Abstract
Quantitative Fitness Analysis (QFA) is an experimental and computational workflow for comparing fitnesses of microbial cultures grown in parallel1,2,3,4. QFA can be applied to focused observations of single cultures but is most useful for genome-wide genetic interaction or drug screens investigating up to thousands of independent cultures. The central experimental method is the inoculation of independent, dilute liquid microbial cultures onto solid agar plates which are incubated and regularly photographed. Photographs from each time-point are analyzed, producing quantitative cell density estimates, which are used to construct growth curves, allowing quantitative fitness measures to be derived. Culture fitnesses can be compared to quantify and rank genetic interaction strengths or drug sensitivities. The effect on culture fitness of any treatments added into substrate agar (e.g. small molecules, antibiotics or nutrients) or applied to plates externally (e.g. UV irradiation, temperature) can be quantified by QFA.
The QFA workflow produces growth rate estimates analogous to those obtained by spectrophotometric measurement of parallel liquid cultures in 96-well or 200-well plate readers. Importantly, QFA has significantly higher throughput compared with such methods. QFA cultures grow on a solid agar surface and are therefore well aerated during growth without the need for stirring or shaking.
QFA throughput is not as high as that of some Synthetic Genetic Array (SGA) screening methods5,6. However, since QFA cultures are heavily diluted before being inoculated onto agar, QFA can capture more complete growth curves, including exponential and saturation phases3. For example, growth curve observations allow culture doubling times to be estimated directly with high precision, as discussed previously1.
Here we present a specific QFA protocol applied to thousands of S. cerevisiae cultures which are automatically handled by robots during inoculation, incubation and imaging. Any of these automated steps can be replaced by an equivalent, manual procedure, with an associated reduction in throughput, and we also present a lower throughput manual protocol. The same QFA software tools can be applied to images captured in either workflow.
We have extensive experience applying QFA to cultures of the budding yeast S. cerevisiae but we expect that QFA will prove equally useful for examining cultures of the fission yeast S. pombe and bacterial cultures.
Keywords: Physiology, Issue 66, Medicine, Robotic, microbial, culture, yeast, array, library, high-throughput, analysis, fitness, growth rate, quantitative, solid agar
Protocol
Manual QFA Protocol (for S. cerevisiae strains)
1. Culturing Yeast Strains
Up to 96 independent yeast strains are cultured in 200 μl of rich liquid media in a 96-well culture dish. The strains are grown to saturation in a temperature controlled incubator.
A 96 pin, 1/8" diameter manual replica plater (SIGMA R-2508) is sterilized by first dipping the replica plater in 100% ethanol, flaming and then leaving to cool.
200 μl of sterile water is arrayed in a 96-well culture plate. The saturated cultures are vortexed (Eppendorf MixMate, 30 sec, 750 rpm) and diluted by dipping the sterilized pin tool into the saturated strains three times then into the plate containing the water once.
The pin tool is re-sterilized as per 1.2.
The sterilized pin tool is used to spot the diluted culture onto sold agar plates by dipping into the plate containing the diluted culture three times then transferring the pin tool to a rectangular solid agar plate.
The rectangular solid agar plates are incubated at an appropriate temperature before being imaged manually using an S&P Robotics spImager. Alternative image capture methods may be also be used such as a FujiFilm LAS4000, or a less expensive digital SLR camera if care is taken to ensure even illumination and consistent plate positioning relative to the imaging camera.
The method is adaptable to smaller numbers of parallel cultures, such as 48, which can be fitted onto round Petri dishes if care is taken to ensure the same angle relative to the imaging camera during each photograph.
Fully Automated QFA Protocol (for S. cerevisiae strains)
2. Culturing Yeast Strains
Start with a rectangular array of independent yeast strains growing on solid agar. In this example the starting strains are the result of crossing a temperature sensitive query mutation with the Yeast Deletion Collection (YDC) by Synthetic Genetic Array (SGA). Final SGA plates are in 1536 format. This is 16 rows by 24 columns of colonies/cultures representing four replicates of 384 independent yeast strains. See Figure 1 for plate layouts.
