Abstract
Combinatorial sequence optimization for protein design requires libraries of discrete side-chain conformations. The discreteness of these libraries is problematic, particularly for long, polar side chains, since favorable interactions can be missed. Previously, an approach to loop remodeling where protein backbone movement is directed by side-chain rotamers predicted to form interactions previously observed in native complexes (termed “motifs”) was described. Here, we show how such motif libraries can be incorporated into combinatorial sequence optimization protocols and improve native complex recapitulation. Guided by the motif rotamer searches, we made improvements to the underlying energy function, increasing recapitulation of native interactions. To further test the methods, we carried out a comprehensive experimental scan of amino acid preferences in the I-AniI protein–DNA interface and found that many positions tolerated multiple amino acids. This sequence plasticity is not observed in the computational results because of the fixed-backbone approximation of the model. We improved modeling of this diversity by introducing DNA flexibility and reducing the convergence of the simulated annealing algorithm that drives the design process. In addition to serving as a benchmark, this extensive experimental data set provides insight into the types of interactions essential to maintain the function of this potential gene therapy reagent.
Keywords: computational modeling and specificity redesign, sequence recovery, LAGLIDADG homing endonucleases, protein–DNA interaction conservation, experimental benchmarks for computation
Introduction
Advances in structural modeling algorithms for protein–DNA complexes lay the groundwork for functional predictions of these classes of interactions and engineering efforts. For example, accurate determination of binding specificity preferences for native complexes1,2 and estimations of the contributions of individual amino acids to the energetics of an interface3 can promote a better understanding of protein–DNA complexes and facilitate the next step: the computational refactoring of these properties for the development of tools for numerous biotechnology applications. 4,5 Improved computational methods have the capability to address the limitations of sampling size and significant experimental effort that constrain traditional combinatorial screening approaches6–8 for engineering novel protein–DNA interactions. Currently, the main focus of protein–DNA interface engineering efforts is the reprogramming of DNA substrate specificity to alter binding or cleavage locations in a genome.9 Promising platforms for generation of genome-specific cleavage reagents are zinc-finger nucleases,10 TALE nucleases,11 and homing endonucleases or meganucleases. 12 While there are a number of diverse experimental protocols to accomplish this engineering goal,6–8 the utilization of computational methods has been shown to complement and improve the efficiency of the experimental methods by guiding library design or providing a starting place for directed evolution.13–15
The ROSETTA macromolecular modeling and design suite16 has been used for developing homing endonucleases with novel specificities.9,17–19 ROSETTA depends on a physically based energy function working in conjunction with a simulated annealing sampling algorithm to identify mutations in a protein that are likely to drive the formation of favorable, sequence-specific protein–DNA interactions. 20 The general method for protein design with a fixed protein and DNA backbone involves a search of protein sequence and rotameric space to identify the predicted lowest-energy set of amino acid identities and conformations. Redesign for a specific DNA sequence change consists of substitution of the nucleotide type in the crystal structure DNA followed by redesign and repacking (search of rotameric, but not sequence space) of the amino acids surrounding this nucleotide change. A recent improvement to the ROSETTA modeling of protein–DNA interactions was the incorporation of backbone flexibility on both sides of the interface, improving specificity predictions.1 Backbone flexibility provides a way to further diversify design results over the standard, fixed-backbone approximation available in release versions of ROSETTA. While the use of ROSETTA has resulted in a number of endonucleases with successfully altered specificities, 9,17–19 consistent recapitulation of experimental data has proven challenging,17,19 suggesting that many potentially successful designs are being overlooked by current algorithms.
In this work, we developed methods for exploring energetically relevant sequence diversity in order to produce designs enriched in amino acids making native-like interactions with the DNA bases. These new methods are potentially valuable for guiding design of libraries for experimental engineering methods, and their success was evaluated by comparison to a newly collected experimental data set. The Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB)21 contains within it a wealth of information in the form of the distances and geometries of protein–DNA interactions (“motifs”) present in native complexes (Fig. 1). This information was incorporated into the ROSETTA design process. Previously, motifs had been used to direct protein backbone sampling,22,23 and in this new implementation, they are used to bias both sampling and energetics of amino acid rotameric states in the context of a fixed protein backbone. Comparisons of designs with and without these native interactions helped guide energy function improvements. New protocols for increased diversity generation included differential energetic and sequence-space biasing for rotamers capable of forming canonical motif contacts, simulations with flexible DNA,1 and reducing the convergence of the simulated annealing algorithm. The resulting predictions were analyzed in the context of sequence recovery benchmarks and a newly generated comprehensive experimental data set that identified the tolerated sequence variation at 44 positions in one protein–DNA interface.
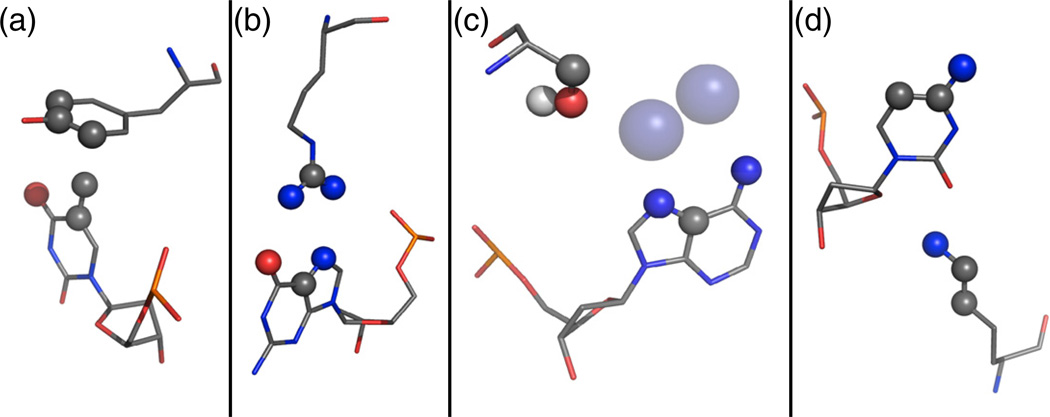
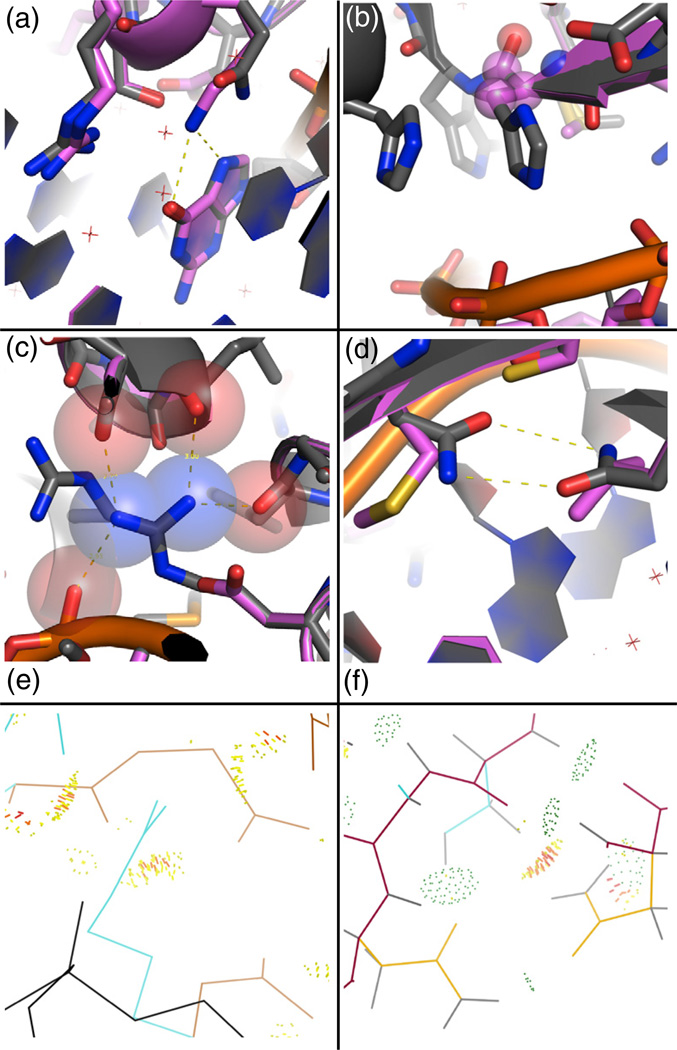
Fig. 1.
Examples of the types of motif interactions included in the motif library. Atoms that define the motif interaction are shown as spheres colored by atom type. (a) Tyrosine residue packing against a thymine methyl group, derived from Tyr25A and Thy317B of 1mow. (b) Bidentate arginine–guanine interaction, derived from Arg274B and Gua418C of 1cyq. (c) Water-mediated interaction identified by placement of waters (transparent blue spheres) on the DNA at canonical locations, derived from Ser47A and Ade516C of 1m5x. (d) Minor groove interaction, derived from Lys116A and Cyt16C of 2np6.
Results
Improving sequence recovery with motifs
A library of canonical amino acid–base interactions, referred to as motifs, was collected from protein–DNA complexes available in the PDB (Fig. 1). Rotameric conformations of amino acid side chains capable of forming interactions seen in that motif library were identified through a newly developed search process (Fig. 2). This process scores the rotamers based on the distance between a canonical base placed in the motif-forming location and the closest base of the same type in the crystal structure. The rotamers that can form motif interactions, identified by a small distance between the canonical base and the crystal structure base, are added, with an energetic bonus, to the rotamer set used by the standard, fixed-backbone ROSETTA design protocol. The size of the rotamer library used in standard design calculations is limited due to computational considerations, and this search process allows assessment of many more rotamers than could normally be included. While only a small fraction of the screened rotamers are added to the rotamer library—the procedure is limited to 100 extra rotamers of each amino acid type at each position—the incorporation of these interaction-biased side chains provides a way to increase exploration in areas of sequence and rotameric space that are most likely to result in the formation of native-like contacts.
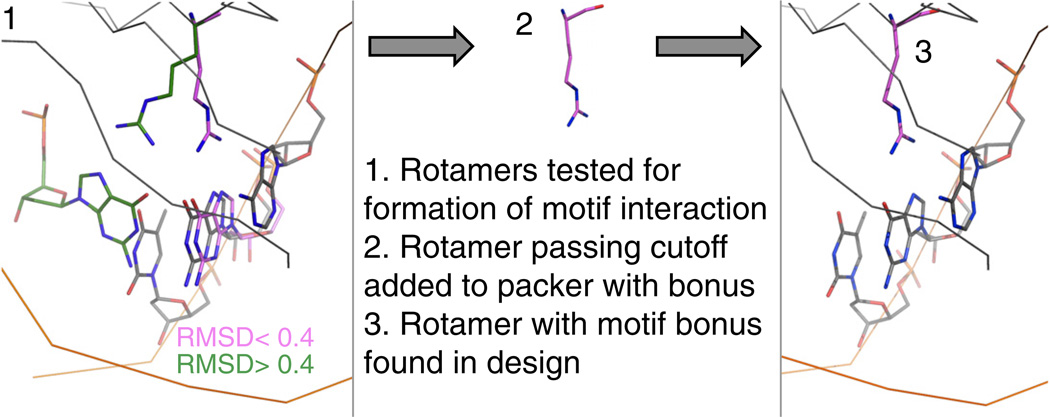
Fig. 2.
