Abstract
Autophagy and autophagy-related processes are fundamentally important in human health and disease. These processes are viewed primarily as cellular degradative pathways that recycle macromolecules and dysfunctional or redundant organelles into amino acids, sugars and lipids, especially during starvation. However, the ubiquitin-like autophagy proteins and other components of the autophagic machinery additionally participate in cellular reprogramming. We highlight these non-autophagic roles of autophagy proteins with the aim of drawing attention to this growing, but unexplored, research topic. We focus on the non-autophagic functions of autophagy proteins in cell survival and apoptosis, modulation of cellular traffic, protein secretion, cell signalling, transcription, translation and membrane reorganization.
Keywords: autophagy, non-autophagic roles, unconventional protein secretion, signalling, Atg protein function
See the Glossary for abbreviations used in this article.
Glossary.
- AMBRA1
activating molecule in beclin 1-regulated autophagy 1
- AP3
adaptor protein complex 3
- Bcl-xL
B-cell lymphoma extra large
- BLOC1
biogenesis of lysosome-related organelles complex 1
- CatK
cathepsin K
- COPII
coat protein complex II
- dsRNA
double-stranded RNA
- EDEM1
endoplasmic reticulum degradation enhancer, mannosidase alpha-like 1
- FADD
Fas-associated death domain
- Grh1
Grasp homolog 1
- HEK
human embryonic kidney
- keap1
kelch-like ECH associated protein 1
- LC3
light chain 3
- MEKK
mitogen-activated protein/ERK kinase
- NFκB
nuclear factor kappa B
- Nrf2
NF-E2 related factor 2
- RIG1
retinoic acid-inducible gene 1
- Sar1
secretion-associated RAS-related protein 1
- SARS
severe acute respiratory syndrome
- SNARE
soluble NSF-attachment protein receptor
- TRAF
tumour necrosis factor receptor-associated factor
- ULK1
Unc-51-like kinase 1
- VAMP7
vesicle-associated membrane protein7
- Vps23
Vacuolar protein sorting 23
Introduction
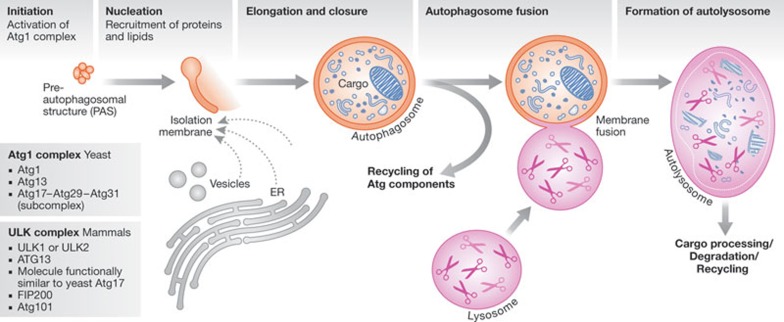
Thirty-six known ATG genes that encode autophagy (Atg) proteins coordinate autophagy-related processes. During autophagy, non-selective or specific cargos are captured in a double membrane vesicle (DMV) known as the autophagosome, which forms by the expansion of an isolation membrane from a phagophore assembly site. The autophagosomes fuse with the lysosome (vacuole in yeast), in which hydrolases degrade the cargo for reuse by the cell. The canonical formation of autophagosomes involves five steps [1]: initiation, nucleation, elongation and closure, recycling, and autophagosome fusion with the lysosome to form an autolysosome (Fig 1).
Figure 1.
Steps in autophagy—initiation occurs by activation of the Atg1 complex (ULK complex I in mammals). In yeast, this complex includes Atg1, Atg13 and the Atg17–Atg29–Atg31 subcomplex. The mammalian Atg1 complex has the yeast Atg1 homologue, Unc-51-like kinases 1 or 2 (ULK1 or ULK2, respectively), the autophagy-related 13 homologue (ATG13), a molecule functionally similar to yeast Atg17, FIP200 (focal adhesion kinase (FAK)-family-interacting protein of 200 kDa) and Atg101 [72]. Nucleation is the step that recruits proteins and lipids for autophagosome biogenesis. It begins with the recruitment of Atg proteins, such as ATG14L and WD-repeat proteins interacting with phosphoinositides (WIPIs), to the PAS—a step requiring the activity of the class 3, PI3K complex, as well as phosphatidylinositol 3-phosphate. Membrane expansion and closure—during autophagosome formation, an isolation membrane expands from the PAS, either by direct flow of membranes from a source such as the endoplasmic reticulum (ER) or by vesicle fusion. The isolation membrane surrounds the cargo, and fusion of the growing edge of the isolation membrane completes autophagosome formation. The two UBL-conjugation systems (Atg8/LC3 and Atg12) are involved in vesicle expansion and closure, and SNAREs are also probably involved. After autophagosome formation, several Atg proteins are recycled in a process involving Atg9. Autophagosome and lysosome fusion—the autophagosome fuses with the lysosome (the vacuole in yeast) to form an autolysosome, in which the cytosolic cargos are degraded and recycled. This process also involves SNAREs. PAS, phagophore assembly site; SNARE, soluble NSF-attachment protein receptor; UBL, ubiquitin-like.