384 strains out of 1536 are transferred robotically using an S&P Robotics Inc BM3-SC robot with a 96 pin, 1-mm pin diameter sterile pintool into four 96-well culture plates containing 200 μl of selective media (Figure 1). Pintool cleanliness and sterility is achieved by sequentially washing in sterile water twice (once with a revolving brush to remove debris), 70% ethanol (with sonication) and finally 100% ethanol.
The cultures are grown to saturation (for three days at 20 °C in this example, see Figure 1).
3. Spotting Yeast Cultures
Using a Beckman Biomek FX, saturated cultures are resuspended by vortexing on a Variomag Teleshake (anticlockwise at 1000 rpm for 20 sec) and diluted approximately 1:70 by pinning into 200 μl sterile water using a sterile 96 pin robotic pin tool (V&P Scientific magnetic mounting pintool, 2 mm pin diameter). Pin tool cleanliness and sterility is achieved by washing in sterile water, washing in 70% ethanol (with a brush to remove debris) and finally dipping in 70% ethanol followed by drying.
Diluted cultures are spotted onto solid agar plates in 384 format using the same robotic pin tool (see Figure 1) after cleaning and sterilisation.
After pinning, plates are transferred to a Cytomat temperature controlled, humidified incubator with automatic carousel and access hatch.
4. Image Capture
An S&P Robotics automated imager repeatedly removes the plates from the Cytomat incubator, removes the plate lid, places them precisely under an enclosed, uniformly illuminated space beneath a Canon EOS Rebel Ti 35mm DSLR, captures an image at 5184 x 3456 px resolution, replaces the plate lid, and returns them to the incubator carousel. One image is captured immediately following initial transfer to the incubator to allow for a zero time point growth measurement (background). Robot handling and photography takes 2 min per plate.
With a fully loaded carousel, the maximum achievable plate image capture frequency is one photograph every 6 hr. Plates are incubated for up to six days (until colony growth saturation. Figure 1 shows three typical images from a time-course at 20 °C.
For tracking purposes, plates are named according to incubator name, temperature, unique sequential batch number and position in the incubator. Image names are a concatenation of plate name and timestamp and are automatically constructed upon image capture (e.g. K000011_030_010_2011-09-22_09-47-54.jpg, File Type FT1, Table1).
5. Capture of Experimental Metadata
The metadata files described in this section are tab-delimited text files which are manually generated (e.g. using spreadsheet software). The Experimental Description file (FT2, Table 1) is unique to each experiment, but the Library Description (FT3, Table 1) can be re-used extensively once constructed.
The experiment is described in the Experimental Description file (FT2, Table1) containing columns for barcode (or automatically generated plate name), experiment starting timestamp, plate treatment, contents of solid agar medium, screen name, library, plate number (for libraries with multiple plates) and a repeat-quadrant number (see Figure 1) for scaling down from 1536 format to 384 format.
The yeast strain library is described with a Library Description file (FT3, Table1) stating the genotype grown in each culture location in each plate of a library. It contains columns for: library name, ORF, plate number, plate row, plate column and an optional notes column.
An optional Standard Gene Name file (FT4, Table 1) describing the standard gene name (e.g. RAD9) associated with each systematic ORF y-number (e.g. YDR217C) identifying strains being screened can be provided. This file contains two columns: ORF & Gene name.
6. Data Analysis
The QFA computational workflow requires access to a reasonably powerful multicore computer workstation (e.g. a Dell Precision T3500 with Xeon quad-core 2.67 GHz processor and 12Gb RAM) on which is installed the Colonyzer image analysis software tool 3,4 and the QFA R package7, both of which are documented online, are freely available and run on a range of operating systems.
