Abstract
The equilibrium unfolding reaction of Ltn, a metamorphic C-class chemokine was monitored by tryptophan fluorescence to determine unfolding free energy values. Measurements revealed that addition of 150mM NaCl stabilized the Ltn chemokine fold by approximately 1 kcal/mol. Specific mutations involving Arg23 and Arg43 also increased stability by 1 kcal/mol suggesting involvement in chloride ion coordination. This interaction was confirmed by NMR salt titrations studies that revealed chemical shift perturbations localized to these residues and backbone amides within the proximal 40’s-loop. The effects of NaCl on the free energy landscape were further verified by ZZ-exchange NMR spectroscopy. Our results suggest that changes in the electrostatic environment modulate the Gibb’s free energy of folding and alter the forward and reverse rates of interconversion. These results demonstrate how solution ions can promote metamorphic folding by adjusting the relative stabilities of two unrelated Ltn native state structures.
Keywords: Chemokine, Lymphotactin, native state, unfolding, metamorphosis, free energy
The interactions between a protein and its surroundings ultimately define its function, which is encoded in a specific 3D structure. The consensus view originally proposed by Anfinsen is that a protein adopts a single native-state fold, within the context of its environment, representing a minimum in the Gibbs free energy (1). This thermodynamic hypothesis implies that a protein’s native-state structure is determined solely by its primary sequence and the composition of the surroundings (e.g. pH, solvent, ions, etc). However, while the vast majority of proteins predictably adopt a single native-state conformation (with varying degrees of flexibility), nothing precludes the existence of primary sequences that populate more than one native-state fold provided their free energies are nearly equal. Interestingly, a small number of naturally occurring proteins have shown the extraordinary ability to actively switch their native-state fold defining a new category of “metamorphic proteins” (2, 3).
Of the several examples now documented (4–7), the chemokine lymphotactin (Ltn/XCL1) undergoes one of the most remarkable native-state transformations. In one state, termed Ltn10, the protein adopts the canonical chemokine fold comprised of a three strand β-sheet and a C-terminal α-helix, which converts into a dimeric β-sheet fold termed Ltn40. Unlike other metamorphic proteins where a majority of substructure is retained throughout the interconversion process, Ltn native-state transformation requires complete restructuring of all hydrogen bonds and tertiary contacts (7). At near-physiological conditions (37 °C, 150mM NaCl) both Ltn conformations are equally populated (Keq ~1) and each is required for proper biological function (7).
Early on it was revealed that the Ltn10-Ltn40 equilibrium could be shifted by adjusting solution conditions (8). For instance, in conditions of high salt and low temperature (200mM NaCl, 10 °C) the Ltn10 chemokine fold is exclusively populated, while at high temperature with no salt (0mM NaCl, 40 °C) the alternate Ltn40 β-strand configuration is strongly favored. Numerous reports have detailed the effects of various salts on protein stability (9–11), but the specific role of solution ionic strength in modulating metamorphic interconversion has not been investigated.
Ltn is rich in basic amino acids (9 Arg and 6 Lys; 93 total residues). Because the protein carries a net positive charge, it is likely that electrostatic interactions contribute to salt dependence of the Ltn10 conformation. In fact, replacement of Arg23 and/or Arg43 with alanine led to an increase in the Ltn10 population relative to Ltn40, even in the absence of salt (12). Given the proximity of positively charged Arg23 and Arg43 side chains in Ltn10 (~10 Å), which are separated by ~20 Å in Ltn40, we speculated that electrostatic repulsion selectively destabilizes the chemokine fold (12). Conversely, Ltn40 contains two potential salt bridges, Arg9 to Asp50 (intramolecular) and Lys25 to Glu31 (intermolecular), which may be weakened by the addition of salt. Interestingly, a molecular dynamic (MD) simulation that investigated the effect of salt concentration on Ltn10 structural stability revealed a chloride ion persistently associated with the Arg23 and Arg43 side chains as well as backbone amides within the 40’s-loop (13). Collectively, these observations suggest that coulombic forces contribute to metamorphic rearrangements in Ltn. However, the effects of salt on the Ltn10-Ltn40 equilibrium have not been quantified, and the participating structural elements remain undefined. Here we use a combination of equilibrium unfolding, NMR binding, and kinetic exchange measurements to quantify the changes in Ltn free energy associated with an increased ionic environment and mutation. Our results indicate that interactions between Arg23 and Arg43 have a destabilizing effect on Ltn10 free energy that can be overcome by increasing the ionic strength of the surroundings. This effect is further corroborated through kinetic analysis revealing that ionic environments beneficial to Ltn10 will destabilize the Ltn40 structure. The consequence of electrostatic tuning of Ltn free energy is discussed in the context of other metamorphic proteins.
Material and Methods
Protein expression, purification, and mutagenesis
All proteins were expressed and purified using established protocols that have been previously reported (14). All mutant Ltn proteins were produced by employing the QuickChange® Site-Directed mutagenesis kit following manufacturers instructions, with no deviation in expression or purification protocols.