Ubiquitin and ubiquitin-like-molecules constitute a broad class of protein and lipid modifiers the diverse cellular roles of which continue to be discovered [2]. Atg8 and Atg12 are two ubiquitin-like proteins that are integral to general and selective autophagy. Atg12 is coupled with Atg5 to form an Atg12–Atg5 conjugate, whereas Atg8—or its mammalian orthologues MAP1LC3, GATE16 and GABARAP, often referred to as LC3—is conjugated to a lipid, phosphatidylethanolamine. Conjugation of Atg12 and Atg8—or its orthologues—to Atg5 or phosphatidylethanolamine, respectively, requires an E1 enzyme, E2 enzymes, and an E3-like ligase activity supplied by the Atg12–Atg5/Atg16L complex [3,4], similar to the conjugation of ubiquitin and its relatives.
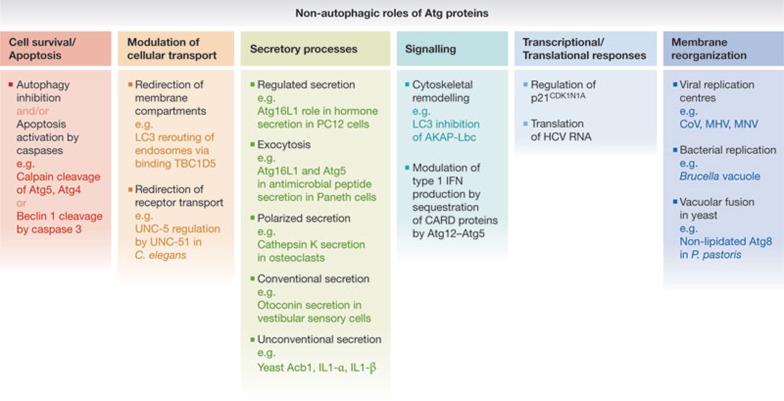
This review focuses on the ever-expanding array of non-autophagic biological functions for Atg proteins (Fig 2). We define these processes as those generally distinct from, or occasionally in addition to, an involvement in autophagosome formation or lysosomal cargo degradation. Our focus is distinct from ‘non-canonical’ autophagy, in which the biogenesis of conventional autophagosomes involved in macroautophagy proceeds either in the absence of key Atg proteins that originally defined the canonical process, or originates from membrane sources that differ from the traditional phagophore assembly site [1]. The growing appreciation for other functions of Atg proteins has paralleled the explosion of research activity in the ubiquitin field, which began with a narrow focus on the role of ubiquitination in protein turnover, but has now broadened to study its tremendous impact on many aspects of cellular physiology, including protein sorting, DNA repair, gene regulation, protein retrotranslocation, apoptosis and the immune response [5].
Figure 2.
Summary of non-autophagic roles of autophagy proteins. See text for details. C. elegans, Caenorhabditis elegans; CARD, caspase recruitment domain; CoV, coronavirus; HCV, hepatitis C virus; IFN, interferon; MHV, mouse hepatitis virus; MNV, murine norovirus; P. pastoris, Pichia pastoris.
We begin by bringing together information on the non-autophagic roles of the ubiquitin-like Atg proteins and then highlight the roles of other Atg proteins.
Non-autophagic roles of ubiquitin-like Atg proteins
Various analyses of the partners of Atg proteins, particularly of Atg8 and LC3, have revealed many new interacting components, including those that affect other cellular pathways. This has raised the question of whether the ubiquitin-like Atg proteins and their conjugates have a broader role that goes beyond autophagy.
As with the interaction of ubiquitin with many protein domains [6,7,8], Atg8 and LC3 bind to many proteins either through Atg8-interaction motifs (AIMs) or LC3-interacting regions (LIRs), respectively [9,10,11]. The six ATG8 orthologues in humans—encoded by the MAP1LC3A, MAP1LC3B and MAP1LC3C, GABARAP, GABARAPL1 and GATE16 genes—interact with at least 67 other proteins. Furthermore, several cryptic AIMs and LIRs are activated by the phosphorylation of serine and threonine residues in the vicinity of the AIM and LIR [12], thereby potentially expanding the number of putative Atg8/LC3-interacting proteins. Among the non-autophagy-related proteins that interact with Atg8 and LC3 are GTPases [13], GTPase-activating proteins (GAPs; [14,15]) and guanine-nucleotide exchange factors (GEFs; [16]). Although these are not direct targets of the autophagic process, they might serve to regulate events involved in the cellular response to autophagy, as illustrated in Table 1.
Table 1. Non-autophagic roles of autophagy proteins.
