Abstract
We recently demonstrated that resveratrol induces caspase-dependent apoptosis in multiple cancer cell types. Whether apoptosis is also regulated by other cell death mechanisms such as autophagy is not clearly defined. Here we show that inhibition of autophagy enhanced resveratrol-induced caspase activation and apoptosis. Resveratrol inhibited colony formation and cell proliferation in multiple cancer cell types. Resveratrol treatment induced accumulation of LC3-II, which is a key marker for autophagy. Pretreatment with 3-methyladenine (3-MA), an autophagy inhibitor, increased resveratrol-mediated caspase activation and cell death in breast and colon cancer cells. Inhibition of autophagy by silencing key autophagy regulators such as ATG5 and Beclin-1 enhanced resveratrol-induced caspase activation. Mechanistic analysis revealed that Beclin-1 did not interact with proapoptotic proteins Bax and Bak; however, Beclin-1 was found to interact with p53 in the cytosol and mitochondria upon resveratrol treatment. Importantly, resveratrol depleted ATPase 8 gene, and thus, reduced mitochondrial DNA (mtDNA) content, suggesting that resveratrol induces damage to mtDNA causing accumulation of dysfunctional mitochondria triggering autophagy induction. Together, our findings indicate that induction of autophagy during resveratrol-induced apoptosis is an adaptive response.
Keywords: Resveratrol, mitochondria, autophagy, apoptosis, mitochondrial DNA
1. Introduction
Resveratrol [3, 4′, 5-trihydroxy-trans-stilbene], a plant polyphenol, is most commonly found in the skin of grapes (Athar et al., 2009; Dong, 2003) and in red wine (Athar et al., 2007; Dong, 2003). Numerous beneficial effects of resveratrol have been reported such as anti-aging effects (Chung et al., 2010; Lagouge et al., 2006; Manna et al., 2000), anti-oxidant and anti-inflammatory activities, inhibition of platelet aggregation and inhibition of growth of a variety of cancer cells (Athar et al., 2007; Cal et al., 2003). Resveratrol affects all three stages of carcinogenesis. For example, resveratrol induces phase II drug metabolizing enzymes, inhibits cyclooxygenase and hydroperoxidase enzymes, and triggers differentiation to target initiation, promotion, and progression, respectively (Jang et al., 1997). Resveratrol is a promising molecule for cancer prevention as well as for anti-cancer therapy (Delmas et al., 2006; Liu et al., 2007). Resveratrol exhibits minimal toxicity to normal cells (Aziz et al., 2005; Baur and Sinclair, 2006; Delmas et al., 2003; Gill et al., 2007; Niles et al., 2003) and targets a wide range of signaling pathways such as apoptosis and autophagy to impair the growth and survival of a variety of cancer cell types (Athar et al., 2009). We recently observed that resveratrol induces p53-independent death of cancer cells (Gogada et al., 2011b), however, whether autophagy could also be considered a key pathway for cancer cell death is still not clearly understood.
Autophagy is initiated by the formation of a double membrane autophagosome, which fuses with the lysosomes causing degradation of engulfed organelles such as mitochondria, cytoplasmic proteins, genomic materials and lipids. The degraded products can be re-directed to form new macromolecules and ATP. Thus autophagy serves dual purpose within cells, damage control and energy efficiency. Several proteins such as Beclin-1, ATG5, and LC3 are involved in various stages of autophagosome formation (Glick et al., 2010; Levine and Yuan, 2005; Scarlatti et al., 2008). Autophagy is regulated by nutrient sensors such as mammalian target of rapamycin (mTOR) kinase (Levine and Yuan, 2005) and by the Bcl-2 family of proteins (Levine et al., 2008). Thus, autophagy is a survival mechanism and can also serve as a form of non-apoptotic programmed cell death in response to multiple stresses including resveratrol (Glick et al., 2010; Levine and Yuan, 2005; Opipari et al., 2004; Scarlatti et al., 2008).
Resveratrol has been shown to induce apoptotic (Filomeni et al., 2007; Gogada et al., 2011b) and autophagic cell death in cancer cells (Opipari et al., 2004; Puissant et al., 2010; Scarlatti et al., 2008). Autophagy contributes to resveratrol mediated cell survival (Morselli et al., 2009; Morselli et al., 2010) and suppresses resveratrol-induced apoptosis (Li et al., 2009). The effects of autophagy on resveratrol-induced caspase activation and cancer cell death are not well defined. A clear understanding of how resveratrol-induced autophagy regulates apoptosis in cancer cells is essential for designing effective chemopreventive and chemotherapeutic strategies. We investigated the effects of autophagy inhibition on resveratrol-mediated caspase activation and cell death. Pharmacological inhibition of autophagy as well as the use of siRNA-mediated ATG5 and Beclin-1 knockdown enhanced resveratrol-mediated caspase activation and cell death. Resveratrol depleted ATPase 8 gene encoded by mtDNA, suggesting that mitochondria are critical for autophagy induction and its crosstalk with apoptosis.
2. Materials and Methods
2.1 Cells and Reagents
HCT116 colon cancer cells, PC3 and LNCaP prostate cancer cells, and MDA-MB231 breast cancer cells were cultured as described previously (Gogada et al., 2011a; Gogada et al., 2011b). The primary antibodies for ATG5, Beclin-1, LC3 (Cell signaling), Caspase-3 (Rabbit polyclonal antibody (Rb pAb); Biomol), Bak, p53 (Santa Cruz Biotechnology), Bax (Rb pAb; BD Pharmingen), Bax N Terminus, Bak N Terminus (Rb pAb; Upstate) and Actin (monoclonal antibody (mAb); ICN) were obtained from the indicated suppliers. Secondary antibodies and ECL reagents were obtained from GE healthcare. Fluorogenic caspase substrates DEVD-AFC, LEHD-AFC, IETD-AFC were acquired from Enzo Life Sciences. All other chemicals not specified were obtained from Sigma.