Equilibrium unfolding experiments and determination of unfolding free energies
Experimental measurements were performed on a Photon Technology International (PTI) spectrofluorometer in a fluorescence cuvette with a 1 cm path length equipped with a stir bar rotating at 240 rpm. An excitation wavelength of 283 nm was used with a bandwidth of 4 nm. The fluorescence emission for WT-Ltn was collected between 310–360 nm (λmax 332 nm) recorded at 1nm increments with signal acquired for 2 sec at each wavelength employing a 6 nm bandwidth. Fluorescence emission spectra for the mutant proteins were collected with identical parameters and all displayed λmax of 332 ± 1nm. Fresh urea stocks were used for all unfolding measurements following standard laboratory practices (15). All data points in the unfolding curve were determined with a separate solution containing 5µM protein in 20mM Na2HPO4 pH 6. Unfolding curves were recorded by monitoring fluorescence intensity at 332 nm as a function of urea concentration. The free energy of each point in the unfolding transition zone was determined from the equation:
| Equation 1) |
where Ku is the equilibrium unfolding value at each urea concentration. Ku was determined using a two-state unfolding model based on the equation:
| Equation 2) |
With Yf and Yu representing the folded and unfolded baselines, respectively and Yurea representing the fluorescence signal as a function of urea concentration. The final ΔGunfold value was obtained by extrapolation to zero molar urea based on the equation:
| Equation 3) |
where ΔGH2O is free energy unfolding value in the absence of denaturant, [urea] is molar concentration of denaturant , and m is slope in units of cal/mol M.
NMR Spectroscopy
All NMR experiments were performed on a Bruker DRX 600 equipped with a 1H, 15N, 13C cyroprobe. To detect NaCl interactions with Ltn10, titration experiments were performed with 100 µM 15N-labeled Ltn in 20mM sodium phosphate pH 6 at 283 K. The salt concentration was incrementally increased to 5, 15, 35, 75, 150, 225, and 365 mM NaCl, with subsequent collection of a 2D 1H-15N-HSQC spectrum. HSQC cross peak movements were quantified as a combined 1H-15N chemical shift perturbation calculated from:
| Equation 4) |
where Ho and No represent reference proton and amide nitrogen ppm values obtained in starting buffer (0 M NaCl), and H and N represent proton/amide nitrogen cross peak ppm values determined at the different NaCl concentrations. Salt binding curves were generated by plotting differences in chemical shift values as a function of NaCl concentration. As NaCl concentrations were increased above 15 mM, chemical shift perturbations were observed for nearly all amide groups indicating non-specific interactions across the Ltn10 backbone. As a result NaCl binding affinities were determined using a ligand interaction model that accounted for non-specific binding when titration curves appeared unsaturable using the equation:
| Equation 5) |
, where ΔCSobs is the observed chemical shift change calculated from equation 4, ΔCSmax is the maximum chemical shift difference, L is ligand concentration (NaCl), Kd is binding affinity and NS accounts for non-specific interactions. Binding affinities were obtained through curve fitting using program pro Fit 6.1. When inclusion of the non-specific interaction term (NS) did not effect the measured Kd value, binding fits were generated using an NS value of zero and are represented as solid lines in Figure 4. In all other instances titration data displayed unsaturable binding warranting the use of equation 5 to accurately determinine Kd values. Binding fits that include the NS term are represented as dashed lines in Figure 4. Normalized ∆CS was determined from the largest recorded chemical shift difference observed in each titration curve generated
Figure 4.
Chloride ion binding curves generated from NMR titration. Kd values were determined by curve fitting to equation 5 with error estimated from fitting shown in insert. Solid and dashed lines representing binding curves denote removal or inclusion of non-specific interaction term, as described in Material and Methods. A) NaCl titration curves generated at 10 °C in 20mM sodium phosphate. B) NaCl titration curves generated at 10 °C in 10mM Bis-Tris.
Ltn interconversion rates were determined by 2D longitudinal 15N ZZ-exchange spectroscopy (16). In these experiments a series of 2D exchange spectra were acquired with variable mixing times of 50, 150, 200, 250, 300, 350, 400, 500, 750, and 900 ms. Sample conditions consisted of 1mM Ltn in 20mM NaH2PO4, pH 6 at 298 K. Comparisons with salt samples were obtained in an identical buffer and temperature with the addition of 150mM NaCl. Nonlinear fitting of cross peak and exchange peak intensities that accounted for two-site exchange and 15N -T1 relaxation was conducted with an in-house MATLAB 2007b script employing the equations described by Tollinger et al (17) that allow extraction of kfor and krev Ltn interconversion rates.