Run Colonyzer with each of the captured plate images as input, generating one Colonyzer Output file (FT5 Table 1) for each image captured. Colonyzer Output files specify culture density estimates, culture area, shape and color for each of the 384 locations on the imaged plate. Output filename is automatically copied from the source image filename (e.g. the plate photograph K000011_030_010_2011-09-22_09-47-54.jpg (FT1, Table 1) corresponds to the Colonyzer output file K000011_030_010_2011-09-22_09-47-54.dat (FT5, Table 1)).
Using the QFA R package, load experimental metadata (FT2, Table1) and Colonyzer output files (FT5 Table 1). These data are merged into R data frames (which can be exported as QFA Raw Data text files (FT6, Table 1)) for further analysis.
The QFA R package contains functions to gather cell density timecourses for each culture, to fit generalized logistic population models to observations and to plot both (see Figures 2 and 3 for examples). Fitted parameter values are written to QFA Logistic Parameter files (FT7 Table 1). See QFA R package documentation for further details.
The QFA R package also contains functions for quality control: edge colonies are discarded due to the greater availability of nutrients at the plate edges and difficulty with image analysis near plate walls, cultures which failed the SGA and genotypes displaying linkage with screen-specific marker genes are stripped from analysis.
Several quantitative fitness measures are derived from logistic population model parameters for each culture. These include maximum population doubling rates (MDR) and number of divisions from inoculation to saturation (MDP). For each set of replicate cultures (of unique genotype for example) and for each fitness definition, several summary statistics of fitness estimates are computed: mean and median fitness, fitness standard deviation and number of replicates observed. Summary statistics are output to QFA Fitness Summary files (FT8, Table 1) for further analysis, e.g. calculation of genetic interaction scores (FT9 Table 1).
7. Representative Results
Figure 2 shows one typical growth curve out of 384 QFA cultures growing at 20 °C on solid agar as quantified by Colonyzer, with increase in cell density first becoming detectable after roughly 2 days. This delay is likely a combined effect of a culture lag phase after inoculation onto solid medium and a lower limit of detection for cell density. Figure 3 shows 308 similar growth curves captured from a single plate. Figure 4 demonstrates the comparison of two genome-wide QFA screens to infer genetic interaction strengths (adapted from 1). The QFA R package 7 also includes functions to produce Figure 3 and to output ranked lists of examined genotypes and estimates of genetic interaction strength (GIS), together with a q-value (False Discovery Rate (FDR) corrected p-value) for the significance of observed GIS.
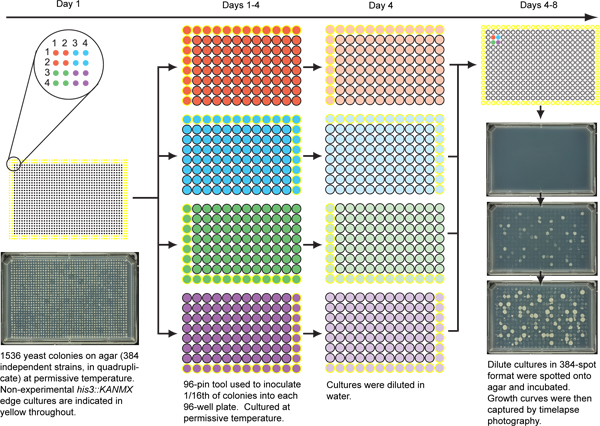 Figure 1. Scheme for robotic inoculation of yeast strains in 384-spot format. This procedure begins with 1536 independent cultures per plate (left). In this typical example, colonies at positions 1,1; 1,2; 2,1 and 2,2 (colored red) are four replicates of the same genotype. his3::KANMX cultures in yellow, growing on the edge of the plate, have a growth advantage due to lack of competition and are therefore not examined by QFA. One of these replicates (e.g. 1,1) is inoculated into liquid growth media in 96-well plates using a 96-pin tool which inoculates 96 out of 1536 colonies each time. In order to inoculate one replicate for each of 384 gene deletions, four different "quadrants" (indicated as red, blue, green and purple) are inoculated into four different 96-well plates containing growth media. After growth to saturation (e.g. 3 days at 20 °C), cultures are diluted in water, then the four quadrants from one repeat are spotted in 384-format onto a solid agar plate (right) in the same pattern as the original SGA plate (as indicated by color). The process can be repeated to test other replicates: 1,2; 2,1 and 2,2. Example time-lapse images on the right were captured 0.5, 2 and 3.5 days after inoculation. Adapted from Supplementary Figure 1