2.2 Clonogenic survival assay
Cells (0.5 × 103/well) in 6-well tissue culture dishes were treated with different doses of resveratrol for 48 h. At the end of this treatment, the resveratrol-containing medium was replaced with drug-free fresh medium. Clones were counted within 1–2 weeks after fixation and staining with Giemsa (Jeter et al., 2009; Patrawala et al., 2006). Cell number in each clone was also counted.
2.3 Preparation of cell lysates, subcellular fractionation, and Western blotting
Preparation of the whole cell lysate, isolation and purification of mitochondrial and cytosolic fractions, Western blotting were performed as described previously (Chandra et al., 2007; Zhang et al., 2011).
2.4 Quantification of cell death and caspase activity measurement
Trypan blue dye exclusion test was used to quantify live and dead cells. DEVDase, LEHDase, and IETDase activities were determined as described previously (Gogada et al., 2011b; Zhang et al., 2011).
2.5 Establishment of MDA-MB231 cells stably expressing ATG5 or Beclin-1 siRNA using shRNA lentiviral vectors
To generate lentiviral particles, green fluorescence protein (GFP)-tagged short hairpin RNAs (shRNAs) specific to ATG5, Beclin-1, and negative control shRNA were cloned into the pGIPZ (Open Biosystems) lentiviral vector. The shRNA sequences for ATG5 and Beclin-1 were 5′-ACCGTGGAATGGAATGAGATTA-3′, and 5′-CCGCTATATCAGGATGAGATAA-3′ respectively. The lentiviral particles specific for ATG5, Beclin-1, and control shRNAs were obtained from the Roswell Park Cancer Institute (RPCI) shRNA core resource and were directly utilized to infect MDA-MB231 cells at a multiplicity of infection or MOI of 3. Puromycin (1 μg/ml) was used to select stable clones and ATG5 or Beclin-1 knockdown was analyzed by Western blotting (Gogada et al., 2011b; Jeter et al., 2009).
2.6 Immunoprecipitation
Immunoprecipitation analysis for Bax, Bak, p53, and Beclin-1 was performed as previously described (Gogada et al., 2011b).
2.7 Analysis of mtDNA content by real-time PCR
Total genomic DNA, containing both mtDNA and nuclear DNA, was isolated from unstimulated and resveratrol treated MDA-MB231 cells using the ZR Genomic DNA II Kit (Zymo Research). After quantification of DNA samples by the NanoDrop8000 (Thermo Scientific), Applied Biosystems 7300 real-time PCR system was used to amplify the GAPDH, β-actin and the mtDNA encoded ATPase 8 gene. Previously described primer sequences (Chen et al., 2007; Xia et al., 2009) were used. GAPDH (forward): 5′-CCC CAC ACA CAT GCA CTT ACC-3′, GAPDH (reverse): 5′ CCT AGT CCC AGG GCT TTG ATT-3′, β-actin (forward): 5′-TCA CCC ACA CTG TGC CCA TCT ACG A-3′, β-actin (reverse): 5′-CAG CGG AAC CGC TCA TTG CCA ATG G-3′, ATPase 8 (forward): 5′-AAT ATT AAA CAC AAA CTA CCA CCT ACC-3′, ATPase 8 (reverse): 5′-TGG TTC TCA GGG TTT GTT ATA-3′. The real-time PCR reaction was carried out in a total reaction volume of 10μl containing 5μl of 2X iTaq SYBR Green Supermix with ROX (Bio-Rad, Cat# 172-5850), 10 ng of template DNA, 500 nM each of forward and reverse primers, and nuclease-free water. Melting curve analysis done at the end of amplification showed the absence of nonspecific amplification or primer dimer formation. Standard curves generated using 10 ng to 10 pg DNA provided PCR efficiency based on the equation (Heid et al., 1996). The threshold cycle number (Ct) values for each reaction were calculated using the 7300 system SDS software. Average threshold cycle number (Ct) values were obtained by amplification of ATPase 8 (mtDNA-specific), GAPDH, and β-actin (nDNA-specific). MtDNA content was determined as 2^ΔCt, or fold difference of mtDNA from nDNA (Kulawiec et al., 2009; Xia et al., 2009).
2.8 Statistical analysis
Results are presented as mean +/− standard deviation of data from three or more independent experiments. Statistical analysis was performed using Sigma Stat. Statistically significant changes with p-values of <0.05 or <0.01 are mentioned in the Figures.
