Abstract
Streptomyces bacteria are known for producing important natural compounds by secondary metabolism, especially antibiotics with novel biological activities. Functional studies of antibiotic-biosynthesizing gene clusters are generally through homologous genomic recombination by gene-targeting vectors. Here, we present a rapid and efficient method for construction of gene-targeting vectors. This approach is based on Streptomyces phage φBT1 integrase-mediated multisite in vitro site-specific recombination. Four ‘entry clones’ were assembled into a circular plasmid to generate the destination gene-targeting vector by a one-step reaction. The four ‘entry clones’ contained two clones of the upstream and downstream flanks of the target gene, a selectable marker and an E. coli-Streptomyces shuttle vector. After targeted modification of the genome, the selectable markers were removed by φC31 integrase-mediated in vivo site-specific recombination between pre-placed attB and attP sites. Using this method, part of the calcium-dependent antibiotic (CDA) and actinorhodin (Act) biosynthetic gene clusters were deleted, and the rrdA encoding RrdA, a negative regulator of Red production, was also deleted. The final prodiginine production of the engineered strain was over five times that of the wild-type strain. This straightforward φBT1 and φC31 integrase-based strategy provides an alternative approach for rapid gene-targeting vector construction and marker removal in streptomycetes.
Introduction
Members of the Gram-positive, spore-producing genus Streptomyces play critical roles in soil ecology and are useful for synthesizing medically and industrially important secondary metabolites. Genomic sequencing of the model organism Streptomyces coelicolor A3(2) was completed in 2002 [1], which dramatically facilitated gene function studies of these bacteria, especially those involved in antibiotic production through targeted modification [2]. Additionally, with the increased data of whole genomic actinomycetes sequences, robust and rapid methods are required to study the functions of a large number of important genes. The gene-targeting approach through homologous recombination was the first step in construction of gene-targeting vectors. The traditional method of constructing vectors is by step-by-step enzymatic digestion and ligation following PCR amplification and assembly of independent DNA segments into a plasmid. However, this method is complicated and time consuming, and often limited by lack of proper restriction enzymatic sites.
Many methodologies have been developed for gene targeting vector construction [3], [4], [5], [6], especially those of recombination-based strategies [4], [5], [7], [8], [9], [10], [11], [12], [13], [14]. Among these, a powerful method, termed REDIRECT© technology (PCR-targeting system) [4], [7], [15] has been widely used in Streptomyces. This procedure is based on phage λ-Red proteins to promote recombination, which employs Redα (exo), Redβ (bet) and Redγ (gam) to mediate recombination when only tens of nucleotides are homologous to the target region [4]. In E. coli with helper plasmids, PCR-amplified selectable markers using primers with only 39 nt homology extensions are used to amplify chromosomal sequences within a genomic cosmid library [15] to generate the desired gene-targeting vectors. Another universal cloning method, referred to as Gateway® technology [8], is based on phage λ site-specific integrase between attB and attP sites and excision between attL and attR sites. Using expanded properties of recombination sites with unique specificities, many segments could be cloned into a vector backbone [16], thereby expediting several simple one-week methods to construct gene-targeting vectors [5], [12], [14]. These two phage λ-recombination-based technologies are simple and efficient, and have been combined together for constructing gene targeting vectors to generate knock-out mice [17]. Furthermore, the μ transposon was developed to allow random insertion of selectable markers and other desired sequences into destination plasmids for rapid generation of gene-targeting vectors [3], [18], [19], which complemented existing recombination-based approaches for generation of gene-targeting constructs [3].
We previously established a highly efficient in vitro site-specific recombination system based on Streptomyces phage φBT1 integrase and identified the minimal sizes of attB and attP sites (36-bp and 48-bp, respectively), which was smaller than that of λ site-specific recombination (25-bp and 200-bp, respectively) [20]. We selected 16 pairs of non-compatible recombination sites, of which the central dinucleotides were not identical, and inhibited DNA strand exchange and religation, thus no recombination could occur between site pairs containing different core sequence mutations [21]. Here, we report a simple and highly efficient system for marker-free gene targeting in S. coelicolor by combining φBT1 integrase-mediated multisite recombination in vitro, homologous recombination and φC31 integrase-mediated site-specific recombination in vivo [22]. This simple strategy should be readily suitable and advantageous for poorly genetically established Streptomyces systems without an ordered cosmid library, coupled with a desire to knockout longer DNA segments, and could be easily adopted to other organisms to construct gene-targeting vectors.