Results and Discussion
Salt stabilization of the Ltn10 conformation
Ltn structural stability was studied by chemical unfolding at 25 °C using urea as a denaturant. As Ltn contains only one tryptophan at position 55 (Trp55), the equilibrium unfolding reaction was monitored by measuring tryptophan fluorescence intensity at 332 nm as a function of urea concentration. To ensure that fluorescence emission detected was sensitive primarily to monomeric Ltn10 and not a mixture of both structural states, the dependence of the unfolding curve on protein concentration was investigated. It has been shown that the denaturation midpoint for dimeric proteins is dependent on protein concentration (18) (19) . To assess the contribution of each structural state to fluorescence emission we collected urea unfolding curves at different Ltn concentrations. It was reasoned that if fluorescence emission contained substantial contributions from Ltn40 a characteristic shift in the unfolding curve would be observed. Figure 1A displays unfolding curves generated at different Ltn concentrations indicating that between 2 and 10 µM Ltn protein, the denaturation midpoint is unaffected by protein concentration, which is consistent with fluorescence reporting mainly on the monomeric Ltn10 state. This is not unexpected considering that Trp55 in Ltn10 is buried in the hydrophobic core resulting in a more intense blue-shifted fluorescence emission spectrum relative to the dimeric Ltn40, where Trp55 is solvent exposed (7). The baseline WT-Ltn10 unfolding value used in our investigation was established at 2 kcal/mol with a 5 µM protein sample in 20mM sodium phosphate (Figure 1B), using the linear extrapolation method (15) shown in Figure 2A. To test the effect of NaCl concentration on Ltn10 stability additional denaturation experiments were conducted in the presence of 70mM and 150mM NaCl. Under both conditions Ltn10 was stabilized by about 1 kcal/mol (Figure 2A and Table 1) indicating that within this range, increasing the ionic strength of the environment had a similar effect on Ltn10 stability. We speculated that the observed salt stabilization resulted from a specific interaction rather than a general effect of ionic strength.
Figure 1.
Urea denaturation unfolding curves monitored by fluorescence. A) Comparison of Ltn unfolding curves collected at 2 and 10 µM in 20 mM Na2HPO4 pH 6 at 25 °C. B) Unfolding curves for WT and mutant Ltn. All data were obtained with 5 µM protein in 20 mM Na2HPO4 pH 6 at 25 °C. Ltn mutants are denoted as single letter amino acid abbreviations, followed by position and identity of substitution in protein sequence. Solid lines connecting data points represent non-linear fitting to a two-state unfolding model where included for clarity and not used in analysis.
Figure 2.
Unfolding free energy (ΔGunfold) of WT and mutant Ltn as a function of urea concentration. All ΔGunfold values were calculated based on equations 1 and 2, with final extrapolated values determined with equation 3. Experiments conducted in the presence of 70mM or 150mM NaCl in buffer are denoted. A) Comparison of WT Ltn unfolding values in the absence and presence of NaCl. B) Comparison of R23A, R43A and R23/43A Ltn mutants, and effect of salt on free energy. C) and D) ΔGunfold vs [urea] for additional arginine and lysine Ltn mutants.
Table 1.
Ltn10 free energy unfolding values determined for WT and mutant constructs.
| Ltn Construct |
ΔGunfold (kcal/mol) |
m-value (kcal/mol M) |
Repulsion pair* |
Distance (Å) |
|---|---|---|---|---|
| WT | 2.1 ± 0.1 | 0.92 ± .04 | NA | NA |
| WT70 | 2.7 ± 0.1 | 0.93 ± .02 | NA | NA |
| WT150 | 2.7 ± 0.2 | 0.85 ± .06 | NA | NA |
| R9A | 2.0 ± 0.3 | 0.72 ± .02 | R9 – K46 | 10.1 ± 2.5 |
| R23A | 2.9 ± 0.1 | 0.73 ± .02 | R23 – R43 | 10.1 ± 2.3 |
| R23A150 | 2.8 ± 0.1 | 0.68 ± .01 | R23 – R43 | 10.1 ± 2.3 |
| K25A | 2.2 ± 0.1 | 0.78 ± .03 | K25 – R70 | 5.8 ± 1.9 |
| R35A | 2.0 ± 0.2 | 0.65 ± .07 | R35 – R57 | 10.5 ± 1.8 |
| R43A | 2.8 ± 0.1 | 0.82 ± .03 | R23 – R43 | 10.1 ± 2.3 |
| K66A | 2.2 ± 0.1 | 0.80 ± .03 | K66 – K42 | 5.5 ± 2.4 |
| R23/43A | 3.0 ± 0.1 | 0.61 ± .06 | R23 – R43 | 10.1 ± 2.3 |
Denotes side chain pairs with potentially unfavorable charge-charge interactions that were disrupted by mutation. Distance represents ensemble average between arginine Cζ or lysine Nζ atoms ± standard deviation (PDB 1J8I). Superscript 70 and 150 refer to NaCl concentration 70mM and 150mM used in unfolding experiments. NA-not applicable. Error in ΔGunfold and m-values values estimated from linear fittings using equation 3.
Effect of charge-change mutants on ΔGunfold
Given the excess of positive charge carried by Ltn, we reasoned that salt ions might increase stability by weakening repulsive forces within the protein. To differentiate between a general ionic strength effect and a specific NaCl interaction within the Ltn10 structure, a series of charge change mutants was generated. Mutants were designed to disrupt potentially destabilizing arginine and lysine side chain configurations by alanine substitution. We hypothesized that if the observed salt stabilization was due to non-specific interactions individual mutations would have little or no effect on fold stability. In contrast, if the increased stability resulted from electrostatic screening of specific repulsive side chain contacts, then mutants at those sites would display a pronounced increase in Ltn10 stability independent of salt. The free energies of unfolding for six Ltn charge-change mutants were determined by linear extrapolation (Figure 2B–D) with results shown in Table 1. In the absence of salt, R23A (Figure 1B) and R43A each stabilize the chemokine fold by approximately 1 kcal/mol. Interestingly introduction of the double mutant R23/43A, or addition of 150 mM NaCl to R23A contributed no additional increase in protein stability. Other mutations investigated (i.e. R9A, K25A, R35A, K66A) showed no significant change in protein stability. Taken together the free energy analysis is consistent with electrostatic repulsion between Arg23-Arg43 side chains destabilizing the Ltn10 fold, which can be counteracted through increased ionic strength or removal of one of the charged side chains.
