Abstract
Objectives
The Xq28 region containing IRAK1 and MECP2 has been identified as a risk locus for systemic lupus erythematosus (SLE) in previous genetic association studies. However, due to the strong linkage disequilibrium between IRAK1 and MECP2, it remains unclear which gene is affected by the underlying causal variant(s) conferring risk of SLE.
Methods
We fine-mapped ≥136 SNPs in a ~227kb region on Xq28, containing IRAK1, MECP2 and 7 adjacent genes (L1CAM, AVPR2, ARHGAP4, NAA10, RENBP, HCFC1 and TMEM187), for association with SLE in 15,783 case-control subjects derived from 4 different ancestral groups.
Results
Multiple SNPs showed strong association with SLE in European Americans, Asians and Hispanics at P<5×10−8 with consistent association in subjects with African ancestry. Of these, 6 SNPs located in the TMEM187-IRAK1-MECP2 region captured the underlying causal variant(s) residing in a common risk haplotype shared by all 4 ancestral groups. Among them, rs1059702 best explained the Xq28 association signals in conditional testings and exhibited the strongest P value in trans-ancestral meta-analysis (Pmeta=1.3×10−27, OR=1.43), and thus was considered to be the most-likely causal variant. The risk allele of rs1059702 results in the amino acid substitution S196F in IRAK1 and had previously been shown to increase NF-κB activity in vitro. We also found that the homozygous risk genotype of rs1059702 was associated with lower mRNA levels of MECP2, but not IRAK1, in SLE patients (P=0.0012) and healthy controls (P=0.0064).
Conclusion
These data suggest contributions of both IRAK1 and MECP2 to SLE susceptibility.
Keywords: Systemic Lupus Erythematosus, Gene Polymorphism, Xq28, IRAK1, MECP2
INTRODUCTION
SLE (OMIM 152700), a chronic multi-organ autoimmune disease, is associated with significant morbidity and mortality. A large body of literature supports a role for genetic, environmental and epigenetic factors in the pathogenesis of SLE.[1–5]
Previously, single nucleotide polymorphisms (SNP) in IRAK1 (interleukin-1 receptor-associated kinase 1) and its adjacent gene MECP2 (methyl CpG binding protein 2), separated by 1.7 kb on Xq28, have been independently associated with risk of SLE, mainly in subjects with European and Asian ancestries.[6–10] Both IRAK1 and MECP2 are strong candidate genes for SLE susceptibility. IRAK1 associates with interleukin-1 receptor, up-regulates transcription factor NF-κB [11] and activates the innate immune system which is important in SLE pathogenesis.[12, 13] IRAK1 deficiency in mice abrogated SLE-associated phenotypes, including IgM and IgG autoantibodies, lymphocytic activation and renal disease, and reversed the dendritic cell “hyperactivity” associated with the Sle3 lupus susceptibility interval.[7] MECP2 plays a role in two epigenetic repression mechanisms, DNA methylation and histone deacetylation, leading to a chromatin configuration inaccessible for transcription, thereby silencing gene expression.[14, 15] In both humans and mice, defects of DNA methylation have been implicated in the pathogenesis of SLE.[2, 3] The strong linkage disequilibrium (LD) between these two genes has led to the hypothesis that only one or the other of IRAK or MECP2 is the SLE risk gene on Xq28,[16] a debate which has not yet been solved. Furthermore, rs2269368 in ARHGAP4 has been associated with SLE in subjects with European ancestry,[17] suggesting that genes located upstream of the IRAK1-MECP2 region may also contribute to SLE susceptibility.
Leveraging different LD patterns among multiple ancestral groups, the trans-ancestral fine-mapping approach has shown its power in identifying underlying causal variants at SLE-associated loci.[18–20] Here, we fine mapped 9 genes in Xq28 using 136–173 SNPs and assessed their association with SLE in subjects from 4 different ancestral groups. After localizing the candidate causal variant, we tested its association with the mRNA level of IRAK1 and MECP2.
METHODS
Sample collection
DNA samples used in the collaborative Large Lupus Association Study 2 (LLAS2) were from subjects recruited by multiple participating institutions and processed at the Oklahoma Medical Research Foundation (OMRF). Each institution had Institutional Review Board (IRB) approval to recruit subjects and the overall study was approved by the IRB of OMRF. Each patient met at least four of eleven 1997 American College of Rheumatology revised criteria for the classification of SLE.[21]
Genotyping and data cleaning
We selected potential functional SNPs, previously reported SLE-associated Xq28 SNPs and tag SNPs based on HapMap datasets (r24) for genotyping. Fifty-five Xq28 SNPs and 347 admixture informative markers (AIMs) were successfully genotyped using an Illumina custom array on the iSCAN instrument (San Diego, CA, USA).
Subjects with genotype missing rate >10% (due to low quality), shared identical by descent >0.4 or showing mismatch between the reported and estimated gender were removed. The global ancestry of each subject was estimated based on genotype of AIMs, using principal components analysis [22] and ADMIXMAP,[23–25] as described in another LLAS2 study.[19] Genetic outliers were removed.
Final clean data from 15,783 subjects were divided into 4 groups according to ancestry, including EA (European Americans), AA (composed of 92.5% African Americans and 7.5% Gullahs), AS (comprised of 74.6% of Koreans, 16.1% of Chinese and subjects from Japan and Singapore) and HA (Hispanics enriched for the Amerindian-European admixture) (Table S5). Some subjects were previously analyzed in two published MECP2/IRAK1 studies (Table S6).[6,7]
Imputation
To obtain genotypes of additional Xq28 SNPs, SNP genotypes of 381 Europeans, 246 Africans, 286 Asians and 181 Americans from the 1000 Genomes Project (June 2011 data release) were used as references in imputation for our EA, AA, AS and HA subjects, respectively. Imputation was performed using IMPUTE 2.1.2;[26] imputed SNPs with an information score >0.9 were included for further analyses.