Using this method, we constructed an S. coelicolor strain for overproduction of prodiginine (Red), which is one of the four main antibiotics produced by S. coelicolor A3(2) with anti-fungal, anti-bacterial, anti-protozoan, anti-malarial, immunosuppressive and anti-cancer activities [23], [24]. The biosynthesis of calcium-dependent antibiotic (CDA) and actinorhodin (Act), which might influence Red production by competition of common precursors, were disrupted by homologous recombination after parts of the key genes of these two biosynthetic clusters were deleted. For CDA, nonribosomal peptide synthetase (NRPS) coding genes cdaPS1, cdaPS2 and part of cdaPS3, were replaced by the apramycin resistance gene aac(3)IV; and for Act, structural genes (actIII to actVB) were deleted. In addition, one of the TetR family protein genes, rrdA, which negatively regulates Red production by controlling the abundance of RedD mRNA [25], was also deleted using our method. The final Red production of the engineered strain was over five times that of the wild-type strain.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli DH5α [26] was used for plasmid propagation. Mannitol soy flour [27] agar was used to generate spores and select for S. coelicolor exconjugants. R2YE agar was used for phenotype screening. R4 liquid medium (100 mg casamino acids, 1 g yeast extract, 3 g proline, 10 g MgCl2·6H2O, 10 g glucose, 4 g CaCl2·2H2O, 5.6 g TES, and 0.2 ml trace element in 1.0 L H2O) was used for antibiotic production. The conjugal transfer from E. coli ET12567/pUZ8002 into S. coelicolor was performed as described previously [27]. Bacillus mycoides Flugge ATCC 6462 was used as indicator strain for CDA production assay [27]. Antibiotics were added at the following final concentrations: ampicillin, 50 µg ml−1; apramycin, 30 µg ml−1; chloramphenicol, 34 µg ml−1; kanamycin, 30 µg ml−1; and thiostrepton, 20 µg ml−1.
Table 1. Strains and plasmids used in this study.
| Strains or plasmids | Genotype or description | Ref. or source |
| S. coelicolor | ||
| M145 | SCP1- SCP2- | [27] |
| ZB1 | M145 with CDA gene cluster disrupted, containing aac(3)IV gene copy in the chromosome | This work |
| ZB2 | ZB1 with the resistance gene removed | This work |
| ZB3 | ZB2 with Act gene cluster disrupted, containing a aphII gene copy in the chromosome | This work |
| ZB4 | ZB3 with the resistance gene removed | This work |
| ZB7 | ZB4 with rrdA gene disrupted, containing a aphII gene copy in the chromosome | This work |
| ZB8 | ZB7 with the resistance gene removed | This work |
| ZB8/pFDZ16-rrdA | ZB8 carrying integrative plasmid pFDZ16-rrdA | This work |
| ZB8/pFDZ16 | ZB8 carrying integrative plasmid pFDZ16 | This work |
| E. coli | ||
| DH5α | F- recA lacZ ΔM15 | [26] |
| ET12567 | dam dcm hsdS | [27] |
| Plasmids | ||
| pMD19-T | 2.7-kb cloning vector; Ampr | Takara |
| pBC-AM | Donor of aac(3)IV; Aprar Cmr | [25] |
| pHZ1358 | E. coli-Streptomyces shuttle vector; Ampr Thior | [25] |
| pXD34-int | E. coli-Streptomyces shuttle vector; derivative obtained from pIJ6021, containing tipA promoter and the φC31 integase gene; Aprar Thior Kanr | This work |
| pFDZ100 | E. coli-Streptomyces shuttle vector; derivative obtained from pHZ1358, containing attP 0 site and attB 15 site; Thior Ampr Cmr | This work |
| pTA0006 | Derivative obtained from pMD19-T, containing attB 0 site and attP 6 site; Aprar | [32] |
| pTA0613 | Derivative obtained from pMD19-T, containing attB 6 site and attP 13 site; Aprar | [32] |
| pTA1315 | Derivative obtained from pMD19-T, containing attB 13 site and attP 15 site; Aprar | This work |
| pFDZ101 | Derivative obtained from pTA0006, containing attB 0-φC31 site | This work |
| pFDZ102 | Derivative obtained from pTA0613, replacing the aac(3)IV resisitance gene with the aphII resistance gene | This work |
| pFDZ103 | Derivative obtained from pTA1315, containing attP 0-φC31 site | This work |
| pFDZ101-CDA-5′arm | Derivative obtained from pFDZ101, containing upstream homologous arm of CDA biosynthetic gene cluster gene-targeting | This work |
| pFDZ103-CDA-3′arm | Derivative obtained from pFDZ103, containing downstream homologous arm of CDA biosynthetic gene cluster gene-targeting | This work |
| pFDZ101-Act-5′arm | Derivative obtained from pFDZ101, containing upstream homologous arm of Act biosynthetic gene cluster gene-targeting | This work |