Effects of NaCl and mutations on m-values
The m-value, reflected in the slopes of the lines in Figure 2, (listed in Table 1) reports on the dependence of the free energy of unfolding on the denaturant concentration. Comparisons of m-values reveal that within experiential error, addition of 70mM or 150mM NaCl had no significant effect on values observed for WT-Ltn10. However all charge-change mutants revealed decreases in the m-value relative to WT-Ltn with the double mutant R23/43A displaying the largest effect. Decreases in m-value can be indicative of an increase population of intermediate states during protein unfolding or formation of a more compact denatured state (20), however our data does not allow us to conclusively differentiate between these two possibilities. Considering Ltn unfolding properties, we have not seen evidence of an intermediate state in kinetic unfolding measurements (21) or observed biphasic denaturation curves (Figure 1). Given the excess of positively charge residues in Ltn it is possible that removal of Arg or Lys sidechains also reduces unfavorable interactions in the denatured state. As changes in m-value have been correlated to differences in accessible surface area between the folded and denatured states (ΔASA) (22), we speculate that decreases in m-values observed for the Ltn mutants may reflect a reduction is ΔASA resulting from increased compactness of the denatured states caused by removal of positively charged amino acids.
Identification of a chloride ion binding site
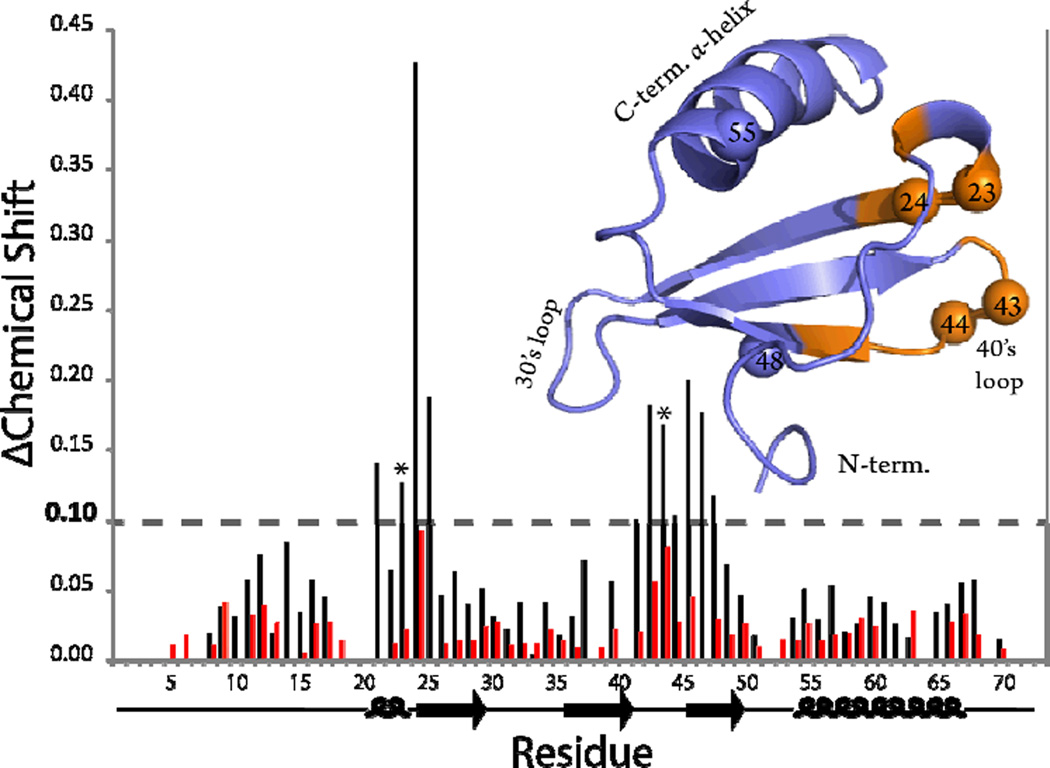
The interaction between Ltn10 and NaCl was further investigated by 1H-15N-HSQC spectroscopy. It was rationalized that if NaCl binding was mediated through the Arg23 /Arg43 side chains then salt-induced chemical shift perturbations would localize to this region of Ltn10. In these experiments 15N-labeled Ltn in 20mM Na2HPO4 at pH6 was titrated against incremental additions of an identically buffered NaCl solution. All HSQC spectra were collected at 10 °C to ensure that Ltn10 cross peak movements resulted from a direct interaction with salt ions and not structural interconversion. At low salt concentration (15mM) the only significant amide chemical shift perturbations observed (≥ 0.1 ppm) were constrained to areas near Arg23 and Arg43 (Figure 3). The changes to Ltn10 chemical shift values were mapped onto the 3D structure and revealed a specific interaction site that was consistent with a chloride ion binding pocket identified through molecular dynamic simulations (13). Interestingly, this same pocket on Ltn was also identified as the binding site for the viral protein M3 chemokine decoy receptor (23). Considering the composition of residues identified in chemical shift mapping (5 positively charged side chains, 5 hydrophobic side chains) the results are consistent with chloride, rather than sodium ion, acting as the ligand.