3. Results
3.1 Resveratrol inhibits colony formation
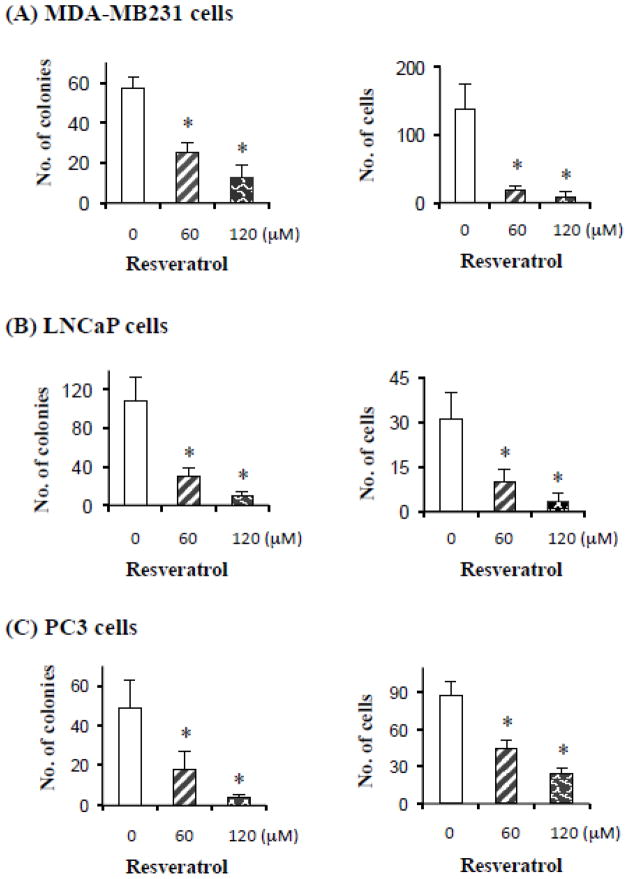
To understand the anticancer properties of resveratrol, we studied the effect of resveratrol on the proliferation capacity of cancer cells using colony-forming assay. Breast (MDA-MB231) and prostate (LNCaP and PC3) cancer cells were assayed in the presence of different concentrations of resveratrol, the number of colonies and the number of cells per colony were assessed. Resveratrol inhibited colony formation in a dose-dependent manner. For example, in control, DMSO-treated MDA-MB231 cells, 57 colonies were observed, whereas in 60 and 120 μM resveratrol treatments, the number of colonies was reduced to 24 and 13, respectively (Figure 1A). The number of cells per colony was also drastically reduced, for example, the average number of cells in vehicle-treated MDA-MB231 cells was 138, whereas with 60 and 120 μM resveratrol, the number of cells was 19 and 8 respectively (Figure 1A). Similarly, in LNCaP and PC3 cells, the number of colonies as well as the number of cells per colony was reduced upon resveratrol treatment (Figure 1B and C). Thus, resveratrol treatment reduces colony formation and number of cells per colony in a dose-dependent manner.
Figure 1. Resveratrol-inhibits colony formation in cancer cells.
Prostate (LNCaP and PC3 cells) and breast (MDA-MB231 cells) cancer cells were plated at a density of 500 cells/well in a 6-well plate and treated with 0, 60 and 120 μM of resveratrol for 48 h. After 48 h, the cells were grown in drug-free medium. When colonies became visible (~ 1–2 weeks), cells were fixed and stained with Giemsa (1:10 in distilled water at room temperature), and counted. Number of cells in individual colony was also counted (right hand side panels). Data are mean ± SD, n=3; *P < 0.05.
3.2 Autophagy occurs in response to resveratrol treatment and inhibition of autophagy enhances resveratrol-mediated cell-death
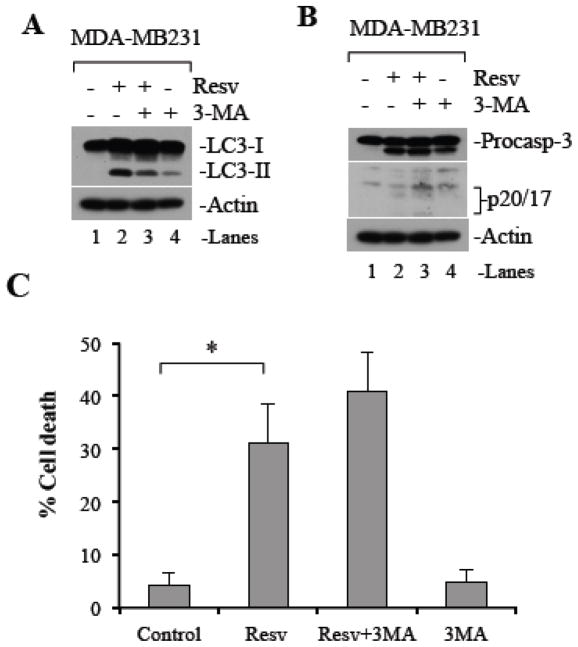
We have previously demonstrated that resveratrol induces mitochondria-dependent caspase activation and apoptosis in cancer cells (Gogada et al., 2011b). Next we evaluated whether cancer cells treated with resveratrol also undergo autophagy. Autophagy is initiated in response to cellular stress by autophagosome formation, which requires the induction of microtubule associated protein 1 light chain 3 (LC3-I) and its conjugation with phosphatidylethanolamine (PE). The cytosolic LC3 (i.e., LC3-I) is converted to the autophagosome associated LC3-II. Therefore, an increase in the levels of LC3-II in response to stress, is a marker for autophagy (Mizushima et al., 2010). To understand if resveratrol also induces autophagy, we determined the levels of LC3-I and LC3-II upon resveratrol treatment by Western blot analysis in MDA-MB231 cells and observed that the level of LC3-II was increased at 24 h upon 120 μM resveratrol treatment (Figure 2A, lanes 1–2) showing that resveratrol induces autophagy.
Figure 2. Resveratrol induces autophagy in cancer cells.
MDA-MB231 cells were pretreated with 3-MA (5 mM) for 1 h followed by treatment with 120 μM resveratrol (Resv) for 24 h or untreated as indicated. (A) Whole cell lysates were subjected to Western blotting for LC3. (B) Whole cell lysates were subjected to Western blotting for determination of caspase 3 processing. Actin serves as a loading control. (C) Percent cell death was quantified using Trypan Blue exclusion assay. Procasp-3, procaspase-3; Resv, resveratrol. Data are mean ± SD, n 3; *P < 0.01.
LY294002 and 3-methyladenine (3-MA) are known to inhibit autophagy by class III phosphatidylinositol 3-kinase inhibition (Blommaart et al., 1997; Seglen and Gordon, 1982). Resveratrol-induced autophagy (i.e., increased LC3-II levels) was reversed upon pretreatment with 3-MA in combination with resveratrol (Figure 2A, lanes 3) in MDA-MB231 cells. However, the level of autophagy was not completely inhibited as a slight background level of LC3-II was detected with 3-MA alone (Figure 2A, lanes 3 and 4). Surprisingly, resveratrol-induced caspase 3 activation was increased in the presence of 3-MA, suggesting that 3-MA may further sensitize cancer cells to undergo apoptotic cell death (Figure 2B, lanes 2–4). To delineate the role of resveratrol-induced autophagy in cancer cell death, we measured viability of MDA-MB231 cells in response to resveratrol (120 μM) treatment for 24 h using trypan blue exclusion assay. In the control condition, we observed 5 % cell death, which was increased to 31% upon resveratrol treatment. Interestingly, the combination of resveratrol and 3-MA further increased the number of dead cells to 41% (Figure 2C). The additive effect of resveratrol and 3-MA on cell death in MDA-MB231 cells indicates that autophagy in response to resveratrol is a cell survival mechanism.