| pFDZ103-Act-3′arm | Derivative obtained from pFDZ103, containing downstream homologous arm of Act biosynthetic gene cluster gene-targeting | This work |
| pFDZ101-rrdA-5′arm | Derivative obtained from pFDZ101, containing upstream homologous arm of rrdA gene | This work |
| pFDZ103-rrdA-3′arm | Derivative obtained from pFDZ103, containing downstream homologous arm of rrdA gene | This work |
| pFDZ100-CDA-tandem | Derivative obtained from pFDZ100, containing two homologous arms of CDA biosynthetic gene cluster gene-targeting; Aprar | This work |
| pFDZ100-Act-tandem | Derivative obtained from pFDZ100, containing two homologous arms of Act biosynthetic gene cluster gene-targeting; Kanr | This work |
| pFDZ100-rrdA-tandem | Derivative obtained from pFDZ100, containing two homologous arms of rrdA gene; Kanr | This work |
| pZB101 | E. coli-Streptomyces shuttle vector; derivative obtained from pHZ1358, containing tipA promoter and the φC31 integase gene; Kanr Thior Ampr | This work |
| pZB102 | E. coli-Streptomyces shuttle vector; derivative obtained from pZB101, replacing the aphII resisitance gene with the aac(3)IV resistance gene | This work |
| pSET152 | Integrative vector for actinomycetes; containing oriT, int, and attP-φC31 site, Aprar | [27] |
| pRT802 | E.coli-Streptomyces shuttle plasmid, encoding φBT1-int and attP, resistant to kanamycin | [28] |
| pFDZ16 | E. coli-Streptomyces integrative shuttle vector containing tipA promoter, Kanr, Thior, Ampr. | [25] |
| pFDZ16-rrdA | Derivative obtained from pFDZ16, containing the rrdA gene located downstream of the tipA promoter, Kanr, Thior, Ampr. | [25] |
Construction of the plasmids
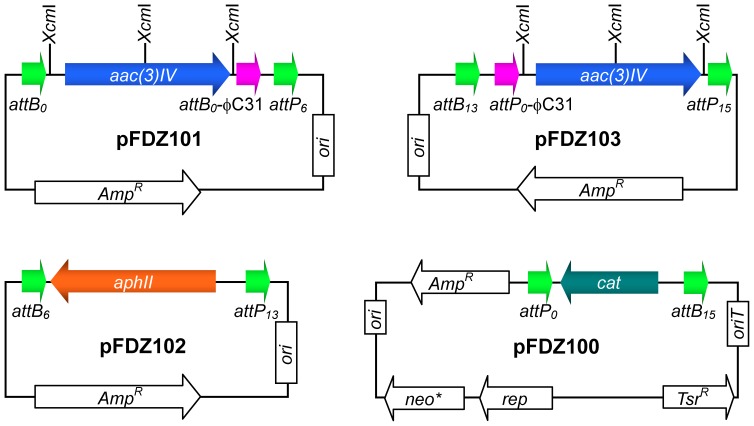
All plasmids used in this study are described in Table 1, and primers are listed in Table S1. To construct the ‘entry vectors’, three 1.0 kb cassettes, attB0-aac(3)IV-attP6, attB6-aac(3)IV-attP13, and attB13-aac(3)IV-attP15, were PCR-amplified using the PrimeStar® PCR Kit (TaKaRa, Kyoto, Japan) and ligated into the pMD19-T vector (TaKaRa) to generate plasmids pTA0006, pTA0613 and pTA1315 (see Table 1). Primers ZB153 and ZB154 (containing attB0-φC31), ZB155 (containing attP0-φC31) and ZB156 were used to amplify the aac(3)IV gene fragment, which was inserted into XcmI-linearized pTA0006 and pTA1315 to generate pFDZ101 and pFDZ103. These two plasmids were used to carry homologous arms after XcmI digestion (see Fig. 1). Plasmid pSET152 was used as a template for the aac(3)IV gene.
Figure 1. Map of four ‘entry vectors’ used in this study.
Plasmids pFDZ101 and pFDZ103 were used to harbor the 5′-arm and 3′-arm respectively, and pFDZ102 contained the selectable marker (aphII gene). pFDZ100 was the backbone of the gene-targeting vector. Each vector contained a pair of non-compatible recombination φBT1 integration sites (arrows in green). The pink arrows represent the recombination φC31 integration sites, where recombination occurred to remove the marker gene.
Primer pairs ZB129/ZB130 and ZB131/132 were used to amplify two 2.1 kb homologous arm fragments of the CDA biosynthetic gene cluster, and the PCR products were inserted into pFDZ101 and pFDZ103 to generate the pFDZ101-CDA 5′arm and pFDZ103-CDA 3′arm, respectively. Primer pairs ZB125/ZB126 and ZB127/128 were used to amplify the 2 kb upstream homologous arm and 2.2 kb downstream homologous arm fragments of the Act biosynthetic gene cluster, and the PCR products were inserted into pFDZ101 and pFDZ103 to generate the pFDZ101-Act 5′arm and pFDZ103-Act 3′arm, respectively. Using the same method, we obtained the pFDZ101-rrdA 5′arm and the pFDZ103-rrdA 3′arm, containing a 1.6 kb upstream homologous arm fragment and a 1.7 kb downstream homologous arm fragment of the rrdA gene, respectively, which were amplified using primer pairs ZB180/ZB181 and ZB182/ZB183.
Primers Oxj128 and Oxj129 were used to amplify the aphII gene fragment from pRT802 [28], which was inserted into the XcmI-linearized pTA0613 to generate pFDZ102. Fragment amplifications containing the cat gene from pBC-AM [25] flanked by attP0 and attB15 were generated using the primer pair PTP00/PTB15 and the product was digested with XbaI/BamHI and inserted into the E. coli-Streptomyces shuttle vector pHZ1358 [27], which contained the plasmid origin of transfer (oriT) to generate pFDZ100. pFDZ100 is very unstable in streptomycetes and is easily lost when not maintained with antibiotic selection.