Figure 3.
Chemical shift mapping of NaCl interaction with the Ltn chemokine fold. Bar plot represents chemical shift difference (ΔChemical shift) in amide backbone resonances collected at 10 °C from 0 and 15mM NaCl calculated from equation 4, where (*) denotes positions of R23 and R43. Residues that displayed ΔCS values greater than 0.1 ppm are mapped onto the Ltn chemokine fold highlighted in orange. Red bars depict shift perturbations generated using Ltn-R23A collected at 10 °C from 0 and 15mM NaCl. Spheres shown on the Ltn chemokine structure denote amino acid positions in the primary sequence described in text.
To confirm the identity of the R23/R43 chloride recognition site, chemical shift perturbations induced by addition of 15 mM NaCl were monitored for the R23A mutant. Significant salt-induced perturbations (i.e. ΔCS < 0.1) are no longer observed, indicating that removal of the Arg23 side chain disrupts a specific chloride binding site within the chemokine fold.
Ion binding affinity and selectivity
For the majority of residues identified through chemical shift mapping binding affinities (Kd) were determined to be approximately 16mM (Figure 4A), with two outliers displaying values ranging between 40 and 70mM. Residues outside the revealed binding site, displayed much weaker affinities ( Kd > 700mM, Supporting information, Figure S1). It is difficult to speculate on the discrepancy of these lower affinities, but based on the consensus of data we estimate a NaCl binding affinity of 16mM for residues within the Arg23 / Arg43 region. This value appears consistent with other proteins (e.g. α-amylase, and Runx1) known to coordinate chloride ion through arginine and lysine side chains demonstrating binding affinities between 6 and 34mM (24) (25). Considering a Kd value of 16mM, one expects at physiological concentrations (20mM phosphate, 150mM NaCl) about 90% of Ltn10 would have an occupied Arg23 / Arg43 binding pocket, with a binding energy of up to 2 kcal/mol. While this exceeds the salt-dependent effect determined by thermodynamic unfolding measurements (WT-Ltn10 ΔΔG ~ 0.6 kcal/mol, Table 1), we conclude that a specific interaction at the R23/R43 pocket can easily account for the salt-induced stabilization of Ltn10.
It was demonstrated previously that addition of 10mM phosphate improved the quality of 2D NMR spectra of Ltn (26), and we considered the possibility that other anions might bind preferentially to the site detected in our titrations with NaCl which were buffered by 20 mM phosphate. A NaCl titration conducted in 10mM Bis-Tris buffer (Figure 4B) yielded Kd values for residues located in the Ltn10 chloride binding pocket (Arg23, Ile24, Thr4, Lys42) in good agreement with the values determined in 20mM phosphate. One notable difference was observed for R43, which required a non-specific binding term in fitting NaCl titration data in the presence of Bis-Tris (Figure 4B). However, these results indicate that phosphate concentrations at or below 20mM have a negligible effect on Ltn10 / chloride interactions. This does not exclude the possibility of phosphate interacting with the binding pocket, but comparisons of the relative affinity of phosphate and chloride ions (data not shown) indicate that chloride binds preferentially to the R23/R43 site.
Effect of NaCl on Ltn10 interconversion kinetics
In order to further verify the effects of salt on Ltn10 stability we investigated the dependence of Ltn interconversion rates on NaCl concentration. We reasoned that increasing the stability of Ltn10 would increase the barrier height (ΔG‡) associated with structural interconversion, and slow the rate at which Ltn10 rearranges to Ltn40 (krev). Therefore we determined the effect of NaCl on Ltn interconversion rates (kfor, krev) using ZZ-exchange NMR spectroscopy. A series of 2D exchange spectra were acquired at 25 °C in 20mM Na2HPO4 pH 6 containing either 0 or 150mM NaCl employing exchange mixing times ranging from 50 to 900 ms. The ZZ-exchange cross peak intensities were measured as a function of time (Figure 5A and B) and Ltn interconversion rates were determined by non-linear fitting using established equations (17) . Measurements involving Gly44 cross peaks revealed that addition of 150mM NaCl decreased the Ltn10 → Ltn40 conversion rate (krev), which is indicated in Figure 5. Additional rates were also determined from Cys48 cross peak intensities (Supporting information, Figure S2) revealing a similar decrease in krev values.
Figure 5.