Statistical analyses
The same quality control criteria were applied to genotyped and imputed SNPs. SNPs with minor allele frequency (MAF) <1% or Hardy Weinberg equilibrium P<0.001 in controls were excluded. SNPs with genotype missing rate >5% or showing significantly different missing rates between cases and controls (missing rate >2% and P<0.05) were also excluded.
In each ancestral group, SNPs were assessed for association with SLE under a logistic regression model adjusting for gender and the first 3 principal components estimated using AIMs. Haplotype-based conditional association testings were also performed by adjusting for gender and the first 3 principal components. The trans-ancestry meta-analysis was conducted across all 4 ancestral groups. For each SNP, if the Cochran's Q statistic showed no evidence of genetic heterogeneity (P>0.05), a fixed effect model was applied. Otherwise, a random effect model was used. All analyses described above were performed using PLINK v1.07.[27] Pairwised LD values shown in Figure S2 were calculated using Haploview 4.2.[28]
Real-time quantitative PCR (RT-PCR)
Total mRNA extracted from peripheral blood mononuclear cells (PBMC) using the All Prep DNA/RNA mini kit (QIAGEN, Valencia, CA, USA) were reverse-transcribed into cDNA (Invitrogen, Carlsbad, CA, USA). Using RT-PCR (Applies Biosystems, Foster City, CA, USA), the expression of IRAK1 and MECP2, including all isoforms, were measured with TaqMan probe Hs01018347_m1 and Hs00172845_m1, respectively. The relative expression levels of IRAK1 and MECP2, normalized to housekeeping gene RPLP0, were calculated using the 2−ΔΔCt method. Log10 transformed IRAK1 and MECP2 levels were compared between individuals carrying different genotypes using Student's t test.
RESULTS
We genotyped 55 SNPs at Xq28, together with 347 AIMs, in 15,783 case-control subjects from 4 ancestral groups including EA, AA, AS and HA. In addition, we imputed genotypes for ungenotyped SNPs using reference data from the 1000 Genomes Project, resulting in 173 (EA), 157 (AA), 157 (AS) and 136 (HA) SNPs with MAF>1% that covers a ≥227kb region in Xq28 containing genes L1CAM, AVPR2, ARHGAP4, NAA10, RENBP, HCFC1, TMEM187, IRAK1 and MECP2 (Figure 1A). SNPs were assessed for the association with SLE under a logistic regression model adjusting for gender and global ancestry. The significance level was defined as Bonferroni-corrected P<0.05/173=2.9×10−4, using the most stringent criterion.
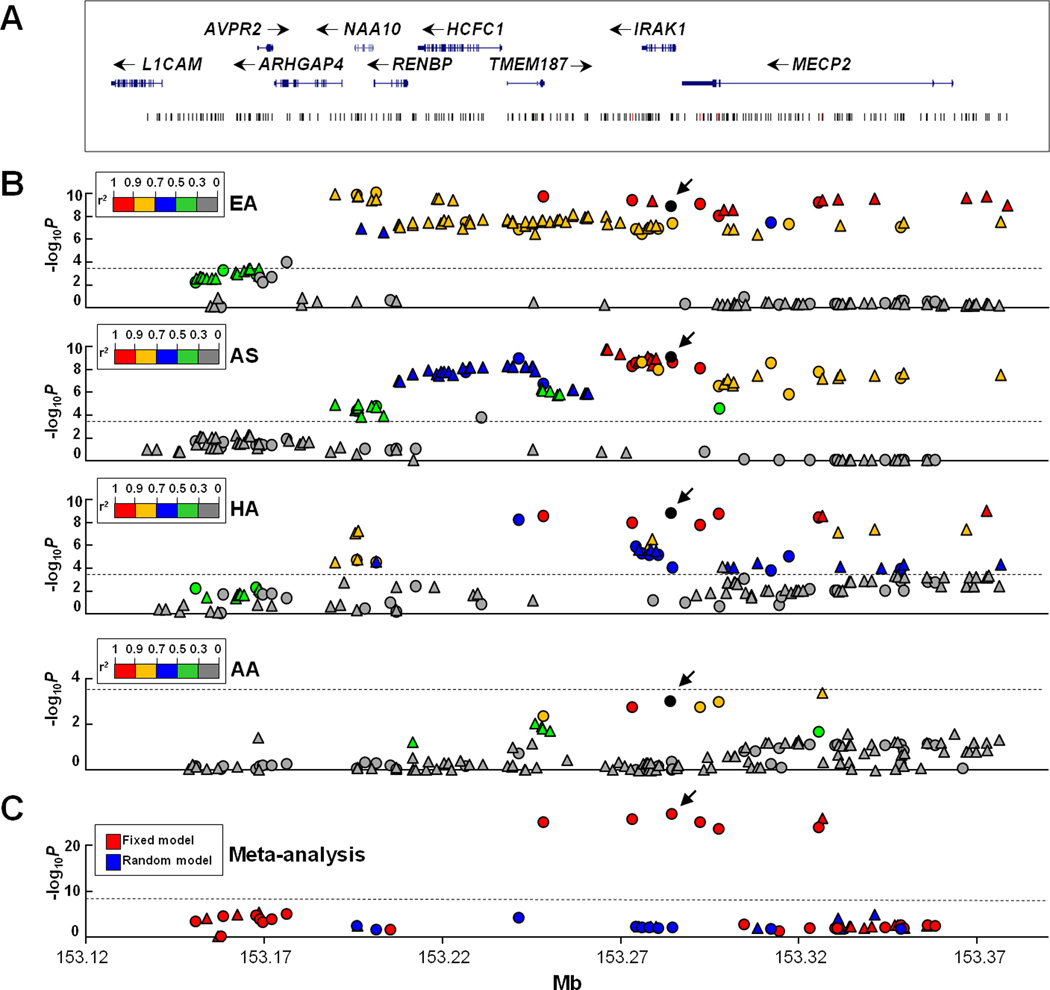
Figure 1. Association of SNPs in the Xq28 region with SLE.