Construction of plasmid pXD34-int is described in the Supplemental Materials section. Plasmid pXD34-int was cut with EcoRV/NheI to generate a 7.1-kb fragment, which contained the tipA promoter and φC31 integrase gene, and then inserted into the corresponding sites in vector pHZ1358, yielding plasmid pZB101. pZB101 was further digested with StuI/Ecl136II and then linked with a 1.5 kb SmaI-linearized fragment containing the aac(3)IV gene from plasmid pBC-AM [25] (Table 1) to generate pZB102. pZB102 was subsequently conjugated from the donor E. coli ET12567/pUZ8002 into the null mutants to remove the resistance gene by φC31 integrase-mediated in vivo site-specific recombination (see Fig. 2B). The exconjugants were selected by growth on MS media supplemented with thiostrepton (20 µg ml−1) and apramycin (30 µg ml−1). The thiostrepton was used to induce the expression of φC31 integrase. As plasmids pZB101 and pZB102 were derived from pHZ1358, which contained the sti DNA region (strong incompatibility locus), they could be easily lost in non-resistance stress condition [27].
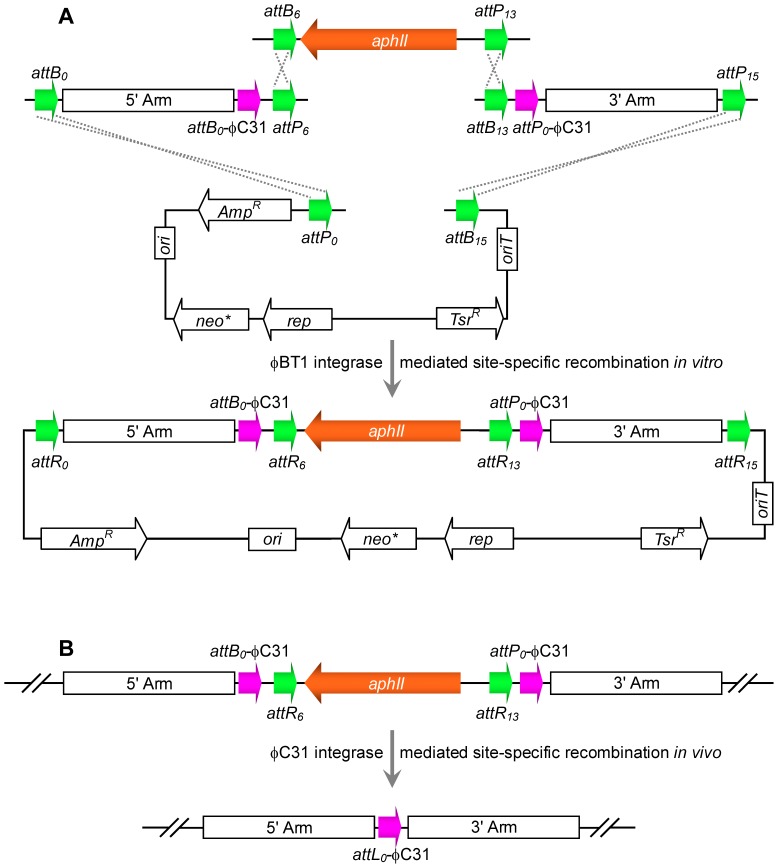
Figure 2. Strategy of tandem assembly in vitro and the marker removing in vivo based on site-specific recombinations.
(A) One step assembly of the gene-targeting vector by φBT1 integrase-mediated site-specific recombination in vitro. Four recombination reactions occurred simultaneously between attB0/attP0, attB6/attP6, attB13/attP13, and attB15/attP15. (B) Deletion of marker by φC31 integrase-mediated site-specific recombination in vivo. The reaction occurred between attB0-φC31 and attP0-φC31 to remove the marker and leave attL0-φC31.
Standard in vitro recombination assays and verification of the positive assembly products
Expression and purification of φBT1 integrase was performed as described previously [20]. The recombination reaction was performed in a reaction mixture (10 µl) containing 10 mM Tris-HCl, pH 8.0, 100 mM KCl, 50 mM NaCl, 5% glycerol, integrase (4 U in 0.5 µl, ∼75 nM) and ∼30 ng DNA reaction substrates [21]. The reaction was incubated at 30°C overnight and treated with proteinase K at 55°C for 30 min and then heated to 80°C for 20 min for inactivation and transformed into E. coli strain DH5α. After selection with antibiotics, plasmids of positive clones were isolated and verified by enzyme digestion.
CDA production assay
The CDA bioassay was carried out as previously described by Kieser et al. [27]. Spores were spotted onto an Oxoid nutrient agar plate and incubated for 2 days, then overlaid with soft nutrient agar containing Bacillus mycoides and calcium nitrate at a concentration to yield 12 mM throughout the plate. CDA produced by S. coelicolor killed the B. mycoides cells, resulting in a clearance zone on the plate.
Quantification of Red production
Red production was assayed according to previous descriptions [27]. A culture grown in 40 ml R4 liquid medium was filtered to separate the supernatant and pellet. The mycelium pellet was collected and dried under a vacuum, then extracted with methanol overnight at room temperature. The methanol was acidified with HCl (to a final concentration of 0.5 M). Optical density was measured at 530 nm. Measurements were always performed for triplicate samples.
