Abstract
Thyroid hormone is essential for normal metabolism and development, and overt abnormalities in thyroid function lead to common endocrine disorders affecting approximately 10% of individuals over their life span. In addition, even mild alterations in thyroid function are associated with weight changes, atrial fibrillation, osteoporosis, and psychiatric disorders. To identify novel variants underlying thyroid function, we performed a large meta-analysis of genome-wide association studies for serum levels of the highly heritable thyroid function markers TSH and FT4, in up to 26,420 and 17,520 euthyroid subjects, respectively. Here we report 26 independent associations, including several novel loci for TSH (PDE10A, VEGFA, IGFBP5, NFIA, SOX9, PRDM11, FGF7, INSR, ABO, MIR1179, NRG1, MBIP, ITPK1, SASH1, GLIS3) and FT4 (LHX3, FOXE1, AADAT, NETO1/FBXO15, LPCAT2/CAPNS2). Notably, only limited overlap was detected between TSH and FT4 associated signals, in spite of the feedback regulation of their circulating levels by the hypothalamic-pituitary-thyroid axis. Five of the reported loci (PDE8B, PDE10A, MAF/LOC440389, NETO1/FBXO15, and LPCAT2/CAPNS2) show strong gender-specific differences, which offer clues for the known sexual dimorphism in thyroid function and related pathologies. Importantly, the TSH-associated loci contribute not only to variation within the normal range, but also to TSH values outside the reference range, suggesting that they may be involved in thyroid dysfunction. Overall, our findings explain, respectively, 5.64% and 2.30% of total TSH and FT4 trait variance, and they improve the current knowledge of the regulation of hypothalamic-pituitary-thyroid axis function and the consequences of genetic variation for hypo- or hyperthyroidism.
Author Summary
Levels of thyroid hormones are tightly regulated by TSH produced in the pituitary, and even mild alterations in their concentrations are strong indicators of thyroid pathologies, which are very common worldwide. To identify common genetic variants associated with the highly heritable markers of thyroid function, TSH and FT4, we conducted a meta-analysis of genome-wide association studies in 26,420 and 17,520 individuals, respectively, of European ancestry with normal thyroid function. Our analysis identified 26 independent genetic variants regulating these traits, several of which are new, and confirmed previously detected polymorphisms affecting TSH (within the PDE8B gene and near CAPZB, MAF/LOC440389, and NR3C2) and FT4 (within DIO1) levels. Gender-specific differences in the genetic effects of several variants for TSH and FT4 levels were identified at several loci, which offer clues to understand the known sexual dimorphism in thyroid function and pathology. Of particular clinical interest, we show that TSH-associated loci contribute not only to normal variation, but also to TSH values outside reference range, suggesting that they may be involved in thyroid dysfunction. Overall, our findings add to the developing landscape of the regulation of thyroid homeostasis and the consequences of genetic variation for thyroid related diseases.
Introduction
Through the production of thyroid hormone (TH), the thyroid is essential for normal development, growth and metabolism of virtually all human tissues. Its critical role in heart, brain, bone, and general metabolism is illustrated by the clinical manifestations of thyroid disease, which affects up to 10% of the population. Low thyroid function (i.e., hypothyroidism) can lead to weight gain, high cholesterol, cognitive dysfunction, depression, and cold intolerance, whereas hyperthyroidism may result in weight loss, tachycardia, atrial fibrillation, and osteoporosis. Mild variation in thyroid function, both subclinical and within the normal range, is associated with these TH-related clinical outcomes as well [1]–[4].
The thyroid gland secretes predominantly the pro-hormone thyroxine (T4), which is converted into the active form triiodothyronine (T3) in peripheral tissues. The production of TH by the thyroid gland is regulated by the hypothalamus-pituitary-thyroid (HPT) axis, via a so-called negative feedback loop. Briefly, low levels of serum TH in hypothyroidism result in an increased release of thyroid stimulating hormone (TSH) by the pituitary, under the influence of hypothalamic thyrotropin releasing hormone (TRH) [5]. TSH, a key regulator of thyroid function, stimulates the synthesis and secretion of TH by the thyroid. When circulating TH levels are high, as in hyperthyroidism, TRH and TSH synthesis and secretion are inhibited.
In healthy (euthyroid) individuals, TSH and free T4 (FT4) levels vary over a narrower range than the broad inter-individual variation seen in the general population, suggesting that each person has a unique HPT axis set-point that lies within the population reference range [6]. Besides environmental factors such as diet, smoking and medication, little is known about the factors that influence this inter-individual variation in TSH and FT4 levels [7]–[9]. The heritability of TSH and FT4 has been estimated from twin and family studies at about 65% and 40%, respectively [10]–[12]. However, the underlying genetic variants are not fully established, and the contribution of those discovered so far to the overall variance is modest. Single nucleotide polymorphisms (SNPs) in the phosphodiesterase type 8B (PDE8B), upstream of the capping protein (actin filament) muscle Z-line, β (CAPZB) and, more recently, of the nuclear receptor subfamily 3, group C, member 2 (NR3C2) and of v-maf musculoaponeurotic fibrosarcoma oncogene homolog (MAF/LOC440389) genes have been implicated in TSH variation by genome-wide association studies (GWAS) [13]–[15], whereas SNPs in the iodothyronine deiodinase DIO1 have been associated with circulating levels of TH by candidate gene analysis [16]–[18].
To identify additional common variants associated with thyroid function, we performed a meta-analysis of genome-wide association data in 26,420 euthyroid individuals phenotyped for serum TSH and 17,520 for FT4 levels, respectively. In addition, we also assessed gender-specific effects and correlation with subclinical thyroid dysfunction.
Results
To identify common genetic variants associated with serum TSH and FT4 levels, we carried out a meta-analysis of genome-wide association results from 18 studies for TSH and 15 studies for FT4 levels, which assessed the additive effect of ∼2.5 million genotyped and HapMap-imputed SNPs in relation to those traits in individuals of European ancestry (for cohort description see Table 1 and Table S1). In order to avoid bias due to the presence of thyroid pathologies, prior to analysis we excluded all individuals with TSH values outside the normal range (TSH<0.4 mIU/L and TSH>4.0 mIU/L) and those taking thyroid medication for known thyroid pathologies whenever the relevant information was available. Our meta-analysis was thereby carried out in up to 26,420 and 17,520 euthyroid subjects, respectively for TSH and FT4. Additional exclusion criteria used by individual cohorts are detailed in Table S1.
Table 1. Descriptive statistics of all cohorts.
| Cohort | Subjects (N) | Age, mean (SD) | Age (range) | Men (%) | TSH, mean (SD) | FT4, mean (SD) |
| BLSA | 593 | 69.9 (15.4) | 22–98 | 54.8 | 2.1 (0.9) | 1.1 (0.2) |
| CHS | 1,655 | 74.6 (4.9) | 67–94 | 41.8 | 2.1 (0.9) | 1.2 (0.2) |
| FHS | 2,140 | 47.4 (10.0) | 21–77 | 49.9 | 1.6 (0.8) | NA |
| GARP | 290 | 60.3 (7.5) | 42–79 | 20.3 | 1.9 (0.8) | 1.2 (0.2) |
| HBCS | 454 | 60.9 (2.8) | 56–68 | 51.3 | 1.8 (0.8) | 1.1 (0.1) |
| InChianti | 951 | 68.4 (15.4) | 21–102 | 45.3 | 1.5 (0.8) | 1.4 (0.3) |
| LBC1921 | 401 | 79.0 (0.6) | 77–80 | 44.0 | 1.7 (0.8) | 1.1 (0.2) |
| LBC1936 | 834 | 69.5 (0.8) | 67–71 | 54.0 | 1.7 (0.8) | 1.2 (0.2) |
| LifeLines | 1,306 | 45.0 (10.0) | 20–79 | 44.6 | 2.7 (4.1) | 1.3 (0.2) |
| LLS | 736 | 59.1 (6.8) | 30–75 | 45.9 | 1.7 (0.8) | 1.2 (0.2) |
| MICROS | 1,047 | 44.6 (16.5) | 8–94 | 45.7 | 1.9 (0.9) | 1.0 (0.2) |
| NBS | 1,617 | 61.5 (10.3) | 27–78 | 50.7 | 1.6 (1.1) | 1.1 (0.2) |
| OOA | 1,025 | 49.9 (16.7) | 20–97 | 57.8 | 2.2 (0.5) | NA |
| PROSPER | 4,402 | 75.3 (3.4) | 69–83 | 49.1 | 1.9 (0.8) | 1.3 (0.2) |
| RS | 1,346 | 68.7 (7.4) | 55–93 | 40.7 | 1.6 (0.8) | 1.3 (0.2) |
| SardiNIA | 4,087 | 42.5 (17.7) | 14–101 | 46.9 | 1.7 (0.8) | 1.3 (0.2) |
| TwinsUK | 2,133 | 46.6 (12.5) | 18–82 | 0 | 1.4 (0.7) | 1.1 (0.1) |
| ValBorbera | 1,403 | 53.6 (18.3) | 18–102 | 46.9 | 1.5 (0.8) | NA |
The table shows descriptive statistics of all cohorts included in the meta-analysis. TSH is reported in mIU/L and FT4 in ng/dl. SD, standard deviation.
Using the standard genome-wide threshold of 5×10−8, we observed significant associations for SNPs at 23 loci, of which 19 were associated with TSH, and 4 with FT4 (Figure S1). The results are presented in Table 2 and Figure 1, Figure 2, Figure 3, Figure 4, Figure 5. In Table S2 single cohort results for each GW significant SNP are reported.
Table 2. Independent SNPs associated with TSH and FT4 serum levels.
| Gene | SNP | Chr | Position | A1/A2 | Freq A1 | Effect | StdErr | P | N | Het P |
| TSH levels | ||||||||||
| PDE8B | rs6885099 | 5 | 76566105 | A/G | 0.594 | −0.141 | 0.009 | 1.95×10−56 | 26042 | 0.520 |
| PDE10A | rs753760 | 6 | 165966473 | C/G | 0.691 | 0.100 | 0.010 | 1.21×10−24 | 25988 | 0.363 |
| CAPZB | rs10799824 | 1 | 19713761 | A/G | 0.161 | −0.113 | 0.012 | 3.60×10−21 | 26031 | 0.042 |
| MAF/LOC440389 | rs3813582 | 16 | 78306854 | T/C | 0.674 | 0.082 | 0.010 | 8.45×10−18 | 25948 | 0.292 |
| VEGFA | rs9472138 | 6 | 43919740 | T/C | 0.285 | −0.079 | 0.010 | 6.72×10−16 | 25767 | 0.017 |
| VEGFA | rs11755845 | 6 | 44012758 | T/C | 0.266 | −0.065 | 0.010 | 1.68×10−10 | 25710 | 0.417 |
| NR3C2 | rs10032216 | 4 | 149888956 | T/C | 0.781 | 0.087 | 0.011 | 9.28×10−16 | 26053 | 0.504 |
| IGFBP5 | rs13015993 | 2 | 217333768 | A/G | 0.736 | 0.078 | 0.010 | 3.24×10−15 | 26016 | 0.605 |
| SOX9 | rs9915657 | 17 | 67639131 | T/C | 0.541 | −0.064 | 0.009 | 7.53×10−13 | 25692 | 0.349 |
| NFIA | rs334699 | 1 | 61393084 | A/G | 0.052 | −0.141 | 0.021 | 5.40×10−12 | 25757 | 4.05×10−3 |
| FGF7 | rs10519227 | 15 | 47533656 | A/T | 0.245 | −0.072 | 0.011 | 1.02×10−11 | 25988 | 0.098 |
| PRDM11 | rs17723470 | 11 | 45184143 | T/C | 0.279 | −0.065 | 0.010 | 8.83×10−11 | 26054 | 0.833 |
| MIR1179 | rs17776563 | 15 | 86920108 | A/G | 0.322 | −0.060 | 0.010 | 2.89×10−10 | 25758 | 0.452 |
| INSR | rs4804416 | 19 | 7174848 | T/G | 0.569 | −0.057 | 0.009 | 3.16×10−10 | 25632 | 0.438 |
| ABO | rs657152 | 9 | 135129086 | A/C | 0.343 | 0.058 | 0.009 | 4.11×10−10 | 25765 | 1.22×10−4 |
| ITPK1 | rs11624776 | 14 | 92665344 | A/C | 0.660 | −0.064 | 0.011 | 1.79×10−9 | 23482 | 0.845 |
| NRG1 | rs7825175 | 8 | 32535816 | A/G | 0.210 | −0.066 | 0.011 | 2.94×10−9 | 25996 | 0.711 |
| MBIP | rs1537424 | 14 | 35643769 | T/C | 0.608 | −0.052 | 0.009 | 1.17×10−8 | 25478 | 0.333 |
| SASH1 | rs9497965 | 6 | 148562985 | T/C | 0.415 | 0.051 | 0.009 | 2.25×10−8 | 25980 | 0.444 |
| GLIS3 | rs1571583 | 9 | 4257209 | A/G | 0.249 | 0.057 | 0.010 | 2.55×10−8 | 25766 | 0.118 |
| FT4 levels | ||||||||||
| DIO1 | rs2235544 | 1 | 54148158 | A/C | 0.510 | 0.138 | 0.012 | 7.87×10−32 | 17226 | 0.193 |
| LHX3 | rs7860634 | 9 | 138229500 | A/G | 0.530 | 0.102 | 0.013 | 2.30×10−14 | 14529 | 0.067 |
| FOXE1 | rs7045138 | 9 | 99631284 | T/C | 0.553 | 0.098 | 0.015 | 1.50×10−11 | 10997 | 0.457 |
| AADAT | rs11726248 | 4 | 171290094 | A/G | 0.106 | 0.111 | 0.019 | 5.20×10−9 | 17515 | 0.972 |
| LPCAT2/CAPNS2 | rs6499766 | 16 | 54161629 | A/T | 0.478 | 0.056 | 0.012 | 1.18×10−6 | 17489 | 0.269 |
| NETO1/FBXO15 | rs7240777 | 18 | 69318732 | A/G | 0.5632 | −0.049 | 0.012 | 3.13×10−5 | 17146 | 7.84×10−3 |
The table shows the association results for SNPs that reached genome-wide level (p<5×10−08) in the main meta-analysis. SNPs at LPCAT2/CAPNS2 and NETO1/FBXO15 reached the GW threshold in the gender-specific meta analysis (see Table 3), and here the p-value in the main meta-analysis is reported. For each SNP, the best candidate gene is showed, as well as its genomic position in build 36, the effect allele (A1) and the other allele (A2), its combined frequency across studies and its standard error, the effect size and its standard error, the p-value for association, the number of samples analyzed, and the p-values for heterogeneity of effects across the cohorts meta-analyzed. Effect sizes are standardized, so they represent the estimated phenotypic change, per each copy of the effect allele, in standard deviation units.