| Atg proteins | Non-autophagic role | References |
|---|---|---|
| Atg8/LC3-PE | GTPases—balancing autophagy with other cellular processesGAPs—many TBC-family proteins (signalling platforms, redirection of endosomal components to autophagosomes, recruitment of autophagy regulators)GEF—LC3 regulates Rho–GEF activity of AKAP-Lbc | [17,18,19][15][16] |
| Non-lipidated Atg8 | Component of double-membrane vesicles involved in replication of certain virusesVacuole fusion in yeast | [20,21][22] |
| Atg5 or Atg12–Atg5 | Regulation of isRNA-generated type I IFN production through Atg12–Atg5 interactions with CARDs of helicases RIG1, MDA5Unknown functions—Atg5 interactions with other CARD proteins | [26,27][26,27] |
| Calpain-cleaved Atg5 | Apoptosis—mitochondrial insertion, Bcl-xL activation, cytochrome c release | [54] |
| Worm Unc-51 (Atg1) | Axon guidance | [28] |
| Atg16L | Interaction with Rab33A is required for hormone secretion in PC12 neuroendocrine cells; required for granule exocytosis pathway in intestinal Paneth cells | [29,31,32] |
| Atg5, Atg7, Atg4B and LC3 | Required for cathepsin K secretion in bone osteoclasts | [33] |
| Atg4B and Atg5 | Required for maintenance of balance (equilibrioception) and indirectly for secretion of otoconins by vestibular sensory epithelial cells of the inner ear in mice | [40,41] |
| Many Atg proteins but not vacuolar hydrolases or proteins necessary for autophagosome fusion with vacuole | Necessary for unconventional secretion of certain proteins, such as Acb1, lacking classical signal peptides | [44,45,46,47,48] |
| Atg5, LC3, Atg16L and autophagosomal membranes | Caspase-8-dependent apoptosis | [56] |
| Atg4, beclin 1 | Cleavage by caspases suppresses autophagy and induces apoptosis | [55,57,58] |
| Atg5, Atg7 | Needed for adipogenesis in mice | [59,60] |
| LC3, Atg2 | Lipid droplet formation in mice | [62,63] |
| Atg12–Atg5, Atg7 and Atg16L1 | Required for IFNγ-mediated host defence against murine norovirus replication | [65] |
| Beclin 1, LC3, Atg4B, Atg5, Atg7 and Atg12 | Crucial for HepC viral RNA translation, virus replication and egress from cells | [64] |
| ULK1, beclin 1, Atg14L and PI3K activity | Membrane compartment needed for Brucella abortus replication | [66] |
| Atg7 | Cell cycle arrest via p53 interaction and expression of p21CDK1N1A | [68] |
AKAP, A-kinase anchoring protein; Bcl-xL, B-cell lymphoma extra large; CARD, caspase recruitment domain; GAP, GTPase-activating protein; GEF, guanine-nucleotide exchange factor; HepC, hepatitis C; IFN, interferon; isRNA, immunostimulatory RNA; MDA5, melanoma differentiation-associated protein 5; PI3K, phosphatidyl-inositol 3 kinase; RIG1, retinoic acid inducible gene 1; TBC, Tre2, Bub2 and Cdc16; ULK1, Unc-51-like kinase 1.
Atg8 and LC3 interactions with GTPases. The Rho family of small GTPases regulate diverse signalling pathways that affect cytoskeletal dynamics, cell cycle progression, gene expression, cell polarity, migration and cell transformation [5]. These and other GTPases behave as two-state molecular switches, comprising an active GTP-bound and an inactive GDP-bound form, and controlled by many proteins including GEFs, GAPs and guanine-nucleotide dissociation inhibitors. The ubiquitin–proteasome pathway regulates several Rho GTPase family members and modulates regulators of the cycling of Rho between its GTP- and GDP-bound states. Although many Rab GTPases regulate autophagy [17,18], the impact of their recruitment by autophagy components on the other cellular processes that they affect, as well as the coordination of the use of these regulators for autophagy and cellular membrane trafficking events, remain to be elucidated.
Atg8–GAP interaction as a regulatory signalling platform. The TBC (Tre2, Bub2 and Cdc16) domain-containing family of proteins affect membrane trafficking through their action as Rab–GAPs to inhibit Rab–GTPases. A screen for TBC proteins that inhibit starvation-induced autophagy identified 11 candidates that are likely to have an effect on autophagosome formation indirectly through a membrane trafficking step [19]. At least 14 Rab–GAPs of the TBC domain-containing family interact through LIRs with LC3 [15]. One such member, TBC1D5, interacts with Vps29—a component of the retromer complex on endosomes that modulates the recycling of transmembrane receptors from endosomes to the trans-Golgi network [15]. TBC1D5 also interacts with LC3 through two LIRs, but the amino-terminal LIR of TBC1D5 has a dual role by interacting directly with Vps29 in a step that is necessary for the transport of cargo in retromer vesicles. A second carboxy-terminal LIR of TBC1D5 interacts with LC3 on autophagosomes and LC3 binding displaces Vps29, indicating that the redirection of TBC1D5 from endosomal trafficking to autophagsomes is a form of cellular reprogramming.
LC3 as a regulator of GEF activity. LC3 interacts with a GEF called AKAP-Lbc, a member of the A-kinase anchoring protein (AKAP) family [16]. This interaction inhibits the Rho–GEF activity of AKAP-Lbc in HEK293 cells, thus preventing it from activating RhoA, a GTP-binding protein that controls many cellular processes, including actin cytoskeleton remodelling. LC3 overexpression strongly reduces the ability of AKAP-Lbc to interact with RhoA and impairs the Rho–GEF activity of the anchoring protein. Consequently, the ability of RhoA to promote cytoskeletal rearrangements associated with the formation of actin stress fibres is impaired. Conversely, AKAP-Lbc mutants that do not interact with LC3 have a higher basal Rho–GEF activity compared with the wild-type protein. This suggests that LC3 binding maintains AKAP-Lbc in an inactive state, thereby impairing its ability to promote downstream signalling.
Non-lipidated LC3 as a regulator of viral replication. Coronaviruses (CoVs), including SARS and mouse hepatitis virus (MHV), are enveloped RNA viruses that trigger the formation of DMVs used for viral replication and transcription. MHV uses the host cell machinery for COPII-independent vesicular export, from the endoplasmic reticulum, of a protein called EDEM1 to produce cellular membranes for its replication. MHV infection results in the accumulation of EDEM1 and OS9, an endoplasmic reticulum chaperone, in the DMVs coated with the non-lipidated LC3-I and Atg8 autophagy marker. The downregulation of LC3, but not the inactivation of host cell autophagy, inhibits CoV infection [20]. Nidoviruses and rotaviruses also replicate in DMVs decorated with LC3 [21].