Results and Discussion
Rapid construction of the gene-targeting vectors by site-specific recombination in vitro
Streptomyces phage φBT1 integrase is capable of catalyzing accurate and efficient site-specific in vitro recombination between attB and attP sites [20]. In our previous work, 16 pairs of non-compatible sites containing different core sequences were identified, which confirmed that no recombination could occur between site pairs containing different core sequence mutations [20]. Thus, these sites could be used for multisite cloning [21], [29]. We established a multiple DNA fragment assembly method, termed site-specific recombination-based tandem assembly (SSRTA). SSRTA details and principles were described in our previous report [4], [7], [15]. As shown in Fig. 1, plasmids pFDZ101 and pFDZ103 were digested with XcmI to produce 3′-T overhangs on both ends. Homologous arms could be inserted by TA cloning to generate the pFDZ101 5′-arm and pFDZ103 3′-arm. Plasmid pFDZ102 containing the aphII gene was used for clone selection in E.coli and Streptomyces. Plasmid pFDZ100 was used for propagation of the assembled gene-targeting vector in E.coli and transformed into Streptomyces by conjugation. Fig. 2A demonstrated the one-step tandem assembly of the four DNA fragments into a circular plasmid by φBT1 integrase-mediated in vitro site-specific recombination. Each fragment was flanked by a pair of non-compatible recombination sites, and after incubation with φBT1 integrase in proper buffer conditions (see Materials and Methods), the four fragments were assembled together by four simultaneous recombination events.
Using this method, we successfully constructed three gene-targeting vectors. Vectors pFDZ100-CDA-tandem and pFDZ100-Act-tandem were used to disrupt the biosynthesis of CDA and Act, respectively, and pFDZ100-rrdA-tandem was used for rrdA gene deletion. Construction details of the three vectors are in the Supplemental Information. This tandem assembly method for gene-targeting vector construction was efficient and convenient, and the resistance genes (aac(3)IV and aphII in this study) could be easily replaced by any others in pTA0613 and pFDZ102. Plasmids pFDZ101∼pFDZ103 could be XcmI-digested to produce 3′-T overhangs on both ends and the homologous PCR-amplified arm fragments can be indirectly inserted by TA cloning to generate the pFDZ101 5′arm and the pFDZ103 3′arm, which avoided the limitation of available restriction recognition sites and restriction recognition sites contained in the homologous arms. Thus the construction procedure was condensed to approximately one week by our method.
Markerless disruption of CDA, Act biosynthetic gene clusters and Deletion of the rrdA gene
Cross-regulation of endogenous gene clusters in S. coelicolor M145 might involve competition for common precursors [25]. Therefore, we deleted parts of two main antibiotic biosynthesis gene clusters, calcium-dependent antibiotic (CDA) and actinorhodin (Act), to disrupt their production in S. coelicolor M145 in order to increase prodiginine (Red) production. After pFDZ100-CDA-tandem conjugation into S. coelicolor M145, parts of the CDA biosynthetic gene cluster containing NRPS-encoding genes cdaPS1, cdaPS2 and part of cdaPS3 (sco3228∼sco3232, ∼42 kb), were replaced by the apramycin resistance gene, aac(3)IV. Double-crossover colonies were obtained by screening the Aprar Thios exconjungants to generate the mutant strain ZB1 (see Table 1). We then transferred the marker-removal plasmid pZB101, which expresses the φC31 integrase, into ZB1 by conjugation. As shown in Fig. 2B, the resistance gene was flanked by attB0-φC31 and attP0-φC31. After in vivo site-specific recombination between these two sites catalyzed by φC31 integrase, the resistance gene in the plasmid pTA0613 would be removed, leaving one attL0-φC31 in the genome. Because the central dinucleotide attP0-φC31 sequence was not identical to the wild type attB site of φC31 integrase, no reaction occurred between attP0 and attB in the genome. Exconjungants were screened in the Thior Kanr MS media, and after the resistance gene was removed, the mutant strain was named ZB2 (see Table 1). We then constructed the CDA and Act biosynthetic double deficient strain ZB3 by further deleting part of the Act biosynthetic gene cluster (including key genes required for ACT biosynthesis, ranging from sco5087 to sco5092, ∼3.6 kb). After deletion of the resistance gene aphII by marker-removal plasmid pZB102, the strain was named ZB4 (see Table 1).
In our previous work, we identified the rrdA gene as a TetR family protein gene, which regulated secondary metabolite production in S. coelicolor by negative regulation of Red biosynthesis and controlling RedD mRNA abundance [25]. Thus, we used the pFDZ100-rrdA-tandem to further knock-out the rrdA gene from the mutant strain ZB4 for Red overproduction. The mutated strains before and after removing the resistance gene aphII were named ZB7 and ZB8 (see Table 1).