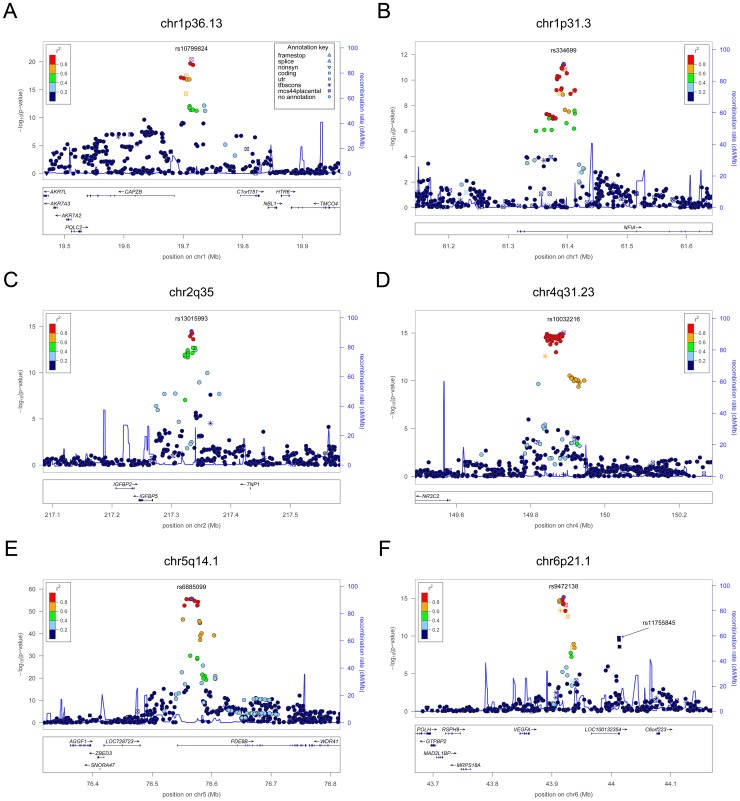
Figure 1. Regional association plots showing genome-wide significant loci for serum TSH.
In each panel (A–F), the most significant SNP is indicated (purple circle). In panel F, an independent signal at the associated locus is indicated with an arrow. The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP as in the inset (taken from pairwise r2 values from the HapMap CEU database build 36/hg18). Symbols reflect genomic functional annotation, as indicated in the legend [61]. Genes and the position of exons, as well as the direction of transcription, are noted in lower boxes. In each panel the scale bar on the Y-axis changes according to the strength of the association.
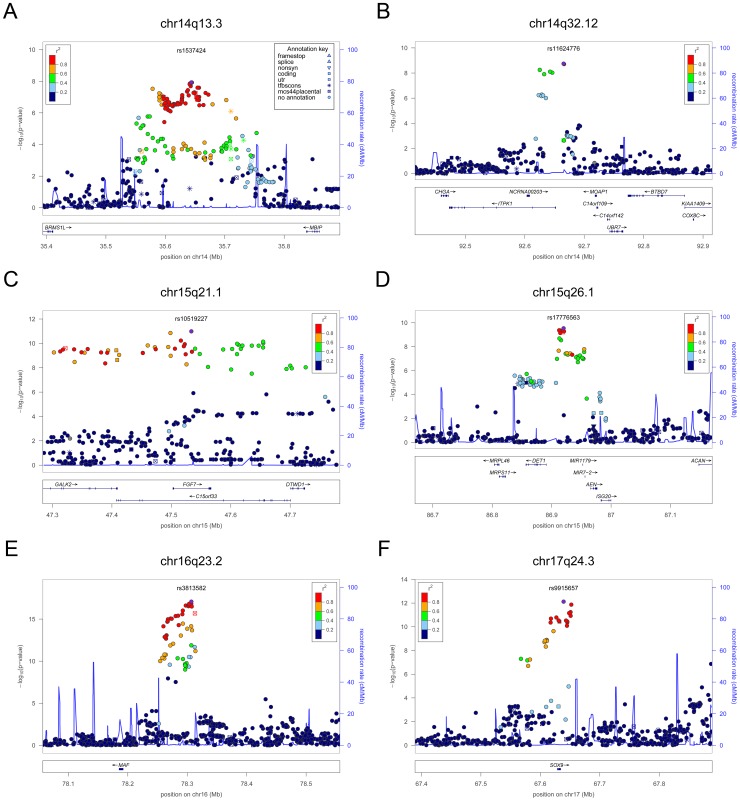
Figure 2. Regional association plots showing genome-wide significant loci for serum TSH.
In each panel (A–F), the most significant SNP is indicated (purple circle). The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP as in the inset (taken from pairwise r2 values from the HapMap CEU database build 36/hg18). Symbols reflect genomic functional annotation, as indicated in the legend [61]. Genes and the position of exons, as well as the direction of transcription, are noted in lower boxes. In each panel the scale bar on the Y-axis changes according to the strength of the association.
Figure 3. Regional association plots showing genome-wide significant loci for serum TSH.
In each panel (A–F), the most significant SNP is indicated (purple circle). The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP as in the inset (taken from pairwise r2 values from the HapMap CEU database build 36/hg18). Symbols reflect genomic functional annotation, as indicated in the legend [61]. Genes and the position of exons, as well as the direction of transcription, are noted in lower boxes. In each panel the scale bar on the Y-axis changes according to the strength of the association.
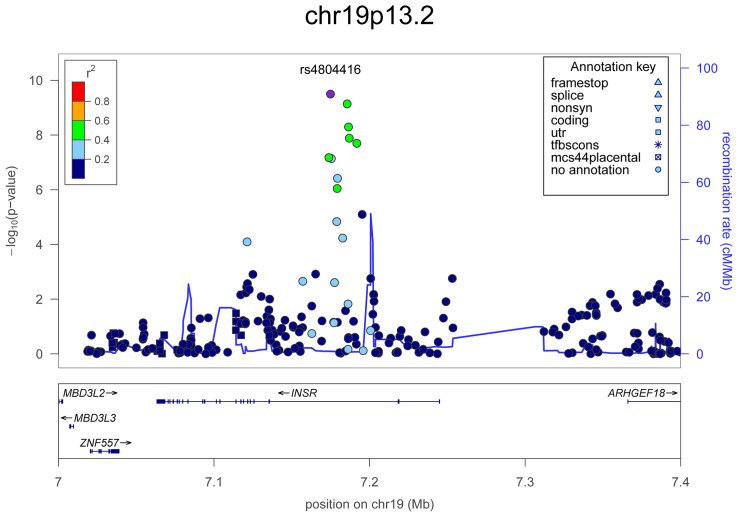
Figure 4. Regional association plots showing genome-wide significant loci for serum TSH.
In the upper panel, the most significant SNP is indicated (purple circle). The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP as in the inset (taken from pairwise r2 values from the HapMap CEU database build 36/hg18). Symbols reflect genomic functional annotation, as indicated in the legend [61]. Genes and the position of exons, as well as the direction of transcription, are noted in the lower box. The scale bar on the Y-axis changes according to the strength of the association.
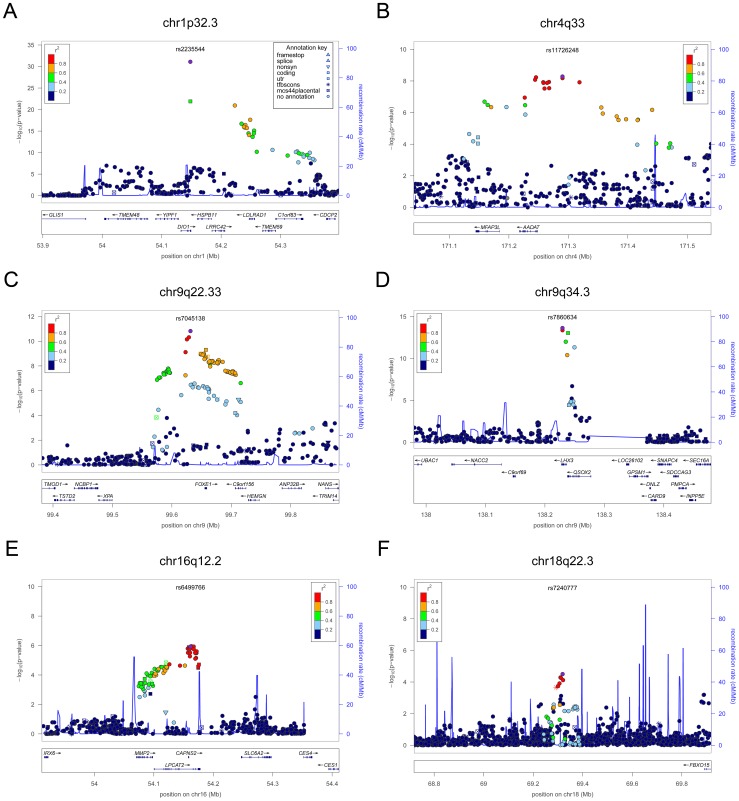
Figure 5. Regional association plots showing genome-wide significant loci for serum FT4.
In each panel (A–F), the most significant SNP is indicated (purple circle). The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP as in the inset (taken from pairwise r2 values from the HapMap CEU database build 36/hg18). Symbols reflect genomic functional annotation, as indicated in the legend [61]. Genes and the position of exons, as well as the direction of transcription, are noted in lower boxes. In each panel the scale bar on the Y-axis changes according to the strength of the association.
For TSH, 4 signals confirmed previously described loci with proxy SNPs at PDE8B (P = 1.95×10−56, r2 = 0.94 with the reported rs4704397), CAPZB (P = 3.60×10−21, r2 = 1 with the reported rs10917469) and NR3C2 (P = 9.28×10−16, r2 = 0.90 with the reported rs10028213), whereas the signal was coincident at MAF/LOC440389 (P = 8.45×10−18) [13]–[15]. The remaining signals were in or near 15 novel loci: PDE10A (phosphodiesterase type 10A, P = 1.21×10−24), VEGFA (Vascular endothelial growth factor, P = 6.72×10−16), IGFBP5 (insulin-like growth factor binding protein 5, P = 3.24×10−15), SOX9 (sex determining region Y-box 9, P = 7.53×10−13), NFIA (nuclear factor I/A, P = 5.40×10−12), FGF7 (fibroblast growth factor 7, P = 1.02×10−11), PRDM11 (PR domain containing 11, P = 8.83×10−11), MIR1179 (microRNA 1179, P = 2.89×10−10), INSR (insulin receptor, P = 3.16×10−10), ABO (ABO glycosyltransferase, P = 4.11×10−10), ITPK1 (inositol-tetrakisphosphate 1-kinase, P = 1.79×10−9), NRG1 (neuregulin 1, P = 2.94×10−9), MBIP (MAP3K12 binding inhibitory protein 1, P = 1.17×10−8), SASH1 (SAM and SH3 domain containing 1, P = 2.25×10−8), GLIS3 (GLIS family zinc finger 3, P = 2.55×10−8), (Figure 1, Figure 2, Figure 3, Figure 4).