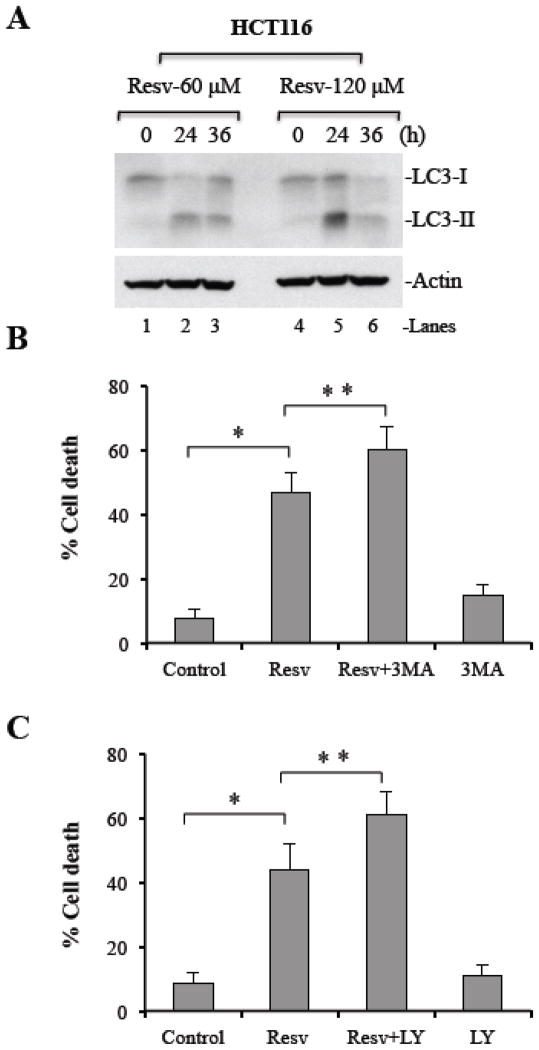
To understand whether resveratrol-induced autophagy is dose-dependent, we treated HCT116 colon cancer cells with both lower (60 μM) and higher (120 μM) doses of resveratrol. We observed that both doses of resveratrol induced LC3-II accumulation in cancer cells at 24 h after treatment (Figure 3A). In addition, we tested whether inhibition of autophagy by LY294002 and 3-MA show additive effect on resveratrol-mediated cell death in HCT116 cells. Similar to MDA-MB231 cells, cell death was increased upon inhibition of autophagy in HCT116 cells (Figure 3B and 3C). Thus, autophagy seems to be a survival mechanism in response to resveratrol treatment of cancer cells and inhibition of autophagy enhanced resveratrol-mediated cell death.
Figure 3. Inhibition of autophagy enhances resveratrol-induced cell death.
(A) HCT116 cells were treated with resveratrol (60 or 120 μM) for indicated times. At the end of treatment, whole cell lysates were subjected to Western blotting for LC3. Actin serves as loading control. HCT116 WT cells were pretreated with 3-MA (B; 5 mM) or LY294002 (C; 10 μM) for 1 h followed by treatment with resveratrol (120 μM for 24 h) or left untreated. Percentage cell death was quantified using Trypan Blue exclusion assay. Resv, resveratrol. Data are mean ± SD, n=3; *P < 0.01; **P < 0.05.
3.3 Inhibition of autophagy enhances resveratrol-mediated caspase activation
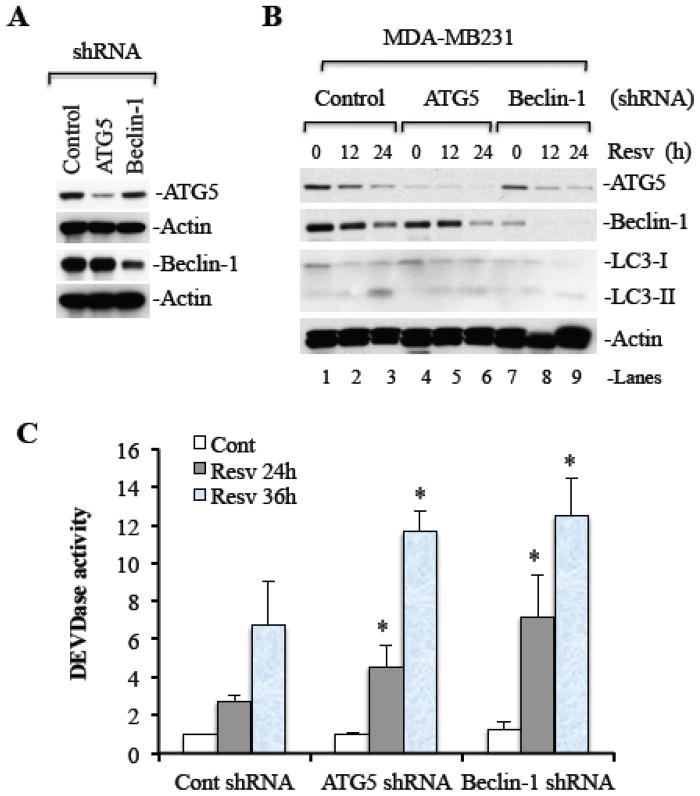
The induction of autophagy is associated with cell survival and may protect cells during apoptosis. If autophagy plays a prosurvival role in cancer cells, then silencing of autophagy related genes should further increase resveratrol-induced caspase activation. We silenced ATG5 or Beclin-1 genes (Figure 4A), which play a crucial role in autophagosome formation and leads to the execution of autophagy. In MDA-MB231 cells, silencing of ATG5 and Beclin-1 by siRNA inhibited resveratrol-induced LC3-II accumulation at 24 h (Figure 4B, lanes 3, 6, and 9). These results clearly demonstrate that LC3-II accumulation occurs in resveratrol treated cells and is dependent on the activation of autophagy. Thus, resveratrol treated cells undergo ATG5 and Beclin-1 dependent autophagy.
