Abstract
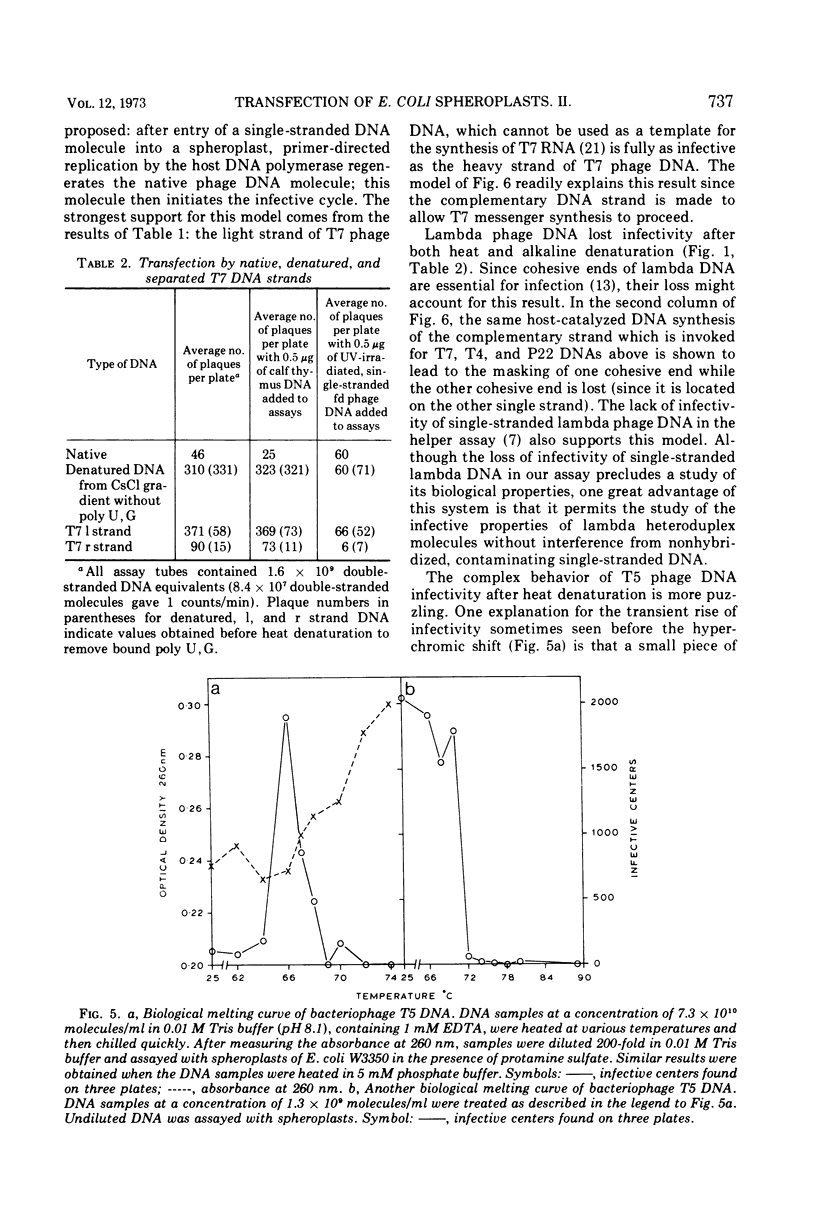
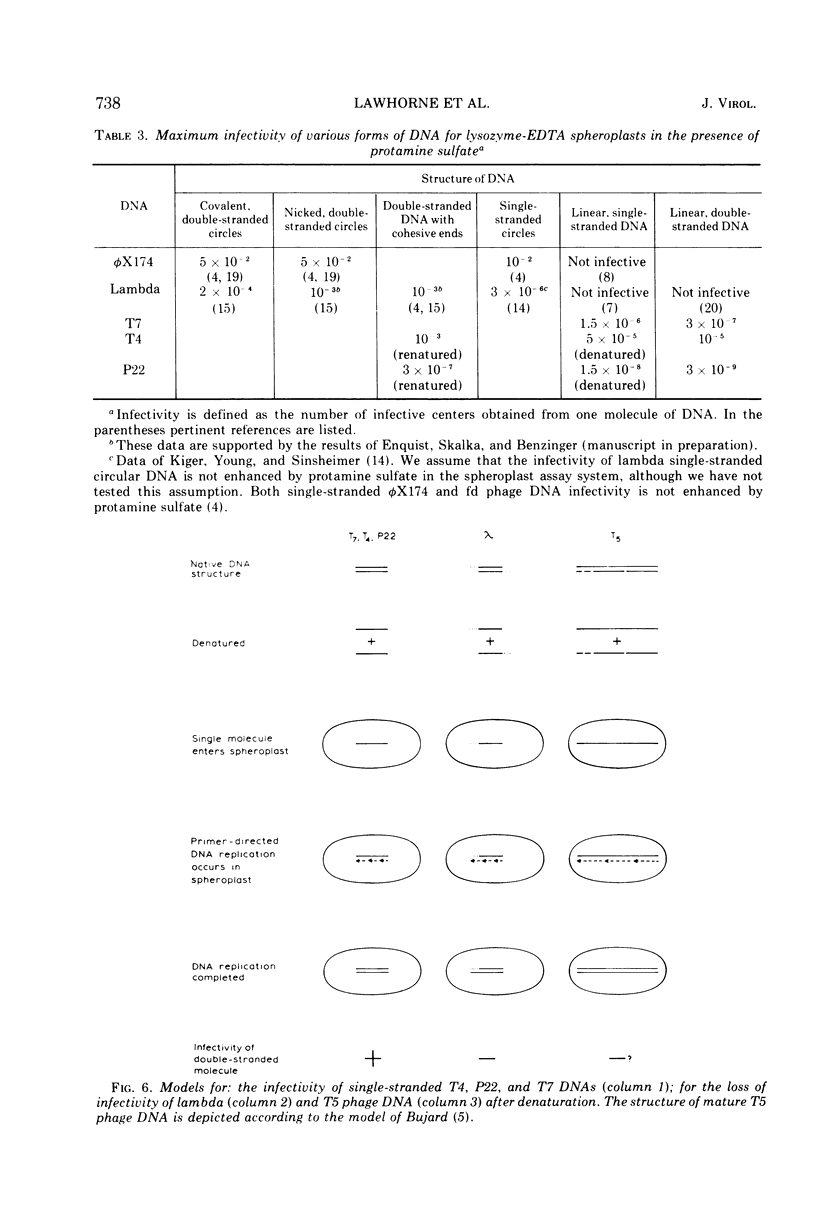
The change of infectivity of phage DNAs after heat and alkali denaturation (and renaturation) was measured. T7 phage DNA infectivity increased 4- to 20-fold after denaturation and decreased to the native level after renaturation. Both the heavy and the light single strand of T7 phage DNA were about five times as infective as native T7 DNA. T4 and P22 phage DNA infectivity increased 4- to 20-fold after denaturation and increased another 10- to 20-fold after renaturation. These data, combined with other authors' results on the relative infectivity of various forms of φX174 and lambda DNAs give the following consistent pattern of relative infectivity. Covalently closed circular double-stranded DNA, nicked circular double-stranded DNA, and double-stranded DNA with cohesive ends are all equally infective and also most highly infectious for Escherichia coli lysozyme-EDTA spheroplasts; linear or circular single-stranded DNAs are about 1/5 to 1/20 as infective; double-stranded DNAs are only 1/100 as infective. Two exceptions to this pattern were noted: lambda phage DNA lost more than 99% of its infectivity after alkaline denaturation; this infectivity could be fully recovered after renaturation. This behavior can be explained by the special role of the cohesive ends of the phage DNA. T5 phage DNA sometimes showed a transient increase in infectivity at temperatures below the completion of the hyperchròmic shift; at higher temperatures, the infectivity was completely destroyed. T5 DNA denatured in alkali lost more than 99.9% of its infectivity; upon renaturation, infectivity was sometimes recovered. This behavior is interpreted in terms of the model of T5 phage DNA structure proposed by Bujard (1969). The results of the denaturation and renaturation experiments show higher efficiencies of transfection for the following phage DNAs (free of single-strand breaks): T4 renatured DNA at 10−3 instead of 10−5 for native DNA; renatured P22 DNA at 3 × 10−7 instead of 3 × 10−9 for native DNA; and denatured T7 DNA at 3 × 10−6 instead of 3 × 10−7 for native DNA.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BENZINGER R., HOFSCHNEIDER P. H. BIOLOGICAL MELTING CURVES FOR THE "REPLICATIVE FORM" OF PHI X 174 DNA. Z Vererbungsl. 1963 Nov 21;94:316–321. doi: 10.1007/BF00894775. [DOI] [PubMed] [Google Scholar]