For FT4, we confirmed the DIO1 locus (P = 7.87×10−32), with the same marker previously reported in candidate gene studies [17], [18], and identified 3 additional novel loci, LHX3 (LIM homeobox 3, P = 2.30×10−14), FOXE1 (forkhead box E1, P = 1.50×10−11) and AADAT (aminoadipate aminotransferase, P = 5.20×10−9) (Figure 5). The most associated SNP at the FOXE1 locus, rs7045138, is a surrogate for rs1443434(r2 = 0.97), previously only suggestively associated with FT4 levels [18], and is also correlated with SNPs recently reported to be associated with both low serum TSH and FT4 levels (r2 = 0.59 with rs965513) [19], as well as with hypothyroidism (r2 = 0.59 with rs7850258) [20].
At each locus, a single variant was sufficient to explain entirely the observed association, except for the VEGFA locus, which contained an independent signal located 150 kb downstream of the gene, detected by conditional analyses (Figure 1F and Table 2).
Of all 24 independent markers, significant evidence for heterogeneity (P<0.002, corresponding to a Bonferroni threshold of 0.5/24) was only observed at ABO (P = 1.22×10−4). Iodine nutrition, which may profoundly affect thyroid function, is quite different in some of the cohorts under study (i.e., Europe vs North America). To test whether the observed heterogeneity could be attributable to different iodine intake, we combined cohorts from South Europe (an iodine-deficient region) and compared effect sizes with those observed in a meta-analysis of North American samples (an iodine-replete region). Interestingly, the effect size of the top marker at ABO was three times larger in Europeans vs North American, and this difference remained significant after Bonferroni correction (P = 7.0.9×10−4) (Table S3). However, the relation of the ABO SNP, a tag for the blood group O, to iodine intake remains to be determined.
Gender-specific analyses
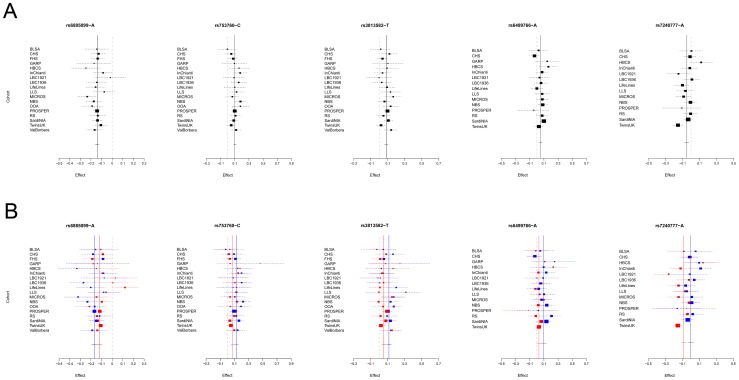
Given the reported clinical differences in thyroid function in males and females [21]–[23], we searched for gender-specific loci by whole-genome sex-specific meta-analysis, analyzing males and females separately in each cohort. Some of the loci detected in the main meta-analysis were seen at genome-wide significance level only in females (NR3C2, VEGFA, NRG1 and SASH1) or in males (MAF/LOC440389, FGF7, SOX9, IGFBP5) with either the same top SNP or one surrogate, but effect sizes at their variants were significantly gender-specific only at PDE8B, PDE10A and MAF/LOC440389, considering a false discovery rate of 5% [24]. In addition, effects at MAF/LOC440389 were significantly different also at the more stringent Bonferroni threshold of 1.9×10−3 ( = 0.05/26), and close to significance at PDE8B and PDE10A (Table 3). At these latter loci, the TSH-elevating alleles showed a stronger impact on trait variability in males compared to females (Figure 6). In addition, the gender specific meta-analysis for FT4, revealed a novel female-specific locus on chromosome 18q22, and a novel male-specific locus on chromosome 16q12.2, that had not been detected in the main meta-analysis (Table 3, Figure 6 and Figure S2). The female-specific signal (rs7240777, P = 3.49×10−8) maps in a “gene desert” region, with the nearest genes NETO1 (neuropilin (NRP) and tolloid (TLL)-like 1), located, about 550 kb upstream and FBXO15 (F-box only protein 15) 500 kb downstream (Figure 5D). The male-specific association is located in intron 11 of the LPCAT2 (lysophosphatidylcholine acyltransferase 2) gene, and near CAPNS2 (calpain, small subunit 2) (rs6499766, P = 4.63×10−8), a gene which may play a role in spermatogenesis [25]. The FT4-elevating alleles in the NETO1/FBXO15 and LPCAT2/CAPNS2 were fully gender-specific, i.e. there was no effect in males and in females, respectively (P>0.01).
Table 3. Top associated SNPs and their effect on males and females separately.
| Females | Males | |||||||||||
| Gene | SNP | A1/A2 | Effect | StdErr | P | N | Effect | StdErr | P | N | Het P | FDRs |
| TSH levels | ||||||||||||
| PDE8B | rs6885099 | A/G | −0,120 | 0.012 | 6.09×10−24 | 14509 | −0.168 | 0.013 | 2.70×10−38 | 11533 | 7.12×10−3 | 0.037 |
| PDE10A | rs753760 | C/G | 0.076 | 0.013 | 4.64×10−9 | 14473 | 0.131 | 0.014 | 5.85×10−20 | 11515 | 5.40×10−3 | 0.035 |
| CAPZB | rs10799824 | A/G | −0.123 | 0.016 | 2.69×10−14 | 14504 | −0.099 | 0.017 | 1.17×10−8 | 11527 | 0.309 | 0.618 |
| MAF/LOC440389 | rs3813582 | T/C | 0.055 | 0.013 | 1.75×10−5 | 14449 | 0.115 | 0.014 | 6.17×10−17 | 11499 | 1.50×10−3 | 0.019 |
| VEGFA | rs9472138 | T/C | −0.090 | 0.013 | 6.30×10−12 | 14291 | −0.065 | 0.014 | 4.89×10−6 | 11476 | 0.208 | 0.450 |
| VEGFA | rs11755845 | T/C | −0.058 | 0.014 | 1.98×10−5 | 14250 | −0.076 | 0.015 | 2.98×10−7 | 11460 | 0.368 | 0.683 |
| NR3C2 | rs10032216 | T/C | 0.106 | 0.014 | 1.72×10−13 | 14519 | 0.069 | 0.016 | 1.12×10−5 | 11534 | 0.092 | 0.294 |
| IGFBP5 | rs13015993 | A/G | 0.069 | 0.013 | 1.71×10−7 | 14491 | 0.095 | 0.015 | 7.60×10−11 | 11525 | 0.193 | 0.450 |
| SOX9 | rs9915657 | T/C | −0.063 | 0.012 | 1.46×10−7 | 14241 | −0.068 | 0.013 | 2.39×10−7 | 11451 | 0.793 | 0.896 |
| NFIA | rs334699 | A/G | −0.143 | 0.027 | 1.80×10−7 | 14253 | −0.149 | 0.030 | 5.93×10−7 | 11504 | 0.874 | 0.909 |
| FGF7 | rs10519227 | A/T | −0.051 | 0.014 | 3.80×10−4 | 14462 | −0.095 | 0.015 | 6.09×10−10 | 11526 | 0.036 | 0.156 |
| PRDM11 | rs17723470 | T/C | −0.069 | 0.013 | 2.92×10−7 | 14519 | −0.056 | 0.015 | 1.45×10−4 | 11535 | 0.519 | 0.710 |
| MIR1179 | rs17776563 | A/G | −0.053 | 0.013 | 3.70×10−5 | 14305 | −0.069 | 0.014 | 6.16×10−7 | 11453 | 0.395 | 0.684 |
| INSR | rs4804416 | T/G | −0.058 | 0.012 | 1.76×10−6 | 14205 | −0.058 | 0.013 | 1.12×10−5 | 11427 | 0.969 | 0.969 |
| ABO | rs657152 | A/C | 0.054 | 0.013 | 1.31×10−5 | 14290 | 0.067 | 0.014 | 1.01×10−6 | 11475 | 0.498 | 0.710 |
| ITPK1 | rs11624776 | A/C | −0.053 | 0.015 | 3.29×10−4 | 12255 | −0.069 | 0.015 | 2.61×10−6 | 11227 | 0.453 | 0.693 |
| NRG1 | rs7825175 | A/G | −0.084 | 0.015 | 1.64×10−8 | 14475 | −0.049 | 0.016 | 2.36×10−3 | 11521 | 0.113 | 0.294 |
| MBIP | rs1537424 | T/C | −0.054 | 0.012 | 1.26×10−5 | 14091 | −0.050 | 0.013 | 1.59×10−4 | 11387 | 0.848 | 0.909 |
| SASH1 | rs9497965 | T/C | 0.067 | 0.012 | 3.36×10−8 | 14462 | 0,031 | 0.013 | 0.023 | 11518 | 0.046 | 0.171 |
| GLIS3 | rs1571583 | A/G | 0.041 | 0.014 | 2.86×10−3 | 14290 | 0.074 | 0.015 | 9.82×10−7 | 11476 | 0.104 | 0.294 |
| FT4 levels | ||||||||||||
| DIO1 | rs2235544 | A/C | 0.130 | 0.015 | 2.62×10−18 | 10019 | 0.143 | 0.018 | 4.59×10−15 | 7201 | 0.605 | 0.786 |
| LHX3 | rs7860634 | A/G | 0.098 | 0.018 | 5.01×10−8 | 7665 | 0.108 | 0.019 | 1.72×10−8 | 6864 | 0.715 | 0.845 |
| FOXE1 | rs7045138 | T/C | 0.093 | 0.020 | 3.10×10−6 | 5801 | 0.105 | 0.021 | 4.96×10−7 | 5196 | 0.679 | 0.840 |
| AADAT | rs11726248 | A/G | 0.123 | 0.024 | 4.03×10−7 | 10252 | 0.093 | 0.029 | 1.47×10−3 | 7263 | 0.440 | 0.693 |
| LPCT2/CAPNS2 | rs6499766 | A/T | 0.030 | 0.015 | 0.040 | 10231 | 0.099 | 0.018 | 4.63×10−8 | 7258 | 3.42×10−3 | 0.029 |
| NETO1/FBXO15 | rs7240777 | A/G | −0.083 | 0.015 | 3.49×10−8 | 9963 | −0.001 | 0.018 | 0.950 | 7183 | 5.64×10−4 | 0.014 |
The table shows the association results in males and females separately for all independent SNPs associated with TSH and FT4 in the main meta-analysis (Table 2), as well as for the marker found to be associated only in females in the gender-specific meta-analysis. The last two columns report the p-value (Het P) and the false discovery rates (FDRs) for differences of effect sizes. SNPs with significantly different effect sizes at 5% FDRs and/or Bonferroni threshold (p = 1.9×10−3) are highlighted in bold. SdtErr, standard error; A1, effect allele; A2 other allele.
Figure 6. Forest plot of SNPs with gender-specific effects.
Squares represent the estimated per-allele beta-estimate for individual studies (a) and in males and females separately (b). The area of the square is inversely proportional to the variance of the estimate. Diamonds represent the summary beta estimates for the subgroups indicated. Horizontal lines represent 95% confidence intervals. In b, red and blue dotted lines represent, respectively, females and males.
Overall, the 20 TSH and the 6 FT4 associations account, respectively, for 5.64% and 2.30% of total trait variance.
Common loci regulating TSH and FT4 levels
To explore overlap between TSH- and FT4-associated loci and their involvement in the HPT-negative feedback loop, we assessed the associations of the top TSH-associated SNPs on FT4 levels, and vice versa. For the SNPs in or near PDE8B, MAF/LOC440389, VEGFA, IGFBP5, NFIA, MIR1179, MBIP and GLIS3 the TSH-elevating allele appeared to be associated with decreasing FT4 levels (P<0.05, Table S4). However, after application of Bonferroni correction (threshold for FT4 association of TSH SNPs, P = 2.5×10−3), none of these reciprocal associations remained significant.
By contrast, a positive relationship was seen for one of the FT4 associated loci, since the variant at the LHX3 locus was significantly associated with higher levels of both FT4 and TSH (P = 5.25×10−3, with Bonferroni threshold 0.05/6 = 0.008).
As the presence of reciprocal associations between TSH and FT4 regulating SNPs would be expected from physiology, we tested the power of our study to detect such a relationship. Power calculation for the top SNP at PDE8B, which has the largest effect on TSH levels, revealed that our meta-analysis only has 9% power to detect an association of FT4 at a Bonferroni P = 2.5×10−3. We also carried out a bivariate analysis in the SardiNIA study using poly software to estimate specific contributions [26]. This analysis showed that most of the observed negative feedback correlation is due to environmental factors (environmental correlation = −0.130, genetic correlation = −0.065).