Non-lipidated Atg8 as a regulator of yeast vacuole fusion. In the yeast Pichia pastoris, the non-lipidated form of Atg8 is important in fusion of the vacuole [22]. Septated vacuoles fuse to become a single spherical vacuole during adaptation from glucose to methanol. Atg8 is necessary for vacuolar fusion during adaptation of P. pastoris cells to methanol. Although vacuole fusion requires the processing of an inhibitory C-terminal peptide of Atg8 by Atg4, it does not require the lipidation of Atg8 to phosphatidylethanolamine on membranes. Neither Atg7, nor the Gly 116 residue of Atg8 involved in conjugation to phosphatidylethanolamine, are necessary for this process. Atg8 localizes to vacuolar membranes and its previously described tethering and fusion activities are necessary for vacuole fusion [23]. Vacuole fusion has been studied in vivo and recapitulated in vitro from yeast. It requires multiple SNAREs, but Atg8 was not identified in this analysis [24]. It is therefore probable that Atg8 localized at the vacuolar membrane recruits SNAREs and/or other fusion components through its ability to interact with AIMs on many yeast proteins [11], rather than serving as the direct fusogen [25].
Non-autophagic roles of Atg5 and Atg12–Atg5 conjugates
Atg12–Atg5 as a suppressor of innate antiviral immune signalling. Atg12 and its conjugation partner, Atg5, are also involved in non-autophagic signalling processes. The infection of mammalian cells by many RNA viruses produces virus-derived immunostimulatory RNA (isRNA) structures. In cells infected by vesicular stomatitis virus (VSV), Newcastle disease virus, influenza or Japanese encephalitis virus, 5′-triphosphorylated RNA produced during the virus life cycle is recognized by a cytoplasmic RNA helicase protein, RIG1. Other RNA viruses, such as encephalomyocarditis virus, generate dsRNA that is recognized by an alternative RNA helicase, MDA5. These helicases have N-terminal caspase recruitment domains (CARDs) that interact with other CARD proteins, such as the signalling protein interferon-β promoter stimulator 1 (IPS1). The homotypic interaction between the CARDs of IPS1 and either RIG1 or MDA5 is crucial for subsequent signal transmission, which in turn activates NFκB, interferon regulatory factor 3 (IRF3) and IRF7, leading to their translocation into the nucleus and transcriptional activation of type I interferon (IFN) and other pro-inflammatory genes. Secreted type I IFNs act in an autocrine or paracrine manner to induce viral RNA degradation by activating RNase L activity and also by blocking viral protein synthesis by inhibiting eukaryotic translation initiation factor 2 activity [26].
The Atg12–Atg5 conjugate negatively regulates isRNA-generated type I IFN production (Table 1; [26,27]). The conjugate interacts with the RNA helicases RIG1 and MDA5 thereby interfering with the ability of the helicases to engage in their normal CARD interactions with IPS1, consequently impairing type I IFN production. This is reflected by the increase in type I IFN production in VSV-infected Atg5-knockout fibroblasts and a reduction in viral replication. Overexpression of Atg5 or Atg12 alone in HEK293 cells increases the formation of the Atg12–Atg5 conjugates and suppresses RIG1 or MDA5-mediated signals, leading to the reduced production of type I IFNs. Atg7-knockout fibroblasts, in which the formation of the Atg12–Atg5 conjugate is impaired, also show hyper-responsiveness to isRNA stimulation.
Atg5 also interacts with the CARDs of interleukin-β converting enzyme (also known as caspase 1, a key regulator of apoptosis) and the nucleotide-binding oligomerization domain protein 1 (an intracellular receptor for peptidoglycan), suggesting additional unexplored roles of Atg5 and the Atg12–Atg5 conjugate.
In summary, as with ubiquitin-signalling pathways, ubiquitin-like Atg proteins in their conjugated and unconjugated forms function as signalling scaffolds to modulate many cellular pathways in addition to their traditional roles in autophagy.
Non-autophagic roles of other Atg proteins
Several modulators of the ubiquitin-like Atg proteins, as well as other non-ubiquitin-like Atg proteins, are crucial for a variety of cellular processes (listed in Table 1).
Axon guidance. In Caenorhabditis elegans, UNC-51, the homologue of yeast Atg1 and human ULK1, regulates axon guidance in many neurons. As such, worms with mutated UNC-51 have uncoordinated movement [28]. This serine/threonine kinase modulates the subcellular transportation of UNC-5, a growth cone receptor for the dorsoventral axon guidance protein UNC-6/Netrin. UNC-51 also interacts with LET-92, the catalytic subunit of protein phosphatase 2A (PP2A), which is important for cellular functions and the dephosphorylation of UNC-51 targets. Thus, the PP2A–UNC-51 complex regulates axon guidance.
Regulated secretion. In neuroendocrine PC12 cells, Atg16L1 is localized on hormone-containing dense-core vesicles by interaction with Rab33A. Knockdown of Atg16L1 causes a marked reduction in the level of hormone secretion in PC12 cells, independently of autophagic activity [29].
Secretion granule exocytosis. Interestingly, coding variants of ATG16L1 are genetically linked to susceptibility to Crohn disease—a chronic inflammatory condition of the intestinal tract [30]. Atg16L1- and Atg5-deficient Paneth cells of the mouse ileal epithelium have significant abnormalities in the exocytosis of granules containing antimicrobial peptides [31]. Similarly, Paneth cell abnormalities are apparent in Crohn disease patients homozygous for an ATG16L1 risk allele that predisposes these patients to the disease [32].