A) The genomic structure of the Xq28 region and the location of all SNPs are indicated. B) Association signals (−log10P) are plotted against the position of each SNP in EA, AS, HA and AA, respectively. Genotyped and imputed SNPs are indicated as circles and triangles, respectively. SNPs are highlighted using different colors according to their LD strength (r2) with rs1059702 (shown as a black circle). An arrowhead is used to indicate the position of rs1059702. The dashed line represents the significance level after Bonferroni correction. C) Trans-ancestry meta-analysis P value generated using fixed and random model are highlighted as red and blue, respectively. The dashed line represents the significance level of 5×10−8.
Xq28 SNPs were associated with SLE in four different ancestral groups
To confirm the previously reported association of Xq28 region with SLE,[6,7,17] we firstly performed association testing in the largest EA dataset (3,915 cases and 3,462 controls). Eighty-six SNPs located in the region containing ARHGAP4, NAA10, RENBP, HCFC1, TMEM187, IRAK1 and MECP2 were significantly associated with SLE, of which 61 SNPs had P<5.0×10−8 exceeding the genome-wide association study (GWAS) significance level and rs5945377 in RENBP exhibited the strongest association signal (P=8.4×10−11, OR=1.38) (Figure 1B, Table S1). These data confirmed that Xq28 is a risk locus for SLE in EA.
Association of Xq28 with SLE was also confirmed in our AS (1,262 cases and 1,256 controls) and HA (1,487 cases and 807 controls) datasets. In total, 85 and 40 SNPs were significantly associated with SLE in AS and HA, respectively, of which 48 and 10 SNPs had P<5.0×10−8 (Figure 1B, Table S2 and S3). Both datasets showed the strongest association signal in the IRAK1-MECP2 region. SNPs in the upstream ARHGAP4-NAA10-RENBP region did not reach the GWAS significance level.
In the AA dataset (1,674 cases and 1,920 controls), 16 SNPs showed modest association with SLE (P<0.05) (Figure 1B, Table S4), of which SNPs in the IRAK1-MECP2 region exhibited peak association signals but none of them reached the Bonferroni-corrected significance level.
Comparing across EA, AS and HA, 34 SNPs located in a ~187 kb region spanning from ARHGAP4 to MECP2 were significantly associated with SLE in all 3 datasets (Table 1). Of them, 7 SNPs (rs13397, rs4898375, rs1059702, rs2734647, rs2075596, rs1734787 and rs1616369) were consistently associated with SLE in AA at P<0.05 (Table 1), had no genetic heterogeneity (P>0.05) across 4 ancestral groups, and generated a combined Pmeta<5×10−8 in trans-ancestral meta-analysis (Figure 1C, Table 1). We performed association testing in females and males, respectively, which yielded no evidence for a gender-specific association with SLE. Consistent association detected in EA, AS, HA and AA indicated that Xq28 is a risk locus of SLE in all these 4 ancestral groups.
Table 1.
Significant association of SNPs at Xq28 with SLE
| Tested | EA | AS | HA | AA | Meta-analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | SNP | Gene | Annotation | allele | P | OR | Pc | P | OR | Pc | P | OR | Pc | P | OR | Pc | P | OR |
| I | rs2269368c | ARHGAP4 | intronic | T | 1.3E-10 | 1.38[1.25-1.52] | 0.98 | 1.3E-05 | 1.32[1.17-1.49] | 0.06 | 3.4E-05 | 1.32[1.16-1.50] | 0.05 | M | M | -- | M | M |
| G | rs2071129 | NAA10 | intronic | G | 1.3E-10 | 1.38[1.25-1.52] | 0.87 | 4.1E-05 | 1.29[1.14-1.46] | 0.02 | 1.9E-05 | 1.32[1.16-1.50] | 0.19 | 0.818 | 1.01[0.92-1.12] | -- | 4.7E-03 | 1.24 |
| I | rs2071130 | NAA10 | intronic | C | 2.0E-10 | 1.37[1.25-1.52] | 0.99 | 2.7E-05 | 1.30[1.15-1.47] | 0.03 | 1.9E-05 | 1.32[1.16-1.51] | 0.11 | 0.773 | 1.02[0.92-1.12] | -- | 3.9E-03 | 1.24 |