Association of loci with hypothyroidism and hyperthyroidism
To assess possible clinical implications, we investigated whether the variants identified in individuals without overt thyroid pathologies (i.e., with TSH levels within the normal range and not taking thyroid medication) were also associated in individuals with abnormal TSH values (i.e., outside the reference range), who were not included in the initial meta-analysis as potentially affected by thyroid pathology. Towards this, we first assessed the global impact of TSH- and FT4-associated SNPs on the risk of increased or decreased TSH levels by comparing weighted genotype risk score (GRS) quartiles in the individuals with abnormal TSH values that were discarded for the GWAS analyses. For the TSH-associated SNPs, the odds of increased TSH levels were 6.65 times greater in individuals with a GRS in the top quartile compared to individuals in the bottom quartile (P = 3.43×10−20) (Table 4, top panel, lower vs upper tail). When we compared subjects with high TSH values with subjects within the normal TSH reference range, subjects with a GRS in the top quartile had odds of an elevated TSH 2.37 times greater than for subjects in the bottom quartile (P = 1.06×10−17) (Table 4). With regard to low TSH values versus the normal range, the odds ratio was 0.26 (P = 5.43×10−13) (Table 4, top panel, lower vs normal tail). By contrast, with the FT4-associated SNPs we found no significant associations for any of the tested comparisons (data not shown).
Table 4. TSH associated SNPs in extreme phenotype categories.
| Quartile – based analysis | UPPER vs LOWER | UPPER vs NORMAL | LOWER vs NORMAL | |||||||||
| OR | StdErr | P value | N (cases/controls) | OR | StdErr | P value | N (cases/controls) | OR | StdErr | P value | N (cases/controls) | |
| Quartile1 | – | – | – | 141/169 | – | – | – | 215/2699 | – | – | – | 143/1842 |
| Quartile2 | 2.16 | 0.17 | 7.09×10−6 | 194/108 | 1.31 | 0.10 | 0.010 | 280/2635 | 0.52 | 0.15 | 9.74×10−6 | 77/1913 |
| Quartile3 | 3.20 | 0.18 | 1.51×10−10 | 219/86 | 1.43 | 0.11 | 7.00×10−4 | 311/2595 | 0.48 | 0.15 | 8.31×10−7 | 71/1913 |
| Quartile4 | 6.65 | 0.21 | 3.43×10−20 | 256/49 | 2.37 | 0.10 | 1.06×10−17 | 447/2467 | 0.26 | 0.19 | 5.43×10−13 | 38/1947 |
The table shows results for the quartile-based GRS scores (top panel) and single marker (bottom panel) analyses in extreme phenotype categories, defined as TSH >4 mIU/L (UPPER) or TSH <0.4 mIU/L (LOWER). NORMAL, individuals with TSH within the normal range. OR, odds ratio; StdErr, standard error. A1, effect allele; A2 other allele. SNPs reaching the Bonferroni significance threshold are highlighted in bold.
We also assessed the 20 independent TSH SNPs individually in relation to the risk of abnormal TSH levels by case-control meta-analysis in subjects with high (cases) versus low (controls) TSH values. This analysis showed that variants at PDE8B, CAPZB, FGF7, PDE10A, NFIA and ITPK1 loci are significantly associated (Bonferroni threshold P = 2.5×10−3) with abnormal TSH levels (Table 4, bottom panel). PDE8B, CAPZB and FGF7 were also strongly associated with the risk of decreased TSH levels in an analysis of individuals with low (cases) versus normal range TSH (controls). In addition, variants at VEGFA were also significantly associated in this comparison. Finally, when individuals with high TSH values were analyzed versus controls, the NR3C2 locus appeared significantly associated in addition to PDE8B and CAPZB.
Association of TSH lead SNPs in pregnant women
Normal thyroid function is particularly important during pregnancy and elevated TSH levels are implicated in a number of adverse outcomes for both mother and offspring. We therefore assessed whether the TSH lead SNPs were also associated with elevated TSH during pregnancy, when increased TH production is necessary. We tested 9 of the 20 lead TSH variants (or their proxies, see Text S1) in a cohort of 974 healthy pregnant women at 28 weeks gestation [27] and found, as expected, that mean TSH levels were correlated with the number of TSH-elevating alleles (P = 3.0×10−12, Table S5). Effect size estimates in pregnant women were not significantly different when compared to those of women in the main gender-specific meta-analysis (heterogeneity P value>0.05), suggesting that the effects of the TSH-elevating alleles are no greater during pregnancy (data not shown). However, there was evidence of association between the number of TSH-raising alleles and subclinical hypothyroidism in pregnancy, both in the whole sample (OR per weighted allele: 1.18 [95%CI: 1.01, 1.37], P = 0.04) and in TPO antibody-negative women (1.29 [95%CI: 1.08, 1.55], P = 0.006) (Table S6).
Discussion
We report 26 independent SNPs associated with thyroid function tests in euthyroid subjects, 21 of which represent novel signals (16 for TSH and 5 for FT4). Overall they explain 5.64% and 2.30% of the variation in TSH and FT4 levels, respectively.
We observed that carriers of multiple TSH-elevating alleles have increased risk of abnormal TSH levels, and also found association between the number of TSH-elevating alleles and subclinical hypothyroidism in pregnancy. These results are potentially clinically relevant, because abnormal TSH values are the most sensitive diagnostic markers for both overt and subclinical thyroid disease [4]. The variants identified in the current study, or those in LD with them, may thus contribute to the pathogenesis of thyroid disease. Of note, we found eight loci significantly associated with abnormal TSH levels (PDE8B, PDE10, CAPZB, VEGFA, NR3C2, FGF7, NFIA and ITPK1), of which two were specifically associated with either abnormally low (VEGFA) or elevated (NR3C2) TSH values, suggesting differential mechanisms for the contribution of these variants to hyper- and hypothyroidism, respectively. Interestingly, the mineralocorticoid receptor NR3C2 gene has recently been found to be up-regulated in adult-onset hypothyroidism [28], and PDE8B and CAPZB have been suggestively associated with hypothyroidism by GWAS [29]. Alternatively, it may be that carriers of these alleles are healthy individuals who may be misdiagnosed as having thyroid disease because their genetically determined TSH concentrations fall outside the population-based reference range. More research is required to determine which of these interpretations is correct, and the relevance of these variants as markers for thyroid dysfunction or thyroid-related clinical endpoints.
The evidence for gender-specific differences at several TSH and FT4 regulatory loci is intriguing. They included variants at PDE8B, PDE10A, and MAF/LOC440389, which showed significantly stronger genetic effects with pituitary-thyroid function in males, and variants at NETO1/FBX015 and LPCAT2/CAPNS2 which seems to have an effect only in females and males, respectively. Sex differences in the regulation of thyroid function have generally been linked to the influence of sex hormones and autoimmune thyroid disease, resulting in a higher prevalence of thyroid dysfunction in women, without clear understanding of underlying molecular mechanisms [21]–[23]. Our study suggests that differential genes and mechanisms are potentially implicated in the regulation of thyroid function in men and women. Given the impact of thyroid function on several disease outcomes as well as male and female fertility and reproduction, clarifying the underlying associations may provide additional insight for future interventions.
Although it is well known that TSH and FT4 levels are tightly regulated through a negative feedback loop involving the HPT axis, we detected significant overlap between TSH and FT4 signals only at the LHX3 locus, which was primarily associated in our study with FT4. The LHX3 allele is associated with an increase of both TSH and FT4, which is consistent with the essential role of this transcription factor in pituitary development. Inactivating mutations in LHX3 cause the combined pituitary hormone deficiency-3 syndrome [CPHD3 (MIM#221750)] [30], [31], characterized by low TSH and FT4 levels. The positive association of the LHX3 variant with both TSH and FT4 suggests an effect of this allele at the level of the HPT-axis, resulting in an increased exposure to thyroid hormone throughout life. In contrast, although several of the TSH-elevating alleles appeared to be associated with decreasing FT4 levels, none of these reciprocal associations remained significant after Bonferroni correction. Lack of loci associated in a reciprocal manner with both TSH and FT4 is somewhat puzzling, as their presence would be expected from physiology. However, these findings are consistent with initial reports by Shields et al. [27] and more recent findings by Gudmundsson et al. [32]. A power analysis showed that our study – in spite of being one of the larger conducted so far on these traits – is underpowered to detect an inverse relationship between TSH and FT4 variants, considering a Spearman rank correlation of −0.130 between these traits [12]. As a consequence, contrasting studies on smaller sample sizes may also lack power and cannot be considered robust when testing this relationship [33]. In addition, we estimated that most of the observed negative feedback correlation is due to environmental factors; so it is unlikely that negative feedback is controlled by a genetic locus with large effect. This observation can rationalize the lack of reciprocal, significant associations detected for both TSH and FT4 in this and other studies, and further supports the crucial role of the HPT-axis in maintaining normal levels of thyroid hormone.
At present the relationship between the associated variants and specific mechanisms involved in regulating TSH and FT4 levels has not been established, but we have identified strong candidates at the majority of the loci by literature-mining approaches, as detailed below and in Table 5.
Table 5. Candidate genes at newly discovered loci for TSH and FT4 levels.