Polarized cathepsin secretion in osteoclasts. In osteoclasts, autophagy proteins, such as Atg5, Atg7, Atg4B and LC3, participate in the polarized secretion of lysosomal contents [33]. Osteoclasts secrete lysosomal enzymes such as cathepsin K (CatK) and hydrochloric acid into an extracellular resorptive space (resorptive lacuna). Upon attachment to the bone, osteoclasts become polarized, the ruffled border is formed and an actin ring confines and seals the contact site between the osteoclasts and the bone. The secretion of demineralizing and degradative enzymes is confined to the resorption lacuna [33]. Bone resorption causes the formation of bone pits or cavities. Atg5 and Atg7 are required for the localization of secretory lysosomes within the actin ring and for bone resorption, but not for osteoclast development or actin ring formation. These processes require LC3 and its conversion to LC3-II. Whilst it is clear that these autophagy proteins are important in CatK secretion at the ruffled border membrane in osteoclasts, the mechanism of secretion is not understood. Lysosomes containing the degradative enzymes were proposed to fuse with the plasma membrane, in which LC3 is localized in an Atg5-dependent manner.
LC3 localizes to many cellular membranes to orchestrate autophagosome formation [34], but how its localization to the plasma membrane in osteoclasts allows lysosome fusion with the plasma membrane is unclear. LC3 might assemble SNAREs that allow this selective fusion event to occur [35]. Indeed, SNAREs such as the v-SNARE VAMP7 are commonly required for lysosome exocytosis by fusion with the plasma membrane [36], as well as for autophagosome formation from membranes derived from the cell surface [35]. Previously, Ca2+-triggered lysosomal exocytosis events involving a subset of vesicles containing VAMP7 and synaptotagmin VII were implicated in plasma membrane wound healing and neurite outgrowth [37,38]. Interestingly, synaptotagmin VII and VAMP7 are also important in osteoclast function, CatK secretion and bone resorption [39], suggesting that LC3 isoforms and other Atg proteins [33] might be involved more broadly in processes such as neurite outgrowth and plasma membrane wound healing.
Balance disorders. Additional evidence for the role of Atg proteins in protein secretion comes from the analysis of Atg4 isoforms in mice, which have four orthologues of yeast Atg4, the protease that activates LC3 orthologues and also deconjugates LC3-II to free LC3-I. A significant proportion—approximately 25%—of Atg4B−/− mice have a motor coordination defect manifested by abnormal tilting of the head, a tendency to walk backwards and a marked inability to swim, which are all characteristics of balance-related disorders caused by inner ear otoconial abnormalities [40,41]. These otoconia consist of organic calcium carbonate crystals embedded in a proteinaceous matrix composed of proteins called otoconins. The defects in the Atg4B−/− and Atg5−/− mice have been traced to a deficiency in the secretion of otoconins by vestibular sensory cells of the inner ear. This process depends on appropriate mRNA and protein levels of BLOC1 and AP3 subunits, both of which are adaptors in the coat-mediated vesicular transport of otoconial proteins from the Golgi apparatus to the plasma membrane. Similar balance defects have also been noted in mice deficient in BLOC1 or AP3 subunits.
How Atg protein defects impair the mRNA and protein levels of BLOC1 and AP3 is unclear, although p62, a key signalling regulator that coordinates cell-fate decisions and transcriptional programmes that allow cells to cope with redox insults [42], might be involved. A block in autophagy causes levels of p62 to increase. The functions of p62 are mediated through its interaction with partners such as keap1, TRAF6, MEKK3 and caspase 8. When p62 protein levels increase in cells as a consequence of a block in autophagy, other cellular pathways are dysregulated. In particular, p62 sequesters keap1, an E3 ligase for the transcriptional activator Nrf2. Keap1 itself is downregulated by autophagy. Thus, as p62 levels increase, free keap1 might decrease, allowing Nrf2 levels to increase, perhaps indirectly having an impact on BLOC1 and AP3 mRNA and protein expression [43].
Role of Atg proteins in unconventional protein secretion. Eukaryotic cells release proteins into the extracellular space by two main routes. The first is the conventional secretion pathway for proteins that are synthesized at the endoplasmic reticulum and contain a signal for translocation into the endoplasmic reticulum, followed by their vesicular transport to the Golgi membranes and subsequent export from the cell. Examples of molecules that follow this pathway include insulin, neurotransmitters, hormones, mucins and collagens. The second pathway is the unconventional secretion process of proteins synthesized in the cytoplasm that lack a signal for entry into the endoplasmic reticulum–Golgi membrane pathway. Examples of proteins that use this pathway include insulin-degrading enzyme, acyl-CoA-binding protein (Acb1), fibroblast growth factor 2, interleukin 1-β (IL-1β) and many galectins. The mechanism of unconventional protein secretion remains poorly understood, but there is good evidence that Acb1, and probably IL-1β, are secreted by an autophagosome-like vesicular intermediate, requiring many but not all proteins of the normal degradative autophagic pathway [44,45,46,47,48]. Unlike autophagy, this vesicular intermediate captures cytosolic cargo and delivers it to the exterior of the cell, bypassing the normal process of autophagosome and lysosome fusion [49]. Data from studies of Saccharomyces cerevisiae suggests that this process begins at a new compartment for unconventional protein secretion (CUPS) that contains the proteins Grh1, Vps23, Atg8 and Atg9, as well as phosphatidylinositol-3-phosphate [50]. The mechanism of formation of the CUPS and secretory autophagosome-like vesicles from this compartment are unknown. Several autophagy proteins that produce degradative autophagosomes are probably involved in the formation of secretory autophagosomes, but how these vesicles avoid fusion with the lysosome and vacuole is not understood. Intriguingly, this unconventional secretory pathway might be used by many pathogens to evade destruction in the lysosomes and instead used for their release from the cells. New evidence suggests that caspases, particularly caspase 3, the death regulator caspase, regulate unconventional protein secretion. A significant proportion of the caspase-dependent secretome of nutrient-starved human apoptotic endothelial cells comprises proteins lacking conventional secretion signals, suggesting an association between caspase activation and unconventional secretion pathways [51].