| G | rs5945377 | RENBP | intronic | C | 8.4E-11 | 1.38[1.25-1.52] | 0.86 | 1.7E-05 | 1.31[1.16-1.48] | 0.02 | 3.2E-05 | 1.31[1.16-1.49] | 0.22 | 0.465 | 0.96[0.87-1.07] | -- | 2.2E-02 | 1.23 |
| I | rs5945378 | RENBP | intronic | G | 3.6E-10 | 1.37[1.24-1.51] | 0.79 | 2.0E-05 | 1.31[1.16-1.48] | 0.03 | 2.9E-05 | 1.32[1.16-1.50] | 0.19 | M | M | -- | M | M |
| G | rs4898374 | TMEM187 | intronic | T | 1.3E-07 | 1.28[1.17-1.40] | 0.42 | 1.2E-09 | 1.53[1.34-1.76] | 0.32 | 5.6E-09 | 1.48[1.30-1.69] | 0.03 | 0.184 | 1.09[0.96-1.25] | -- | 6.2E-05 | 1.33 |
| G | rs13397 | TMEM187 | T245T | A | 1.9E-10 | 1.37[1.24-1.51] | * | 1.8E-07 | 1.39[1.23-1.58] | * | 3.0E-09 | 1.48[1.30-1.69] | * | 4.2E-03 | 1.41[1.12-1.79] | * | 8.4E-26 | 1.40F |
| G | rs4898375 | Intergenic | A | 3.9E-10 | 1.36[1.23-1.49] | * | 5.2E-09 | 1.52[1.32-1.75] | * | 1.2E-08 | 1.46[1.28-1.67] | * | 1.7E-03 | 1.44[1.15-1.82] | * | 1.5E-26 | 1.42F | |
| G | rs633 | Intergenic | C | 1.5E-07 | 1.26[1.16-1.37] | 0.94 | 2.6E-09 | 1.55[1.34-1.79] | 0.34 | 1.3E-06 | 1.37[1.21-1.56] | 0.53 | 0.950 | 1.00[0.91-1.11] | -- | 6.0E-03 | 1.27 | |
| I | rs12400188 | Intergenic | G | 1.2E-07 | 1.26[1.16-1.38] | 0.82 | 1.7E-09 | 1.56[1.35-1.80] | 0.30 | 2.3E-06 | 1.37[1.20-1.55] | 0.42 | 0.998 | 1.00[0.99-1.11] | -- | 6.7E-03 | 1.27 | |
| G | rs3027898 | Intergenic | C | 3.4E-07 | 1.25[1.15-1.36] | 0.77 | 2.3E-09 | 1.55[1.34-1.79] | 0.28 | 5.5E-06 | 1.35[1.19-1.54] | 0.43 | 0.948 | 1.00[0.99-1.19] | -- | 7.9E-03 | 1.26 | |
| I | rs731642 | IRAK1 | intronic | A | 1.0E-07 | 1.27[1.16-1.39] | 0.92 | 8.7E-10 | 1.58[1.36-1.82] | ND | 1.9E-06 | 1.37[1.21-1.57] | 0.34 | M | M | -- | M | M |
| G | rs2239673a | IRAK1 | intronic | C | 1.3E-07 | 1.26[1.16-1.38] | 0.96 | 1.3E-09 | 1.57[1.35-1.81] | 0.35 | 7.3E-06 | 1.34[1.18-1.53] | 0.46 | 0.976 | 1.00[0.90-1.19] | -- | 7.5E-03 | 1.27 |
| I | rs763737a | IRAK1 | intronic | G | 7.0E-08 | 1.27[1.16-1.39] | 0.99 | 1.5E-09 | 1.56[1.35-1.81] | 0.29 | 2.5E-06 | 1.36[1.20-1.55] | 0.57 | 0.999 | 1.00[0.90-1.11] | -- | 6.7E-03 | 1.28 |
| I | rs1059703 | IRAK1 | L532S/intronic | G | 4.9E-10 | 1.35[1.23-1.48] | ND | 4.3E-09 | 1.53[1.33-1.77] | ND | 2.8E-07 | 1.41[1.24-1.60] | 0.73 | M | M | -- | M | M |
| I | rs5945174a | IRAK1 | intronic | G | 6.9E-08 | 1.27[1.16-1.39] | 0.98 | 1.1E-09 | 1.57[1.36-1.82] | 0.29 | 3.4E-06 | 1.36[1.19-1.55] | 0.53 | 0.945 | 1.00[0.91-1.11] | -- | 6.0E-03 | 1.28 |
| G | rs7061789a | IRAK1 | intronic | G | 1.3E-07 | 1.26[1.16-1.38] | 0.96 | 1.0E-08 | 1.53[1.32-1.77] | 0.22 | 6.8E-06 | 1.34[1.18-1.53] | 0.28 | 0.650 | 0.98[0.88-1.08] | -- | 1.3E-02 | 1.26 |
| G | rs1059702 | IRAK1 | S196F | A | 1.2E-09 | 1.35[1.22-1.48] | * | 8.2E-10 | 1.56[1.35-1.79] | * | 1.5E-09 | 1.49[1.31-1.70] | * | 1.0E-03 | 1.48[1.17-1.87] | * | 1.3E-27 | 1.43F |
| G | rs1059701 | IRAK1 | V562V | G | 3.9E-08 | 1.27[1.17-1.39] | 0.75 | 2.5E-09 | 1.55[1.34-1.79] | 0.24 | 9.0E-05 | 1.30[1.14-1.48] | 0.09 | 0.879 | 0.99[0.89-1.11] | -- | 7.3E-03 | 1.26 |
| G | rs2734647 | MECP2 | 3'UTR/intergenic | T | 7.8E-10 | 1.35[1.23-1.48] | * | 7.9E-09 | 1.51[1.32-1.74] | * | 1.7E-08 | 1.46[1.28-1.66] | * | 1.7E-03 | 1.42[1.14-1.78] | * | 7.1E-26 | 1.41F |
| G | rs2075596b | MECP2 | intronic | A | 9.0E-09 | 1.33[1.21-1.46] | * | 3.0E-07 | 1.44[1.25-1.66] | * | 1.8E-09 | 1.50[1.31-1.71] | * | 9.8E-04 | 1.45[1.16-1.80] | * | 2.2E-24 | 1.40F |
| I | rs4898467 | MECP2 | intronic | G | 1.4E-07 | 1.27[1.16-1.39] | 0.93 | 8.0E-08 | 1.48[1.28-1.71] | ND | 9.0E-05 | 1.30[1.14-1.48] | 0.03 | M | M | -- | M | M |
| I | rs1734790 | MECP2 | intronic | C | 1.4E-07 | 1.27[1.16-1.39] | 0.93 | 1.5E-07 | 1.47[1.27-1.70] | ND | 9.0E-05 | 1.30[1.14-1.48] | 0.02 | M | M | -- | M | M |