Overview of the motif-biased design protocol. In step 1, a series of rotamers and motifs are tested to see if they are compatible with the crystal structure undergoing design. These rotamers and motifs are subject to a series of cutoffs: distance of C1*, how parallel the placed base is to the crystal structure DNA, and RMSD of nucleobase atoms. In this example, two arginine rotamers (green and pink) are tested with a bidentate arginine–guanine motif, and the pink rotamer passes a nucleobase RMSD cutoff of < 0.4 when an ideal guanine base is placed in a motif-compatible position and compared to the nearest guanine base in the crystal structure. This pink arginine rotamer is then added to the standard rotamer sets used by the ROSETTA packer. The rotamer is given an energy bonus over other rotamers and is found in a design completed for this guanine base.
In order to analyze the effect on design of adding these motif-biased rotamers and determine the optimal bonus value for them, we carried out calculations for a set of 112 protein–DNA co-crystal structures. This set was divided into a training set of 48 proteins and a test set of 64 proteins for assessing the validity of protocol optimizations found to improve results for the training set. The sequence recovery for this test set, analyzed by two metrics (“weighted” and “unweighted” recovery), is shown in Fig. 3a for a range of motif bonus values. The addition of motif rotamers was found to improve the sequence recovery for both recovery metrics, across multiple variants of the ROSETTA energy function (Fig. 3a). Examining sequence recovery as a function of the motif bonus term revealed that low bonuses generally give the best results. Values of −1.25 or −2.50 ROSETTA energy units (REUs; most closely correlated with kilocalories per mole24), depending on the other scoring parameters and the recovery metric, resulted in optimal recovery. Higher bonus values have reduced recovery due to the incorporation of motif rotamers without regard to other energy function terms. The motif bonus resulting in the highest sequence recovery for the weighted metric was slightly less than that for the unweighted metric. The unweighted metric counts every designed position equally and is thus subject to a bias favoring incorporation of the amino acid types most commonly found in protein–DNA interfaces (such as those types in the motif library). The weighted metric is an average over the recoveries for each amino acid type and free from biases in the amino acid composition of the interface positions. Accordingly, the very high motif bonus values were less detrimental to unweighted recovery, which benefited from biases toward abundant amino acid types, than to the weighted metric.
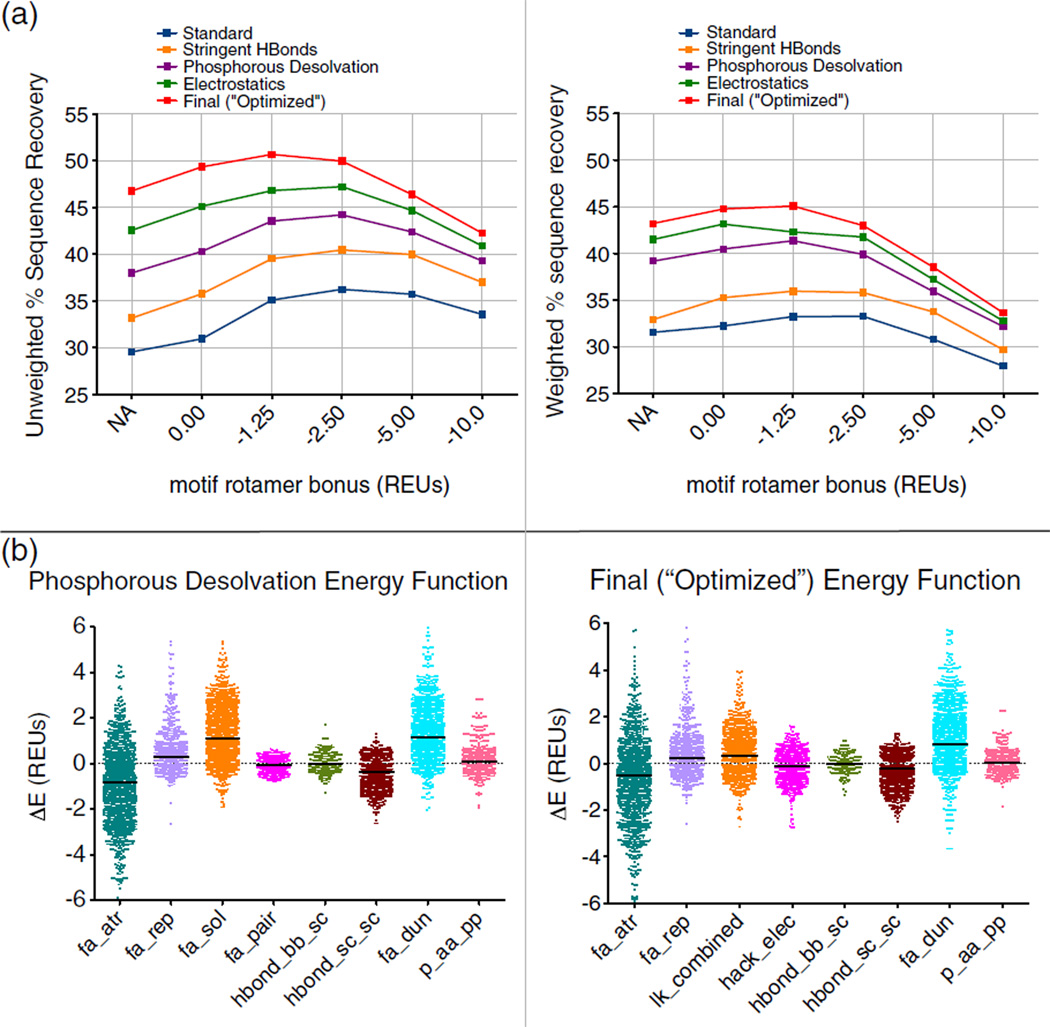
Fig. 3.
Optimization of ROSETTA energy function. Abbreviations for energy function terms are as follows: fa_atr, attractive; fa_rep, repulsive; fa_sol, solvation; fa_pair, distance-dependent atom pair potential; hbond_bb_sc, hydrogen bonds between backbone and side-chain atoms; hbond_sc_sc, hydrogen bonds between side-chain atoms; fa_dun, rotamer probability; p_aa_pp, probability of amino acid given backbone conformation; hack_elec, simple electrostatics; lk_combined, combination of terms for orientation-dependent desolvation model. (a) A comparison to two metrics of sequence recovery over several motif rotamer bonuses and several iterations of energy function optimization (Figs. S2–S5). The “Standard” energy function was the starting point for the optimization. The “Standard” energy function was improved by the addition of motifs, increasing the stringency of the hydrogen-bonding model (“Stringent HBonds”), modification of the phosphorous desolvation penalty (“Phosphorous Desolvation”), and the addition of a coulombic electrostatics term1 for the “Electrostatics” energy function. The “Final (“Optimized”)” energy function includes multiple additional changes detailed further in the text and in the supplement. (b) Energy differences, separated out by energy term, between incorrectly designed rotamers and rotamers with a motif bonus that match the native amino acid type, or more correctly match the native rotamer, than a designed rotamer with no bonus. The units for these energy differences are in REUs. The differences collected with the “Standard” energy function reveal that the solvation term (fa_sol) and the rotamer probability term26 (fa_dun) are the two energy terms that are being offset by the motif bonus. As a part of the energy function optimization, the solvation term was replaced with an orientation-dependent solvation model1 (lk_combined), and changes were made to the atom-specific desolvation parameters for several amino acid types.
Optimization of the ROSETTA energy function
We next used the motif-biased design results to guide optimization of the ROSETTA energy function, improving sequence recovery significantly over “Standard” scoring. The complete set of modifications to the energy function resulted in a high unweighted recovery of 50.7% with motifs added, an increase of 20% over the initial “Standard” recovery of 29.6% with no motif rotamers or optimization (Fig. 3a and Table S1). The recovery pattern and the magnitude of the differences in recovery observed for this test set are similar to those changes seen for the training set, over the same iterations of the energy function (Fig. S1).
Of these scoring improvements, many were implemented specifically for modeling of protein–DNA interactions, such as increase in the stringency of the hydrogen-bonding model and correction of the ROSETTA phosphorous desolvation25 parameter (Fig. 3a).1 The combination of this corrected solvation model and the increased hydrogen bond stringency provides over 8% of the total 20% improvement in unweighted recovery. The change having the next largest effect was the replacement of the database-derived, residue-pair potential (the fa_pair term) with a simple, short-range explicit electrostatics term. 1 Recoveries with only this “Electrostatics” modification are shown in Fig. 3a. Both the electrostatics model and the motif bonus favor charged interactions—charged residues are overrepresented in the motif library due to their abundance at protein–DNA interfaces—thus a higher motif weight is less beneficial in the presence of the electrostatics model (Fig. 3a, comparing “Phosphorous Desolvation” to “Electrostatics”). The “Final” optimized scoring function garners further improvements in recovery of over 4% unweighted (1.7% weighted). This finalized scoring function is a composite of several smaller improvements, the individual effects of which are detailed in the supplement (Figs. S2–S5). These changes are (1) a modification to the solvation model (lk_ball), introduced by Yanover and Bradley,1 in which desolvation contributions for polar atoms are dependent on the relative orientation of the desolvating atom; (2) the modification of desolvation parameters for atom types found in asparagine, glutamine, lysine, and arginine amino acids; (3) an increased weight of the attractive (fa_atr) scoring term; (4) an increased positive charge for the lysine NH3 group as a proxy for an inability in ROSETTA to differentially weight hydrogen-bonding types; and (5) an optimization of the amino-acid-specific reference energies.
This optimization of the ROSETTA energy function was guided in part by analyzing the biases in the sequence recovery results. Examining the ratio of the number of times an amino acid was designed to the number of times it is found in the initial population reveals amino acid types that are underrepresented and overrepresented by the design process. All modifications to the desolvation terms, as well as the increased positive charge of lysine, were prompted by a low recovery of those amino acid types and a corresponding low representation of these types in the designs completed using the energy function with only the electrostatics term added. The sequence recoveries and amino acid ratios leading to and resulting from each modification are detailed in Figs. S2–S5. Optimization of the amino-acid-specific reference energies, representing the average energy of the residue in the unfolded state, was also guided by looking for biases in the distribution of designed amino acids.