Cleavage of Atg proteins by caspases and regulation of apoptosis. There is both in vitro and in vivo evidence that caspases cleave Atg proteins and regulate autophagy, and that this also activates apoptosis [52,53]. A calpain-mediated (calpains 1 and 2) N-terminal cleavage product of Atg5 makes human neutrophils and other cell types more responsive to apoptotic stimuli. Apoptosis is associated with the translocation of this Atg5 fragment from the cytosol to mitochondria, in which it associates with the anti-apoptotic molecule Bcl-xL and triggers cytochrome c release and caspase activation, without activating autophagy [54]. Similar results have also been observed in several other cell types [55].
Autophagosomal membrane as a platform for apoptosis. During apoptosis triggered by the extrinsic pathway, extracellular signals activate plasma membrane-localized death receptors of the tumour necrosis factor receptor (TNFR) family and cause them to multimerize. This multimerization recruits FADD and the initiator caspase 8 to form a death-inducing signalling complex (DISC), which in turn causes caspase 8 oligomerization and autoactivation through self-cleavage. Interestingly, there is complex crosstalk between autophagy and apoptosis pathways through both caspase-dependent and caspase-independent mechanisms [56]. For example, human Atg4 processing by caspase 3 [55], and beclin 1 cleavage by caspase 3 or caspase 8, can stimulate apoptosis whilst inhibiting autophagy [57,58].
The anti-tumour compound SKI-1 is a sphingosine kinase inhibitor that induces apoptosis in SV40 T-antigen transformed mouse embryonic fibroblasts (MEFs) and also triggers autophagy [56]. In these cells, the formation of the apoptotic DISC depends on the presence of Atg5 and the autophagosomal membrane containing LC3 and Atg16L. Caspase 8 is recruited to the autophagosomal membrane through the association of Atg5 with FADD, and its activation within the DISC requires these Atg proteins and the autophagosomal membrane as a platform.
Atg proteins required for adipogenesis. Atg5 and Atg7, in their autophagic roles, are necessary for adipogenesis in mice [59,60]. In mutant animals lacking one of these proteins, adipose tissue deposits are smaller in mass than those observed in wild-type mice, and mutant adipocytes have unusual morphological characteristics including compartmentalized lipid droplets and many more mitochondria. The animals are also resistant to high-fat diet-induced obesity. The requirement of Atg5 and Atg7 in adipogenesis might be an indirect consequence of their roles in autophagy. One reflection of this is the fact that a form of autophagy, lipophagy and lipid-droplet-associated lipases are important in the lysosomal degradation of lipid droplets, and defective autophagy causes the accumulation of lipid droplets in hepatocytes [61]. However, there is evidence for the direct involvement of Atg proteins in lipid droplet formation. As examples, Atg2 and LC3 are involved in lipid droplet formation in mouse hepatocytes and cardiac myocytes [62,63].
Host–pathogen responses. The Atg12–Atg5/Atg16L1 protein complex, along with other autophagy proteins, is actively involved in host defence mechanisms against pathogens such as bacteria and viruses. Both host cells and viruses exploit subsets of the autophagy machinery, as well as autophagy-related membranous structures, to either restrict or enhance viral replication, independent of the conventional autophagy pathways [64]. For example, a subset of the autophagic machinery is required, in a non-degradative capacity, for IFNγ-mediated host defence against murine norovirus (MNV—a plus strand, single-stranded RNA virus responsible for human epidemics of gastroenteritis). Essentially, IFNγ-activated macrophages co-opt some of the cellular Atg proteins to block norovirus infection by inhibiting formation of the replication complex. Importantly, the direct antiviral activity of IFNγ against MNV in macrophages required Atg5–Atg12, Atg7 and Atg16L1, but not the induction of autophagy, the degradative activity of lysosomal proteases, the fusion of autophagosomes with lysosomes or Atg4B [65]. The mechanism by which the autophagy proteins described above interfere with membrane alterations and membrane-associated viral replication is not known.
Viral protein translation. In contrast to the example above, is the case of hepatitis C (HepC) virus, another subset of autophagy proteins, specifically beclin 1, LC3, Atg4B, Atg5, Atg7 and Atg12, which are crucial for the translation of HepC RNA, virus replication and egress from cells, rather than interfering with the viral life cycle [64].