Figure 4. Inhibition of autophagy by ATG5 or Beclin-1 silencing enhances caspase activation.
MDA-MB231 cells were transduced using control (cont) or ATG5 or Beclin-1 targeted shRNAs in lentiviral vectors. Whole cell lysates were subjected to Western blot analysis for ATG5 and Beclin-1 (A). Stably transduced cells were treated with 120 μM resveratrol for 0, 12, 24, and 36 h. Whole cell lysates were prepared and equal amounts of protein were subjected to Western blotting to detect autophagy related proteins (B), and for caspase-3 activity measurement (C). Cont, control; Resv, resveratrol. Data are mean ± SD, n=3; *P < 0.05 as compared to control shRNA cells treated with resveratrol.
To investigate whether inhibition of autophagy causes increased levels of apoptosis, ATG5 or Beclin-1 silenced MDA-MB231 cells were treated with resveratrol and caspases-3 activity was determined. As shown in Figure 4C, silencing of ATG5 or Beclin-1 resulted in increased caspase-3 activation as compared to control shRNA infected cells. These results confirm the data in Figures 2 and 3 and reiterate the principle/phenomenon that resveratrol-induced autophagy is a prosurvival mechanism.
3.4 Resveratrol treatment induces interaction between p53 and Beclin-1
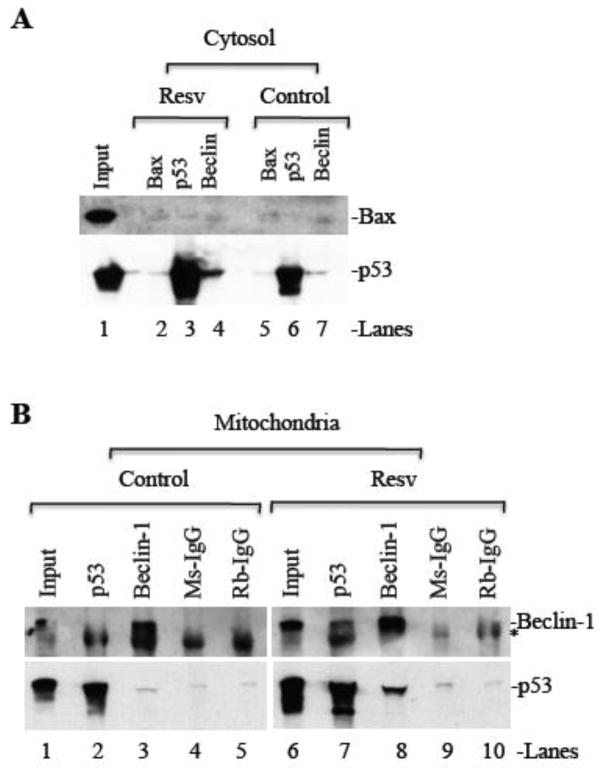
In order to investigate the mechanism of crosstalk between autophagy and apoptosis in response to resveratrol treatment in cancer cells, we performed immunoprecipitation experiments to determine the interaction between various proapoptotic proteins such as Bax, Bak, and p53 with autophagy regulator protein Beclin-1. In the cytosol, resveratrol treatment induced interaction between Beclin-1 and p53 (Figure 5A), but Beclin-1 does not interact with Bax (Figure 5A). Similarly, p53 IP pulled down Beclin-1 and Beclin-1 precipitated p53 (Figure 5B) in mitochondria isolated from resveratrol-treated cells. However, Bax and Bak did not interact with Beclin-1 in purified mitochondria from resveratrol treated cells (data not shown). Thus, it is likely that resveratrol-mediated autophagy involves Beclin-1 interaction with p53 in the cytosol and mitochondria.
Figure 5. Beclin-1 interacts with p53 in the cytosol and mitochondria.
MDA-MB231 cells were treated with either DMSO or resveratrol (120 μM for 24 h) and subjected to subcellular fractionation. (A) Equal amounts of cytosolic protein were immunoprecipitated with anti- Bax or p53 or Beclin-1 antibodies, followed by Western blot analysis for Bax and p53. (B) Equal amounts of mitochondrial proteins were subjected to immunoprecipitation with anti-p53 or Beclin-1 antibodies followed by Western blot analysis for p53 and Beclin-1. Rabbit-IgG (Rb-IgG) and Mouse-IgG serves as negative controls. Resv, resveratrol; * non-specific band.