In addition to correcting biases in amino acid composition, a comparison between designs completed with and without motifs highlighted the energy terms most in need of optimization. The sequence recoveries of designs with a bonus on motifs were higher than those without the added motif rotamers. Determination of those energy terms that were offset by the motif bonus helped guide our energy function optimization. If a motif rotamer of the native amino acid type is incorporated in a design and more closely matches the wild-type rotamer than an incorrectly designed rotamer without a motif bonus, the differences in energy terms between the motif rotamer and the incorrect rotamer can illuminate what terms are responsible for favoring the incorrect rotamer. This analysis was completed over the entire set of 112 designed interfaces, and the results for the “Phosphorous Desolvation” and “Final (“Optimized”)” weight sets are shown in Fig. 3b. Energy differences with a positive value are the ones being offset by the motif bonus for the more correct rotamer choice. For the starting energy function, the two energy terms that are positively shifted are the solvation (fa_sol) and rotamer probability26 (fa_dun). The final energy function indicates that the design failures associated with a solvation penalty were significantly corrected by a combination of the modifications to desolvation terms and the addition of the orientation-dependent solvation model. Ways to correct the remaining penalty associated with the rotamer probability term are currently under study. These findings correlate with the shift toward a preference for lower motif weights in concert with higher sequence recovery as the energy function was optimized. This result indicates that more successful motif-like interactions were being made without the aid of such significant motif favoring as energy function improvements were incorporated.
Sequence optimality of a wild-type endonuclease
A designed amino acid that does not match the native sequence is not necessarily a failure of the computational methods. Depending on the physiological role of a DNA-binding protein, the wild-type amino acid may not be the most energetically favorable. Some regions of a protein–DNA interface may require low specificity and hence few direct nucleotide contacts in order to accommodate multiple DNA bases—such as transcription factors that must bind to multiple promoters.27 While some protein positions in an interface require the wild-type amino acid for activity or binding, other positions can tolerate multiple amino acid types. Without knowing the role and importance of each amino acid in an interface, it is insufficient to use sequence recovery of native interfaces as the sole metric for determining the success of the computational methods. A straightforward way to address this question is to make and characterize protein mutations and to see if they are tolerated or disallowed as computationally predicted. This experiment was carried out for one protein in the benchmark set, the homing endonuclease I-AniI. Full randomization of each of 44 positions in the interface of the homing endonuclease I-AniI and screening of all single-position libraries for activity against the wild-type target site was completed using a bacterial directed evolution system. 28 Sequencing ~20 protein mutants for each library (Table S2) after activity selection showed which positions tolerated only the wild-type amino acid and which positions could accept a number of amino acids.
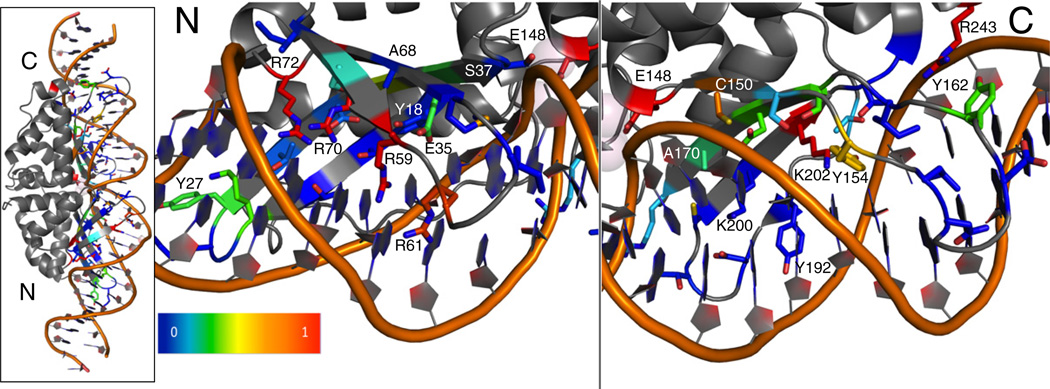
The experimental data revealed that the wild-type amino acid type is not highly favored over other possibilities at many positions in the interface (Fig. 4). The calculated experimental recovery, an average over all wild-type recovery frequencies, is 31%. Only a few positions show very high preservation of the wild-type amino acid. In the N-terminal domain, only four arginine residues are preserved, certainly contributing significant binding energy (R59, R61, R70, and R72). In the C-terminal domain, preserved residues include the position Arg243, stabilizing the position of a C-terminal DNA-contacting loop through interactions with the protein backbone, and interacting amino acids Lys202 and Tyr154, likely key contributors to formation of the catalytic complex.18 The importance of these three C-terminal residues for cleavage of this particular target DNA is underscored by their complete conservation in homologues of I-AniI predicted to cleave a very similar target DNA sequence, even in those with sequence identity of less than 50%.29 The other aromatic residue positions on both sides of the interface display higher conservation in this data set than the majority of positions, with the exception of Tyr192. While these aromatics did not always show a high recovery of the exact native amino acid type, they all displayed a tendency to remain an aromatic. The frequency of recovering the wild-type amino acid at each position is visually presented on the IAniI structure (2qoj30) using a gradient from red to blue; positions that come back as wild type are colored red, and the positions with very little wild type observed in the sequencing results are blue (Fig. 5). The significant number of positions displaying little or no preference indicates that many amino acid substitutions in the I-AniI interface are functionally neutral, at least in the context of this selection system. The ability of the interface to accommodate such neutral drift—the accumulation of non-deleterious mutations with adaptive potential—has been implicated as a mechanism for the acquisition of new substrate specificities.29,31,32 This neutral drift facilitates enzyme adaptations by reducing the number of mutations necessary to acquire new functions in the face of evolutionary pressure and is particularly important for the endonuclease family of proteins. These DNA-cleaving enzymes are parasitic elements, catalyzing transfer of their own gene, and their interface flexibility allows for their continued propagation by facilitating cleavage of a wide range of target sites that are themselves subject to genetic drift.
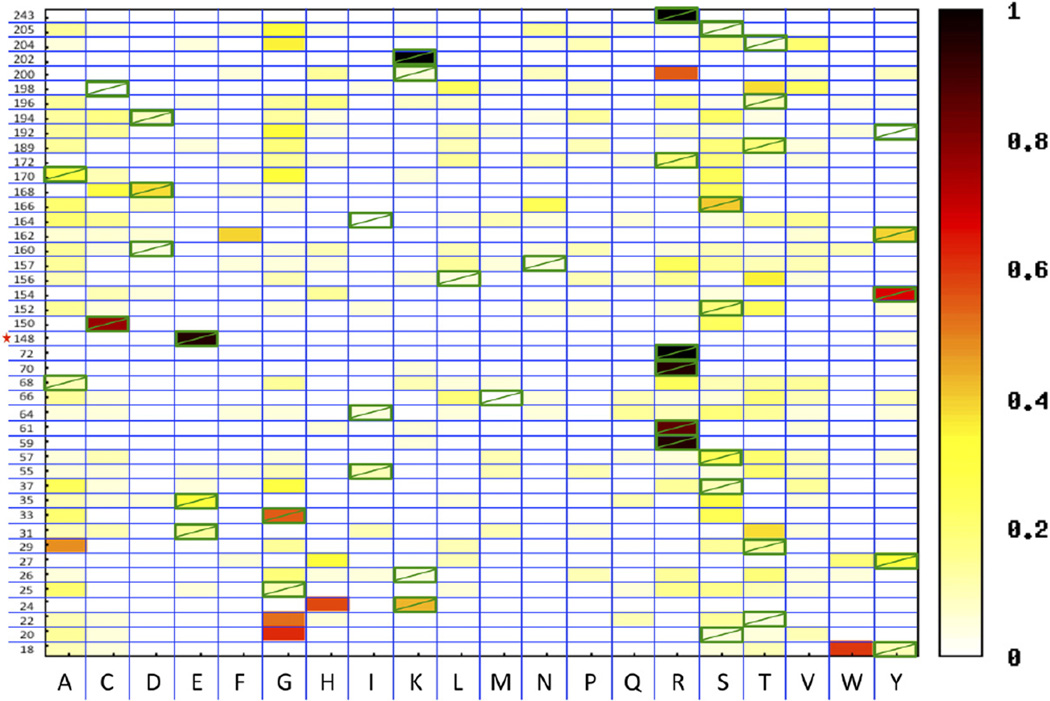
Fig. 4.
Sequence optimality of the interface residues of I-AniI. Heat map displaying the frequencies observed of each amino acid type in a selected pool of sequences at each of 44 positions in the I-AniI interface. The wild-type amino acid is marked with a green box. Each position in the interface was fully randomized, and these single-position libraries were subject to an activity selection.28 A frequency of 1 means that the amino acid with this frequency was the only amino acid type observed at that protein position, whereas a frequency of 0.05 would be an amino acid type observed once from a set of 20 sequences.
Fig. 5.
Visual representation of the interface conservation of I-AniI. The frequency of observing the wild-type amino acid after full randomization and selection (Fig. 4) is summarized on the structure of I-AniI. Only the 44 residues that were randomized are shown in this representation. Blue corresponds to a frequency of 0 or non-conserved positions. Red corresponds to positions that are highly conserved as the wild-type amino acid. The overall protein–DNA complex is shown on the leftmost panel, and the N- and C-terminal domains are separated in the other panels to allow for a closer examination of the conserved contacts. Four arginine residues are most conserved in the N-terminal domain and are likely essential for formation of the initial substrate-bound complex. Lys202 and Tyr154 are conserved in the C-terminal domain, and these interactions likely play an important role in the formation of the catalytic complex.18 This representation is incomplete in that it loses information if the preferred amino acid is not the wild type, but still a conserved type. For example, positions Tyr18, Tyr27, and Tyr162 are strongly conserved as aromatic residues (Fig. 4), but the native aromatic shows up at lower or equivalent frequencies as other aromatic types, resulting in blue or green shading at these positions.
Numerous positions show very low levels of wild-type amino acid in the sequencing results (at or below 5% or 1 of 20 sequences), and understanding how differences in frequency correlate with differences in enzyme activity is important for utilizing this data set. When there is strong selective pressure, the position converged almost completely to the preferred sequence, such as in the case of the magnesium-binding catalytic residue Glu148 that was randomized as a control for the experiment (Figs. 4 and 5). This assay of activity is also sensitive to small differences in activity, as is demonstrated by the data collected for position Lys200. K200R and K200N were previously tested mutants, since they were both observed in homologues of I-AniI and shown to have levels of activity very similar to wild type.29 Both mutants were found to be slightly more active than the wild-type enzyme, and in this current assay, both of them were found in the selected pool with higher frequencies than the wild-type lysine (0.55 for Arg, 0.09 for Asn, and 0.05 for Lys). Given the extremely high activity of both mutants, it was challenging to resolve whether one was more active than the other with previously published enzymatic cleavage assays.29 However, arginine was by far the most common amino acid observed at position 200 in an alignment of homologous enzymes29 (Fig. S6), matching the data here showing that it is observed more frequently than any other amino acid in the selected pool (Fig. 4). While the amino acid frequencies at this particular position match those observed in a multiple sequence alignment of endonucleases predicted to cut a very similar site to I-AniI, the majority of the positions observed experimentally to have high flexibility are significantly less variable in the alignment (Fig. S6). The conditions of the bacterial selection system differ from natural evolution, likely resulting in this divergence between the alignment and the results observed from the described experiments. In particular, the bacterial system is selecting only for activity on the wild-type I-AniI, not for specificity against competing target sites or lack of specificity at areas facilitating new specificity acquisition, and artificial selections allow for full randomization at any interface position, whereas natural evolution generally traverses a pathway constrained by single nucleotide substitutions in the starting codon.