| I | rs909131 | MECP2 | intronic | G | 4.1E-07 | 1.25[1.15-1.37] | 0.91 | 3.6E-08 | 1.50[1.30-1.73] | ND | 3.8E-05 | 1.32[1.16-1.50] | 0.14 | 0.775 | 0.98[0.89-1.09] | -- | 1.0E-02 | 1.24 |
| G | rs17435b | MECP2 | intronic | T | 3.6E-08 | 1.28[1.17-1.39] | 0.67 | 2.7E-09 | 1.53[1.33-1.76] | ND | 1.8E-04 | 1.28[1.13-1.46] | 0.02 | 0.522 | 0.97[0.87-1.07] | -- | 1.8E-02 | 1.24 |
| G | rs1624766b | MECP2 | intronic | C | 5.0E-08 | 1.28[1.17-1.40] | 0.77 | 1.5E-06 | 1.42[1.23-1.64] | ND | 9.9E-06 | 1.35[1.18-1.54] | 0.34 | M | M | -- | M | M |
| G | rs1734787b | MECP2 | intronic | C | 6.4E-10 | 1.35[1.23-1.49] | ND | 1.6E-08 | 1.50[1.31-1.73] | ND | 4.0E-09 | 1.48[1.30-1.69] | ND | 0.021 | 1.22[1.03-1.45] | 0.91 | 9.4E-25 | 1.39F |
| I | rs1616369 | MECP2 | intronic | A | 4.2E-10 | 1.36[1.23-1.49] | * | 6.8E-08 | 1.48[1.28-1.71] | * | 2.9E-09 | 1.49[1.30-1.69] | * | 4.0E-04 | 1.50[1.20-1.88] | * | 1.2E-26 | 1.43F |
| I | rs1734791b | MECP2 | intronic | T | 3.4E-10 | 1.36[1.23-1.49] | ND | 5.7E-08 | 1.49[1.29-1.71] | ND | 7.8E-08 | 1.43[1.26-1.63] | 0.14 | 0.210 | 1.08[0.96-1.21] | -- | 9.1E-05 | 1.32 |
| I | rs1734789 | MECP2 | intronic | G | 6.9E-08 | 1.27[1.16-1.38] | 0.91 | 3.3E-08 | 1.51[1.30-1.74] | ND | 7.6E-05 | 1.30[1.14-1.48] | 0.08 | 0.492 | 0.96[0.87-1.07] | -- | 1.7E-02 | 1.24 |
| I | rs1734792b | MECP2 | intronic | A | 3.1E-10 | 1.36[1.24-1.49] | ND | 3.8E-08 | 1.50[1.30-1.73] | ND | 4.4E-08 | 1.44[1.26-1.64] | 0.14 | 0.134 | 1.10[0.97-1.24] | -- | 1.4E-05 | 1.34 |
| G | rs2239464b | MECP2 | intronic | A | 8.4E-08 | 1.27[1.16-1.39] | 0.79 | 5.3E-08 | 1.49[1.29-1.72] | ND | 1.2E-04 | 1.29[1.13-1.46] | 0.03 | 0.517 | 0.97[0.87-1.07] | -- | 1.8E-02 | 1.23 |
| I | rs5945393 | MECP2 | intronic | G | 3.5E-08 | 1.28[1.17-1.40] | 0.84 | 2.3E-08 | 1.51[1.31-1.74] | ND | 5.1E-05 | 1.31[1.15-1.49] | 0.08 | 0.665 | 0.98[0.88-1.08] | -- | 1.3E-02 | 1.25 |
| I | rs2872736 | Intergenic | C | 3.4E-08 | 1.28[1.17-1.39] | 0.89 | 3.0E-08 | 1.51[1.31-1.75] | ND | 5.3E-05 | 1.31[1.15-1.48] | 0.09 | M | M | -- | M | M | |
Abbreviation: G, genotyped SNP; I, imputed SNP; OR, odds ratio; Pc, P value after conditioning on 6 SNPs shown as "*"; ND, non-distinguishable in conditional testing; F, fixed effect model in meta-analysis; M, missing data. Only SNPs that remained significant association with SLE after correction for multiple comparisons in EA, AS and HA are listed in this table. Position of each SNP is based on GRch37. SNPs that showed consistent association with SLE in all 4 ancestral groups are highlighted in bold. For SNPs that were not tested in conditional testing (P>0.05), the Pc value is denoted as “--”. Previously reported SLE-associated SNPs located in IRAK1,[7] MECP2 [6] and ARHGAP [17] were noted using "a", "b" and "c", respectively, all of which were confirmed to be significantly associated with SLE in EA, AS and HA.
SLE-associated SNPs shared by four different ancestral groups were localized to the TMEM187-IRAK1-MECP2 region
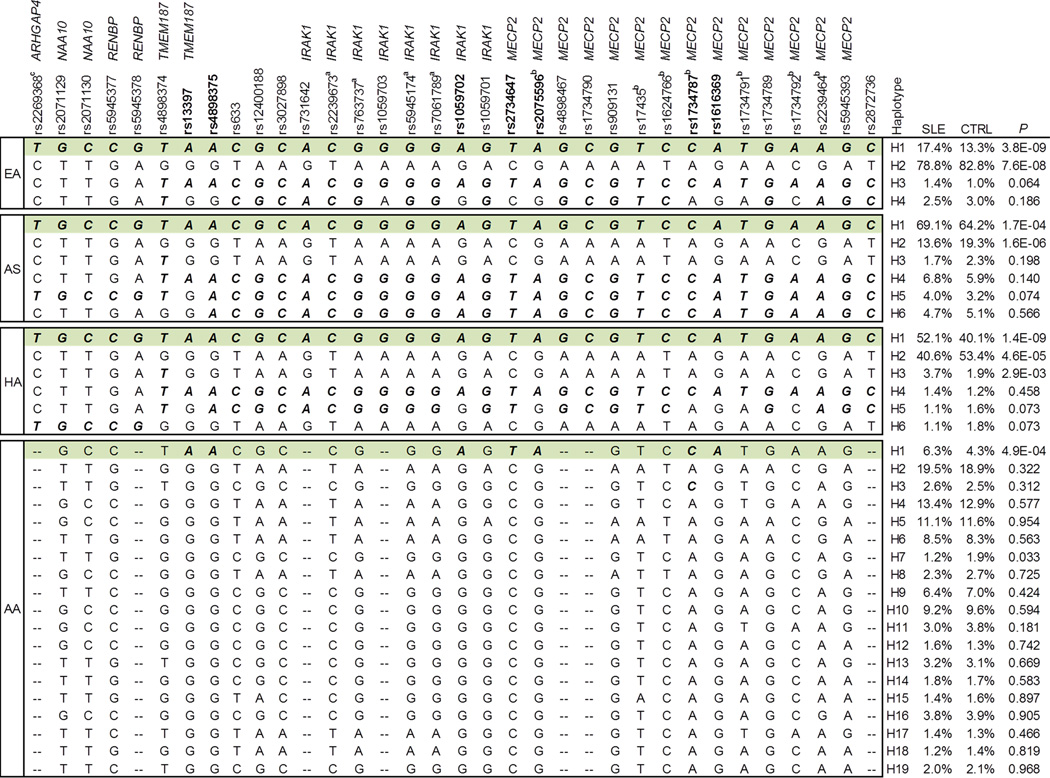
Haplotypes with frequency >1% were constructed using the 34 SNPs that were significantly associated with SLE in EA, AS and HA. Only haplotype H1 showed consistent association with increased SLE risk in EA (frequency of 17.4% in cases vs. 13.3% in controls, P=3.8×10−9), AS (69.1% vs. 64.2%, P=1.7×10−4), HA (52.1% vs. 40.1%, P=1.4×10−9) and AA (6.3% vs. 4.3%, P=4.9×10−4) (Figure 2). H1 shared by these 4 ancestral groups could be perfectly tagged by the risk allele of 6 SNPs (rs13397, rs4898375, rs1059702, rs2734647, rs2075596 and rs1616369) in AA, which suggested that the underlying risk variant(s) of SLE was best captured by these 6 SNPs located in the TMEM87-IRAK1-MECP2 region in this study.