| SNP | Region | Gene | Position | Trait | Function |
| rs753760 | 6q26 | PDE10A | intron 1 | TSH | Encodes a dual specificity phosphodiesterase abundant in the thyroid, which can hydrolyze both cAMP and cGMP to the corresponding nucleoside 5′ monophosphate, but has higher affinity for cAMP, and is more efficient with cAMP as substrate. This gene was previously suggestively associated with TSH levels and hypothyroidism and linkage has been observed over this gene in families with individuals reaching the clinical criteria for sub-clinical and clinical thyroid disorders [13], [34]. The top SNP is only weakly correlated with previously reported variants (r2 = 0.449 with rs2983521 and r2 = 0.184 with rs9347083), but is a perfect proxy of a SNP recently reported in association with TSH [32]. |
| rs9472138; rs11755845 | 6p12 | VEGFA | intergenic | TSH/FT4 | Encodes a growth factor implicated in angiogenesis, which acts as an important regulator of both benign and malignant processes in the thyroid [62]. Angiogenesis is particularly critical for thyroid function, as the local microvasculature exerts an essential role in the continuous supply of iodine, the key element of thyroid hormone synthesis. In response to a reduction in intracellular iodine concentration, thyrocytes rapidly release angiogenic signals, including an increase in VEGFA expression and secretion [63], [64]. Notably, thyroid hormone stimulation in rat brain has been shown to induce VEGFA upregulation [65], which is consistent with the nominal association of the VEGFA locus with FT4 levels observed in this study. The two independent signals, rs9472138 and rs11755845, associated with TSH levels map 40 kb downstream of VEGFA, which is the nearest gene in the region. SNPs in this region were recently reported in association with TSH levels (r2 = 0.874 and r2 = 0.947) [32]. |
| rs13015993 | 2q33-36 | IGFBP5 | intergenic | TSH | It belongs to a protein family that interacts with insulin-like growth factors (IGFs) and plays a major role in regulating cell proliferation, differentiation, apoptosis and transformation. IGFBP5 is significantly over expressed in thyroid papillary carcinoma [54], [66]. Studies in vitro showed that TSH, through cAMP, inhibits IGFBP5 transcription, whereas enhanced production of IGFBP5 is correlated with inhibition of thyroid function [67];[68]. In addition, IFGBP5 has been found up-regulated in response to thyroid hormone in bone, where it interacts with the growth hormone/insulin-like growth factor (GH/IGF) system to contribute to bone formation [69], suggesting that thyroid hormone may potentate the effect of IGF-1 at the receptor level. The top SNP maps about 60 kb upstream of IGFBP5. A proxy of this SNP has been recently found associated with TSH and FT4 levels (rs737308, r2 = 0.927) [32]. |
| rs9915657 | 17q23 | SOX9 | 3′UTR | TSH | Encodes a transcription factor involved in chondrocyte differentiation and male sex determination, although other specific functions are known. The TA domain of SOX9, which is expressed both in the pituitary and in the thyroid, has been reported to interact with a component of the thyroid hormone receptor complex (TRAP230) [70]. How this interaction could affect TSH levels is at present unclear. The top SNP maps 5 kb downstream of SOX9, which is the nearest gene in the region. |
| rs334699 | 1p31.3-p31.2 | NFIA | intron 3 | TSH | Encodes a member of the NF1 (nuclear factor 1) family of transcription factors. NFI proteins have been implicated in regulating developmental processes by their specific expression pattern during embryonic development and by analysis of NFI-deficient mice [71]. In addition, they play crucial roles in the transcription of many cellular genes. Members of this family, including NFIA, have been shown to interact with thyroid transcription factor 1 (TTF1) [72], a transcription factor essential for thyroid-specific gene expression [73]. The top SNP maps in intron 3 of the gene. A proxy of this SNP has been recently found associated with TSH levels (rs334725, r2 = 1) [32]. |
| rs10519227 | 15q21.2 | FGF7 | intron 2 | TSH | Encodes a member of the fibroblast growth factor (FGF) family. FGF family members are involved in a variety of biological processes, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth and invasion. FGF signals play a role in the development of the thyroid gland and mice deficient for corresponding receptors show thyroid agenesis [74]. The top SNP maps in intron 2 of FGF7. A proxy of this SNP has been recently associated by GWAS with thyroid volume and goiter (rs4338740, r2 = 0.874) [36]. |
| rs17723470 | 11p11 | PRDM11 | intron 2 | TSH | Encodes a member of the family of PR-domain genes involved in human cancers [75]. The function of PRDM11 and its correlation with TSH levels is unclear; however the association with low TSH values in the extreme phenotype analysis supports a role of this gene in this trait. The top SNP maps in intron 2 of the gene and is moderately correlated with a SNP associated with TSH levels (rs7128207, r2 = 0.55) [32]. |
| rs17776563 | 15q25.3 | MIR1179 | intergenic | TSH | Encodes a microRNAs (miRNAs), which are short non-coding RNAs involved in post-transcriptional regulation of gene expression in multicellular organisms by affecting both the stability and translation of mRNAs. The associated SNP maps about 30 kb upstream of the gene, which is the nearest gene in the region. |
| rs4804416 | 19p13.3-13.2 | INSR | intron 2 | TSH | Encodes the insulin receptor precursor, which is post-translationally cleaved after removal of the precursor signal peptide into two chains (alpha and beta) that are covalently linked. Binding of insulin to the insulin receptor (INSR) stimulates glucose uptake. Two transcript variants encoding different isoforms have been found for this gene. INSR isoforms appear overexpressed in thyroid tumors, where they interact with insulin homolog IGFs (I and II), which act as potent mitogenic and antiapoptotic factors in a variety of human malignancies, and supporting a specific role of the GH/IGF pathway in thyroid function. The top SNP maps in intron 2 of the INSR, and is moderately correlated (rs10420008, r2 = 0.435) with a variant recently associated with TSH levels [32]. |
| rs657152 | 9q34.2 | ABO | intron 1 | TSH | Encodes proteins related to the blood group system, ABO, which determines the individual blood group. The associated SNP is in intron 1 of the gene and is a tag of the O blood group allele, caused by a deletion of guanine-258 near the N-terminus of the protein which results in a frameshift and translation of an almost entirely different protein. The correlation of TSH with ABO blood groups is currently unclear. ABO has been found associated with several phenotypes, including serum levels of different molecules [76]. It was reported that serum levels of TSH vary in individuals with ABO blood types, and that blood group O may be associated with hyperthyroidism [77]. |
| rs11624776 | 14q31 | ITPK1 | intergenic | TSH | Encodes the enzyme inositol 1,3,4-trisphosphate 5/6-kinase, which catalyzes the rate-limiting step in the formation of higher phosphorylated forms of inositol in mammalian cells. ITPK1 plays a pivotal role in inositol metabolism and mice producing reduced levels of ITPK1 develop neural tube defects [78]. Its role in the regulation of TSH levels is at present unclear. Of note, inositol phosphates/Ca2+ cascades mediates TSH action on thyroid hormone synthesis [79] The associated SNP is located about 15 kb upstream of the gene, which is the nearest gene in the region. |
| rs7825175 | 8p12 | NRG1 | intron 2 | TSH | Encodes neoregulin 1, a glycoprotein that interacts with the NEU/ERBB2 receptor tyrosine kinase to increase its phosphorylation on tyrosine residues. NRG1 is a signaling protein that mediates cell-cell interactions and plays critical roles in the growth and development of multiple organ systems. Its gene dysregulation has been linked to cancer, schizophrenia and bipolar disorder. The associated SNP maps in intron 2 of the gene and is only weakly correlated with a variant recently associated with TSH levels and thyroid cancer risk (rs2439302, r2 = 0.257) [32]. |
| rs1537424 | 14q13.3 | MBIP | intergenic | TSH | Encodes MAP3K12/MUK-binding inhibitory protein, a MAP3K regulator during osmolarity sensing and stress signaling that localizes in either the cytoplasm or nucleus [80]. SNPs in this locus have been recently reported as associated with TSH levels and thyroid cancer risk [32]. The top associated SNP maps at about 190 kb downstream of MBIP, which is the nearest gene in the region. Recently, a long, intergenic, noncoding RNA gene (lincRNA) named Papillary Thyroid Carcinoma Susceptibility Candidate 3 (PTCSC3) has been mapped 3.2 kb downstream of rs944289 (r2 = 0.708 with our top SNP), whose expression is strictly thyroid specific and acts as a PTC tumor suppressor gene [81]. |
| rs9497965 | 6q24.3 | SASH1 | intergenic | TSH | Encodes a member of the SLY-family of signal adapter proteins and is a candidate tumor suppressor in breast and colon cancer. However, the biological function of SASH1 and its involvement in malignant transformation remain largely unknown. Of note, SASH1 has been identified as a downstream target of the insulin/IGF1/PI 3-kinase signaling pathway [82], which appears implicated in TSH levels in the current study. The associated SNP is located about 130 kb upstream of SASH1, which is the nearest gene in the region. |
| rs1571583 | 9p24.2 | GLIS3 | intron 2 | TSH | Encodes a nuclear protein with five C2H2-type zinc finger domains, which is a member of the GLI-similar zinc finger protein family. GLIS3 functions as both a repressor and activator of transcription and is specifically involved in the development of pancreatic beta cells, the thyroid, eye, liver and kidney. Mutations in this gene have been associated with neonatal diabetes and congenital hypothyroidism (NDH) [52]. The top SNP at this locus maps in intron 2 of the gene. |
| rs7860634 | 9q34.3 | LHX3 | intron 6 | FT4/TSH | Encodes a transcription factor with an essential role in pituitary development [30]. Mutations in LHX3 cause the combined pituitary hormone deficiency-3 (CPHD3) syndrome (OMIM #221750), characterized by a complete deficit in growth hormone, prolactin, gonadotropin, and TSH, a rigid cervical spine leading to limited head rotation, as well as an extended spectrum with variable sensorineural hearing loss and ACTH deficiency [31], [42], [43], which is consistent with its association also with TSH levels observed in this study. This locus has been also recently associated with height in Japanese [44] The top SNP maps in intron 6 of the gene. |
| rs7045138 | 9q22 | FOXE1 | intergenic | FT4 | Encodes a transcription factor crucial for thyroid morphogenesis. Mutations in FOXE1 cause the Bamforth-Lazarus syndrome (MIM #241850), characterized by thyroid dysgenesis with neonatal hypothyroidism, cleft palate, choanal atresia and spiky hair [40], [41]. In addition, FOXE1 is a susceptibility locus for thyroid cancer [19], [83]–[85]. The top SNP maps 25 kb upstream of FOXE1 and is also associated in our study with FT4 levels. SNPs in the FOXE1 locus were previously associated with FT4 levels in a recent candidate gene analysis (rs1443434, r2 = 0.776) [18], as well as by GWAS with both low serum TSH and T4 levels (rs965513, r2 = 1) [19], and hypothyroidism (rs7850258, r2 = 0.625) [20]. |
| rs716822 | 4q33 | AADAT | intron 4 | FT4 | AADAT catalyzes the synthesis from kynurenine (KYN) of kynurenic acid (KYNA), which is implicated in the pathophysiology of several diseases of the central nervous system involving inflammation-induced brain injury 46–49. The KYN pathway has been associated with the induction of proinflammatory cytokines in the brain, which are known to activate the hypothalamo-pituitary-adrenal (HPA)-axis, involved in stress response and affecting the HPT-axis and thyroid function, including FT4 levels [45]. The top SNP maps in intron 4 of the gene. |
| rs7240777 | 18q22 | NETO1/FBXO15 | intergenic | FT4 | The top SNP maps in a gene desert region, with NETO1 located about 550 kb upstream and FBXO15 about 500 kb downstream. None of these genes has a clear role in thyroid function. NETO1 is expressed in brain and encodes a predicted transmembrane protein containing two extracellular CUB domains followed by a low-density lipoprotein class A (LDLa) domain. A similar gene in mice plays a critical role in spatial learning and memory by regulating the function of synaptic N-methyl-D-aspartic acid receptor complexes in the hippocampus. FBOX15 encodes a member of the F-box protein family characterized by an approximately 40-amino acid F-box motif. SCF complexes, formed by SKP1 (MIM#601434), cullin (see CUL1; MIM#603134), and F-box proteins, act as protein-ubiquitin ligases. F-box proteins interact with SKP1 through the F box, and they interact with ubiquitination targets through other protein interaction domains. |
| rs6499766 | 16q12.2 | LPCAT2/CAPNS2 | intron 11 | FT4 | LPCAT2 encodes a member of the lysophospholipid acyltransferase family. The enzyme may function to catalyze both the biosynthesis of platelet-activating factor and of glycerophospholipid precursors from arachidonyl-CoA and lysophosphatidylcholine. The encoded protein may function in membrane biogenesis and production of platelet-activating factor in inflammatory cells. The associated SNP maps in intron 11 of the gene, and is also near CAPNS2, which is contained in the LPCAT2 gene. None of these genes has a clear role in thyroid function. Of note, calpain subunit genes (CAPNS1 and CAPNS2) play important functions in mammalian reproduction [25]. |
The table lists genes of interest in the novel associated regions. For each associated region, the reported gene either contains the lead SNP or is in closest physical proximity with the lead SNP.
Most of the 16 novel loci implicated in the regulation of TSH are highly represented in the thyroid with the exception of PRDM11, expressed in brain, ABO, in blood, and MIR1179. PDE10A encodes a cAMP-stimulated phosphodiesterase, which was previously only suggestively associated with TSH levels and hypothyroidism [13], [34], although the tested variants were weakly correlated with our top signal (r2 = 0.55 with rs2983521 and r2 = 0.15 with rs9347083). The presence of linkage at this gene in families reaching accepted clinical criteria of thyroid dysfunction reinforces the observation that variants in this gene may contribute to clinical thyroid disorders [34]. PDE10A, together with PDE8B and CAPZB, emerged in our study as the strongest currently known genetic determinants of this trait. Both PDE8B and PDE10A are implicated in cAMP degradation in response to TSH stimulation of thyrocytes. In addition, the activity of both PDE10A and CAPZB appear modulated by cAMP [35], [36]. These three genes most likely act in a pathway that leads to cAMP-dependent thyroid hormone synthesis and release, thus highlighting a critical role of cAMP levels in thyroid function. For the other TSH-associated loci (VEGFA, IGFBP5, SOX9, NFIA, FGF7, PRDM11, MIR1179, INSR, ABO, ITPK1, NRG1, MBIP, SASH1 and GLIS3), hypotheses can be formulated based on the published literature (see Table 5), but further studies will be necessary to clarify the exact biological mechanisms and the specific genes involved at each locus. The association of TSH levels with IGFBP5, INSR and NR3C2 is, however, an indication of a specific role of the growth hormone/insulin-like growth factor (GH/IGF) pathway in thyroid function. Remarkably, expression of IGFBP5 is tightly regulated by cAMP, again underlying the pivotal role of this second messenger in determining net TSH levels [37].
For FT4, the DIO1, FOXE1 and LHX3 identified loci have strong biological support as potential effectors. While both DIO1 and FOXE1 were previously associated with FT4 levels and hypothyroidism by candidate gene analysis and functional studies [17]–[19], [38]–[41], association at LHX3 is novel and is consistent with the essential role of this transcription factor in pituitary development (see above) [30], [31], [42], [43]. Consistent with the role of pituitary in growth, this locus has also recently been associated with height in Japanese [44]. The associations of AADAT, NETO1/FXBO15 and LPCT2/CAPNS2 with FT4 levels are currently less clear. It may be relevant that AADAT catalyzes the synthesis of kynurenic acid (KYNA) from kynurenine (KYN), a pathway that has been associated with the induction in brain of proinflammatory cytokines that are known to activate the hypothalamo-pituitary-adrenal (HPA) axis, in turn affecting the HPT axis and thyroid function, including FT4 levels [45]–[49].