Experimental data and fitted curves for G44 ZZ-exchange. A) Curves describe the decay of the auto peaks (Ltn10 and Ltn40), with lower two curves showing buildup and decay of exchange peaks in 20 mM Na2HPO4 at 25 °C. Insert shows Gly44 cross peaks for 750 ms mixing time. B) Curves describe the decay of the auto peaks (Ltn10 and Ltn40), with lower two curves showing buildup and decay of exchange peaks in 20 mM Na2HPO4, 150 mM NaCl at 25 °C. Insert shows Gly44 cross peaks for 750 ms mixing time. C) Salt dependence of the Ltn free energy landscape. The terms kfor and krev indicate conversion rate of Ltn as shown by dashed arrows. The NaCl superscript reflects rate measured in the presence of 150 mM NaCl. ΔΔG‡ values were calculated from the equation described in text. D) Intramolecular (R9–D50) and intermolecular (K25–E31) salt-bridge interactions found within the Ltn40 dimer.
Considering the nature of the Ltn energy landscape, it was possible that differences observed in the krev interconversion rates arose from an increased transition state free energy as well as stabilization of the Ltn10 conformation. Therefore we computed the free energy change (ΔΔG‡) reflected in altered krev rates as −RTln(krev / krevNaCl) (27). We reasoned if the free energy changes derived from kinetic measurements were greater than the energy differences revealed by chemical unfolding it would support destabilization of the Ltn interconversion transition-state. Conversely if the two values were similar then any change in kinetics caused by salt could be attributed solely to the protein thermodynamic native-state. ΔΔG‡ values of 0.5 kcal/mol determined from ZZ-exchange experiments (Figure 5C) are in excellent agreement with the ΔΔG value of 0.6 kcal/mol derived from Ltn10 urea unfolding experiments (Table 1) and we concluded that increased ionic strength has little or no effect on the interconversion transition state. This conclusion is consistent with our previous determination that the interconversion transition-state is predominately unfolded (21) , since further destabilization would have been highly unlikely.
Effect of NaCl on Ltn40 interconversion
As ZZ-exchange spectroscopy reports on the relative stability of each structural conformation at equilibrium, the kinetic measurements also had the potential to detect salt-dependent changes in the free energy of Ltn40. Interestingly, the addition of 150mM NaCl accelerated the Ltn40 → Ltn10 conversion rate (kfor), consistent with a salt-dependent destabilization of the Ltn40 conformation (ΔΔG‡ ~ 0.3 kcal/mol; Figure 5). Considering the Ltn40 structure, which contains two salt-bridge interactions (e.g. intramolecular Arg9 to Asp50 and intermolecular Lys25 to Glu31, Figure 5D) this result suggested that increases in the ionic environment could destabilize the Ltn40 fold through disruption of electrostatic interactions.
Assessing salt-bridge contributions to Ltn40 stability
To assess the energetic contribution of electrostatic contacts within the Ltn40 fold, we determined Ltn10↔Ltn40 equilibrium constants from 1H-15N-HSQC peak volumes using the relationship Keq = [Ltn40vol] / [Ltn10vol] (Supporting information, Table S1). As noted earlier, the R9A and K25A substitutions have no effect on free energy of unfolding for Ltn10 (Table 1). Thus, we hypothesized that if a specific coulombic interaction involving either side chain contributed to Ltn40 stability, its disruption would shift the equilibrium toward Ltn10. We found that in the absence of NaCl the Keq of WT-Ltn at 25 °C was 1.2 ± 0.2 consistent with a previous report (12). Measurements conducted under identical conditions using 15N-labeled R9A and K25A produced Keq values of 1.1 ± 0.5 and 0.40 ± 0.03, respectively. From these results we concluded that the Arg9–Asp50 contact has a negligible effect of Ltn40 stability. However disruption of the intermolecular Lys25–Glu31 salt-bridge shifted the equilibrium toward the Ltn10 state, consistent with destabilization of Ltn40 (Figure 5 panels C and D). By comparing the change in Keq values observed for WT and Ltn-K25A, we estimate that the Lys25–Glu31 salt-bridge contributes about 0.6 kcal/mol toward Ltn40 stability. While this value is slightly higher than what is expected from ZZ-exchange measurements this discrepancy may result from the Lys25-Glu31 salt bridge being only partially disrupted in the presence of NaCl, compared to complete removal caused by the K25A mutation.
Conclusions
Our measurements of Ltn thermodynamic stability and interconversion kinetics expand our view into the structural and environmental factors defining its metamorphic folding. Our previous temperature-dependent kinetic investigation was designed to resolve the nature of metamorphic rearrangement, revealing similarities between the transition-states associated with protein interconversion and unfolding (21). This led us to the perspective that Ltn metamorphosis follows a path resembling protein unfolding and that factors affecting the stability of either native state conformation can be defined empirically. Here we have identified specific structural elements and environmental factors that optimize the metamorphic folding of Ltn.
Our current work demonstrates that the Ltn10 fold is destabilized by unfavorable electrostatic interactions between two arginine side chains, which can be counteracted by increasing the ionic strength of the surroundings or specific mutations. These observations support a model where chloride ion coordination increases the thermodynamic stability of Ltn10 by reducing the local net charge and coulombic repulsion between Arg23 and Arg43 slowing the Ltn10 → Ltn40 conversion rate. Conversely, a pair of symmetric intermolecular salt-bridges that stabilize the Ltn40 structure are weakened in solutions of high ionic strength increasing the Ltn40 → Ltn10 conversion rate. These structural elements appear to balance the relative stabilities of each Ltn conformer ensuring that both are thermodynamically accessible in physiological conditions of temperature and ionic strength.