Due to the high efficiency and accuracy of the site-specific recombination reaction, the ratio of positive exconjungants with marker removal was about 50%, and the entire process was very time-efficient. Including the construction of plasmids, the deletion of the gene cluster and removal of selectable markers, the whole process took less than a month. Furthermore, compared to other marker excision systems (Flp, Cre, Dre recombinases) [30], [31], φC31 integrase-mediated recombination is highly directional in the absence of the excisionase. As no resistance marker was preserved in the chromosome, this approach avoided the limitations of selectable resistance markers and was quite convenient for the following gene replacement, especially for multiple gene knock-outs.
PCR verification of the mutants and phenotypes
Each null mutant was verified by PCR amplification using a pair of oligonucleotide primers specific for the flanking chromosomal DNA sequences of the target gene. The results are shown in Fig. 3A. In lane 1, no band was seen because the PCR fragment size was very large (∼42 kb), which almost contained the entire CDA biosynthesis gene cluster, and could not be amplified. The further verification was also carried on by PCR amplification using two different pairs of oligonucleotide primers. One pair of primers was synthesized according to the internal sequences of the deleted gene clusters or gene, and the other one was based on the internal sequences of the resistance gene. The results were shown in Fig. 3B and Fig. 3C. No band was amplified with the former primers and a 500 bp-size band could be seen clearly with the later primers in the double-crossover null mutant. It turned out just the opposite result in the wild type Streptomyces coelicolor M145. The confirmation of removing the resistance gene was seen in Fig. 3A and Fig. 3C. The PCR products had also been sequenced to ensure excision of the antibiotic cassettes occurred precisely (Fig. S1). The aac(3)IV gene was used as a selectable marker in ZB1 while using aphII in ZB3 and ZB7, due to cross-resistance (<50 µg/ml) of the aac(3)IV gene to kanamycin that could have complicated the marker-removal process by false positives. However, almost no cross-resistance (<5 µg/ml) of the aphII gene to apramycin occurred.
Figure 3. PCR analysis and phenotypes of mutants.
(A) PCR verification of gene deletion. Lane 1, a 42,725 bp fragment was amplified using primers ZB287/ZB288. The size of the fragment was too large and could not be PCR-amplified, thus no band was seen in lane 1. Lane 2, a 1981 bp fragment was amplified using primers ZB287/ZB288. The CDA biosynthetic gene cluster was replaced with the aac(3)IV gene. Lane3, a 927 bp fragment was amplified using primers ZB287/ZB288, and the resistance gene was removed. Lane 4, a 4073 bp fragment was amplified with primers ZB289/ZB290. Lanes 5 and 6 showed the bands before and after removing the aphII resistance gene. Lane 7, a 928 bp fragment was amplified with primers ZB285/ZB286. Lanes 8 and 9 showed thebands before and after removing the aphII resistance gene. WT, wild type strain; ZB1∼ZB8, serial mutant strains. (B) PCR verification of gene deletion. Lane 1: the fragment was amplified with primers ZB145/ZB146 and primers were both within sco3229. Lane 2: the fragment was amplified with primers ZB195/ZB196 and primerZB195 was within sco5089 and primer ZB196 was within sco5090. Lane 3: the fragment was amplified with primers ZB184/ZB186 and primerZB184 was within sco1103 and primer ZB184 was within sco1104. Lane 4: the fragment as a positive control was amplified with primers ZB147/ZB148 and primers were both within sco5888. M: 150 bp ladder. (C) PCR verification of the resistance removing. Lane 1: M145; Lane 2: ZB1; Lane 3: ZB2. The three fragments were amplified with primers ZB189/ZB190, primers were both within the acc(3)IV gene. Lane 4: M 145; Lane 5: ZB3; Lane 6: ZB4; Lane 7: ZB7; Lane 8: ZB8. The five fragments were amplified with primers ZB187/ZB188, primers were both within the aphII gene. M: 150 bp ladder. (D) Bioassay of CDA extracts from the WT strain and CDA-null mutant strain ZB2. (E) Phenotypes of wild-type and three mutant strains. The spores were cultured for 2 days (left) and 5 days (right) on R2YE agar.
Mutant phenotypes are shown in Fig. 3D and 3E. CDA production analysis was carried out in line with previous descriptions [25], [27]. B. mycoides was used as an indicative strain to detect CDA activity. First, spores were spotted onto oxoid nutrient agar plates, air-dried and cultured for 36∼48 h in a 30°C incubator. Then, the plate was overlaid with the soft nutrient agar containing B. mycoides and Ca(NO3)2 at a 12 mM final concentration and CDA activity was detected after one day. CDA produced by wide-type M145 could kill B. mycoides cells, thus a clear inhibition zone was seen on the plate, but no inhibition zone was observed during CDA biosynthesis by the mutant (see Fig. 3D).
The phenotypes of the wild-type strain and the mutants ZB2, ZB4 and ZB8 are exhibited in Fig. 3E. The loss of blue pigment production indicated that Act biosynthesis was disrupted. The deletion of the rrdA gene was shown by an increase of red pigment production (at 2 and 5 days).
Analysis of growth and prodiginine (Red) production
To assess mutant effects on the growth and development of the different strains, each mutant and the parent strain were spotted onto the rich R2YE agar medium plate, then spores were incubated at 30°C for 2 to 5 days to test the growth performance. All strains exhibited no apparent differences in aerial mycelia formation or sporulation (Fig. 3E). Strain ZB4 and ZB8 failed to produce Act.