- Benzinger R., Delius H., Janenisch R., Hofschneider P. H. Infectious nucleic acids of Escherichia coli bacteriophages. 10. Preparation and properties of Escherichia coli competent for infectious DNA from bacteriophages phi X 174 and M 13 and RNA from bacteriophage M 12. Eur J Biochem. 1967 Nov;2(4):414–428. doi: 10.1111/j.1432-1033.1967.tb00154.x. [DOI] [PubMed] [Google Scholar]
- Benzinger R., Kleber I., Huskey R. Transfection of Escherichia coli spheroplasts. I. General facilitation of double-stranded deoxyribonucleic acid infectivity by protamine sulfate. J Virol. 1971 May;7(5):646–650. doi: 10.1128/jvi.7.5.646-650.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzinger R., Kleber I. Transfection of Escherichia coli and Salmonella typhimurium spheroplasts: host-controlled restriction of infective bacteriophage P22 deoxyribonucleic acid. J Virol. 1971 Aug;8(2):197–202. doi: 10.1128/jvi.8.2.197-202.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujard H. Location of single-strand interruptions in the DNA of bacteriophage T5. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1167–1174. doi: 10.1073/pnas.62.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W., Hogness D. S. Gene orientation in bacteriophage lambda as determined from the genetic activities of heteroduplex DNA formed in vitro. J Mol Biol. 1968 May 14;33(3):661–678. doi: 10.1016/0022-2836(68)90312-4. [DOI] [PubMed] [Google Scholar]
- FIERS W., SINSHEIMER R. L. The structure of the DNA of bacteriophage phi-X174. I. The action of exopolynucleotidases. J Mol Biol. 1962 Oct;5:408–419. doi: 10.1016/s0022-2836(62)80029-1. [DOI] [PubMed] [Google Scholar]
- HERSHEY A. D., GOLDBERG E., BURGI E., INGRAHAM L. Local denaturation of DNA by shearing forces and by heat. J Mol Biol. 1963 Mar;6:230–243. doi: 10.1016/s0022-2836(63)80072-8. [DOI] [PubMed] [Google Scholar]