2. Click here to view larger figure.
Figure 1. Scheme for robotic inoculation of yeast strains in 384-spot format. This procedure begins with 1536 independent cultures per plate (left). In this typical example, colonies at positions 1,1; 1,2; 2,1 and 2,2 (colored red) are four replicates of the same genotype. his3::KANMX cultures in yellow, growing on the edge of the plate, have a growth advantage due to lack of competition and are therefore not examined by QFA. One of these replicates (e.g. 1,1) is inoculated into liquid growth media in 96-well plates using a 96-pin tool which inoculates 96 out of 1536 colonies each time. In order to inoculate one replicate for each of 384 gene deletions, four different "quadrants" (indicated as red, blue, green and purple) are inoculated into four different 96-well plates containing growth media. After growth to saturation (e.g. 3 days at 20 °C), cultures are diluted in water, then the four quadrants from one repeat are spotted in 384-format onto a solid agar plate (right) in the same pattern as the original SGA plate (as indicated by color). The process can be repeated to test other replicates: 1,2; 2,1 and 2,2. Example time-lapse images on the right were captured 0.5, 2 and 3.5 days after inoculation. Adapted from Supplementary Figure 1
2. Click here to view larger figure.
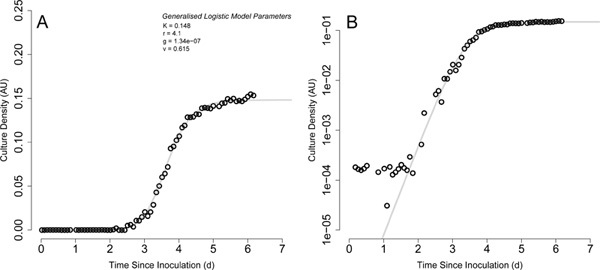 Figure 2. A single robot-captured QFA growth curve. A) QFA growth curve of a yeast strain growing on agar containing galactose at 20 °C. Images were captured robotically approximately every 2 hours. Exponential phase at this temperature was observable for approximately 1.5 days, with culture density increase first becoming detectable approximately 2 days after inoculation. A generalized logistic model was fit to the observed data (grey curve). Model parameters automatically fitted by the QFA R package are presented: K (carrying capacity (AU)), r (growth rate (d-1)), g (inoculum density (AU)), v (growth symmetry). B) As for panel A, plotted with cell density on a log scale. Click here to view larger figure.
Figure 2. A single robot-captured QFA growth curve. A) QFA growth curve of a yeast strain growing on agar containing galactose at 20 °C. Images were captured robotically approximately every 2 hours. Exponential phase at this temperature was observable for approximately 1.5 days, with culture density increase first becoming detectable approximately 2 days after inoculation. A generalized logistic model was fit to the observed data (grey curve). Model parameters automatically fitted by the QFA R package are presented: K (carrying capacity (AU)), r (growth rate (d-1)), g (inoculum density (AU)), v (growth symmetry). B) As for panel A, plotted with cell density on a log scale. Click here to view larger figure.