Additional pathway analyses by MAGENTA [50], GRAIL [51], and IPA (Ingenuity Systems, www.ingenuity.com) to look for functional enrichment of the genes mapping to the regions associated with TSH, FT4 or both, yielded no novel interactions. However, IPA highlighted an over-representation of genes implicated in developmental processes (11/26, P = 6.27×10−6–8.85×10−3) and cancer (16/26 loci, P = 2.44×10−6–9.30×10−3). This is consistent with the notion that a normally developed thyroid gland is essential for both proper function and thyroid hormone synthesis, and that defects in any of the essential steps in thyroid development or thyroid hormone synthesis may result in morphologic abnormalities, impaired hormonogenesis and growth dysregulation. It is also interesting to note that 11 of the 20 TSH signals and 3 of the 6 FT4 signals are connected in a single protein network, underlying the biological interrelationship between genes regulating these traits (Figure S3).
While our manuscript was in preparation, a GWAS of comparable sample size was published on levels of TSH in the general Icelandic population, which confirmed 15 of our reported loci (E. Porcu et al., 2011, ESHG, abstract), and inferred a role for three TSH-lowering variants in thyroid cancer [32]. Four additional TSH loci identified by Gudmundsson and colleagues were also associated in our sample-set of euthyroid individuals with p<0.05 and consistent direction of effects (VAV3, NKX2–3, TPO and FOXA2). Finally, 2 loci (SIVA1, ELK3) could not be tested because the corresponding SNPs or any surrogate (r2>0.5) were not available in our data set (Table S7). Our study shows that most of the loci described in Icelanders are reproducible in other populations of European origin; differences in sample size, phenotype definition (i.e., selection of euthyroid subjects vs general population) and in the genetic map used to detect associations most likely explain non-overlapping genome-wide significant signals. Among them, the reported signals at SOX9, ABO, SASH1, GLIS3 and MIR1179 will need to be confirmed in other studies; but one of them - GLIS3- is a prime candidate, because it is involved in congenital hypothyroidism [52]. Interestingly, despite the use of variants detected through whole-genome sequencing in Icelanders, the top signals at seven overlapping loci (PDE8B, PDE10A, CAPZB, MAF/LOC440389, VEGFA, NR3C2, IGFBP5) were either coincident or in high LD (r2>0.9) with those detected in our HapMap-based meta-analysis. Thus, such variants are likely to be the causative ones.
In conclusion, our study reports the first GWAS meta-analysis ever carried out on FT4 levels, adds to the existing knowledge novel TSH- and FT4-associated loci and reveals genetic factors that differentially affect thyroid function in males and females. Several detected loci have potential clinical relevance and have been previously implicated both in Mendelian endocrine disorders (LHX3 [MIMM#221750], FOXE1 [MIMM#241850], PDE8B [MIMM#614190], NR3C2 [MIMM#177735], INSR [MIMM#609968], GLIS3 [MIMM#610199]) and thyroid cancer (FOXE1 [19], VEGFA [53], IGFBP5 [54], INSR [55], NGR1 [32], MBIP [32], FGF7 [56]). Furthermore, the TSH-associated variants were found to contribute to TSH levels outside the reference range. Overall, our findings add to the developing landscape of the regulation of hypothalamic-pituitary-thyroid axis function and the consequences of genetic variation for hypo- or hyperthyroidism.
Methods
Ethics statement
All human research was approved by the relevant institutional review boards, and conducted according to the Declaration of Helsinki.
Cohort details
Cohort description, genotyping and statistical methods for individual study cohorts are reported in Text S1 and Table S1.
Statistical analyses
We carried out a meta-analysis including up to 26,523, individuals from 18 cohorts for TSH and up to 17,520 individuals from 15 cohorts for FT4 (see Table 1). FT4 measures were not available for all 21,955 individuals with TSH levels of the 15 participating cohorts. We combined evidence of associations from single GWAS using an inverse variance meta-analysis, where weights are proportional to the squared standard error of the beta estimates, as implemented in METAL [57]. Prior to GWAS, each study excluded individuals with known thyroid pathologies, taking thyroid medication, who underwent thyroid surgery, and with out-of-range TSH values (<0.4 mIU/L and >4 mIU/L), and an inverse normal transformation was applied to each trait (Table S1). Age, age-squared, and gender were fitted as covariates, as well as principal components axes or additional variables, as required (Table S1). Family-based correction was applied if necessary (see Table S1). Uniform quality control filters were applied before meta-analysis, including MAF <0.01, call rate <0.9, HWE P<1×10−6 for genotyped SNPs and low imputation quality (defined as r2<0.3 or info <0.4 if MACH [58] or IMPUTE [59], [60] were used, respectively) for imputed SNPs.
Genomic control was applied to individual studies if lambda was >1.0. The overall meta-analysis showed no significant evidence for inflated statistics (lambda for TSH, FT4 and were 1.05 and 1.03 respectively). To evaluate for heterogeneity in effect sizes across populations, we used a chi-square test for heterogeneity, implemented in METAL [57]. The same test was used to evalute heterogeneity related to iodine intake, by comparing effect sizes obtained in a meta-analysis of studies assessing individuals from South Europe (InChianti, MICROS, Val Borbera, SardiNIA, totaling up to 7,488 subjects) with those estimated in a meta-analysis of studies assessing individuals from North America (BLSA, CHS, FHS, OOA, totaling up to 5,407 subjects). Finally, the main meta-analysis was carried out independently by two analysts who obtained identical results.
Conditional analysis
To identify independent signals, each study performed GWA analyses for both TSH and FT4 by adding the lead SNPs found in the primary analysis (19 for TSH, and 4 for FT4, see Table 2) as additional covariates to the basic model, and removing those from the test data set. When lead SNPs were not available, the best proxies (r2>0.8) were included. We then performed a meta-analysis on the conditional GWAS results, using the same method and filters as described above. We used the standard genome-wide significance cutoff (P<5×10−8) to declare a significant secondary association.
Gender-specific analysis
To identify sex-specific effects, each study performed GWA analyses for each gender separately, using the same covariates and transformation as in the basic model (with the exception of gender covariate). We then performed a meta-analysis on association results using the same method and filters described for the primary analysis. To evaluate sex-specific differences we tested heterogeneity between effect sizes as described above. False-discovery rates (FDRs) on the 26 associated SNPs were calculated with R's p.adjust() procedure via the method of Benjamini and Hochberg [24].
Variance explained
The variance explained by the strongest associated SNPs was calculated, for each trait and in each cohort, as the difference of R2 adjusted observed in the full and the basic models, where the full model contains all the independent SNPs in addition to the covariates. The estimates from each cohort were combined using a weighted average, with weights proportional to the cohort sample size.
Extreme phenotype analysis
To evaluate the impact of the detected variants with clinically relevant TSH levels, we compared the allele frequencies observed in different categories of individuals in a case-control approach. Specifically, we compared individuals in the upper and lower TSH tails (individuals with TSH >4 mIU/L and TSH <0.4 mIU/L, respectively, whom were excluded for the GWAS analyses), as well as individuals in each tail with those in the normal TSH range. In the first case, individuals in the lower tail were considered controls and those in the upper tail cases. In the other two cases, we defined individuals in the normal range as controls and individuals on the two tails cases. To avoid sources of bias, individuals taking thyroid medication and/or with thyroid surgery were excluded. Only unrelated individuals were selected from the family-based cohort SardiNIA, while GEE correction was applied to the TwinsUK dataset. Results from single cohorts were then meta-analyzed. We first assessed the global impact of the 20 TSH- and 6 FT4-associated variants by defining a genotype-risk score (GRS) for each individual as the weighted sum of TSH- and FT4-elevating alleles, with weights proportional to the effect estimated in the meta-analysis. For each comparison, we then calculated quartiles from the global distribution (cases+controls) of the genotype score and used quartile 1 as the baseline reference to compare the number of cases and controls in the other quartiles. In addition, for TSH-associated variants we conducted single SNP comparisons. GRS quartile and single SNP analyses were performed by each study separately. Cohort specific results were then meta-analyzed for both the GRS score and single SNP results only if they had at least 50 cases and 50 controls. Specifically, cohorts included were: CHS, Lifelines, PROSPER, RS, SardiNIA and TwinsUK.
Bivariate analysis
Bivariate analysis was carried out with the software poly [26] in the SardiNIA cohort using the same individuals included in the GWAS and considering the same covariates and transformation for TSH and FT4 levels.
Web resources
The URLs for data presented herein are as follows:
METAL, http://www.sph.umich.edu/cgs/abecasis/metal
MACH, http://www.sph.umich.edu/csg/abecasis/MACH/
IMPUTE, https://mathgen.stats.ox.ac.uk/impute/impute.html
LocusZoom, http://csg.sph.umich.edu/locuszoom/
HapMap, http://www.hapmap.org
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
Supporting Information
Manhattan plots from meta-analysis results of serum TSH (panel A) and FT4 (panel B) levels. SNPs are plotted on the x axis according to their position (build 36) on each chromosome against association with TSH (A) and FT4(B) on the y axis (shown as –log10 P value) in. The loci highlighted in green are those that reached genome-wide significance (P<5×10−8). In each panel, quantile-quantile plots obtained with all SNPs (red dots) and after removal of SNPs within associated regions (blue dots) are also shown. The gray area corresponds to the 90% confidence region from a null distribution of P values (generated from 100 simulations).
(TIF)
Panel A. Manhattan plots from meta-analysis results of serum TSH and FT4 levels for men and women separately are shown as indicated. SNPs are plotted on the x axis according to their position (build 36) on each chromosome; association with TSH and FT4 is indicated on the y axis (as –log10 P value). Signals reaching genome-wide statistical significance in the gender specific analysis are shown in green. Panel B. Quantile-quantile plots are shown for all SNPs (red dots) and after removal of SNPs within associated regions (blue dots).
(TIF)
Ingenuity pathway analysis (IPA) results for candidate genes in the TSH and FT4 associated loci. A single protein network connects most of the identified loci.
(PDF)
Analysis details and methods for individual GWAS studies.
(XLS)
GWAS results of top SNPs for single cohorts. In the row named “G/I” we report the imputation quality (RSQR or INFO, according to study specific imputation method) or indicate with the letter “G” if the SNP was genotyped (G).
(XLSX)
Heterogeneity analysis of South European vs North American cohorts. The table shows the results of the two meta-analyses carried out in studies of South Europe and North America individuals, respectively. For each meta-analysis, we report the frequency of the effect allele (FreqA1), the effect size and the corresponding standard error (Effect, StdErr), the combined association pvalue (P), the pvalue for heterogeneity between studies (Het P), and the number of samples analyzed (N). The last two columns report the pvalue for differences of the effect size estimated in the two groups, and the total number of samples analyzed.
(DOC)
Association results for TSH and FT4 overlapping loci and their involvement in the negative feedback loop. The table shows the association results for all independent TSH (top panel) and FT4 (bottom panel) associated SNPs with FT4 and TSH levels, respectively. Effect sizes are standardized, so they represent the estimated phenotypic change, per each copy of the effect allele, in standard deviation units.
(DOC)
Genotype risk score for TSH alleles in pregnant women.
(DOC)
Association between TSH genetic risk score in pregnant women and subclinical hypothyroidism in pregnancy. Genotype risk score (GRS) was calculated in women with TSH level above reference range (>4.21 mIU/L) versus TSH level ≤4.21 mIU/L. Notably, excluding the 2 women with overt hypothyroidism (TSH>4.21 mIU/L and FT4 <9.13 pg/L), the results were essentially unchanged: Effect = 0.163, StdErr = 0.080, P = 0.043, OR = 1.18. StdErr, standard error, OR, odds ratio.
(DOC)
Association of SNPs reported by Gudmundsson and colleagues in our dataset. The table shows the association results for SNPs reported by Gudmundsson and colleagues [32] with TSH and FT4 levels available in our data-set. When the same marker was not available, we reported a proxy (r2>0.8) and the relative r2. StdErr, standard error. Loci reaching genome-wide significance in our data set with either the same SNP or a proxy are highlighted in bold.
(DOC)
Supplemental acknowledgments and funding information, cohort description, phenotyping, genotyping, analysis methods, supplemental references.
(DOC)
Acknowledgments
We thank all study participants, volunteers, and study personnel that made this work possible. We also thank Dr. Dawood Dadekula for running Ingenuity Pathway Analysis (IPA) on candidate genes. A detailed list of acknowledgements for individual study cohorts is available in Text S1.