3.5 Resveratrol treatment causes depletion of ATPase 8 gene encoded by mitochondrial DNA
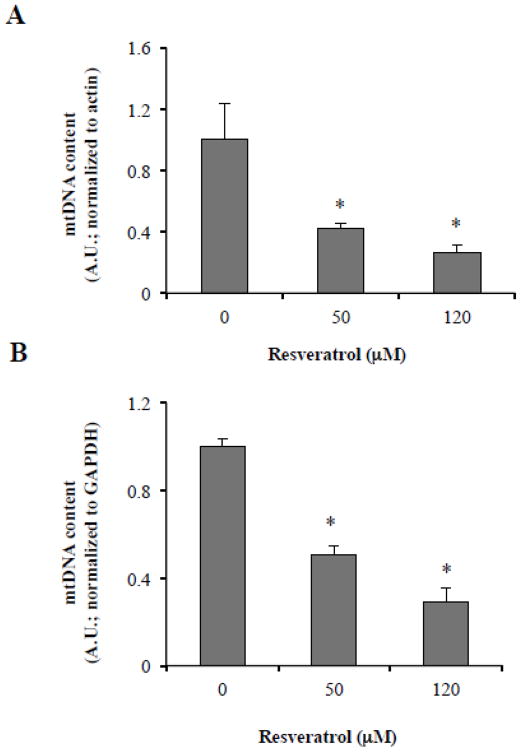
ROS production upon resveratrol treatment of cancer cells (Miki et al., 2012) could damage mtDNA leading to the accumulation of damaged mitochondria due to decreased efficiency of mtDNA repair enzymes (Kujoth et al., 2007), thus triggering autophagy to remove damaged mitochondria could be a pro-survival mechanism. To directly test whether resveratrol treatment modulates mtDNA content, we used real time PCR approach to quantitate the levels of mtDNA encoded ATPase 8 gene. In MDA-MB231 cells, we observed a decrease in the content of mtDNA at 24 h in response to resveratrol treatment compared to control cells (Figure 6). This indicates that cancer cells induce autophagy in order to cope with the stress in response to resveratrol treatment.
Figure 6. Resveratrol depletes mitochondrial DNA.
ATPase-8 gene encoded by the mitochondrial genome was amplified and quantified by real-time PCR using the SYBR green chemistry on the Applied Biosystems 7300 real-time PCR system. Total DNA was extracted from MDA-MB231 cells treated with DMSO or resveratrol (0, 50 or 120 μM) for 24 h. Mitochondrial ATPase-8 gene specific primers were used to amplify, and values were normalized to actin (A) or GAPDH (B). Data are mean ± SD, n=4; *P < 0.01 as compared to DMSO-treated cells.
4. Discussion
Previously, we observed that resveratrol induces mitochondrial dysfunction leading to the loss of mitochondrial membrane potential, cytochrome c release, and apoptosis. Here we demonstrate that resveratrol causes depletion of the mtDNA encoded ATPase-8 gene causing accumulation of defective mitochondria, which induces autophagy to restore mitochondria homeostasis in cancer cells. Inhibition of autophagy could lead to the accumulation of damaged mitochondria, which may enhance resveratrol-induced caspase activation and apoptotic cell death.
We have shown that resveratrol inhibits the clonal expansion and cell proliferation of breast cancer (MDA-MB231) and prostate cancer (PC3 and LNCaP) cells. These biological effects are consistent with the earlier findings and may be associated with cell cycle arrest and/or induction of apoptosis (Aziz et al., 2006; Hsieh et al., 2011). We previously demonstrated that resveratrol induces p53-independent, XIAP-mediated apoptosis in some cancer cells (Gogada et al., 2011b). Here we show that resveratrol induces autophagy in cancer cells, suggesting that in addition to apoptosis, autophagy may also play a role in the regulation of clonal expansion and cancer cell proliferation.
Our findings are consistent with previous reports that resveratrol-induces autophagy in multiple cancer cell types (Li et al., 2009; Opipari et al., 2004; Puissant and Auberger, 2010; Trincheri et al., 2008). Although previous findings suggest that resveratrol induces autophagy as a form of cell death, our data along with others (Filippi-Chiela et al., 2011) indicate that resveratrol-induced autophagy may represent a prosurvival mechanism in some types of cancer cells. Multiple pieces of evidence support our conclusions. For example, pharmacological inhibition of autophagy enhances caspase activation and cell death in resveratrol treated cells; and silencing of key regulators of autophagy such as ATG5 and Beclin-1 significantly enhanced resveratrol-induced caspase activation.
Our findings support the prosurvival role of autophagy during resveratrol-induced cell death. Indeed, inhibition of autophagy has been shown to enhance cytotoxic effects of resveratrol in glioma cells (Li et al., 2009), and inhibition of autophagy is also known to enhance therapy-induced apoptosis in lymphoma cells (Amaravadi et al., 2007). However, other studies suggest that inhibition of autophagy by its inhibitors suppresses apoptosis (Miki et al., 2012). Additionally, inhibition of autophagy has also been reported in cancer cells upon resveratrol treatment (Armour et al., 2009; Lin et al., 2012). For example, resveratrol enhances the efficacy of temozolomide chemotherapy in malignant glioma both in vitro and in vivo by inhibiting prosurvival autophagy signaling (Lin et al., 2012). These studies indicate that resveratrol-induced autophagy could be regulated by multiple factors exerting prosurvival or proapoptotic functions in multiple cancer cell types.
How inhibition of autophagy enhances apoptosis? It is known that p53 interacts with Bax triggering Bax translocation to mitochondria, which induces Bax oligomerization, cytochrome c release, and thus apoptosis (Chipuk and Green, 2004; Chipuk et al., 2004). Our study suggests that interaction of p53 with Beclin-1 in the cytosolic compartment may reduce efficient Bax translocation to mitochondria. Thus, inhibition of autophagy could induce p53 interaction with Bax leading to increase in cytochrome c release and apoptosis. Proapoptotic BH3-only proteins disrupt Beclin-1 interaction with antiapoptotic proteins Bcl-2/Bcl-xL (Lomonosova and Chinnadurai, 2008). Therefore, Beclin-1 silencing will allow BH3-only proteins to activate Bax/Bak or inhibition of autophagy may lead to the sequestration of Bcl2/Bcl-xL, thus may efficiently activate Bax/Bak to enhance cytochrome c release and apoptosis.