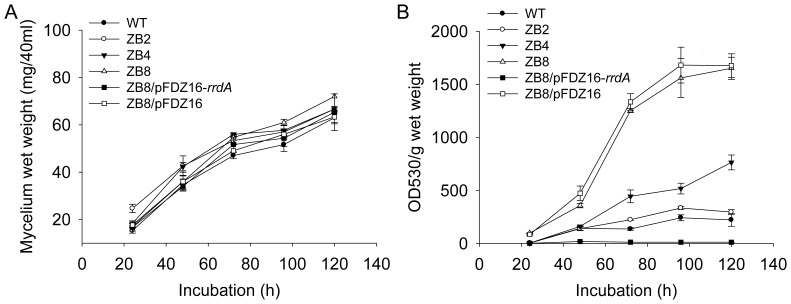
The wild-type and mutated strains were cultured in 40 ml of R4 liquid medium to investigate the growth rate and prodiginine production quantitatively. Spores were inoculated in a reciprocal shaker (180 rpm) at 30°C with an inoculation of 2×107spores per ml. The mycelia were harvested at different time points and treated as described in the Materials and Methods section. As shown in Fig. 4A, no statistical significance in growth rate was observed among all the strains, and the mutated strains (ZB2, ZB4 and ZB8) produced more prodiginine than the wild-type strain (Fig. 4B). Red overproduction might be attributable to competition for common precursors of CDA, Act and Red biosynthesis [25]. Red production of the double null mutant strain ZB4(ΔCDA ΔAct) was approximately 3.5 times that of the wild-type strain, and this further suggested competition for common precursors of the three biosynthetic pathways. Furthermore, our previous study [25] showed that Red production of the rrdA-null mutant was two-fold higher than that of the wild-type strain by releasing the negative control of redD mRNA abundance. In the present study, Red production in strain ZB8 was over five-fold higher than the wild-type strain (Fig. 4B). In order to illustrate the important role of rrdA more clearly, we tried to overexpress rrdA in ZB8 strain by introducing the plasmid pFDZ16-rrdA and red production showed much lower level than wild-type strain. However, the complementation strain harboring the empty vector pFDZ16 produced almost the same prodiginine production as ZB8 strain (Fig. 4B). Thus, the rrdA as a negative regulator of Red production played an important role. The Red-overproducing strain ZB8 could be rapidly constructed using our method.
Figure 4. Growth curves and prodiginine production of wild-type strain, three mutant strains and complemented strains.
Growth curves (A) and prodiginine production (B) of M145, ZB2, ZB4, ZB8, ZB8/pFDZ16 and ZB8/pFDZ16-rrdA growth in 40 ml of R4 liquid medium. Incubation was carried out at 30°C. The symbols indicate the averages of three independent determinations and the error bars indicate the standard errors. OD530, optical density at 530 nm.
In conclusion, we described a rapid and efficient method for marker-free S. coelicolor mutagenesis by site-specific recombination. This approach was time-saving and facilitated multiple deletion rounds in the Streptomyces genome. The utility of this method was clearly demonstrated by construction of a prodiginine (Red) overproducing strain, ZB8, which produced five times the amount of Red compared to the wild-type strain. This strategy provided an alternative approach for rapid markerless modification of the actinobacterial genome.
Supporting Information
Primers used in this study.
(DOCX)
Construction of plasmids pXD34-int, pFDZ100-CDA-tandem, pFDZ100-Act-tandem and pFDZ100- rrdA -tandem.
(DOCX)
Sequence analysis of PCR products for verification of the markerless mutants. Primers ZB469 (for ZB2), ZB472 (for ZB4) and ZB473 (for ZB8) were used for sequencing the PCR products.
(TIFF)
Acknowledgments
We would like to thank Prof. Meifeng Tao (Laboratory of Microbial Metabolism, Shanghai Jiaotong University, China) for providing the strain Bacillus mycoides.