Funding Statement
This work was supported by Intramural Research Program of the NIH (grants AG-023629, AG-15928, AG-20098, AG-027058, 263 MD 9164, 263 MD 821336,_NO1-AG-1-2109, U01 HL72515, R01 AG18728) and NIH General Clinical Research Centers Program, National Center for Research Resources (NCRR); NHLBI contracts (N01-HC-85239, N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HHSN268201200036C, and NHLBI grants HL080295, HL087652, HL105756, contracts N01-HC-25195 and N02-HL-6-4278); R&D contract with MedStar Research Institute; NINDS contract; National Center of Advancing Translational Technologies CTSI grant UL1TR000124; National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491; Robert Dawson Evans Endowment; Italian Ministry of Health grants ICS110.1/RF97.71 and Ricerca Finalizzata 2008; Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano; European Union's Seventh Framework Programme (grant agreements FP7/2007-2011 and FP7/2007-2013, 259679, 223004 and ENGAGE project grant agreement HEALTH-F4-2007-201413, EPIGENESYS grant agreement 257082, BLUEPRINT grant agreement HEALTH-F5-2011-282510); Dutch Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007); Netherlands Organization for Scientific Research (NWO) (grants 050-060-810, 175.010.2005.011, 911-03-012, 050-060-810, MW 904-61-095, 911-03-016, 917 66344, 911-03-012, 175.010.2007.006); South Tyrolean Sparkasse Foundation; Radboud University Nijmegen Medical Centre; University of Maryland General Clinical Research Center grant M01 RR 16500; Johns Hopkins University General Clinical Research Center, grant M01 RR 000052; Baltimore Veterans Administration Geriatric Research and Education Clinical Center (GRECC); Netherlands Research Institute for Diseases in the Elderly (grant 014-93-015; RIDE2); Erasmus Medical Center and Erasmus University, Rotterdam; Netherlands Organization for the Health Research and Development (ZonMw); Dutch Ministry for Health, Welfare and Sports; European Commission (DG XII); Municipality of Rotterdam; German Bundesministerium fuer Forschung und Technology grants 01 AK 803 A-H, 01 IG 07015 G; Wellcome Trust (grants WT089062, WT098051, WT091310,085541/Z/08/Z); English Department of Health, National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre; Canadian Institutes of Health Research, Canadian Foundation for Innovation; Fonds de la Recherche en Santé Québec, Ministère du Développement Économique, de l'Innovation et de l'Exportation; Lady Davis Institute of the Jewish General Hospital (JBR); Australian National Health and Medical Research Council (grants 1010494, 1031422); Sir Charles Gairdner Hospital Research Fund; Italian “Compagnia di San Paolo”; Italian “Fondazione Cariplo”; Leiden University Medical Centre; Dutch Arthritis Association; Pfizer, Groton, CT, USA; Dutch Centre of Medical System Biology; Netherlands Genomics Initiative (NGI), Netherlands Consortium of Healthy Aging” (grant 050-060-810); Academy of Finland; Finnish Diabetes Research Society; Folkhälsan Research Foundation; Novo Nordisk Foundation; Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation; University of Helsinki; European Science Foundation (EUROSTRESS); Finnish Ministry of Education; Ahokas Foundation; Emil Aaltonen Foundation; Juho Vainio Foundation; Biotechnology and Biological Sciences Research Council (BBSRC); Engineering and Physical Sciences Research Council (EPSRC); Economic and Social Research Council (ESRC); Medical Research Council (MRC), as part of the cross-council Lifelong Health and Wellbeing Initiative; AXA Research Fund; Research Into Ageing and programme grants from Help the Aged/Research Into Ageing (Disconnected Mind); Economic Structure Enhancing Fund (FES) of the Dutch government; Dutch Ministry of Economic Affairs; Dutch Ministry of Education, Culture and Science; Northern Netherlands Collaboration of Provinces (SNN); Province of Groningen; University of Groningen; Dutch Kidney Foundation and Dutch Diabetes Research Foundation; Bristol-Myers Squibb; Netherlands Heart Foundation grant 2001 D 032; National Computing Facilities Foundation (NCF), Netherlands, for the use of supercomputer facilities; Endocrine Research Fund; http://www.chs-nhlbi.org/pi.htm. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Toft AD (2001) Clinical practice. Subclinical hyperthyroidism. N Engl J Med 345: 512–516. [DOI] [PubMed] [Google Scholar]
- 2. Cooper DS (2001) Clinical practice. Subclinical hypothyroidism. N Engl J Med 345: 260–265. [DOI] [PubMed] [Google Scholar]
- 3. Fernandez-Real JM, Lopez-Bermejo A, Castro A, Casamitjana R, Ricart W (2006) Thyroid function is intrinsically linked to insulin sensitivity and endothelium-dependent vasodilation in healthy euthyroid subjects. J Clin Endocrinol Metab 91: 3337–3343. [DOI] [PubMed] [Google Scholar]
- 4. Biondi B, Cooper DS (2008) The clinical significance of subclinical thyroid dysfunction. Endocr Rev 29: 76–131. [DOI] [PubMed] [Google Scholar]
- 5. Chiamolera MI, Wondisford FE (2009) Minireview: Thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology 150: 1091–1096. [DOI] [PubMed] [Google Scholar]
- 6. Andersen S, Pedersen KM, Bruun NH, Laurberg P (2002) Narrow individual variations in serum T(4) and T(3) in normal subjects: a clue to the understanding of subclinical thyroid disease. J Clin Endocrinol Metab 87: 1068–1072. [DOI] [PubMed] [Google Scholar]
- 7. Bulow Pedersen I, Knudsen N, Jorgensen T, Perrild H, Ovesen L, et al. (2002) Large differences in incidences of overt hyper- and hypothyroidism associated with a small difference in iodine intake: a prospective comparative register-based population survey. J Clin Endocrinol Metab 87: 4462–4469. [DOI] [PubMed] [Google Scholar]
- 8. Franklyn JA, Ramsden DB, Sheppard MC (1985) The influence of age and sex on tests of thyroid function. Ann Clin Biochem 22 (Pt 5) 502–505. [DOI] [PubMed] [Google Scholar]
- 9. Bartalena L, Bogazzi F, Brogioni S, Burelli A, Scarcello G, et al. (1996) Measurement of serum free thyroid hormone concentrations: an essential tool for the diagnosis of thyroid dysfunction. Horm Res 45: 142–147. [DOI] [PubMed] [Google Scholar]
- 10. Hansen PS, Brix TH, Sorensen TI, Kyvik KO, Hegedus L (2004) Major genetic influence on the regulation of the pituitary-thyroid axis: a study of healthy Danish twins. J Clin Endocrinol Metab 89: 1181–1187. [DOI] [PubMed] [Google Scholar]
- 11. Samollow PB, Perez G, Kammerer CM, Finegold D, Zwartjes PW, et al. (2004) Genetic and environmental influences on thyroid hormone variation in Mexican Americans. J Clin Endocrinol Metab 89: 3276–3284. [DOI] [PubMed] [Google Scholar]
- 12. Panicker V, Wilson SG, Spector TD, Brown SJ, Falchi M, et al. (2008) Heritability of serum TSH, free T4 and free T3 concentrations: a study of a large UK twin cohort. Clin Endocrinol (Oxf) 68: 652–659. [DOI] [PubMed] [Google Scholar]
- 13. Arnaud-Lopez L, Usala G, Ceresini G, Mitchell BD, Pilia MG, et al. (2008) Phosphodiesterase 8B gene variants are associated with serum TSH levels and thyroid function. Am J Hum Genet 82: 1270–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panicker V, Wilson SG, Walsh JP, Richards JB, Brown SJ, et al. (2010) A locus on chromosome 1p36 is associated with thyrotropin and thyroid function as identified by genome-wide association study. Am J Hum Genet 87: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rawal R, Teumer A, Volzke H, Wallaschofski H, Ittermann T, et al. (2012) Meta-analysis of two genome-wide association studies identifies four genetic loci associated with thyroid function. Hum Mol Genet [DOI] [PubMed] [Google Scholar]
- 16. Peeters RP, van Toor H, Klootwijk W, de Rijke YB, Kuiper GG, et al. (2003) Polymorphisms in thyroid hormone pathway genes are associated with plasma TSH and iodothyronine levels in healthy subjects. J Clin Endocrinol Metab 88: 2880–2888. [DOI] [PubMed] [Google Scholar]
- 17. Panicker V, Cluett C, Shields B, Murray A, Parnell KS, et al. (2008) A common variation in deiodinase 1 gene DIO1 is associated with the relative levels of free thyroxine and triiodothyronine. J Clin Endocrinol Metab 93: 3075–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Medici M, van der Deure WM, Verbiest M, Vermeulen SH, Hansen PS, et al. (2011) A large-scale association analysis of 68 thyroid hormone pathway genes with serum TSH and FT4 levels. Eur J Endocrinol 164: 781–788. [DOI] [PubMed] [Google Scholar]
- 19. Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, et al. (2009) Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet 41: 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, et al. (2011) Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 89: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boucai L, Hollowell JG, Surks MI (2011) An approach for development of age-, gender-, and ethnicity-specific thyrotropin reference limits. Thyroid 21: 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McGrogan A, Seaman HE, Wright JW, de Vries CS (2008) The incidence of autoimmune thyroid disease: a systematic review of the literature. Clin Endocrinol (Oxf) 69: 687–696. [DOI] [PubMed] [Google Scholar]
- 23. Hollowell JG, Staehling NW, Flanders WD, Hannon WH, Gunter EW, et al. (2002) Serum TSH, T(4), and thyroid antibodies in the United States population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J Clin Endocrinol Metab 87: 489–499. [DOI] [PubMed] [Google Scholar]
- 24. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57: 289–300. [Google Scholar]
- 25. Ben-Aharon I, Brown PR, Etkovitz N, Eddy EM, Shalgi R (2005) The expression of calpain 1 and calpain 2 in spermatogenic cells and spermatozoa of the mouse. Reproduction 129: 435–442. [DOI] [PubMed] [Google Scholar]
- 26. Pilia G, Chen WM, Scuteri A, Orru M, Albai G, et al. (2006) Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet 2: e132 doi:10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shields BM, Freathy RM, Knight BA, Hill A, Weedon MN, et al. (2009) Phosphodiesterase 8B gene polymorphism is associated with subclinical hypothyroidism in pregnancy. J Clin Endocrinol Metab 94: 4608–4612. [DOI] [PubMed] [Google Scholar]
- 28. Montero-Pedrazuela A, Fernandez-Lamo I, Alieva M, Pereda-Perez I, Venero C, et al. (2011) Adult-onset hypothyroidism enhances fear memory and upregulates mineralocorticoid and glucocorticoid receptors in the amygdala. PLoS ONE 6: e26582 doi:10.1371/journal.pone.0026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, et al. (2012) Novel Associations for Hypothyroidism Include Known Autoimmune Risk Loci. PLoS ONE 7: e34442 doi:10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sheng HZ, Zhadanov AB, Mosinger B Jr, Fujii T, Bertuzzi S, et al. (1996) Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science 272: 1004–1007. [DOI] [PubMed] [Google Scholar]
- 31. Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, et al. (2000) Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet 25: 182–186. [DOI] [PubMed] [Google Scholar]
- 32. Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Masson G, et al. (2012) Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet 44: 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taylor PN, Panicker V, Sayers A, Shields B, Iqbal A, et al. (2011) A meta-analysis of the associations between common variation in the PDE8B gene and thyroid hormone parameters, including assessment of longitudinal stability of associations over time and effect of thyroid hormone replacement. Eur J Endocrinol 164: 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Volpato CB, De Grandi A, Gogele M, Taliun D, Fuchsberger C, et al. (2011) Linkage and association analysis of hyperthyrotropinaemia in an Alpine population reveal two novel loci on chromosomes 3q28–29 and 6q26–27. J Med Genet 48: 549–556. [DOI] [PubMed] [Google Scholar]
- 35. Kitazawa M, Yamakuni T, Song SY, Kato C, Tsuchiya R, et al. (2005) Intracellular cAMP controls a physical association of V-1 with CapZ in cultured mammalian endocrine cells. Biochem Biophys Res Commun 331: 181–186. [DOI] [PubMed] [Google Scholar]
- 36. Teumer A, Rawal R, Homuth G, Ernst F, Heier M, et al. (2011) Genome-wide association study identifies four genetic loci associated with thyroid volume and goiter risk. Am J Hum Genet 88: 664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duan C, Clemmons DR (1995) Transcription factor AP-2 regulates human insulin-like growth factor binding protein-5 gene expression. J Biol Chem 270: 24844–24851. [DOI] [PubMed] [Google Scholar]
- 38. de Jong FJ, Peeters RP, den Heijer T, van der Deure WM, Hofman A, et al. (2007) The association of polymorphisms in the type 1 and 2 deiodinase genes with circulating thyroid hormone parameters and atrophy of the medial temporal lobe. J Clin Endocrinol Metab 92: 636–640. [DOI] [PubMed] [Google Scholar]
- 39. Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, et al. (2008) Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev 29: 898–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Felice M, Ovitt C, Biffali E, Rodriguez-Mallon A, Arra C, et al. (1998) A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat Genet 19: 395–398. [DOI] [PubMed] [Google Scholar]