Two methods for sequence diversity generation
The high sequence diversity tolerated at many positions in the I-AniI interface points to the need for computational protocols that generate multiple, energetically reasonable solutions rather than a single design. Algorithms that produce only a lowest-energy solution are constrained by sampling and the quality of the energy function guiding the design process. Methods are needed to generate diverse structures, thus enabling new local minima to be found. Diversity in design is valuable for comparison to experimental data, as library-screening experiments rarely produce a single best protein sequence for a given target and instead provide several solutions. Multiple low-energy solutions can also be screened concurrently in directed evolution experiments.
Two methods, DNA backbone flexibility and reducing the convergence of the simulated annealing algorithm (“the packer”16) used by the ROSETTA, were developed and assessed in the context of a computational benchmark and experimental data. The DNA flexibility consisted of a 3-base-pair pocket of movement surrounding the target design base pair (Fig. 6a, “DNA-Rebuild”), and the convergence of the packer was reduced by increasing the low temperature of the simulated annealing procedure and removal of the quenching step that drives the packer to identify the sequence with the lowest possible energy (“HighTemp-Packer”). Out of the full set of 112 proteins, a complete set of interface designs was collected with both of these new protocols for 78 that were compatible with the DNA-Rebuild methods in their current state. All data were collected with the “Optimized” energy function. No motif rotamers were added for these computational experiments. A total of 56 designs were completed for every design pocket (DNA base pair and surrounding protein positions) that was previously designed a single time with the standard design protocols. The frequencies of amino acids observed at each designable position were calculated over these 56 designs and compared to frequencies from 56 designs completed with the standard method.
Fig. 6.
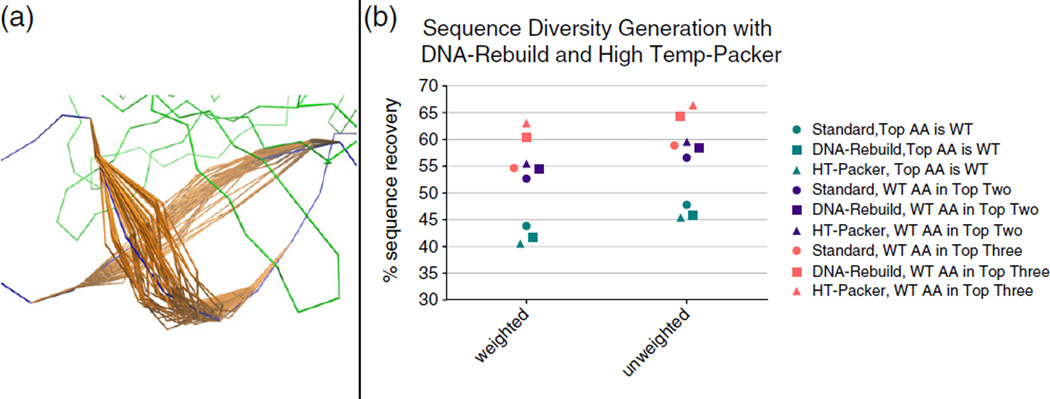
Limited degeneracy increases sampling of the native sequence for two methods of diversity generation. (a) Illustrative example of the level of DNA movement in the DNA rebuilding simulations. (b) Both methods developed for sampling diverse sequences were tested, and compared to the “Standard” method, for a benchmark set of 78 proteins. The frequencies of amino acids observed at each position were calculated from 56 trajectories for each method. If only the highest frequency amino acid is incorporated in the sequence recovery calculation (cyan), the recovery shows a slight decrease for both weighted and unweighted metrics. If the top two (purple) or top three (pink) amino acids are both considered in the recovery calculation, and observing that the wild-type amino acid in any of these top positions counts as correct, then the sequence recoveries are significantly increased.
The results of both protocols on the two sequence recovery metrics revealed that the diversity produced often contains the wild-type amino acid, even if it is not the most frequently observed type at a particular position. If the top two amino acids by frequency were considered when calculating recovery, the chance of correctly identifying the wild type is increased over 12% for both recovery metrics (Fig. 6b). However, while the sequence variation is much less for the 56 design runs with the standard protocol, recovery with this original method also improves by 8% when the top two amino acids are counted, achieving a high of only about 2% lower than the two new methods. Looking at the top three most frequent amino acids drastically increases the recovery gap between the original method and these new methods that generate significant sequence diversity. The HighTemp-Packer achieves a highest unweighted recovery of 66.4%, a 7% improvement over taking only the top two amino acids. The DNA-Rebuild performs slightly less well, achieving only 64.3% unweighted recovery, but still significantly outperforms the original method that only shows a 2% gain to 58.9% unweighted recovery. Computational results that produce possible amino acid choices rather than a single lowest-energy choice are essential for building libraries to guide experimental engineering projects. However, the success of building libraries based on this expanded sequence pool requires that the added information increases the chance of finding a native-like or low-energy state rather than simply diluting the good sequences with inaccurately produced diversity. The result that both of these new protocols significantly improved sequence recovery when the second or third highest frequency amino acids were added to the recovery calculation argues that both protocols could add valuable diversity to a designed library. Comparisons to experimental data conducted in the next section further explore the merits and limitations of both methods.
Computational recapitulation of experimental data
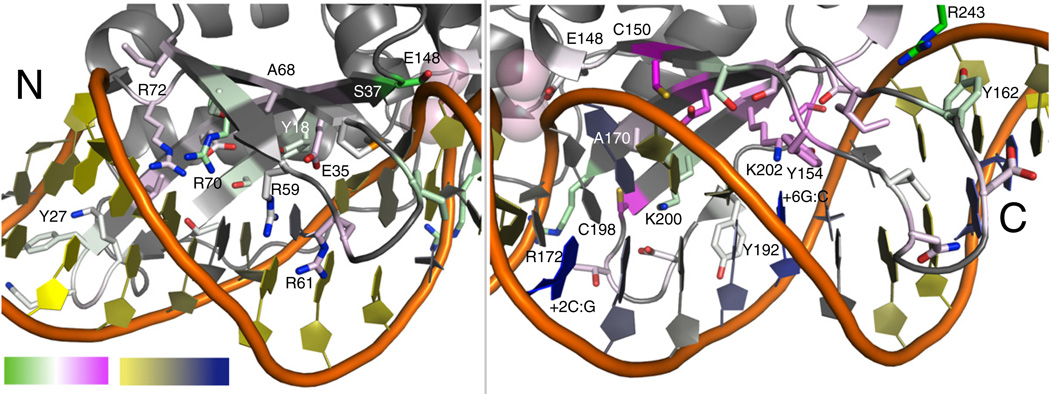
Comparison of the experimental data with the previously described computational protocols indicates that neither of the new protocols stands out as superior and that each method has different strengths (Fig. 7 and Figs. S7 and S8). Both protocols better recapitulate the experimental data than the “Standard” design method (Table 133). The amino acid frequencies observed at some positions better matched the frequencies from the DNA-Rebuild simulations, and others better matched the results of protocol utilizing the HighTemp-Packer. Both computational protocols result in higher sequence convergence, for wild-type amino acids as well as incorrect amino acid types, than the experimental selection. The two different methods of diversity generation are able to drive escape from the converged energy well for different positions in the interface, indicating that they can each overcome different types of protocol limitations (Fig. 7 and Figs. S7 and S8). For example, positions Ala68 and Ala70 are converged in the DNA-Rebuild simulations, likely due to the conformation of the protein backbone structure. The HighTemp-Packer method was able to generate significant diversity at both these positions that better matched the experimental data. Some positions near the DNA backbone benefited more from the DNA-Rebuild simulation. Positions 37 and 172 show very high convergence in the HighTemp-Packer results, and the experimental data indicate that there should be minimal amino acid preferences here. Both these positions are directly interacting with the DNA backbone in the crystal structure of the complex, and the DNA-Rebuild method was able to reproduce this experimental variation by allowing DNA backbone movement.
Fig. 7.
Recovery of experimental data with computational methods. A comparison between the two methods of sequence diversity generation, DNA-Rebuild and HighTemp-Packer, is summarized on the structure of I-AniI. The frequency distributions at each of the I-AniI interface positions were compared to the experimental data (Fig. 4) by both Euclidean distance and Jensen–Shannon divergence measures (Table 1 and Fig. S8). For this illustration, the Jensen–Shannon divergence measure33 calculated for the DNA-Rebuild method was subtracted from the same calculation completed for the HighTemp-Packer. White is designated as a value of 0, indicating that neither computational method better matched the experimental frequency distribution; green is negative values, indicating that the DNA-Rebuild performed better than the HighTemp-Packer; and pink is positive values, indicating that the HighTemp-Packer performed better than the DNA-Rebuild method. The DNA is colored based on the average RMSD between the DNA-Rebuild simulations and the crystal structure DNA, where yellow is the lowest average RMSD and where blue is the highest. The DNA moved farthest away from the crystal structure DNA in the same area that the DNA-Rebuild method performs well much less than the HighTemp-Packer, indicating that the DNA location has a significant effect on the design results.
Table 1.
Comparison of computational protocols to experimental data
| Computational method |
Jensen–Shannon divergence |
Euclidean distance |
|---|---|---|
| Standard | 0.472 | 0.839 |
| DNA-Rebuild | 0.409 | 0.670 |
| HighTemp-Packer | 0.399 | 0.695 |
Divergence between experimentally observed and computationally predicted amino acid frequency distributions at 44 positions of the I-AniI protein–DNA interface was assessed using two standard metrics for comparing probability distributions: the Jensen–Shannon divergence33 and the Euclidean distance. A lower divergence value indicates that the probability distributions better match one another.