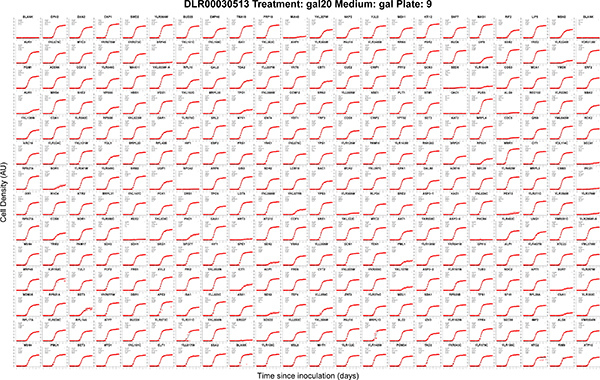 Figure 3. Automatically generated growth curves captured in parallel from a single 384-format plate. Each panel shows cell density estimates (red crosses) and model fit (black curves) for independent cultures labeled by gene name or ORF y-number grown in a QFA procedure. This example plate was imaged by robot and grown in an automated incubator at 20 °C. Experimental metadata are automatically included in the figure title: plate name, plate treatment, agar medium and library plate number. Fitted model parameter values are also automatically printed on each panel (see Figure 2). Edge cultures were stripped from QFA analysis, leaving the 308 cultures in this plot (see QFA Protocol 5.4). Since edge cultures are not analyzed in QFA, these culture locations are typically filled with neutral control strains (pGAL-HIS3 in this case). The original version of this figure, output by the QFA R package, is in .pdf format and is therefore infinitely zoomable and searchable by text queries. Click here to view larger figure.
Figure 3. Automatically generated growth curves captured in parallel from a single 384-format plate. Each panel shows cell density estimates (red crosses) and model fit (black curves) for independent cultures labeled by gene name or ORF y-number grown in a QFA procedure. This example plate was imaged by robot and grown in an automated incubator at 20 °C. Experimental metadata are automatically included in the figure title: plate name, plate treatment, agar medium and library plate number. Fitted model parameter values are also automatically printed on each panel (see Figure 2). Edge cultures were stripped from QFA analysis, leaving the 308 cultures in this plot (see QFA Protocol 5.4). Since edge cultures are not analyzed in QFA, these culture locations are typically filled with neutral control strains (pGAL-HIS3 in this case). The original version of this figure, output by the QFA R package, is in .pdf format and is therefore infinitely zoomable and searchable by text queries. Click here to view larger figure.
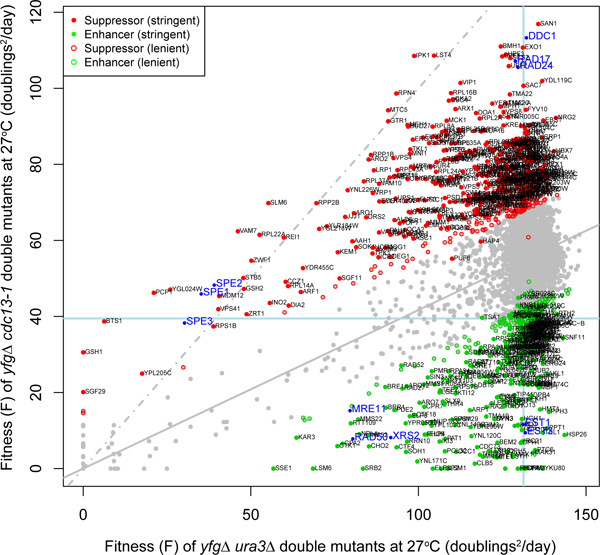 Figure 4. Comparing fitnesses from two QFA screens to deduce genetic interaction. This plot shows comparison of the fitness of budding yeast deletions (yfgΔ) combined with the neutral ura3Δ mutation or the temperature sensitive cdc13-1 mutation and grown at the semi-permissive temperature for cdc13-1 (27 °C). Fitness was calculated as the product of maximum doubling rate and maximum doubling potential 1. Most deletion strains grow well when combined with the neutral ura3Δ mutation, as expected and have high fitness, but deletions combined with cdc13-1 have a wide range of fitnesses. The light blue lines cross at the location of the neutral his3Δ mutation, the solid line is a linear regression model of genetic independence (expected fitness of cdc13-1 yfgΔ mutants given fitness of ura3Δ yfgΔ mutants) and the dashed line is the line of equal fitness. Deletions colored green significantly enhance the fitness defect of cdc13-1 strains. Deletions colored red significantly suppress the same defect. Vertical distance of any mutation cdc13-1
yfgΔ from the solid line indicates the strength of genetic interaction between cdc13-1 and yfgΔ under the plate conditions. Note that some of the strongest suppressors of cdc13-1 are previously identified checkpoint genes, RAD17, DDC1 and RAD24 and the nuclease EXO1 near the top right. Genes whose deletion reduces fitness include YKU80 (encoding the DNA repair and telomere capping protein Yku80) near the bottom right. Also note that some groups of genes that function together tend to be co-located on this plot. Three example clusters are highlighted in blue SPE1, SPE2 and SPE3: spermidine synthesis genes, RAD17, DDC1 and RAD24: encoding components of the checkpoint sliding clamp and checkpoint clamp loader, MRE11, RAD50 and XRS2: the MRX complex, involved in DDR and EST1 & EST3: genes involved in telomere length regulation. Adapted from Supplementary Figure 1B
1, raw data from which this plot was generated can be downloaded from here: http://research.ncl.ac.uk/colonyzer/AddinallQFA/. Click here to view larger figure.