Membrane reorganization events in bacterial replication and egress. A variation on this theme of membrane sequestration by pathogens comes from studies on the intracellular bacterium Brucella abortus that causes brucellosis [66]. This bacterium resides in a vacuole called the brucella-containing vacuole (BCV), which moves in a Sar1- and Rab2-dependent manner from an endocytic compartment to fuse with the endoplasmic reticulum, where the bacterium replicates (rBCV). During the post-replication stages of B. abortus infection in macrophages and epithelioid cells, the rBCV is transformed into a compartment with autophagic properties (aBCV). The aBCV formation and transport of Brucella to the endoplasmic reticulum is unaffected in autophagy-deficient cells obtained either by knockdown of specific autophagy components in HeLa cells or by using macrophages carrying null mutations in Atg proteins. However, BCV formation is dependent on autophagy proteins, such as ULK1, beclin 1, Atg14L and PI3K activity, but independent of other factors, for example Atg5, Atg16L1, Atg4B, Atg7 and LC3B that are required for membrane expansion steps in autophagosome formation. Although the mechanisms involved are unclear, some parallels have been drawn to other ‘non-canonical’ pathways of autophagy that are dependent on beclin 1 and ULK1, but independent of Atg5 and Atg7, as well as LC3 recruitment [67]. However, whilst the pathway described in reference [67] also depends on the small GTPase Rab9, aBCV formation does not require Rab9 in HeLa cells [66]. The authors suggest the possibility that aBCVs are important in bacterial release and cell-to-cell spread by an egress mechanism. Whether this egress mechanism is related to the role of Atg proteins in unconventional protein secretion as described earlier [45,47,48] remains unexplored.
Gene transcriptional control. A recent study shows how autophagy and cell cycle arrest are coordinated during nutrient deprivation. In starved MEFs, Atg7—independent of its E1 activity—Atg5 and Atg6 are required for interaction with the tumour suppressor p53 and for transcriptional activation of the cell cycle inhibitor p21CDK1N1A [68].
Summary
It is evident that the autophagic machinery is far more versatile in coordinating cellular activities than was previously appreciated. Indeed, attention has been drawn to other processes possibly involving non-autophagic roles of the Atg machinery [69], such as the exencephaly caused in mice lacking AMBRA1, a protein that interacts with beclin 1. These new roles point to a complex interplay between autophagy, cell proliferation and cell death during neural development in mammals [70]. Additionally, the knockout of ATG genes in mice yields distinct phenotypes ranging from early embryonic to perinatal lethality, or an increased incidence of tumours in adulthood, which are phenomena we do not yet understand, but which also suggest roles for Atg proteins in tumour suppression [71]. The emerging themes that hold great promise for future exploration are the roles played by the autophagy machinery in the reprogramming of signalling, membrane transport, cell cycle arrest and in cell survival, death and differentiation (Sidebar A).
Sidebar A | In need of answers.
Is the non-autophagic effect caused directly or indirectly by an impact on a canonical or non-canonical autophagy-related pathway? There should be one or more ATG genes that block canonical and non-canonical autophagy that do not affect the non-autophagic pathway being studied. As illustrated in several of the examples cited, it is often necessary to assess the roles of several early- and late-acting ATG genes to understand what part of the autophagy machinery is being diverted to the non-autophagic function.
As with ubiquitin, can the ubiquitin-like proteins Atg8 and Atg12 serve as a general lipid/protein modification for non-autophagic functions? This area needs to be explored further in yeast and mammalian models to define the variety of roles.
What are the proteins that recognize Atg8/LC3 and Atg12 as protein modifiers involved in non-autophagic roles? This would require studying the cellular functions of more Atg8/LC3-interacting partners, as well as Atg12-interacting partners. The effect of mutating or knocking down the genes encoding these interacting partners on autophagy-related pathways should be studied directly, and hopefully there would be little or no effect, but some non-autophagic pathway must be impaired. In general, deeper mechanistic insights are necessary to appreciate the non-autophagic roles of Atg proteins in signalling, membrane transport, cell cycle arrest, and in cell survival, death and differentiation.

Suresh Subramani

Vivek Malhotra
Acknowledgments
S.S. was supported by a National Institutes of Health grant (GM 069373). V.M. is an Institucio Catalana de Recerca I Estudis Avancats professor at the Center for Genomic Regulation in Barcelona and the work in his lab is funded by grants from Plan Nacional (BFU2008-00414), Consolider (CSD2009-00016), Agència de Gestió d'Ajuts Universitaris i de Recerca, Grups de Recerca Emergents (SGR2009-1488; AGAUR-Catalan Government) and the European Research Council (268692), We thank our respective lab members for their comments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Codogno P, Mehrpour M, Proikas-Cezanne T (2011) Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol 13: 7–12 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Hochstrasser M (2003) A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci 28: 321–328 [DOI] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y (2007) The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302 [DOI] [PubMed] [Google Scholar]
- Ding F, Yin Z, Wang HR (2011) Ubiquitination in Rho signaling. Curr Top Med Chem 11: 2879–2887 [DOI] [PubMed] [Google Scholar]
- Fu QS, Song AX, Hu HY (2012) Structural aspects of ubiquitin binding specificities. Curr Protein Pept Sci 13: 482–489 [DOI] [PubMed] [Google Scholar]
- Harper JW, Schulman BA (2006) Structural complexity in ubiquitin recognition. Cell 124: 1133–1136 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G (2006) Ubiquitin-binding domains. Biochem J 399: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW (2010) Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Wild P et al. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z, Huang X (2012) Rab32 is important for autophagy and lipid storage in Drosophila. PLoS ONE 7: e32086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M (2011) OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol 192: 839–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I (2012) Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 32: 1733–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baisamy L, Cavin S, Jurisch N, Diviani D (2009) The ubiquitin-like protein LC3 regulates the Rho-GEF activity of AKAP-Lbc. J Biol Chem 284: 28232–28242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua CE, Gan BQ, Tang BL (2011) Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci 68: 3349–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T (2010) FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 188: 253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA (2012) TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 197: 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M (2010) Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7: 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CA, Reggiori F (2008) Are nidoviruses hijacking the autophagy machinery? Autophagy 4: 276–279 [DOI] [PubMed] [Google Scholar]