The failures of the DNA-Rebuild method are focused on the (+) half of the DNA target site. The interactions with this DNA half-site are implicated in the formation of the catalytic complex;18 thus, it is likely that preservation of the DNA conformation observed in the crystal structure is essential for maintaining activity. Many crystallized protein–DNA complexes contain DNA that is perturbed away from canonical B-form, presumably with a functional purpose. The current implementation of DNA energetics and rebuilding is not yet adequate for capturing the subtleties of these more strained DNA conformations. The DNA-Rebuild method results in low recovery at several I-AniI positions making (+) half-site interactions that do not show significant variation in the experimental data. For example, position Cys150 is maintained as a cysteine or a serine in the experimental data, and the HighTemp-Packer simulation almost exactly produces the frequencies observed experimentally for these two amino acids. The DNA-Rebuild simulation allows numerous amino acids to be incorporated at this position, as the DNA moves away from the crystal structure conformation. The experimental data for position 150 indicates that maintaining the conformation of the bases in this area is likely critical to catalysis. Additionally, the two most conserved residues in the (+) half-site, Lys202 and Tyr154, are lost in most of the DNA-Rebuild simulations. Figure 7 shows that the DNA is rebuilt in such a way that it moves away from the crystal structure conformation. This nonnative DNA conformation allows alternative amino acids to be designed in this area. It is likely that contributions of the DNA conformational state to catalysis in I-AniI are the cause of these inaccurate computational rebuilds. A loss in recovery with the DNA-Rebuild method for other proteins in the benchmark set may similarly be attributable to discrepancies between real and modeled DNA conformational preferences, providing an avenue for improvement of ROSETTA's modeling of DNA flexibility.
Escaping energetic minima with motif-based sequence constraints
Both of the new protocols for diversity generation fail to recover the experimentally preferred amino acid at some I-AniI positions. One of the essential arginine residues in the N-terminal domain, position 61, is highly conserved as the wild-type amino acid and is not observed as arginine with any protocol. Position 24 is a lysine in the native enzyme, and the enzyme tolerates a lysine or a histidine. Neither the DNA-Rebuild nor the HighTemp-Packer recapitulates either of these two possibilities. The previously discussed position 200 is known to be highly active as a lysine (native), asparagine, or arginine, yet none of these amino acids are observed in the computational results.
In order to understand the factors responsible for these mis-designed residues in I-AniI, as well as others in the full sequence recovery set, a modification was made to the previously described protocol for design with motif rotamers. This modified protocol forces amino acid types at each designable protein position to all of the types seen in motifs selected for that position. For example, if both arginine and lysine motifs passed the search procedure for a particular position, the protocol would produce a set of designs with the lysine amino acid type fixed, but not any particular rotamer, at that position, as well as a set with the arginine amino acid type fixed. This sequence constraint can result in sampling of higher-energy alternative structures that better match the wild-type protein sequence, and energetic analysis of these forced amino acids has the potential to reveal why those positions are incorrectly designed without the constraint. In addition, this protocol can be used to generate diverse sequences, revealing many potential native-like interactions instead of only the lowest energy one, for seeding experimental libraries.
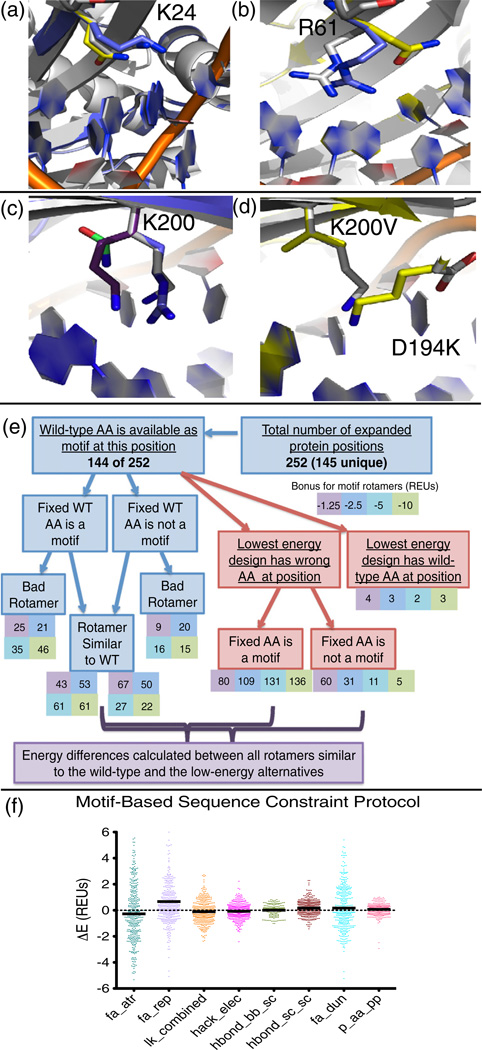
The motif-based sequence constraint method revealed that there is a motif found for every one of the described I-AniI failures. When position 24 is forced to be a lysine, a motif rotamer is incorporated into the design with a very similar conformation to the native lysine (Fig. 8a). The competing low-energy glutamine type is never seen in the experimental interface screen. The difference in total energy between the designs with the lysine and the glutamine is only 0.6 REUs, and when compared to all forced motifs, the design with the forced lysine is the second lowest in energy. The dominant energy term disfavoring arginine at position 61 (Fig. 8b) is the probability of the amino acid given the backbone conformation (p_aa_pp), having a value of 2.46 REUs for the arginine that is forced with the sequence constraint protocol and −0.92 REUs for the lower-energy glutamine type. At position 200, all three of the known, high-activity amino acid types (lysine, arginine, and asparagine) are found to be motifs (Fig. 8c). However, none of these types is designed with the standard motif protocols due to a competing alternative design that incorporates a valine at position 200 and a lysine at the nearby position 194 (Fig. 8d).
Fig. 8.
Motif-based sequence constraints. (a) Lys24 in the I-AniI interface (native rotamer, white) is mis-designed to a glutamine (yellow). The motif-based sequence constraint protocol revealed that position 24 can be a lysine motif, and the motif residue (blue) very closely matches the native lysine. (b) Arg61 in the I-AniI interface (native rotamer, white) is mis-designed to a glutamine (yellow). The motif-based sequence constraint protocol revealed that position 61 can be an arginine motif (blue). (c) The motif-based sequence constraint protocol showed that position Lys200 in the I-AniI interface (native rotamer, white) can be a motif of any of the three amino acid types previously identified to be active at this position (arginine, blue; lysine, purple; and asparagine, green). (d) The alternative low-energy design that disallows any of the motifs in (c) to be designed at position 200. The native structure is shown in white, and the design with K200V and D194K is shown in yellow. (e) Abbreviations: WT, wild type; AA, amino acid. Flowchart summarizing the results of the protocol that generates designs with forced amino acid types for each type of motif identified by the motif search. The protocol was completed only for protein positions that were considered to be true failures of the computational methods by a series of analyses. The chart summarizes the motif status, energetics, and rotameric state of the designs at each of these failed positions. Rotamers are considered similar to the wild-type amino acid if they have an RMSD of <0.8. (f) Energy differences calculated between rotamers that resemble the wild-type amino acid that has a motif rotamer incorporated with a bonus and between the incorrectly designed amino acid observed at this same protein position in the lowest-energy design, as marked on the flowchart in (e). The repulsive energy term (fa_rep) stands out at the biggest contributor to the energy difference between these rotamers.
It was first necessary to determine which interface positions are likely to be the most important for wild-type activity in the absence of experimental data in order to test this motif-biased sequence constraint protocol on proteins other than I-AniI. Given the comprehensive and computationally intensive nature of this protocol, it was additionally necessary to limit its use to a subset of designs. The training set was analyzed to determine the residues that are true failures of the design protocol using a set of metrics described in Materials and Methods. These mis-designed positions are characterized as failures because they are likely important amino acids, as they are amino acids with significant interaction energy, which are designed to a chemically very different amino acid type. The protocol identified 284 of the 3421 designed protein positions from the training set to be failures, which was further reduced to 252 when additional computational constraints due to protein size were taken into account. These design failures were subjected to the described protocol in which the motif residue types are forced at each designable position. This procedure revealed that, for 108 of the 252 positions, a motif of the same type as the wild-type amino acid is not even available (Fig. 8e). For the 144 of these positions where the wild-type amino acid is present in themotifs selected for that position, the number of times that the design actually contains the motif rotamer when the amino acid type is fixed as wild type was found to range from 68 to 107, depending on the motif scoring bonus. The rotameric state of the amino acid making the motif contact was additionally assessed.
For essentially all of the 144 designed positions where a wild-type motif is available, an alternative design sequence that lacked the wild-type amino acid at that position was found to have a lower energy. These designs with the total lowest-energy scores were analyzed to determine the motif status of the mis-designed position. Even for the lowest motif scoring bonus, over half of the positions had a motif rotamer incorporated at the failed position. The components of the energy function were again dissected for each failed protein position by comparing each component from the lowest-energy design and from the design with the forced wild-type amino acid, restricting to positions in which the motif rotamer from the forced wild-type simulation was similar to the native rotamer (Fig. 8f). The results were significantly different from the previous analyses of this type, as the repulsive score (fa_rep) was found to be responsible for the majority of the energy differences between the forced wild-type amino acid and the alternative low-energy designed rotamer. The rotamer probability term is no longer a major component of these differences. These results suggest that the energy function is favoring side chains that are less tightly packed, alleviating the clashes recognized in the high repulsive score.
Visual assessment of design failures suggests future improvements
Human intuition is a valuable tool for assessments of protein interactions.34 Visual analysis of the designs in the training set was used as an additional metric guiding the process of energy function improvement. A large number of the true failures, as determined by analysis described in earlier sections and in Materials and Methods, were visually evaluated in order to gain ideas for the necessary next steps in computational method optimization. While there are many reasons that a design procedure may result in a nonnative amino acid at a protein position, visual analysis of these designs revealed recurrent themes. Four representative design examples are shown in Fig. 9a–d. Of these four examples, one is included to demonstrate how not all mis-designs of the wild-type sequence should be considered failures (Fig. 9a), one was corrected with the HighTemp-Packer sampling strategy described in this work (Fig. 9b), and the remaining two are the result of the fixed-backbone approximation and not optimizing the starting crystal structure in the ROSETTA energy function prior to design (Fig. 9c and d).
Fig. 9.