Figure 4. Comparing fitnesses from two QFA screens to deduce genetic interaction. This plot shows comparison of the fitness of budding yeast deletions (yfgΔ) combined with the neutral ura3Δ mutation or the temperature sensitive cdc13-1 mutation and grown at the semi-permissive temperature for cdc13-1 (27 °C). Fitness was calculated as the product of maximum doubling rate and maximum doubling potential 1. Most deletion strains grow well when combined with the neutral ura3Δ mutation, as expected and have high fitness, but deletions combined with cdc13-1 have a wide range of fitnesses. The light blue lines cross at the location of the neutral his3Δ mutation, the solid line is a linear regression model of genetic independence (expected fitness of cdc13-1 yfgΔ mutants given fitness of ura3Δ yfgΔ mutants) and the dashed line is the line of equal fitness. Deletions colored green significantly enhance the fitness defect of cdc13-1 strains. Deletions colored red significantly suppress the same defect. Vertical distance of any mutation cdc13-1
yfgΔ from the solid line indicates the strength of genetic interaction between cdc13-1 and yfgΔ under the plate conditions. Note that some of the strongest suppressors of cdc13-1 are previously identified checkpoint genes, RAD17, DDC1 and RAD24 and the nuclease EXO1 near the top right. Genes whose deletion reduces fitness include YKU80 (encoding the DNA repair and telomere capping protein Yku80) near the bottom right. Also note that some groups of genes that function together tend to be co-located on this plot. Three example clusters are highlighted in blue SPE1, SPE2 and SPE3: spermidine synthesis genes, RAD17, DDC1 and RAD24: encoding components of the checkpoint sliding clamp and checkpoint clamp loader, MRE11, RAD50 and XRS2: the MRX complex, involved in DDR and EST1 & EST3: genes involved in telomere length regulation. Adapted from Supplementary Figure 1B
1, raw data from which this plot was generated can be downloaded from here: http://research.ncl.ac.uk/colonyzer/AddinallQFA/. Click here to view larger figure.
| File ID | File Type | Manually Constructed | Update Frequency | Prerequisite |
| FT1 | Plate photograph | No | Per plate & timepoint | Yes |
| FT2 | Experimental Description | Yes | Per experiment | Yes |
| FT3 | Library Description | Yes | Per library | Yes |
| FT4 | Standard Gene Names | Yes | Per library | No |
| FT5 | Colonyzer Output | No | Per timepoint per plate | Yes |
| FT6 | QFA Raw Data | No | Per experiment | - |
| FT7 | QFA Logistic Parameters | No | Per experiment | - |
| FT8 | QFA Fitness Summary | No | Per experiment | - |
| FT9 | QFA Genetic Interaction Hitlist | No | Per GIS study (2 expts.) | - |
Table 1. Electronic QFA Files. All files listed (excepting FT1) are tab-delimited text files and can be constructed or read using any text editor or spreadsheet software. For further details about constructing QFA metadata files and precise interpretation of QFA output please see the QFA R package documentation 7 and the Colonyzer software documentation 3.