Low doses of resveratrol induce mitochondrial biogenesis and causes increase of mtDNA content (Park et al., 2012; Price et al., 2012), whereas we observed a depletion of mtDNA-encoded ATPase-8 gene suggesting that a higher dose of resveratrol induces ROS production, which may damage/deplete mtDNA encoded ATPase 8 gene. Damage to mtDNA may lead to accumulation of damaged mitochondria, which may be responsible for increased ROS production. Removal of damaged mitochondria will reduce the oxidative burden and extend cancer cell survival. Thus, induction of autophagy in response to resveratrol treatment in cancer cells may promote survival and prevent/delay apoptosis. Since autophagy results in the engulfment of stressed mitochondria that otherwise may lead to release of cytochrome c release and caspase activation, inhibiting this process may lead to increased caspase activation, and thus, apoptosis in cancer cells (Abedin et al., 2007; Boya et al., 2005). These findings strongly suggest that similar to cardiac myoblast cells (Gurusamy et al., 2010), induction of autophagy in cancer cells is a survival response.
Acknowledgments
This work was supported by National Institutes of Health K01 award CA123142 to Dhyan Chandra and National Cancer Institute Center Support Grant CA016056 to Roswell Park Cancer Institute. Dhyan Chandra was supported by a Research Scholar Grant, RSG-12-214-01 – CCG from the American Cancer Society. We thank Drs. Bert Vogelstein for providing HCT116 cells. Varun Prabhu was a MS student at University at Buffalo. We apologize to all colleagues whose work could not be cited due to space constraints.
Footnotes
Conflict of interest statement: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. The Journal of clinical investigation. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour SM, Baur JA, Hsieh SN, Land-Bracha A, Thomas SM, Sinclair DA. Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging (Albany NY) 2009;1:515–528. doi: 10.18632/aging.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athar M, Back JH, Kopelovich L, Bickers DR, Kim AL. Multiple molecular targets of resveratrol: Anti-carcinogenic mechanisms. Arch Biochem Biophys. 2009;486:95–102. doi: 10.1016/j.abb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athar M, Back JH, Tang X, Kim KH, Kopelovich L, Bickers DR, Kim AL. Resveratrol: a review of preclinical studies for human cancer prevention. Toxicol Appl Pharmacol. 2007;224:274–283. doi: 10.1016/j.taap.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz MH, Nihal M, Fu VX, Jarrard DF, Ahmad N. Resveratrol-caused apoptosis of human prostate carcinoma LNCaP cells is mediated via modulation of phosphatidylinositol 3′-kinase/Akt pathway and Bcl-2 family proteins. Molecular cancer therapeutics. 2006;5:1335–1341. doi: 10.1158/1535-7163.MCT-05-0526. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Reagan-Shaw S, Wu J, Longley BJ, Ahmad N. Chemoprevention of skin cancer by grape constituent resveratrol: relevance to human disease? FASEB J. 2005;19:1193–1195. doi: 10.1096/fj.04-3582fje. [DOI] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. European journal of biochemistry/FEBS. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cal C, Garban H, Jazirehi A, Yeh C, Mizutani Y, Bonavida B. Resveratrol and cancer: chemoprevention, apoptosis, and chemo-immunosensitizing activities. Curr Med Chem Anticancer Agents. 2003;3:77–93. doi: 10.2174/1568011033353443. [DOI] [PubMed] [Google Scholar]
- Chandra D, Choy G, Tang DG. Cytosolic accumulation of HSP60 during apoptosis with or without apparent mitochondrial release: evidence that its pro-apoptotic or pro-survival functions involve differential interactions with caspase-3. J Biol Chem. 2007;282:31289–31301. doi: 10.1074/jbc.M702777200. [DOI] [PubMed] [Google Scholar]
- Chen J, Kadlubar FF, Chen JZ. DNA supercoiling suppresses real-time PCR: a new approach to the quantification of mitochondrial DNA damage and repair. Nucleic Acids Res. 2007;35:1377–1388. doi: 10.1093/nar/gkm010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Green DR. Cytoplasmic p53: bax and forward. Cell Cycle. 2004;3:429–431. [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- Chung S, Yao H, Caito S, Hwang JW, Arunachalam G, Rahman I. Regulation of SIRT1 in cellular functions: Role of polyphenols. Arch Biochem Biophys. 2010;501:79–90. doi: 10.1016/j.abb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas D, Lançon A, Colin D, Jannin B, Latruffe N. Resveratrol as a chemopreventive agent: a promising molecule for fighting cancer. Curr Drug Targets. 2006;7:423–442. doi: 10.2174/138945006776359331. [DOI] [PubMed] [Google Scholar]
- Delmas D, Rébé C, Lacour S, Filomenko R, Athias A, Gambert P, Cherkaoui-Malki M, Jannin B, Dubrez-Daloz L, Latruffe N, Solary E. Resveratrol-induced apoptosis is associated with Fas redistribution in the rafts and the formation of a death-inducing signaling complex in colon cancer cells. J Biol Chem. 2003;278:41482–41490. doi: 10.1074/jbc.M304896200. [DOI] [PubMed] [Google Scholar]
- Dong Z. Molecular mechanism of the chemopreventive effect of resveratrol. Mutat Res. 2003;523–524:145–150. doi: 10.1016/s0027-5107(02)00330-5. [DOI] [PubMed] [Google Scholar]