We speculate that electrostatic ‘tuning’ of conformational stability may be a common feature in other metamorphic proteins. For example, chloride intracellular channel protein 1 (CLIC1) may also exploit electrostatic interactions in bringing about structural transitions as it inserts into the cell membrane. CLIC1 is a large two-domain protein that is comprised of a thioredoxin-like fold (domain1) and an all α-helical fold (domain2). Hydrogen-deuterium exchange mass spectrometry indicated that conformational stability of CLIC1 domain1 is substantially diminished in acidic environments (28). It is speculated that decreased stability results from a reduction of electrostatic attraction caused by protonation of acidic side chains. As CLIC1 approaches the acidic cell membrane, modulation of electrostatic interactions may destabilize the thioredoxin-like domain1 structure and promote the conversion to a transmembrane helix that inserts into the lipid bilayer (28).
Electrostatic effects may also play a contributing role in metamorphic interconversion of the spindle check point protein Mad2 that actively transforms between open and closed states denoted as O-Mad2 and C-Mad2 (PDB 1DUJ, 1S2H respectively). Conversion of O-Mad2 requires translocation of a C-terminal β-hairpin across the protein while simultaneously rearranging to form a new β-hairpin with a different hydrogen bonding network (6). This transition eliminates potentially unfavorable side chain interactions in O-Mad involving Lys80-Lys113 (~7 Å) and Lys161-Lys194 (~8 Å), separating each pair by ~20 Å, similar to the Arg23–Arg43 separation achieved by the Ltn10→Ltn40 rearrangement. Transformation into C-Mad2 is completed by the new β-hairpin displacing a short N-terminal β-strand that incorporates into a central helix. Displacement of the short β-strand appears to require rearrangement of a salt-bridge formed between Arg16–Glu107 that is replaced by Lys194–Glu107 upon metamorphic transition. Although the exact role of these residues in Mad2 structural transitions is not known, we speculate that attractive and repulsive electrostatic forces contribute to the Mad2 interconversion process. The available structural and thermodynamic data collectively indicate that metamorphic interconversion can be potentiated by coulombic forces unique to each native state conformation.
The existence of metamorphic proteins demonstrates that it is possible to encode more than one ordered structure in a single primary sequence. Yet within the framework of the thermodynamic hypothesis there must exist a balance of free energy between alternate native-state folds that permits structural transition while maintaining energetic equipoise. Ltn may provide additional insight into a distinct energy signature that may help better identify metamorphic systems. Considering the energy landscape, we find the relatively shallow well of thermodynamic folding stability observed for Ltn10 (~2–3 kcal/mol) to be of particular interest, because it permits increased sensitivity to changes in temperature and ionic strength. Moreover, a folded conformation of marginal stability allows greater access to the unfolded state, which is likely required for Ltn structural interconversion (21). For example, the folding equilibrium of Ltn10 (ΔGunfold ~ 2 kcal/mol) populates the unfolded state to a level of ~4%. An additional 1 kcal/mol of stability would reduce the unfolded population to 0.6%. In contrast, a typical protein with an unfolding free energy of 4–5 kcal/mol would have a negligible unfolded state population and a folding equilibrium that is relatively insensitive to a 1 kcal/mol change in ΔGunfold. Thus, marginal thermodynamic stability may be a hallmark of metamorphic proteins that permits access to the unfolded state and sensitizes the native state equilibrium to environmental changes.
As more metamorphic proteins are discovered, it will be important to understand the factors that define each native state structure and govern interconversion. By identifying unique structural and thermodynamic signatures associated with these systems it may be possible to encode metamorphic folding into other scaffolds or detect other naturally occurring examples. Molecular switches and reversibly self-assembling nanomaterials with sensitivity to salinity, pH, temperature or other triggers could be envisioned. The results presented here suggest that electrostatic interactions can be manipulated to direct metamorphic structural transitions, but as more of these systems are elucidated the rules governing this unique class of protein will likely expand.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. Evgenii Kovrigin of Marquette University for assistance in the analysis of ZZ-exchange data.
Funding Sources
This work was supported by NIH grant R01 063325.