- 41. Clifton-Bligh RJ, Wentworth JM, Heinz P, Crisp MS, John R, et al. (1998) Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat Genet 19: 399–401. [DOI] [PubMed] [Google Scholar]
- 42. Pfaeffle RW, Savage JJ, Hunter CS, Palme C, Ahlmann M, et al. (2007) Four novel mutations of the LHX3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab 92: 1909–1919. [DOI] [PubMed] [Google Scholar]
- 43. Rajab A, Kelberman D, de Castro SC, Biebermann H, Shaikh H, et al. (2008) Novel mutations in LHX3 are associated with hypopituitarism and sensorineural hearing loss. Hum Mol Genet 17: 2150–2159. [DOI] [PubMed] [Google Scholar]
- 44. Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, et al. (2010) A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum Mol Genet 19: 2303–2312. [DOI] [PubMed] [Google Scholar]
- 45. Helmreich DL, Parfitt DB, Lu XY, Akil H, Watson SJ (2005) Relation between the hypothalamic-pituitary-thyroid (HPT) axis and the hypothalamic-pituitary-adrenal (HPA) axis during repeated stress. Neuroendocrinology 81: 183–192. [DOI] [PubMed] [Google Scholar]
- 46. Vamos E, Pardutz A, Klivenyi P, Toldi J, Vecsei L (2009) The role of kynurenines in disorders of the central nervous system: possibilities for neuroprotection. J Neurol Sci 283: 21–27. [DOI] [PubMed] [Google Scholar]
- 47. Goh DL, Patel A, Thomas GH, Salomons GS, Schor DS, et al. (2002) Characterization of the human gene encoding alpha-aminoadipate aminotransferase (AADAT). Mol Genet Metab 76: 172–180. [DOI] [PubMed] [Google Scholar]
- 48. de Souza FR, Fontes FL, da Silva TA, Coutinho LG, Leib SL, et al. (2010) Association of kynurenine aminotransferase II gene C401T polymorphism with immune response in patients with meningitis. BMC Med Genet 12: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Han Q, Cai T, Tagle DA, Li J (2009) Structure, expression, and function of kynurenine aminotransferases in human and rodent brains. Cell Mol Life Sci 67: 353–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Segrè AV, Groop L, Mootha VK, Daly MJ, Altshuler D (2010) Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 6: e1001058 doi:10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, et al. (2009) Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet 5: e1000534 doi:10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Senee V, Chelala C, Duchatelet S, Feng D, Blanc H, et al. (2006) Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet 38: 682–687. [DOI] [PubMed] [Google Scholar]
- 53. Salajegheh A, Smith RA, Kasem K, Gopalan V, Nassiri MR, et al. (2011) Single nucleotide polymorphisms and mRNA expression of VEGF-A in papillary thyroid carcinoma: potential markers for aggressive phenotypes. Eur J Surg Oncol 37: 93–99. [DOI] [PubMed] [Google Scholar]
- 54. Stolf BS, Carvalho AF, Martins WK, Runza FB, Brun M, et al. (2003) Differential expression of IGFBP-5 and two human ESTs in thyroid glands with goiter, adenoma and papillary or follicular carcinomas. Cancer Lett 191: 193–202. [DOI] [PubMed] [Google Scholar]
- 55. Vella V, Pandini G, Sciacca L, Mineo R, Vigneri R, et al. (2002) A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. J Clin Endocrinol Metab 87: 245–254. [DOI] [PubMed] [Google Scholar]
- 56. Braunschweig T, Kaserer K, Chung JY, Bilke S, Krizman D, et al. (2007) Proteomic expression profiling of thyroid neoplasms. Proteomics Clin Appl 1: 264–271. [DOI] [PubMed] [Google Scholar]
- 57. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39: 906–913. [DOI] [PubMed] [Google Scholar]
- 60. Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529 doi:10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Klein M, Catargi B (2007) VEGF in physiological process and thyroid disease. Ann Endocrinol (Paris) 68: 438–448. [DOI] [PubMed] [Google Scholar]
- 63. Gerard AC, Poncin S, Caetano B, Sonveaux P, Audinot JN, et al. (2008) Iodine deficiency induces a thyroid stimulating hormone-independent early phase of microvascular reshaping in the thyroid. Am J Pathol 172: 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamada E, Yamazaki K, Takano K, Obara T, Sato K (2006) Iodide inhibits vascular endothelial growth factor-A expression in cultured human thyroid follicles: a microarray search for effects of thyrotropin and iodide on angiogenesis factors. Thyroid 16: 545–554. [DOI] [PubMed] [Google Scholar]
- 65. Zhang L, Cooper-Kuhn CM, Nannmark U, Blomgren K, Kuhn HG (2010) Stimulatory effects of thyroid hormone on brain angiogenesis in vivo and in vitro. J Cereb Blood Flow Metab 30: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yu H, Rohan T (2000) Role of the insulin-like growth factor family in cancer development and progression. J Natl Cancer Inst 92: 1472–1489. [DOI] [PubMed] [Google Scholar]
- 67. Eggo MC, King WJ, Black EG, Sheppard MC (1996) Functional human thyroid cells and their insulin-like growth factor-binding proteins: regulation by thyrotropin, cyclic 3′,5′ adenosine monophosphate, and growth factors. J Clin Endocrinol Metab 81: 3056–3062. [DOI] [PubMed] [Google Scholar]
- 68. Backeljauw PF, Dai Z, Clemmons DR, D'Ercole AJ (1993) Synthesis and regulation of insulin-like growth factor binding protein-5 in FRTL-5 cells. Endocrinology 132: 1677–1681. [DOI] [PubMed] [Google Scholar]
- 69. Pepene CE, Kasperk CH, Pfeilschifter J, Borcsok I, Gozariu L, et al. (2001) Effects of triiodothyronine on the insulin-like growth factor system in primary human osteoblastic cells in vitro. Bone 29: 540–546. [DOI] [PubMed] [Google Scholar]
- 70. Zhou R, Bonneaud N, Yuan CX, de Santa Barbara P, Boizet B, et al. (2002) SOX9 interacts with a component of the human thyroid hormone receptor-associated protein complex. Nucleic Acids Res 30: 3245–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, et al. (1999) Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci U S A 96: 11946–11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nakazato M, Chung HK, Ulianich L, Grassadonia A, Suzuki K, et al. (2000) Thyroglobulin repression of thyroid transcription factor 1 (TTF-1) gene expression is mediated by decreased DNA binding of nuclear factor I proteins which control constitutive TTF-1 expression. Mol Cell Biol 20: 8499–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moeller LC, Kimura S, Kusakabe T, Liao XH, Van Sande J, et al. (2003) Hypothyroidism in thyroid transcription factor 1 haploinsufficiency is caused by reduced expression of the thyroid-stimulating hormone receptor. Mol Endocrinol 17: 2295–2302. [DOI] [PubMed] [Google Scholar]
- 74. Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, et al. (2001) Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol 231: 47–62. [DOI] [PubMed] [Google Scholar]
- 75. Jiang GL, Huang S (2000) The yin-yang of PR-domain family genes in tumorigenesis. Histol Histopathol 15: 109–117. [DOI] [PubMed] [Google Scholar]
- 76. Naitza S, Porcu E, Steri M, Taub DD, Mulas A, et al. (2012) A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet 8: e1002480 doi:10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Carmel R, Spencer CA (1982) Clinical and subclinical thyroid disorders associated with pernicious anemia. Observations on abnormal thyroid-stimulating hormone levels and on a possible association of blood group O with hyperthyroidism. Arch Intern Med 142: 1465–1469. [PubMed] [Google Scholar]
- 78. Majerus PW, Wilson DB, Zhang C, Nicholas PJ, Wilson MP (2010) Expression of inositol 1,3,4-trisphosphate 5/6-kinase (ITPK1) and its role in neural tube defects. Adv Enzyme Regul 50: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Grasberger H, Van Sande J, Hag-Dahood Mahameed A, Tenenbaum-Rakover Y, Refetoff S (2007) A familial thyrotropin (TSH) receptor mutation provides in vivo evidence that the inositol phosphates/Ca2+ cascade mediates TSH action on thyroid hormone synthesis. J Clin Endocrinol Metab 92: 2816–2820. [DOI] [PubMed] [Google Scholar]
- 80. Fukuyama K, Yoshida M, Yamashita A, Deyama T, Baba M, et al. (2000) MAPK upstream kinase (MUK)-binding inhibitory protein, a negative regulator of MUK/dual leucine zipper-bearing kinase/leucine zipper protein kinase. J Biol Chem 275: 21247–21254. [DOI] [PubMed] [Google Scholar]
- 81. Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, et al. (2012) The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A 109: 8646–8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dubois F, Vandermoere F, Gernez A, Murphy J, Toth R, et al. (2009) Differential 14-3-3 affinity capture reveals new downstream targets of phosphatidylinositol 3-kinase signaling. Mol Cell Proteomics 8: 2487–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Takahashi M, Saenko VA, Rogounovitch TI, Kawaguchi T, Drozd VM, et al. The FOXE1 locus is a major genetic determinant for radiation-related thyroid carcinoma in Chernobyl. Hum Mol Genet 19: 2516–2523. [DOI] [PubMed] [Google Scholar]
- 84. Landa I, Ruiz-Llorente S, Montero-Conde C, Inglada-Perez L, Schiavi F, et al. (2009) The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genet 5: e1000637 doi:10.1371/journal.pgen.1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Takahashi M, Saenko VA, Rogounovitch TI, Kawaguchi T, Drozd VM, et al. (2010) The FOXE1 locus is a major genetic determinant for radiation-related thyroid carcinoma in Chernobyl. Hum Mol Genet 19: 2516–2523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Manhattan plots from meta-analysis results of serum TSH (panel A) and FT4 (panel B) levels. SNPs are plotted on the x axis according to their position (build 36) on each chromosome against association with TSH (A) and FT4(B) on the y axis (shown as –log10 P value) in. The loci highlighted in green are those that reached genome-wide significance (P<5×10−8). In each panel, quantile-quantile plots obtained with all SNPs (red dots) and after removal of SNPs within associated regions (blue dots) are also shown. The gray area corresponds to the 90% confidence region from a null distribution of P values (generated from 100 simulations).
(TIF)
Panel A. Manhattan plots from meta-analysis results of serum TSH and FT4 levels for men and women separately are shown as indicated. SNPs are plotted on the x axis according to their position (build 36) on each chromosome; association with TSH and FT4 is indicated on the y axis (as –log10 P value). Signals reaching genome-wide statistical significance in the gender specific analysis are shown in green. Panel B. Quantile-quantile plots are shown for all SNPs (red dots) and after removal of SNPs within associated regions (blue dots).
(TIF)
Ingenuity pathway analysis (IPA) results for candidate genes in the TSH and FT4 associated loci. A single protein network connects most of the identified loci.
(PDF)
Analysis details and methods for individual GWAS studies.
(XLS)
GWAS results of top SNPs for single cohorts. In the row named “G/I” we report the imputation quality (RSQR or INFO, according to study specific imputation method) or indicate with the letter “G” if the SNP was genotyped (G).
(XLSX)
Heterogeneity analysis of South European vs North American cohorts. The table shows the results of the two meta-analyses carried out in studies of South Europe and North America individuals, respectively. For each meta-analysis, we report the frequency of the effect allele (FreqA1), the effect size and the corresponding standard error (Effect, StdErr), the combined association pvalue (P), the pvalue for heterogeneity between studies (Het P), and the number of samples analyzed (N). The last two columns report the pvalue for differences of the effect size estimated in the two groups, and the total number of samples analyzed.
(DOC)
Association results for TSH and FT4 overlapping loci and their involvement in the negative feedback loop. The table shows the association results for all independent TSH (top panel) and FT4 (bottom panel) associated SNPs with FT4 and TSH levels, respectively. Effect sizes are standardized, so they represent the estimated phenotypic change, per each copy of the effect allele, in standard deviation units.
(DOC)
Genotype risk score for TSH alleles in pregnant women.
(DOC)
Association between TSH genetic risk score in pregnant women and subclinical hypothyroidism in pregnancy. Genotype risk score (GRS) was calculated in women with TSH level above reference range (>4.21 mIU/L) versus TSH level ≤4.21 mIU/L. Notably, excluding the 2 women with overt hypothyroidism (TSH>4.21 mIU/L and FT4 <9.13 pg/L), the results were essentially unchanged: Effect = 0.163, StdErr = 0.080, P = 0.043, OR = 1.18. StdErr, standard error, OR, odds ratio.
(DOC)
Association of SNPs reported by Gudmundsson and colleagues in our dataset. The table shows the association results for SNPs reported by Gudmundsson and colleagues [32] with TSH and FT4 levels available in our data-set. When the same marker was not available, we reported a proxy (r2>0.8) and the relative r2. StdErr, standard error. Loci reaching genome-wide significance in our data set with either the same SNP or a proxy are highlighted in bold.
(DOC)
Supplemental acknowledgments and funding information, cohort description, phenotyping, genotyping, analysis methods, supplemental references.
(DOC)