral cancer. Int J Oral Maxillofac Surg. 2006;35 (2:101–108. doi: 10.1016/j.ijom.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Shaw R, Robinson M. The increasing clinical relevance of human papillomavirus type 16 (HPV-16) infection in oropharyngeal cancer. Br J Oral Maxillofac Surg. 2011;49 (6:423–429. doi: 10.1016/j.bjoms.2010.06.023. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Akufo-Tetteh EK, Risk JM, Field JK, Liloglou T. Methylation enrichment pyrosequencing: combining the specificity of MSP with validation by pyrosequencing. Nucleic Acids Res. 2006a;34 (11:e78. doi: 10.1093/nar/gkl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Hall GL, Lowe D, Bowers NL, Liloglou T, Field JK, Woolgar JA, Risk JM. CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol. 2007;43 (9:878–886. doi: 10.1016/j.oraloncology.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Hall GL, Lowe D, Liloglou T, Field JK, Sloan P, Risk JM. The role of pyrosequencing in head and neck cancer epigenetics: correlation of quantitative methylation data with gene expression. Arch Otolaryngol Head Neck Surg. 2008;134 (3:251–256. doi: 10.1001/archoto.2007.50. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, Field JK, Risk JM. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer. 2006b;94 (4:561–568. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Lowe D, Woolgar JA, Brown JS, Vaughan ED, Evans C, Lewis-Jones H, Hanlon R, Hall GL, Rogers SN. Extracapsular spread in oral squamous cell carcinoma. Head Neck. 2010;32 (6:714–722. doi: 10.1002/hed.21244. [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314 (5797:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Sodek J, Ganss B, McKee MD. Osteopontin. Crit Rev Oral Biol Med. 2000;11 (3:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 2008;268 (2:177–186. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96 (15:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viet CT, Schmidt BL. Methylation array analysis of preoperative and postoperative saliva DNA in oral cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17 (12:3603–3611. doi: 10.1158/1055-9965.EPI-08-0507. [DOI] [PubMed] [Google Scholar]
- Weber GF. The metastasis gene osteopontin: a candidate target for cancer therapy. Biochim Biophys Acta. 2001;1552 (2:61–85. doi: 10.1016/s0304-419x(01)00037-3. [DOI] [PubMed] [Google Scholar]
- Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, Shi H, Ng SW, Yan PS, Nephew KP, Brown R, Huang TH. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res. 2002;8 (7:2246–2252. [PubMed] [Google Scholar]
- Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YH, Kuo HK, Chang KW. The evolving transcriptome of head and neck squamous cell carcinoma: a systematic review. PLoS One. 2008;3 (9:e3215. doi: 10.1371/journal.pone.0003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.