ABBREVIATIONS
- CLIC1
Chloride intercellular channel protein1
- HSQC
Heteronuclear single quantum coherence
- Mad2
Mitotic arrest deficient 2
- MD
Molecular dynamics
- NMR
Nuclear magnetic resonance
- WT
wild-type
Footnotes
Supporting Information. Supplementary Figure S1displays NMR titration curves generated for residues outside the identified binding site. Figure S2 shows additional ZZ-exchange data resulting from C48 peak intensities. Supplementary Table S1 details HSQC peak volumes and Keq values determined for WT and mutant Ltn proteins as cited in text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 3.Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20:482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Littler DR, Harrop SJ, Fairlie WD, Brown LJ, Pankhurst GJ, Pankhurst S, DeMaere MZ, Campbell TJ, Bauskin AR, Tonini R, Mazzanti M, Breit SN, Curmi PM. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. The Journal of biological chemistry. 2004;279:9298–9305. doi: 10.1074/jbc.M308444200. [DOI] [PubMed] [Google Scholar]
- 5.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 6.Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci U S A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuloglu ES, McCaslin DR, Markley JL, Volkman BF. Structural rearrangement of human lymphotactin, a C chemokine, under physiological solution conditions. J Biol Chem. 2002;277:17863–17870. doi: 10.1074/jbc.M200402200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pace CN, Grimsley GR. Ribonuclease T1 is stabilized by cation and anion binding. Biochemistry. 1988;27:3242–3246. doi: 10.1021/bi00409a018. [DOI] [PubMed] [Google Scholar]
- 10.Courtenay ES, Capp MW, Saecker RM, Record MT., Jr Thermodynamic analysis of interactions between denaturants and protein surface exposed on unfolding: interpretation of urea and guanidinium chloride m-values and their correlation with changes in accessible surface area (ASA) using preferential interaction coefficients and the local-bulk domain model. Proteins Suppl. 2000;4:72–85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Pegram LM, Record MT., Jr Thermodynamic origin of hofmeister ion effects. J Phys Chem B. 2008;112:9428–9436. doi: 10.1021/jp800816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volkman BF, Liu TY, Peterson FC. Chapter 3. Lymphotactin structural dynamics. Methods Enzymol. 2009;461:51–70. doi: 10.1016/S0076-6879(09)05403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Formaneck MS, Ma L, Cui Q. Effects of temperature and salt concentration on the structural stability of human lymphotactin: insights from molecular simulations. Journal of the American Chemical Society. 2006;128:9506–9517. doi: 10.1021/ja061620o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson FC, Elgin ES, Nelson TJ, Zhang F, Hoeger TJ, Linhardt RJ, Volkman BF. Identification and characterization of a glycosaminoglycan recognition element of the C chemokine lymphotactin. J Biol Chem. 2004;279:12598–12604. doi: 10.1074/jbc.M311633200. [DOI] [PubMed] [Google Scholar]
- 15.Shirley BA. Urea and guanidine hydrochloride denaturation curves. Methods Mol Biol. 1995;40:177–190. doi: 10.1385/0-89603-301-5:177. [DOI] [PubMed] [Google Scholar]
- 16.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. Journal of biomolecular NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 17.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. Journal of the American Chemical Society. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 18.Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- 19.Maity H, Mossing MC, Eftink MR. Equilibrium unfolding of dimeric and engineered monomeric forms of lambda Cro (F58W) repressor and the effect of added salts: evidence for the formation of folded monomer induced by sodium perchlorate. Arch Biochem Biophys. 2005;434:93–107. doi: 10.1016/j.abb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Hammack B, Attfield K, Clayton D, Dec E, Dong A, Sarisky C, Bowler BE. The magnitude of changes in guanidine-HCl unfolding m-values in the protein, iso-1-cytochrome c, depends upon the substructure containing the mutation. Protein science : a publication of the Protein Society. 1998;7:1789–1795. doi: 10.1002/pro.5560070814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyler RC, Murray NJ, Peterson FC, Volkman BF. Native-state interconversion of a metamorphic protein requires global unfolding. Biochemistry. 2011;50:7077–7079. doi: 10.1021/bi200750k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein science : a publication of the Protein Society. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander-Brett JM, Fremont DH. Dual GPCR and GAG mimicry by the M3 chemokine decoy receptor. J Exp Med. 2007;204:3157–3172. doi: 10.1084/jem.20071677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feller G, Bussy O, Houssier C, Gerday C. Structural and functional aspects of chloride binding to Alteromonas haloplanctis alpha-amylase. The Journal of biological chemistry. 1996;271:23836–23841. doi: 10.1074/jbc.271.39.23836. [DOI] [PubMed] [Google Scholar]
- 25.Wolf-Watz M, Backstrom S, Grundstrom T, Sauer U, Hard T. Chloride binding by the AML1/Runx1 transcription factor studied by NMR. FEBS Lett. 2001;488:81–84. doi: 10.1016/s0014-5793(00)02390-5. [DOI] [PubMed] [Google Scholar]
- 26.Marcaurelle LA, Mizoue LS, Wilken J, Oldham L, Kent SB, Handel TM, Bertozzi CR. Chemical synthesis of lymphotactin: a glycosylated chemokine with a C-terminal mucin-like domain. Chemistry. 2001;7:1129–1132. doi: 10.1002/1521-3765(20010302)7:5<1129::aid-chem1129>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 27.Gardino AK, Villali J, Kivenson A, Lei M, Liu CF, Steindel P, Eisenmesser EZ, Labeikovsky W, Wolf-Watz M, Clarkson MW, Kern D. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139:1109–1118. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoychev SH, Nathaniel C, Fanucchi S, Brock M, Li S, Asmus K, Woods VL, Jr, Dirr HW. Structural dynamics of soluble chloride intracellular channel protein CLIC1 examined by amide hydrogen-deuterium exchange mass spectrometry. Biochemistry. 2009;48:8413–8421. doi: 10.1021/bi9010607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.