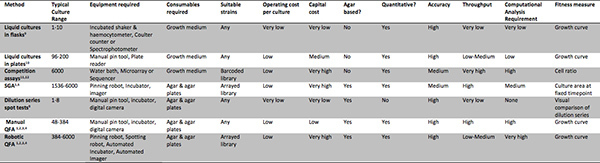 Table 2. Methods for assessing culture fitness. A summary of the features and requirements of a number of methods for assessing microbial culture fitness. A review by Blomberg 8 contains more detailed comparison of some of these. Click here to view larger figure.
Table 2. Methods for assessing culture fitness. A summary of the features and requirements of a number of methods for assessing microbial culture fitness. A review by Blomberg 8 contains more detailed comparison of some of these. Click here to view larger figure.
Discussion
QFA is in many senses a direct descendent of three established bench-scale microbiological techniques: culture dilution series spot tests 9, growth curve determination in aerated liquid cultures 9 and replica plating 13. These three methods are summarized and compared with QFA and other high-throughput techniques in Table 2. Whereas dilution series spot tests define a strain's fitness as its ability to form colonies in a series of cultures grown from a range of inoculum dilutions, QFA measures strain fitness by repeated observations of a single culture to construct a growth curve. Quantifying fitness from single cultures allows many more strains to be examined simultaneously, under identical conditions. Replicating arrays of cultures allows repeated testing of strain collections, most usefully when testing for culture fitness differences in different environments or genetic backgrounds. The QFA protocol presented here uses expensive robotic equipment to replicate, inoculate, incubate and image agar plates, appropriate for observing growth of thousands of independent cultures (robotic QFA, Table 2). However, since QFA is based on established, traditional lab techniques, it can also be carried out much more cheaply by replacing robot assistance with manual steps (manual QFA, Table 2). Manual QFA involves replication and inoculation of cultures onto agar plates using a manual pin tool and manual transfer of plates to and from an incubator for photography. The same computational analysis workflow can be applied to generate growth curves from either experimental design.
QFA fitness estimates seem to be comparatively precise. In Figure 4, the mean fitnesses of four example clusters of functionally related gene deletions are highlighted. The close proximity of members of each functionally related gene class in two independent genetic backgrounds (ura3Δ and cdc13-1) indicates the reproducibility of QFA fitness estimates. For example, only 3 gene deletions (out of a possible 4,300) separate the three members of the conserved MRX complex.
High-throughput alternatives to QFA for testing growth characteristics of microbial strains include SGA 5,6, competition screens in barcoded libraries 11,12 and screens capturing optical density kinetics in spectrophotometric plate readers 10 which are summarized in Table 2 and described more fully elsewhere 8. QFA and SGA plates, where cultures grow on solid agar surfaces (agar based methods, Table 2) can be handled quickly and easily by robot. Solid agar surface cultures are well aerated throughout growth and cells can grow in fixed cultures, allowing sociable microbes to grow in a community 14. Left intact, microbial communities affect their own micro-environments, secreting toxins such as ethanol, and possibly signaling between cells. However, continuous mixing of liquid cultures, necessary to achieve adequate aeration, artificially disrupts microbial communities and their micro-environments which may affect their modes of growth. Cross-contamination is less of a concern in solid agar methods since there is less opportunity for liquid splashes carrying cells between cultures. If contamination occurs by foreign air-borne microbes in solid agar assays, it can often be detected by visual inspection of solid agar plates and accounted for or removed.