Figure 2. A SLE-risk haplotype share by all four ancestral groups.
Haplotypes were constructed using 34 SNPs shown in Table 1. Haplotype H1 (highlighted in green) was consistently associated with SLE in all 4 ancestral groups. Allele conferring risk of SLE is bolded and italicized.
In conditional haplotype-based association testing, after conditioning on rs13397, rs4898375, rs1059702, rs2734647, rs2075596 and rs1616369, association signals of all other SNPs were completely eliminated or reduced to baseline in EA, AS, HA and AA (Table S1–S4), which supported that association signals detected in Xq28 could be attributed to these 6 SNPs.
Of note, in EA, rs2269368, rs2071129, rs2071130, rs5945377 and rs5945378 (named as group 1) in the ARHGAP4-NAA10-RENBP region exhibited even stronger association with SLE than rs13397, rs4898375, rs1059702, rs2734647, rs2075596 and rs1616369 (named as group 2) in the TMEM187-IRAK1-MECP2 region (Table 1). Genetic effects of these two regions could not be distinguished in EA using conditional testing, in which association signals detected at either group of SNPs were completely eliminated when conditioning on another group (Figure S1). In contrast to EA, a stronger association with SLE was detected at group 2 rather than group 1 SNPs in AS and HA (Table 1). In these two datasets, when conditioning on group 2 SNPs, association signals detected at group 1 SNPs were completely eliminated or reduced to baseline (Figure S1). Whereas, when conditioning on group 1 SNPs, group 2 SNPs retained strong residual association signals. Thus, in AS and HA, association signals detected at group 1 SNPs might be attributed to group 2 SNPs. In AA, SNPs in the ARHGAP4-NAA10-RENBP region were not associated with SLE (Table 1). LD analysis showed that these two groups of SNPs were in strong LD in EA and HA (r2>0.7), modest LD in AS (r2<0.5) and low LD in AA (r2<0.3). Taken together, these data suggest that association signals detected in the ARHGAP4-NAA10-RENBP region are driven by SLE-associated SNPs in the TMEM187-IRAK1-MECP2 region.
IRAK1 SNP rs1059702 could best explain association signals detected in the Xq28 region
To further localize underlying causal variant(s) in the TMEM187-IRAK1-MECP2 region, we performed conditional testing among rs13397, rs4898375, rs1059702, rs2734647, rs2075596 and rs1616369. In EA and AA, genetic effects of these 6 SNPs could not be distinguished (Figure S2), probably because they were in strong LD in EA and had too low MAFs in AA. In HA, when conditioning on rs13397, rs1059702, rs2075596 or rs1616369, association signals of the other 5 SNPs were completely eliminated. In contrast, conditioning on rs4898375 or rs2734647 showed residual signals at rs13397, rs1059702, rs2075596 and rs1616369. Thus, association signals detected at rs4898375 and rs2734647 could be attributed to rs13397, rs1059702, rs2075596 and rs1616369. In AS, rs4898375 and rs1059702 explained association signals detected at the other 4 SNPs. Taken together, only rs1059702 could explain the association signals of the other 5 SNPs in EA, AA, HA and AS, suggesting that rs1059702 was the most likely causal variant among the 6 SLE-associated SNPs. In meta-analysis, rs1059702 exhibited the strongest P value (Pmeta=1.3×10−27, OR=1.43) (Table 1). Furthermore, the risk minor allele of rs1059702 (S196F in exon 5) of IRAK1 leads to increased NF-κB activity,[29] suggesting it may confer risk of SLE by affecting the biological function.