- Filippi-Chiela EC, Villodre ES, Zamin LL, Lenz G. Autophagy interplay with apoptosis and cell cycle regulation in the growth inhibiting effect of resveratrol in glioma cells. PloS one. 2011;6:e20849. doi: 10.1371/journal.pone.0020849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomeni G, Graziani I, Rotilio G, Ciriolo MR. trans-Resveratrol induces apoptosis in human breast cancer cells MCF-7 by the activation of MAP kinases pathways. Genes Nutr. 2007;2:295–305. doi: 10.1007/s12263-007-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill C, Walsh SE, Morrissey C, Fitzpatrick JM, Watson RW. Resveratrol sensitizes androgen independent prostate cancer cells to death-receptor mediated apoptosis through multiple mechanisms. Prostate. 2007;67:1641–1653. doi: 10.1002/pros.20653. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogada R, Amadori M, Zhang H, Jones A, Verone A, Pitarresi J, Jandhyam S, Prabhu V, Black JD, Chandra D. Curcumin induces Apaf-1-dependent, p21-mediated caspase activation and apoptosis. Cell Cycle. 2011a;10:4128–4137. doi: 10.4161/cc.10.23.18292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogada R, Prabhu V, Amadori M, Scott R, Hashmi S, Chandra D. Resveratrol induces p53-independent, X-linked inhibitor of apoptosis protein (XIAP)-mediated Bax protein oligomerization on mitochondria to initiate cytochrome c release and caspase activation. J Biol Chem. 2011b;286:28749–28760. doi: 10.1074/jbc.M110.202440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurusamy N, Lekli I, Mukherjee S, Ray D, Ahsan MK, Gherghiceanu M, Popescu LM, Das DK. Cardioprotection by resveratrol: a novel mechanism via autophagy involving the mTORC2 pathway. Cardiovascular research. 2010;86:103–112. doi: 10.1093/cvr/cvp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hsieh TC, Wong C, John Bennett D, Wu JM. Regulation of p53 and cell proliferation by resveratrol and its derivatives in breast cancer cells: an in silico and biochemical approach targeting integrin alphavbeta3. International journal of cancer. 2011;129:2732–2743. doi: 10.1002/ijc.25930. [DOI] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- Jeter CR, Badeaux M, Choy G, Chandra D, Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ, Tang DG. Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells. 2009;27:993–1005. doi: 10.1002/stem.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Bradshaw PC, Haroon S, Prolla TA. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:e24. doi: 10.1371/journal.pgen.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Qin Z, Liang Z. The prosurvival role of autophagy in Resveratrol-induced cytotoxicity in human U251 glioma cells. BMC cancer. 2009;9:215. doi: 10.1186/1471-2407-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Lee CC, Shih YL, Lin TY, Wang SH, Lin YF, Shih CM. Resveratrol enhances the therapeutic effect of temozolomide against malignant glioma in vitro and in vivo by inhibiting autophagy. Free radical biology & medicine. 2012;52:377–391. doi: 10.1016/j.freeradbiomed.2011.10.487. [DOI] [PubMed] [Google Scholar]
- Liu BL, Zhang X, Zhang W, Zhen HN. New enlightenment of French Paradox: resveratrol’s potential for cancer chemoprevention and anti-cancer therapy. Cancer Biol Ther. 2007;6:1833–1836. doi: 10.4161/cbt.6.12.5161. [DOI] [PubMed] [Google Scholar]
- Lomonosova E, Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27(Suppl 1):S2–19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna SK, Mukhopadhyay A, Aggarwal BB. Resveratrol suppresses TNF-induced activation of nuclear transcription factors NF-kappa B, activator protein-1, and apoptosis: potential role of reactive oxygen intermediates and lipid peroxidation. J Immunol. 2000;164:6509–6519. doi: 10.4049/jimmunol.164.12.6509. [DOI] [PubMed] [Google Scholar]
- Miki H, Uehara N, Kimura A, Sasaki T, Yuri T, Yoshizawa K, Tsubura A. Resveratrol induces apoptosis via ROS-triggered autophagy in human colon cancer cells. Int J Oncol. 2012;40:1020–1028. doi: 10.3892/ijo.2012.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, Madeo F, Kroemer G. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–970. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, Michaud M, Madeo F, Tavernarakis N, Kroemer G. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell death & disease. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles RM, McFarland M, Weimer MB, Redkar A, Fu YM, Meadows GG. Resveratrol is a potent inducer of apoptosis in human melanoma cells. Cancer Lett. 2003;190:157–163. doi: 10.1016/s0304-3835(02)00676-6. [DOI] [PubMed] [Google Scholar]
- Opipari AW, Jr, Tan L, Boitano AE, Sorenson DR, Aurora A, Liu JR. Resveratrol-induced autophagocytosis in ovarian cancer cells. Cancer research. 2004;64:696–703. doi: 10.1158/0008-5472.can-03-2404. [DOI] [PubMed] [Google Scholar]
- Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, Chung JH. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, Hubbard BP, Varela AT, Davis JG, Varamini B, Hafner A, Moaddel R, Rolo AP, Coppari R, Palmeira CM, de Cabo R, Baur JA, Sinclair DA. SIRT1 Is Required for AMPK Activation and the Beneficial Effects of Resveratrol on Mitochondrial Function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Auberger P. AMPK- and p62/SQSTM1-dependent autophagy mediate Resveratrol-induced cell death in chronic myelogenous leukemia. Autophagy. 2010;6:655–657. doi: 10.4161/auto.6.5.12126. [DOI] [PubMed] [Google Scholar]
- Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, Auberger P. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer research. 2010;70:1042–1052. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincheri NF, Follo C, Nicotra G, Peracchio C, Castino R, Isidoro C. Resveratrol-induced apoptosis depends on the lipid kinase activity of Vps34 and on the formation of autophagolysosomes. Carcinogenesis. 2008;29:381–389. doi: 10.1093/carcin/bgm271. [DOI] [PubMed] [Google Scholar]
- Xia P, An HX, Dang CX, Radpour R, Kohler C, Fokas E, Engenhart-Cabillic R, Holzgreve W, Zhong XY. Decreased mitochondrial DNA content in blood samples of patients with stage I breast cancer. BMC cancer. 2009;9:454. doi: 10.1186/1471-2407-9-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gogada R, Yadav N, Lella RK, Badeaux M, Ayres M, Gandhi V, Tang DG, Chandra D. Defective molecular timer in the absence of nucleotides leads to inefficient caspase activation. PloS one. 2011;6:e16379. doi: 10.1371/journal.pone.0016379. [DOI] [PMC free article] [PubMed] [Google Scholar]