- Henner W. D., Kleber I., Benzinger R. Transfection of Escherichia coli spheroplasts. 3. Facilitation of transfection and stabilization of spheroplasts by different basic polymers. J Virol. 1973 Oct;12(4):741–747. doi: 10.1128/jvi.12.4.741-747.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz G., Mauser R. Infectious DNA from coliphage T1. I. Some properties of the spheroplast assay system. Mol Gen Genet. 1969;104(2):178–194. doi: 10.1007/BF00272800. [DOI] [PubMed] [Google Scholar]
- Jacquemin-Sablon A., Richardson C. C. Analysis of the interruptions in bacteriophage T5 DNA. J Mol Biol. 1970 Feb 14;47(3):477–493. doi: 10.1016/0022-2836(70)90316-5. [DOI] [PubMed] [Google Scholar]
- Kaiser A. D., Inman R. B. Cohesion and the biological activity of bacteriophage lambda DNA. J Mol Biol. 1965 Aug;13(1):78–91. doi: 10.1016/s0022-2836(65)80081-x. [DOI] [PubMed] [Google Scholar]
- Kiger J. A., Jr, Young E. T., 2nd, Sinsheimer R. L. Infectivity of single-stranded rings of bacteriophage lambda DNA. J Mol Biol. 1967 Aug 28;28(1):157–160. doi: 10.1016/s0022-2836(67)80084-6. [DOI] [PubMed] [Google Scholar]
- Melechen N. E., Hudnik-Plevnik T. A., Pfeifer G. S. Increased stability and reproducibility of Escherichia coli spheroplasts in the transfection assay of phage lambda DNA with polyethylene glycol instead of sucrose. Virology. 1972 Mar;47(3):610–617. doi: 10.1016/0042-6822(72)90550-8. [DOI] [PubMed] [Google Scholar]
- Rhoades M., MacHattie L. A., Thomas C. A., Jr The P22 bacteriophage DNA molecule. I. The mature form. J Mol Biol. 1968 Oct 14;37(1):21–40. doi: 10.1016/0022-2836(68)90071-5. [DOI] [PubMed] [Google Scholar]
- Rhoades M., Rhoades E. A. Terminal repetition in the DNA of bacteriophage T5. J Mol Biol. 1972 Aug 21;69(2):187–200. doi: 10.1016/0022-2836(72)90224-0. [DOI] [PubMed] [Google Scholar]
- STRACK H. B., KAISER A. D. ON THE STRUCTURE OF THE ENDS OF LAMBADA DNA. J Mol Biol. 1965 May;12:36–49. doi: 10.1016/s0022-2836(65)80280-7. [DOI] [PubMed] [Google Scholar]
- THOMAS C. A., Jr, MACHATTIE L. A. CIRCULAR T2 DNA MOLECULES. Proc Natl Acad Sci U S A. 1964 Nov;52:1297–1301. doi: 10.1073/pnas.52.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]