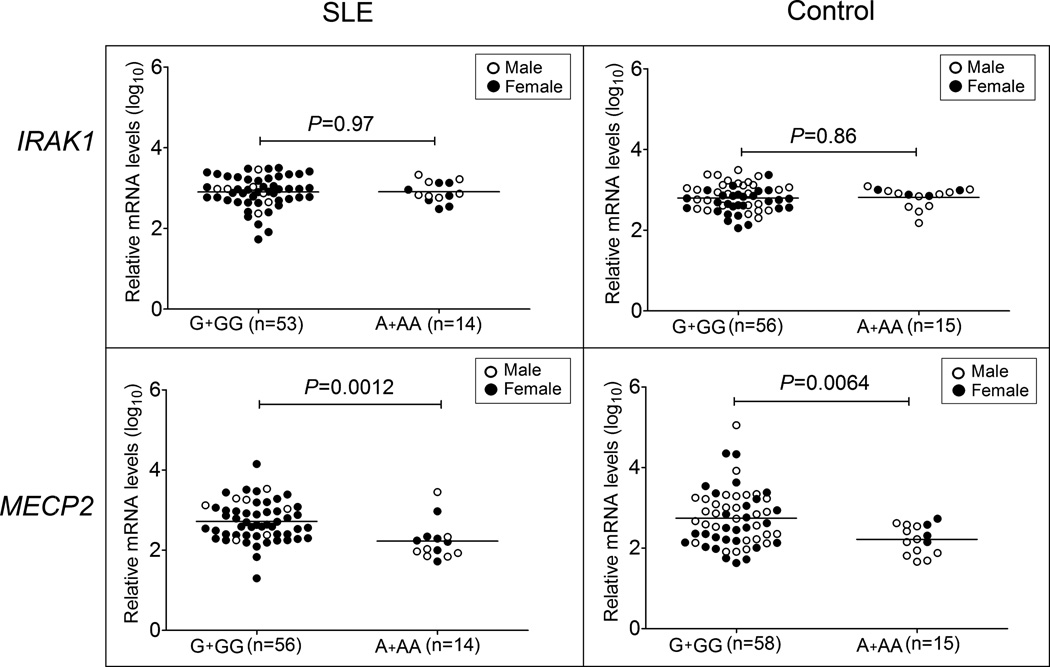
The risk allele of rs1059702 was associated with lower mRNA levels of MECP2
Using RT-PCR, we measured mRNA levels of IRAK1 and MECP2 in PBMCs from 70 SLE cases (56 females and 14 males) and 73 healthy controls (32 females and 41 males) and assessed their association with rs1059702 genotypes. To exclude the influence of X-inactivation, EA males or females carrying a homozygous rs1059702 genotype were used. Compared to those carrying the non-risk GG or G genotype, subjects carrying the SLE-risk AA or A genotype had decreased MECP2, but not IRAK1, levels in both cases (P=0.0012) and controls (P=0.0064) (Figure 3). These data suggested that rs1059702, or other SLE-risk variants tagged by rs1059702, may confer risk of SLE by affecting expression of MECP2.
Figure 3. Association of rs1059702 genotype with IRAK1 and MECP2 mRNA levels.
Expression levels of IRAK1 and MECP2 (total level of all isoforms) were measured in PBMCs of SLE patients and healthy controls with European ancestry using real-time quantitative PCR. The expression level of housekeeping gene RPLP0 was used as an endogenous control. Log10 value of relative mRNA levels of IRAK1 and MECP2 were compared between different genotypes of rs1059702 (G+GG vs. A+AA) in SLE and control groups, respectively, using t test. Females are highlighted as black.
DISCUSSION
We comprehensively investigated the genetic association between Xq28 and SLE susceptibility. In addition to previously reported IRAK1, MECP2 and ARHGAP4, 6 other genes on Xq28 were assessed. We identified SLE-associated SNPs in the TMEM187-IRAK1-MECP2 region in 4 different ancestral groups, and identified rs1059702 (S196F) in IRAK1 as the most likely causal variant. Furthermore, we showed that the SLE-risk genotype of rs1059702 was associated with lower mRNA levels of MECP2. Thus, our data suggested that both IRAK1 and MECP2 are SLE risk genes on Xq28.
The successful localization of a causal variant in our study should be attributed to conducting fine-mapping using high-density SNP markers and performing association testing in subjects with African ancestry. Using fine-mapping, we identified multiple Xq28 SNPs that were strongly associated with SLE in EA, AS and HA. However, these SNPs spanned a ~187 kb region from ARHGAP4 to MECP2 and their independent effects were difficult to distinguish due to strong LD. Compared to EA, AS and HA, the weaker LD at Xq28 in AA helped us localize association signals to a narrower region. Based on the findings that 6 SNPs in the TMEM187-IRAK1-MECP2 region were associated with SLE in EA, AS, HA and AA with similar odds ratios and they could explain association signals of other Xq28 SNPs, we concluded that these 6 SNPs captured the underlying risk variant(s) shared by 4 ancestral groups. Because the risk allele frequency of these 6 SNPs are much lower in AA (~5%) than in EA (~15%), HA (40%) and AS (~75%), the association signals in AA did not reach the Bonferroni-corrected significance level.
IRAK1 plays a pivotal role in the activation of NF-κB. We identified the minor allele of rs1059702 on IRAK1, resulting in a serine to phenylalanine substitution at amino acid 196, as a likely causal variant for SLE. Previous functional study has shown that 196F IRAK1 variant confers increased NF-κB activity in vitro,[29] which is consistent with abrogation of all SLE-associated phenotypes in a IRAK1 deficient mouse lupus model.[7] Of note, the minor allele of rs1059703 (L532S) in exon 12 of IRAK1 also confers increased NF-κB activity in vitro,[29] and minor alleles of both rs1059702 and rs1059703 have been associated with worse outcomes in sepsis [30] and increased risk of systemic sclerosis [31] in European-derived subjects. In this study, rs1059703 was associated with SLE in EA, AS and HA (Table 1), but not in AA (MAF of 34.7% in cases vs. 34.6% in controls, P=0.897. This P value was not shown in Table 1 and S4, because rs1059703 had a genotype missing rate of 6.7% in AA which exceeded our threshold of 5%). LD analysis showed that rs1059703 and rs1059702 were in strong LD in EA (r2=0.94), AS (r2=0.92) and HA (r2=0.86), but in low LD in AA (r2=0.10), suggesting that the association of rs1059703 with SLE in EA, AS and HA might be attributed to rs1059702. IRAK regulates signal transduction of IL-1R and toll-like receptors (TLR), playing a pivotal role in innate immunity and autoimmunity. Of interest, IRAK-M, which mediates suppression of TLR7 signalling, has also been shown as a genetic risk for murine lupus.[32]
It is well recognized that rare variants in MECP2 cause neurodevelopmental disorder Rett syndrome.[33] In this study, the SLE-risk genotype of rs1059702 was associated with lower mRNA levels of MECP2 but not IRAK1 in both cases and controls. Consistent with our results, the risk minor allele of rs1059702 was associated with lower mRNA levels of MECP2 in an eQTL study using peripheral blood from 1,469 unrelated European subjects,[34] in which the minor allele of rs1059702 was also associated with lower levels of RENBP and TMEM187 but at less significant levels. The finding suggests that lower MECP2 levels may have consequences similar to hypomethylation at CpG islands in genes that are regulated epigenetically leading to dysregulated expression of SLE-risk genes. The SLE-risk haplotype tagged by the minor allele of rs1059702 has been associated with the up-regulation of 13 interferon signature genes in B cell lines from female SLE patients.[10] The biological mechanism by which rs1059702 may alter the expression of MECP2 is not known at present. Due to the strong LD in the Xq28 region, it is possible that rs1059702 tags a functional SNP that affects the expression of MECP2, but whether that SNP predisposes to SLE awaits confirmation in subjects from non-EA ancestral groups.