Representative failures of the computational methods. Native structure, gray; designed structure, pink. (a) The designed lysine, making a canonical contact with the guanine nucleotide, is calculated to interact more strongly with the DNA than the wild-type glutamine (Gln39, 1zs4), and no interactions with neighboring protein positions are lost from this substitution. (b) A histidine (His97, 2fl3) is redesigned to an alanine, and energetic analysis revealed that the rotamer probability term was mainly responsible for the alanine preference. The High-Temp-Packer method corrects this failure, as the histidine is regained in 71% of the design trajectories, compared to 19% with the DNA-Rebuild method. (c) An arginine residue (Arg432, 1j1v), making multiple contacts to both the protein and DNA backbone atoms, is redesigned to a smaller aspartate residue that makes no favorable interactions. The atoms in the starting crystal structure are very close to each other, and the repulsive clashes cannot be relieved without backbone movement or minimization. (d) A bidentate asparagine–asparagine hydrogen bond is lost (Asn70–Asn90, 2ex5). This failure is also due to repulsive clashes with the nearby protein backbone. (e) Amount of atomic overlap Arg432 in the 1j1v crystal structure calculated using MolProbity.36–38 The atomic overlap is shown with yellow and red dots, DNA is black, side chains are cyan, and the protein backbone is brown. This analysis indicates that the protein backbone and neighboring side-chain residues are clashing with Arg432. Backbone optimization would be required to relieve the clash with the backbone. (f) Atomic overlap (yellow and red dots) between an asparagine residue (yellow) and a hydrogen atom (gray) of the beta-carbon of a neighboring serine residue shown in cyan (2ex5, Asn90–Ser68). Hydrogen bonding between this same serine residue and the other asparagine (Asn70–Ser68) of the bidentate asparagine pair is shown with green dots.
For the three representative cases (Fig. 9b–d) where the redesigned sequence is clearly suboptimal to the wild-type sequence, small movements of the backbone of the protein and DNA prior to design would most likely correct the failures. The histidine that was redesigned to an alanine (Fig. 9b) was lost because of an excessively high penalty from the rotamer probability term. The energetic contribution of the rotamer probability is dependent on the backbone structure; thus, subtle movement of the protein backbone would likely correct this failure. For the remaining two cases (Fig. 9c and d), the residues being incorrectly designed are all making interactions with the surrounding protein residues. It is possible that these positions provide protein structural stability and thus binding-site pre-organization for these interfaces.35 The atoms making the primary protein–protein interactions are clashing, as determined by MolProbity,36–38 and constrained on multiple sides by the backbone of the protein or DNA, thus prohibiting repacking and instead favoring redesign to relieve repulsion (Fig. 9e and f). The findings for these two examples match the results of the motif-based sequence constraint protocol that the repulsive term was the major source of the higher energy of the designs containing the forced wild-type amino acid type (Fig. 8f). Optimizing the crystal structures in the ROSETTA energy function prior to design is one potential solution to this issue, although this protocol would need to be thoroughly assessed to ensure that it was not generating a bias in the designed sequences for the wild-type amino acids. One way to avoid this artificially generated bias would be to optimize the structures with a different energy function from an external program.
Discussion
In this work, a number of optimizations to ROSETTA have been thoroughly characterized, including energy function improvements and new protocols for sampling diverse design sequences. Limitations of the computation were illuminated, some of which were addressed and others of which still need to be corrected, and a series of methods and analysis tools were developed to increase the ease of such future endeavors. The question of reliability of sequence recovery as a sole metric for energy function improvement was explored in the context of a particularly well-studied enzyme scaffold. Recapitulation of experimental data is a more relevant metric of protein sequence redesign success than sequence recovery, as it removes the biases of potentially overtraining for recovery of the amino acid states observed in crystal structures and is a more direct measure of the functional effect of allowing a protein sequence to vary. There are many factors contributing to the activity and specificity of DNA-binding or DNA-cleaving proteins, such as the transition between the bound and unbound states and the role of neighboring DNA in the formation of the active complex. A crystal structure reveals one state of the interaction complex, and a computational design tool meant to predict sequence changes required to confer certain activities should be assessed with corresponding experimental data, rather than recapitulation of this single, fixed state. Utilizing this combination of experimental and computational benchmarks has revealed several avenues for continuing improvements of the design methodologies. Additionally, the extensive experimental scan completed in this work provides a better understanding of a class of enzymes being actively engineered as gene therapy reagents, and knowledge on the mutability of each position in this particular enzyme will inform future specificity redesign projects.
The ROSETTA force field integrates physicochemical energy terms and database-derived potentials in order to guide sampling and selection of low-energy amino acid sequences. Similarly, the incorporation of interaction-biased motif rotamers into the standard design process provides a way to integrate the information available in the PDB with the energetic guidance of the ROSETTA force field. The collection of motifs can be considered as a step toward formulating a recognition code39,40 for protein–DNA interactions. The interactions in protein–DNA interfaces are complex and shaped by the local environment, suggesting that the information contained in motifs is best utilized in combination with a tool for assessing the likelihood of a given motif in the context of the entire interaction complex. The method described in this work builds on a previous approach in which the motif interaction is held constant as the protein backbone is remodeled to stabilize the desired contact.22,23 Temiz and Camacho have recently described an alternative computational method for investigating this recognition code that combines homology modeling and molecular dynamics simulations to predict changes in binding affinity for zinc-finger mutants. 41 One significant advantage of this approach over the current ROSETTA methods is that explicit waters were simulated at the interface, allowing for improved modeling of water-mediated interface contacts. The incorporation of explicit water into the ROSETTA protein–DNA interface design calculations is currently under study.
While the addition of the motif rotamers improved the results of the ROSETTA design protocol, the optimization of the force field resulted in an even more significant improvement. Indeed, as the force field was iteratively improved, the optimal value for the motif bonus term decreased, suggesting that the new and modified energy terms were able to preferentially reward native-like protein–DNA interactions. While encouraging, these improvements—when applied in the context of the standard, fixed-backbone design simulation—did not enable successful recapitulation of the variability seen in our I-AniI experimental data set. To explore the potential role of DNA backbone flexibility, we integrated a recently described method1 for generating diverse DNA conformations into our design protocols. Most other programs for protein–DNA interface design, such as FoldX,42 use a fixed-backbone model of the DNA. While preliminary DNA minimization was available in older versions of ROSETTA,2 this new implementation of DNA flexibility is significantly more flexible and provides for greater DNA backbone movement (due to the fact that Monte Carlo fragment rebuilding simulations sample a much larger conformational space than gradient-based minimization initiated at crystal structure conformations). Both this new method of sequence diversity generation and the HighTemp-Packer method, defined by an increase in the final temperature used by the simulated annealing algorithm, improve recapitulation of the experimental data set over standard ROSETTA methods (Fig. 7).
In contrast to protein sequences generated by computational design, the primary function of the amino acids in a protein–DNA interface is not always the stabilization of the lowest-energy state or the tightest possible binding. There also may be a range of binding affinities tolerated for maintaining interface functionality. The wild-type amino acid sequence may not always be the most energetically optimal sequence position at the designed position (Fig. 9a). It is challenging to determine whether the seemingly native-like interactions in the design are really compatible with the activity of the protein–DNA complex. Native complexes are evolved for many functions other than tight binding. The only way to fully assess the viability of the mis-designed amino acids is through experimental characterization. There are several positions in the I-AniI interface where the wild-type amino acid is not the most optimal (Fig. 4). For example, position 18 has a significant preference for tryptophan over the wild-type tyrosine, and the previously discussed position 200 shows high experimental recovery of arginine instead of the wild-type lysine. In these two cases, the preferred amino acid likely confers an increased selective advantage through tighter substrate binding or catalytic complex formation. While these positions are somewhat tolerant of substitutions, they differ from the many highly tolerant positions in the I-AniI interface in that they display a significant preference for a particular amino acid type, rather than allowing all amino acid types equally. A successful computational design tool would capture these nonnative energetic preferences while predicting a lack of preference at the most flexible positions. While it is currently challenging to determine which classes of interface mutations are systematically mis-predicted due to the limited size of our experimental data set, we expect that recent work combining next-generation sequencing technology with protein selection43 will revolutionize studies of this sort that attempt to correlate protein mutations with functional characteristics.
The goal of our work is to develop protocols with clear utility for future design projects. Minimizing the starting structure into the native energy well to alleviate predicted clashes in starting structures (Fig. 9) is likely to artificially enhance sequence recovery by biasing toward the wild-type state. Without proper benchmarks, preferably experimental data, it would be challenging to ensure that this over-optimization of the native state was not biasing the results. In light of the experimental data collected for I-AniI that revealed that a number of interface positions tolerated multiple amino acid types, it is likely that the relatively high sequence recovery of 50% is due to an over-optimization for the native sequence in the context of the rigid, fixed-backbone sequence design simulations. While native sequence recovery has proven to be a powerful metric for optimization of protein design scoring functions, its use as the sole benchmark for protein design sampling algorithms would likely penalize the greater exploration of backbone diversity necessary for successful design toward novel DNA target sites. The experimental data are even an underestimate of the acceptable sequence diversity, since only one position is being allowed to change at a time. Varying multiple positions simultaneously would likely show even less conservation of the wild-type sequence due to correlated changes. Computational protocols producing 100% recovery of the wild-type sequences would almost certainly be useless for design purposes. Instead, it would be best to perfectly recover the amino acids forming essential interactions in the protein–DNA interface and have low recovery and multiple solutions generated for the more malleable positions.
Developing a way to perturb the starting crystal structure on both the protein and the DNA side, without biasing toward the native energy minima, will be important for correcting the failures identified from the sequence recovery benchmarks (Fig. 9). There are a number of possible methods to potentially adapt to provide an alternative method of DNA movement that is less extreme than the fragment insertion protocol tested here.24,44 Both the loss in recovery when using the DNA-Rebuild method and the comparisons to experimental data indicate that less conformational freedom of the DNA is likely to produce higher sequence recovery. However, DNA movement is essential for design of new DNA sequences and for predictions of energetics and specificity involving indirect readout;45,46 thus, it is important to develop a reliable method for accomplishing this goal. Adding protein backbone flexibility will also be necessary for improving recapitulation of experimental data and generating diverse designed sequences.47,48 Flexible loop regions of protein–DNA interfaces could benefit from combining the motif-based approach described here with the previously published method that rebuilds protein backbones to accommodate rotamers that can form motif interactions.22 The results of the simulations completed with the HighTemp-Packer showed promising recapitulation of the variation observed in experimental data. However, the loss of some of the strong motif-like interactions of I-AniI when using this approach suggests that incorporation of the motif information could further enhance the method. One potential way to increase the ease of utilizing the motif information, especially for systems other than protein–DNA interfaces, is to incorporate the data about distances and angles of interactions into a knowledge-based contact potential scoring function.49 For current design applications, we suggest an approach that combines subtler DNA backbone optimization with the HighTemp-Packer and motif rotamers. We hope that these proposed improvements, in conjunction with the newly developed methodologies and analysis tools, will accelerate the progress of future design projects.