Funding Statement
This work was supported by grants from the National High Technology Research and Development Program of China (863 Program) (grant number 2007AA021206), National Basic Research Program of China (973 Program) (grant number 2012CB721102), the China Postdoctoral Science Foundation funded project (grant numbers 2012T50444 and 2012M520947) and the National Natural Science Foundation of China (grant numbers 30600009, 30830002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) . Nature 417: 141–147. [DOI] [PubMed] [Google Scholar]
- 2. Raynal A, Karray F, Tuphile K, Darbon-Rongere E, Pernodet JL (2006) Excisable cassettes: New tools for functional analysis of Streptomyces genomes. Appl and Environ Microbiol 72: 4839–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vilen H, Eerikainen S, Tornberg J, Airaksinen MS, Savilahti H (2001) Construction of gene-targeting vectors: A rapid Mu in vitro DNA transposition-based strategy generating null, potentially hypomorphic, and conditional alleles. Transgenic Res 10: 69–80. [DOI] [PubMed] [Google Scholar]
- 4. Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100: 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iiizumi S, Nomura Y, So S, Uegaki K, Aoki K, et al. (2006) Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques 41: 311–316. [DOI] [PubMed] [Google Scholar]
- 6. Frandsen RJN, Andersson JA, Kristensen MB, Giese H (2008) Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. Bmc Mol Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang YM, Buchholz F, Muyrers JPP, Stewart AF (1998) A new logic for DNA engineering using recombination in Escherichia coli . Nat Genet 20: 123–128. [DOI] [PubMed] [Google Scholar]
- 8. Hartley JL, Temple GF, Brasch MA (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10: 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu PT, Jenkins NA, Copeland NG (2003) A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cotta-De-Almeida V, Schonhoff S, Shibata T, Leiter A, Snapper SB (2003) A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res 13: 2190–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu CL, Wehr DR, Edwards J, Hauge B (2008) Rapid one-step recombinational cloning. Nucleic Acids Research 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan W, Costantino N, Li RX, Lee SC, Su Q, et al. (2007) A recombineering based approach for high-throughput conditional knockout targeting vector construction. Nucleic Acids Res 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inoue K, Sone T, Oneyama C, Nishiumi F, Kishine H, et al. (2009) A versatile nonviral vector system for tetracycline-dependent one-step conditional induction of transgene expression. Gene Ther 16: 1383–1394. [DOI] [PubMed] [Google Scholar]
- 15.Gust B, Kieser T, Chater KF (2002) REDIRECT technology: PCR targeting system in Streptomyces coelicolor A3(2). John Innes Centre, Norwich Research Park, Colney, Norwich NR47UH, UK. [Google Scholar]
- 16. Cheo DL, Titus SA, Byrd DRN, Hartley JL, Temple GF, et al. (2004) Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: Functional analysis of multi-segment expression clones. Genome Res 14: 2111–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu S, Ying GX, Wu Q, Capecchi MR (2008) A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. Nat Protoc 3: 1056–1076. [DOI] [PubMed] [Google Scholar]
- 18. Zhang CF, Kitsberg D, Chy H, Zhou Q, Morrison JR (2005) Transposon-mediated generation of targeting vectors for the production of gene knockouts. Nucleic Acids Res 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turakainen H, Saarimaki-Vire J, Sinjushina N, Partanen J, Savilahti H (2009) Transposition-Based Method for the Rapid Generation of Gene-Targeting Vectors to Produce Cre/Flp-Modifiable Conditional Knock-Out Mice. PLoS ONE 4: e4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L, Ou XJ, Zhao GP, Ding XM (2008) Highly efficient in vitro site-specific recombination system based on Streptomyces phage φBT1 integrase. J Bacteriol 190: 6392–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang L, Wang L, Wang J, Ou X, Zhao G, et al. (2010) DNA cleavage is independent of synapsis during Streptomyces phage φBT1 integrase-mediated site-specific recombination. J Mol Cell Biol 2: 264–275. [DOI] [PubMed] [Google Scholar]
- 22. Thorpe HM, Smith MCM (1998) In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci U S A 95: 5505–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williamson NR, Fineran PC, Leeper FJ, Salmond GPC (2006) The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4: 887–899. [DOI] [PubMed] [Google Scholar]
- 24. Williamson NR, Simonsen HT, Ahmed RAA, Goldet G, Slater H, et al. (2005) Biosynthesis of the red antibiotic, prodigiosin, in Serratia: identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces . Mol Microbiol 56: 971–989. [DOI] [PubMed] [Google Scholar]
- 25. Ou XJ, Zhang B, Zhang L, Zhao GP, Ding XM (2009) Characterization of rrdA, a TetR Family Protein Gene Involved in the Regulation of Secondary Metabolism in Streptomyces coelicolor . Appl Environ Microbiol 75: 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 27.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. Norwich, United Kingdom: The John Innes Foundation. [Google Scholar]
- 28. Gregory MA, Till R, Smith MCM (2003) Integration site for streptomyces phage φBT1 and development of site-specific integrating vectors. J Bacteriol 185: 5320–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turan S, Bode J (2011) Site-specific recombinases: from tag-and-target- to tag-and-exchange-based genomic modifications. FASEB J 25: 4088–4107. [DOI] [PubMed] [Google Scholar]
- 30. Herrmann S, Siegl T, Luzhetska M, Petzke L, Jilg C, et al. (2012) Site-specific recombination strategies for engineering actinomycete genomes. Appl Environ Microbiol 78: 1804–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fedoryshyn M, Petzke L, Welle E, Bechthold A, Luzhetskyy A (2008) Marker removal from actinomycetes genome using Flp recombinase. Gene 419: 43–47. [DOI] [PubMed] [Google Scholar]
- 32. Zhang L, Zhao G, Ding X (2011) Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination. Sci Rep 1: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.
(DOCX)
Construction of plasmids pXD34-int, pFDZ100-CDA-tandem, pFDZ100-Act-tandem and pFDZ100- rrdA -tandem.
(DOCX)
Sequence analysis of PCR products for verification of the markerless mutants. Primers ZB469 (for ZB2), ZB472 (for ZB4) and ZB473 (for ZB8) were used for sequencing the PCR products.
(TIFF)