- Tamura N, Oku M, Sakai Y (2010) Atg8 regulates vacuolar membrane dynamics in a lipidation-independent manner in Pichia pastoris. J Cell Sci 123: 4107–4116 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Wickner W (2010) Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol 26: 115–136 [DOI] [PubMed] [Google Scholar]
- Nair U et al. (2011) SNARE proteins are required for macroautophagy. Cell 146: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F, Kobiyama K, Miyawaki A, Jounai N, Okuda K (2008) The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy 4: 67–69 [DOI] [PubMed] [Google Scholar]
- Jounai N et al. (2007) The Atg5–Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA 104: 14050–14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Okada T, Mitani S, Gengyo-Ando K, Baillie DL, Kohara Y, Goshima Y (2010) Protein phosphatase 2A cooperates with the autophagy-related kinase UNC-51 to regulate axon guidance in Caenorhabditis elegans. Development 137: 1657–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Uemura T, Waguri S, Fukuda M (2012) Atg16L1, an essential factor for canonical autophagy, participates in hormone secretion from PC12 cells independently of autophagic activity. Mol Biol Cell 23: 3193–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J et al. (2007) A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 39: 207–211 [DOI] [PubMed] [Google Scholar]
- Cadwell K et al. (2008) A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Komatsu M, Virgin HWt, Stappenbeck TS (2009) A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 5: 250–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deselm CJ et al. (2011) Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell 21: 966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Rubinsztein DC (2012) The plasma membrane as a control center for autophagy. Autophagy 8: 861–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC (2012) Autophagosome precursor maturation requires homotypic fusion. Cell 146: 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaineau M, Danglot L, Galli T (2009) Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett 583: 3817–3826 [DOI] [PubMed] [Google Scholar]
- Arantes RM, Andrews NW (2006) A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J Neurosci 26: 4630–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idone V, Tam C, Andrews NW (2008) Two-way traffic on the road to plasma membrane repair. Trends Cell Biol 18: 552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ito Y, Chappel J, Andrews NW, Teitelbaum SL, Ross FP (2008) Synaptotagmin VII regulates bone remodeling by modulating osteoclast and osteoblast secretion. Dev Cell 14: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera S, Mariño G, Fernandez AF, Lopez-Otin C (2010) Autophagy, proteases and the sense of balance. Autophagy 6: 961–963 [DOI] [PubMed] [Google Scholar]
- Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C (2007) Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem 282: 18573–18583 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kageyama S, Ichimura Y (2012) p62/SQSTM1/A170: physiology and pathology. Pharmacol Res 66: 457–462 [DOI] [PubMed] [Google Scholar]
- Till A, Subramani S (2010) A balancing act for autophagin. J Clin Invest 120: 2273–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral M, Anjard C, Malhotra V, Loomis WF, Kuspa A (2010) Unconventional secretion of AcbA in Dictyostelium discoideum through a vesicular intermediate. Eukaryot Cell 9: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V (2010) Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, Malhotra V (2007) The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell 130: 524–534 [DOI] [PubMed] [Google Scholar]
- Manjithaya R, Anjard C, Loomis WF, Subramani S (2010) Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 188: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Subramani S (2010) Autophagy: a broad role in unconventional protein secretion? Trends Cell Biol 21: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 30: 4701–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V (2011) Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol 195: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois I, Groleau J, Pallet N, Brassard N, Hamelin K, Londono I, Pshezhetsky AV, Bendayan M, Hebert MJ (2012) Caspase activation regulates the extracellular export of autophagic vacuoles. Autophagy 8: 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM (2008) Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol 182: 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JM, Cohen GM, Bampton ET (2010) The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy 6: 1042–1056 [DOI] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU (2006) Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 8: 1124–1132 [DOI] [PubMed] [Google Scholar]
- Betin VM, Lane JD (2009) Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 122: 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MM et al. (2012) Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem 287: 12455–12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E et al. (2010) Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 1: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X, Jin H, Xu H, Chen Q (2010) Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 1: 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerga R, Zhang Y, Chen PH, Goldman S, Jin S (2009) Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 5: 1118–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S (2009) Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA 106: 19860–19865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ (2009) Autophagy regulates lipid metabolism. Nature 458: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M et al. (2010) LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem Biophys Res Commun 393: 274–279 [DOI] [PubMed] [Google Scholar]
- Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N (2012) Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 23: 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Chisari FV (2011) Impact of the autophagy machinery on hepatitis C virus infection. Viruses 3: 1342–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S et al. (2012) Nondegradative role of Atg5–Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, Celli J (2012) Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y et al. (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461: 654–658 [DOI] [PubMed] [Google Scholar]
- Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, Cao L, Finkel T (2012) Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336: 225–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Levine B (2009) Autophagy genes in immunity. Nat Immunol 10: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F et al. (2007) A novel role for autophagy in neurodevelopment. Autophagy 3: 506–508 [DOI] [PubMed] [Google Scholar]
- Liang C, Jung JU (2010) Autophagy genes as tumor suppressors. Curr Opin Cell Biol 22: 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ (2011) The regulation of autophagy - unanswered questions. J Cell Sci 124: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]