Materials and Methods
Computational tools
All protocols were implemented within the ROSETTA molecular modeling package and will be available for free academic use through the ROSETTA Commons. They are currently available to institutions participating in ROSETTA Commons (or upon request), and the code revision numbers are 44353 for trunk ROSETTA and 44354 for the version with the energy function optimized here and the DNA-Rebuild method (source/workspaces/blab/mini). The energy function was similarly optimized for trunk ROSETTA; however, the orientation-dependent desolvation is not available, and the reference energies differed (Fig. S9). These two code versions and energy functions will be integrated in a future release of Rosetta. The executables currently available in both code versions are dna_motif_collector for the generation of motif libraries and motif_dna_packer_design for designing with a motif bias. The flexible DNA simulations are currently limited to the workspaces branch, and the executable that rebuilds the DNA and designs with motifs is called dna_fragment_rebuild_with_motifs. The designs completed with an increased temperature for the low temperature of the simulated annealing algorithm and removal of the final quenching step for the packer are based in the motif_dna_packer_design but require the modification of two lines prior to compilation. These changes are detailed in Supplementary Data. An additional executable, failure_analyzer, for analysis of the design data (failure identification, energy differences between designs) is available in a later revision (source/workspaces/blab/mini, revision 45873). Many parameters of all methods are modifiable via the command line, and all currently available options are discussed in Supplementary Methods. Other data available upon request include, but is not limited to, the final list of PDB codes used to generate the library, the complete motif library either in a single file or in the form of two-residue PDB files, and python analysis scripts (also available in /source/workspaces/sthyme/scripts).
Structural data for training and test sets
A set of 112 largely nonredundant, crystallized protein–DNA complexes all with a resolution of lower than 2.5 Å was downloaded from the RCSB PDB.21 This set split into one group of 48 complexes and another group of 64 complexes; the group containing 48 PDBs was used for training the energy function, and the group containing 64 PDBs was used for testing and analyzing improvements identified from the training procedure. All PDBs were downloaded as the biological assemblies, and several required small modifications for compatibility with the subsequent Rosetta protocols and analysis scripts.
Training set: 1a1f, 1a3q, 1az0, 1bc8, 1bdt, 1bl0, 1ckq, 1d02, 1dc1, 1e3o, 1f4k, 1gd2, 1gu4, 1hcq, 1iaw, 1ig7, 1ign, 1j1v, 1jnm, 1lmb, 1lq1, 1m5x, 1mjo, 1mnm, 1mnn, 1nkp, 1ozj, 1pp7, 1puf, 1r4o, 1r71, 1r7m, 1skn, 1tc3, 1ubc, 1w0u, 1wte, 1zs4, 2bam, 2d5v, 2ex5, 2ezv, 2fl3, 2h27, 2hdd, 2oaa, 2qoj, 3pvi.
Test set: 1a1h, 1a73, 1aay, 1am9, 1b3t, 1b94, 1dfm, 1dmu, 1dp7, 1egw, 1g2f, 1g9y, 1hcr, 1hwt, 1i3j, 1jey, 1jft, 1k61, 1mey, 1mow, 1mus, 1nvp, 1oe5, 1oup, 1qpi, 1r0o, 1sa3, 1tup, 1xbr, 2bop, 2c9l, 2dgc, 2e52, 2fqz, 2o4a, 2odi,2or1, 2wt7, 2x6v, 2xqc, 2xsd, 2z3x, 3bm3, 3bs1, 3c25, 2co6, 3fc3, 2fdq, 3h0d, 3iag, 3igm, 3jtg, 3jxb, 3jy1, 3lnq, 3m4a, 3mln, 3mqy, 3mx4, 3n7q, 3o9x, 3pvv, 3qqy, 6pax.
Generation of motif library
A motif is defined as the spatial arrangement of six atoms. In the case of a protein–DNA motif, three of these atoms are located on a DNA base that interacts with a protein residue, and the other three are derived from that protein residue (Fig. 1). This geometric relationship is expressed as a translation vector and a set of Euler angles, as previously described.22 The atoms that define motifs are currently fixed for different amino acid and DNA residues. Motifs were collected from protein–DNA complexes with a resolution of better than 2.8 Å that were downloaded from the RCSB PDB on August 9, 2011. The set initially consisted of 1459 complexes, which was reduced to 1375 complexes after removal of PDBs that were not compatible with Rosetta without manipulation of the PDB files or modification of Rosetta.
The motif library used for this work includes both major and minor groove interactions, as well as water-mediated contacts. The collection algorithm is defined by iteration over every protein residue in each of the protein–DNA complexes and the identification of up to two DNA bases that have the greatest amount of ROSETTA interaction energy with that protein residue. This interaction energy between the protein and the DNA residue is defined as a packing score (combined attractive and repulsive energies), a direct side-chain–side-chain hydrogen-bonding score, and a water-mediated hydrogen-bonding score, if a theoretical water can be placed at a canonical location on the DNA base.50 The protein–DNA pair must have either a packing score of less than −0.5 REUs, a direct hydrogenbonding score of less than −0.3 REUs, or a water-mediated hydrogen-bonding score of less than −0.3 REUs in order to count as a motif interaction.
Redundancy in the motif library arises mainly from the inclusion of multiple crystal structures of the protein– DNA complex or from equivalent monomers of homo-oligomeric complexes. The amino acid and DNA residue pairs are all placed in the same coordinate frame, based around the motif atoms of the DNA base, for all interactions involving that type of DNA residue in order to reduce this redundancy. Any DNA residue that has less than 0.2 RMSD over the heavy atoms with any other DNA residue is eliminated from the motif library.
Removal of homologous motifs from the motif library
Prior to identifying motif interactions that can be made in a particular protein–DNA complex, it is necessary to remove motifs derived from that same PDB entry or from one of a homologous protein. The inclusion of such motifs would result in artificial biases toward the native sequence. The protocol developed for the removal consists of a BLAST51 run against the PDB database that identified all structures with an e-value of less than 0.05 to the starting structure and a python script to parse the output of the BLAST run and to remove homologous motifs from the library.
Identification of rotamers forming motif interactions
The utilization of motifs in fixed-backbone protein design requires the identification of amino acid rotamers that are capable of forming a motif interaction in a given protein–DNA complex. Backbone-dependent rotamers derived from the Dunbrack rotamer library,26 included with the ROSETTA software, are built at protein positions in a protein–DNA interface in order to accomplish this goal. Interface positions are identified using a previously described protocol17 that builds a set of arginine rotamers at each protein position and checks whether any nucleotide base atom is within 3.8 Å of these arginine side chains. For this motif search protocol, the level of rotamer sampling was set to include extra sampling at χ1–4, as well as an additional four half-step deviations from the bin of the rotamer. Each rotamer is screened against all nearby DNA bases to test whether a motif interaction can be made, and it must pass several cutoffs to be considered a successful rotamer. First, a single atom from a canonical DNA base defined by the motif being tested, currently the C1*, is placed via the defined motif orientation. A distance between this atom and every nearby C1* in the crystal structure DNA is calculated. Passing a defined distance cutoff, set to be 2.0 Å for these experiments, allows the rotamer to be subject to further testing. The next test screens for how parallel a motif-placed canonical base is to the closest crystal structure base by the calculation of a dot product for vectors perpendicular to the plane of the six atoms of a placed nucleobase and the crystal structure nucleobase. The dot product for these experiments was set to be greater than 0.97 to be considered for a final test of the RMSD over the same six atoms of the nucleobase compared with the nearby crystal structure nucleobase. For these experiments, the RMSD had to be less than 1.0 in order for the rotamer to be able to make a successful motif contact. Both the distance and RMSD cutoffs are automatically reduced for motifs with longer side chains that have many more rotamers. Cutoffs for arginine are cut twofold, and cutoffs for methionine, lysine, glutamate, and glutamine are cut by a third. All rotamers passing the cutoffs are then sorted, dependent on a combined score of the RMSD and dot product (RMSD divided by dot product), and the lowest scored rotamers are preferentially considered to be successful if the user indicates a limit on the number of rotamers to be utilized by further design protocols. The default limit is set to be 100 rotamers of each amino acid type at each protein position being designed, and this default was maintained in the experiments described here.
Motif-biased design
Rotamers identified to make motif interactions with the search procedure described in the preceding section are incorporated into the standard design procedure by adding them to the rotamer set being used by the packer. For these experiments, the initial rotamer set included extra sampling of χ1 and χ2 and three one-third step additional deviation samples for χ1 and χ2 of aromatic residues. The packer provides the core functionality for ROSETTA design, utilizing a Monte Carlo simulated annealing algorithm, guided by a physically based atomic-level force field.16 These motif rotamers are flagged and can be given an energy bonus over other rotamers in the rotamer set. The flag is implemented as a residue patch called SpecialRotamer, and the energy term special_rot allows for the user to implement differential bonuses for these rotamers. Alternatively, there are input options that support the definition of a starting motif bonus and a subsequent number of steps of twofold reduction of that bonus, producing multiple designs each with a different bias toward inclusion of these rotamers. The designs completed in this work cover the range of bonuses from −10 to −1.25. Additional designs where motif rotamers are added with no weight and where motif rotamers are left out of the rotamer set are produced by default. Identification of protein positions where mutation of the protein sequence is allowed is described in the section on collection of motif rotamers, as it occurs by the same method. An additional shell of residues surrounding these designable residues is allowed to change rotamer conformation, but not protein sequence.
For the sequence recovery work, individual design runs were done at every single base pair in the interface, simulating the approach used for specificity redesign where only a small group of amino acids are designed simultaneously. Energy function analysis and optimization was guided by sequence recovery calculations. Two metrics, weighted and unweighted recovery, were calculated for each set of design calculations. The unweighted metric counts every designed position equally, and the weighted metric is an average over the recoveries for each amino acid type and free from biases in the amino acid composition of the interface positions. The inclusion of the weighted metric during optimization is necessary to avoid artificial improvements in overall recovery due to biasing the energy function toward recovery of amino acids that are overrepresented in protein–DNA interfaces, namely, lysine and arginine, at the expense of the less abundant types. A previously improved weight set17 that was optimized without consideration of the weighted metric contains this particular bias (Fig. S3).
Flexible DNA interface design
The use of the flexible DNA interface design protocol was limited to computationally tractable PDBs that were compatible with the DNA movement portions of the protocol without any modification or reformatting. This method consists of a previously described1 DNA rebuilding step followed by a motif-biased design run. For each targeted DNA design, that base pair and the two surrounding base pairs were allowed to move. Unpaired DNA base pairs, DNA strands containing chain internal chain breaks, or base pairs on the end or one away from the end of DNA chain were not included because they are not compatible with the DNA rebuilding portion of the protocol. After each design calculation, the rebuilt DNA was allowed to minimize prior to the next design iteration (between each round of lowering the motif bonus).