QFA throughput is significantly higher than that of parallel liquid methods in two ways. First of all, on QFA (and SGA) plates, inoculated cultures are packed together more densely, giving more independent cultures per plate. In QFA there are typically 308 cultures, not counting non-experimental edge cultures (Figure 1) compared to parallel liquid cultures with 96 or 100 cultures per plate. Secondly, QFA experiments can be scaled to a much larger number of plates. Whereas the number of plates analyzed in a single QFA experiment is limited only by the space available in an incubator or warm room, the minimum allowable image capture frequency and the maximum achievable capture frequency, the number of plates in a liquid growth experiment is strongly limited by the capacity the plate reader (typically one or two plates), or of the stacker attached to the plate reader (typically 25-50 plates). We have recently carried out a fully automated QFA experiment on 123 plates, each with 308 experimental cultures, giving 37,884 simultaneous cultures. We estimate that the maximum achievable number of independent cultures growing in liquid is 96 (cultures/plate) x 50 (plates/stacker) = 4800, which is around ten fold less than our QFA throughput. An alternative for liquid culture screens is to utilise many automated growth devices in parallel (Table 1 from the review by Blomberg 8), each having a capacity of one or two plates. Temperature control in each device is independent, and so conditions are not identical, but assuming that they are, this workflow would require at least 190 such devices to match QFA throughput (assuming 200 liquid cultures per device).
In summary, QFA is a high-quality workflow that can be usefully applied to gather quantitative growth phenotypes in both small scale focussed experiments and high-throughput screens. It is flexible enough to be applied effectively with varying requirements for robotic equipment. The computational component of the QFA workflow is based on freely available, open source code.
Disclosures
No conflicts of interest declared.
Acknowledgments
We gratefully acknowledge all members of our laboratory and the Centre for Integrated System Biology of Ageing and Nutrition (CISBAN) for support and helpful discussions. This study was supported by Biotechnology and Biological Sciences Research Council (BBSRC) (BB/C008200/1) and the Wellcome Trust (075294, 093088).
References
- Addinall SG, Holstein E, Lawless C, Yu M, Chapman K, et al. Quantitative Fitness Analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects. PLoS Genetics. 2011;7(4):e1001362. doi: 10.1371/journal.pgen.1001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Downey M, Yu M, Zubko MK, Dewar J, et al. A genomewide suppressor and enhancer analysis of cdc13-1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics. 2008;180:2251–2266. doi: 10.1534/genetics.108.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C, Wilkinson D, Young A, Addinall S, Lydall D. Colonyzer: automated quantification of micro-organism growth characteristics on solid agar. BMC Bioinformatics. 2010;11:287–28. doi: 10.1186/1471-2105-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonyzer - Image analysis software [Internet] Newcastle, UK: Newcastle University; 2012. [cited 2012 Jan 1]. Available from: http://research.ncl.ac.uk/colonyzer/ [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- qfa - An R Package for Quantiative Fitness Analysis [Internet] Vienna, Austria: University of Vienna; 2012. [cited 2012 Jan 1]. Available from: http://qfa.r-forge.r-project.org/ [Google Scholar]
- Blomberg A. Measuring growth rate in high-throughput growth phenotyping. Curr. Opin. Biotechnol. 2011;22(1):94–102. doi: 10.1016/j.copbio.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δmutants. Genes and Dev. 2002;16(15):1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J, Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer M, Fung E, Wildenhain J, Pierce S, Hoon S, Lee W, Proctor M, St. Onge R, Tyers M, Koller D, Altman R, Davis R, Nislow C, Giaever G. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Durbic T, Oh J, Urbanus M, Proctor M, Heisler LE, Giaever G, Nislow C. Competitive Genomic Screens of Barcoded Yeast Libraries. J. Vis. Exp. 2011. p. e2864. [DOI] [PMC free article] [PubMed]
- Lederberg J, Lederberg EM. Replica plating and indirect selection of bacterial mutants. J. Bacteriol. 1952;63:399–406. doi: 10.1128/jb.63.3.399-406.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller C. Behavioural ecology: The social side of wild yeast. Nature. 2008;456:589–590. doi: 10.1038/456589a. [DOI] [PubMed] [Google Scholar]