Rebuild set: 1a1f, 1a1h, 1a3q, 1aay, 1az0, 1bc8, 1bdt, 1bl0, 1ckq, 1d02, 1dc1, 1e3o, 1egw, 1f4k, 1g2f, 1gd2, 1gu4, 1hcq, 1hwt, 1i3j, 1ig7, 1ign, 1j1v, 1jnm, 1lq1, 1m5x, 1mey, 1mnm, 1mnn, 1nkp, 1oe5, 1ozj, 1pp7, 1puf, 1r0o, 1r71, 1r7m, 1sa3, 1skn, 1tc3, 1ubd, 1w0u, 1wte, 1xbr, 1zs4, 2bam, 2c9l, 2d5v, 2e52, 2ex5, 2ezv, 2fl3, 2h27, 2hdd, 2o4a, 2oaa, 2qoj, 2wt7, 2xsd, 2z3x, 3c25, 2co6, 3fc3, 3fdq, 3h0d, 3iag, 3jtg, 3jxb, 3lnq, 3m4a, 3mln, 3mx4, 3n7q, 3o9x, 3pvi, 3pvv, 3qqy, 6pax.
Identification of failed design pockets
The metrics designating an incorrectly designed position as not being a true failure are as follows: (1) the correct amino acid type being seen for over 25% of the design runs from the set of designs completed with a varying motif weight, indicating that the wild type is favorable in the context of a motif bonus; (2) the wild-type amino acid making very little contact to any protein or DNA residue, as defined by a total ROSETTA interaction energy with all nearby residues of no more than −2 REUs; (3) the wild-type amino acid being one of the smallest amino acid types because native protein–DNA interfaces are not always optimized for the tight binding and high specificity that the computational methods are programmed to produce and a small amino acid type being redesigned to a larger one with more contacts is potentially an acceptable change that could increase interface affinity; and (4) the designed amino acid being chemically related to the wild-type amino acid and likely to be making a similar contact, such as a glutamate being redesigned to a glutamine. Future implementations could utilize atom-type-specific analyses for a more accurate assessment of contact success.
Bacterial screen
A bacterial screen for active variants of I-AniI was completed as previously described,28 albeit with minor modifications. Electrocompetent Escherichia coli cells, the DH12S strain from Invitrogen, were transformed with a pCCDb plasmid containing two adjacent copies of the IAniI LIB4 target site,30 a variant of the wild-type target site containing two activating substitutions. This pCCDb-containing strain was prepared for the selection using a standard procedure for electro-competent cell preparation. Each of the 44 libraries, corresponding to the 44 interface positions, was ligated, and the pCCDb-containing electro-competent cells were transformed with the purified ligation products. Transformants were recovered in terrific broth media for a half-hour at 37 °C. The selection procedure was completed for 4 h in 2 mL liquid culture at 30 °C. Following liquid selection, 1 µL was plated on each of minimal selection (100 µg/mL carbenicillin, 1 mM IPTG, and 0.02% L-arabinose) and control (100 µg/mL carbenicillin) plates (1.5% agar, M9 salt, 1% glycerol, 0.8% tryptone, 0.2% thiamine, 1 mM MgSO4, and 1 mM CaCl2) and grown for ca 36 h at 30 °C. Approximately 20 colonies were picked from each selection plate for each of the 44 positions, grown overnight in 96-well culture plates, and submitted for sequencing as 96-well-plate glycerol stocks to the GENEWIZ sequencing facility.
Construction of plasmids and libraries
The pCCDb plasmid containing the I-AniI LIB430 target sites was built by phosphorylating and annealing oligo-nucleotides from Integrated DNA Technologies to form a duplex with sticky ends compatible with the NheI and SacII restriction sites in the pCCDb vector.28 An amino acid library was built for each of the 44 protein interface positions, using assembly PCR52 with oligonucleotides containing an NNS codon (Integrated DNA Technologies) at the randomized position. These libraries were ligated into pEndo vector28 between the NcoI and NotI restriction sites and screened for activity in the bacterial selection system. All C-terminal I-AniI libraries (starting at position 148) were built in the context of the activating M58 mutations, and all N-terminal mutations (from position 18 to position 72) were built in the context of M4, which is M5 without the I55V mutation.
Supplementary Material
Acknowledgements
The authors would like to thank Jim Havranek and Justin Ashworth for previous code development specifically related to DNA design, as well as the entire ROSETTA Commons community for contributions to the ROSETTA code base. Florian Richter and Matthew D. Smith provided helpful discussion with regard to, respectively, the project design and writing of this manuscript. This work was supported by a National Science Foundation graduate research fellowship to S.B.T., the US National Institutes of Health (grants #GM084433 and #RL1CA133832 to D.B.; grant #GM088277 to P.B.), the Foundation for the National Institutes of Health through the Gates Foundation Grand Challenges in Global Health Initiative, and the Howard Hughes Medical Institute.
Abbreviations used
- RCSB
Research Collaboratory for Structural Bioinformatics
- PDB
Protein Data Bank
- REU
ROSETTA energy unit
Footnotes
Supplementary Data
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2012.03.005
References
- 1.Yanover C, Bradley P. Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2zinc fingers. Nucleic Acids Res. 2011;39:4564–4576. doi: 10.1093/nar/gkr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morozov AV, Havranek JJ, Baker D, Siggia ED. Protein–DNA binding specificity predictions with structural models. Nucleic Acids Res. 2005;33:5781–5798. doi: 10.1093/nar/gki875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashworth J, Baker D. Assessment of the optimization of affinity and specificity at protein– DNA interfaces. Nucleic Acids Res. 2009;37:e73. doi: 10.1093/nar/gkp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–215. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Pâques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarjour J, West-Foyle H, Certo MT, Hubert CG, Doyle L, Getz MM, et al. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37:6871–6880. doi: 10.1093/nar/gkp726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urnov FD, Rebar EJ, Holmes MC, Zhang S, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev., Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 11.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotech. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 12.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Guchateau P, Pâques F. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Jr, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 14.Voigt CA, Mayo SL, Arnold FH, Wang Z. Computational method to reduce the search space for directed protein evolution. Proc. Natl Acad. Sci. USA. 2001;98:3778–3783. doi: 10.1073/pnas.051614498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 16.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, et al. ROSETTA3: an object-oriented software suite for simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashworth J, Taylor GK, Havranek JJ, Quadri SA, Stoddard BL, Baker D. Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs. Nucleic Acids Res. 2010;38:5601–5608. doi: 10.1093/nar/gkq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, et al. Exploitation of binding energy for catalysis and design. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulge UY, Baker DA, Monnat RJ., Jr Comprehensive computational design of mCreI homing endonuclease cleavage specificity for genome engineering. Nucleic Acids Res. 2011;39:4330–4339. doi: 10.1093/nar/gkr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havranek JJ, Duarte CM, Baker D. A simple physical model for the prediction and design of protein–DNA interactions. J. Mol. Biol. 2004;344:59–70. doi: 10.1016/j.jmb.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havranek JJ, Baker D. Motif-directed flexible backbone design of functional interactions. Protein Sci. 2009;18:1293–2205. doi: 10.1002/pro.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy PM, Bolduc JM, Gallaher JL, Stoddard BL, Baker D. Alteration of enzyme specificity by computational loop modeling and design. Proc. Natl Acad. Sci. USA. 2009;106:9215–9220. doi: 10.1073/pnas.0811070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutationinduced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 26.Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1668. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel AD, Kim PS. Modular structure of transcription factors: implications for gene regulation. Cell. 1991;165:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- 28.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 29.Szeto MD, Boissel SJ, Baker D, Thyme SB. Mining endonuclease cleavage determinants in genomic sequence data. J. Biol. Chem. 2011;286:32617–32627. doi: 10.1074/jbc.M111.259572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amitai G, Gupta RD, Tawfik DS. Latent evolutionary potentials under the neutral mutational drift of an enzyme. HFSP J. 2007;1:67–78. doi: 10.2976/1.2739115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloom JD, Romero PA, Lu Z, Arnold FH. Neutral drift can alter promiscuous protein functions, potentially aiding functional evolution. Biol. Direct. 2007;2:17. doi: 10.1186/1745-6150-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J. Divergence measures based on the Shannon entropy. IEEE Trans. Inform. Theory. 1991;37:145–151. [Google Scholar]
- 34.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, et al. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleishman SJ, Khare SD, Koga N, Baker D. Restricted sidechain plasticity in the structures of native proteins and complexes. Protein Sci. 2011;20:753–757. doi: 10.1002/pro.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: allatom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen VB, Davis IW, Richardson DC. KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 2009;18:2403–2409. doi: 10.1002/pro.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews BW. Protein–DNA interaction. No code for recognition. Nature. 1988;335:294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- 40.Pabo CO, Nekludova L. Geometric analysis and comparison of protein–DNA interfaces: why is there no simple code for recognition? J. Mol. Biol. 2000;301:597–624. doi: 10.1006/jmbi.2000.3918. [DOI] [PubMed] [Google Scholar]
- 41.Temiz NA, Camacho CJ. Experimentally based contact energies decode interactions responsible for protein–DNA affinity and the role of molecular waters at the binding interface. Nucleic Acids Res. 2009;37:4076–4088. doi: 10.1093/nar/gkp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alibes A, Serrano L, Nadra AD. Structure-based DNA-binding prediction and specificity. Methods Mol. Biol. 2010;649:77–88. doi: 10.1007/978-1-60761-753-2_4. [DOI] [PubMed] [Google Scholar]
- 43.Araya CL, Fowler DM. Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol. 2011;9:435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith CA, Kortemme T. Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant sidechain prediction. J. Mol. Biol. 2008;380:742–756. doi: 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steffen NR, Murphy SD, Tolleri L, Hatfield GW, Lathrop RH. DNA sequence and structure: direct and indirect recognition in protein– DNA binding. Bioinformatics. 2002;18:S22–S30. doi: 10.1093/bioinformatics/18.suppl_1.s22. [DOI] [PubMed] [Google Scholar]
- 46.Becker NB, Wolff L, Everaers R. Indirect readout: detection of optimized sequences and calculation of relative binding affinities using different DNA elastic potentials. Nucleic Acids Res. 2006;34:5638–5649. doi: 10.1093/nar/gkl683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith CA, Kortemme T. Predicting the tolerated sequences for proteins and protein interfaces using RosettaBackrub flexible backbone design. PLoS One. 2011;6:e20451. doi: 10.1371/journal.pone.0020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu X, Apgar JR, Keating AE. Modeling backbone flexibility to achieve sequence diversity: the design of novel α-helical ligands for Bcl-xL . J. Mol. Biol. 2007;371:1099–1117. doi: 10.1016/j.jmb.2007.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kono H, Sarai A. Structure-based prediction of DNA target sites by regulatory proteins. Proteins. 1999;35:114–131. [PubMed] [Google Scholar]
- 50.Jiang L, Kuhlman B, Kortemme T, Baker D. A “solvated rotamer” approach to modeling water-mediated hydrogen bonds at protein–protein interfaces. Proteins. 2005;58:893–904. doi: 10.1002/prot.20347. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Stemmer WPC, Crameri A, Ha KD, Brennan TM, Heyneker HL. Singlestep assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.