SNPs in the ARHGAP4-NAA10-RENBP region exhibited peak association with SLE in EA, although this strong association pattern was not replicated in AS, HA and AA. Based on our data, we favor the explanation that association signals detected in the ARHGAP4-NAA10-RENBP region are driven by SLE-associated SNPs in the TMEM187-IRAK1-MECP2 region. However, it is also possible that SNPs used in this study failed to capture underlying causal variant(s) in the ARHGAP4-NAA10-RENBP region in AS, HA and AA due to different LD patterns in these ancestral groups. ARHGAP4 is a hematopoietic specific gene that belongs to the RhoGAP familiy. A deletion spanning AVPR2 and ARHGAP4 causes congenital nephrogenic diabetes insipidus and has been associated with severe immumodeficiency.[35] NAA10 encodes the catalytic subunit of the major human N-terminal acetyltrasferase.[36] NAA10 knockdown reduced the growth rate in human cancer cell lines.[37] NAA10 variant Ser37Pro results in an X-linked lethal disorder of infancy due to N-terminal acetyltransferase deficiency.[38] RENBP inhibits the activity of renin, [39] and the renin-angiotensinogen system has been implicated in SLE susceptibility.[40] Whether ARHGAP4, NAA10 and RENBP are SLE susceptibility genes will need further investigation.
In conclusion, by taking advantage of the power of trans-ancestral mapping, we identified rs1059702 as the likely causal variant predisposing to SLE susceptibility in 4 different ancestral groups. This SNP leads to an amino acid change on IRAK1 (S196F), with known function of increasing NF-κB activity, and is associated with lower levels of MECP2, suggesting both IRAK1 and MECP2 are SLE susceptibility genes.
Supplementary Material
Acknowledgments
The BIOLUPUS network is composed of Johan Frostegård, MD, PhD (Huddinge, Sweden), Lennart Truedsson, MD, PhD (Lund, Sweden), Enrique de Ramón, MD PhD (Málaga, Spain), José M. Sabio, MD, PhD (Granada, Spain), María F. González-Escribano, PhD (Sevilla, Spain), Javier Martin, MD, PhD (Granada, Spain), Norberto Ortego-Centeno (Granada, Spain), José Luis CAllejas MD (Granada, Spain), Julio Sánchez-Román, MD (Sevilla, Spain), Sandra D’Alfonso, PhD (Novara, Italy), Sergio Migliarese MD (Napoli, Italy), Gian-Domenico Sebastiani MD (Rome, Italy), Mauro Galeazzi MD (Siena, Italy), Torsten Witte, MD, PhD (Hannover, Germany), Bernard R. Lauwerys, MD, PhD (Louvain, Belgium), Emoke Endreffy, PhD (Szeged, Hungary), László Kovács, MD, PhD (Szeged, Hungary), Carlos Vasconcelos, MD, PhD (Porto, Portugal), Berta Martins da Silva, PhD (Porto, Portugal).
The Argentine Collaborative Group is composed of H.R. Scherbarth, P.C. Marino, E.L. Motta, S. Gamron, C. Drenkard, E. Menso, A. Allievi, G.A. Tate, J.L. Presas, S.A. Palatnik, M. Abdala, M. Bearzotti, A. Alvarellos, F. Caeiro, A. Bertoli, S. Paira, S. Roverano, C.E. Graf, E., Bertero, C. Guillerón, S. Grimaudo, J. Manni, L.J. Catoggio, E.R. Soriano, C.D. Santos, C. Prigione, F.A. Ramos, S.M. Navarro, G.A. Berbotto, M. Jorfen, E.J. Romero, M.A. Garcia, J.C. Marcos, A.I. Marcos, C.E. Perandones, A. Eimon, C.G. Battagliotti, C. Garcilazo, J. Musuruana, C. Castel and M. Busajm.
Some of the samples used in this study were provided by the Lupus Family Registry and Repository (LFRR, www.lupus.omrf.org).
Funding: Support for this work was obtained from the US National Institutes of Health grants: R01AR43814 (B.P.T.), R01AR043274 (K.L.M.), R01AI063274 (P.M.G.), N01AR62277 (J.B.H.), R3724717 (J.B.H.), AR042460 (J.B.H.), P01AI083194 (J.B.H.), P20RR020143 (J.B.H.), P01AR49084 (R.P.K. and E.E.B.), R01AR33062 (R.P.K.), P30AR055385 (E.E.B.), 5UL1RR025777 (R.P.K. and J.C.E.), R01AR33062 (R.P.K.), P01AR49084 (R.P.K., J.C.E., E.E.B., G.S.A., J.D.R., R.R.G., L.M.B. and M.A.P.), P30048311 (E.E.B.), K08AI083790 (T.B.N.), LRPAI071651 (T.B.N.), R01CA141700 (M.E.A.R.), RC1AR058621 (M.E.A.R.) and UL1RR024999 (T.B.N.), K24AR002138 (R.R.G.), P602AR30692 (R.R.G.), P01AR49084 (R.R.G.), UL1RR025741 (R.R.G.), R01AR051545-01A2 (A.M.S.), P30AR053483 (J.A.J and J.M.G.), P30GM103510 (J.A.J.), U19AI082714 (J.A.J. and J.M.G.), R21AI070304 (S.A.B.), P01AR052915 (J.D.R.), U01AI090909 (J.D.R.), P60AR053308 (L.A.C.), M01RR-00079 (L.A.C.), P60AR049459 (D.L.K.) and UL1RR029882 (D.L.K.). This study was also supported by a grant from Lupus Research Institute (B.P.T., T.B.N.), the Merit Award from the US Department of Veterans Affairs (J.B.H. and G.S.G.), The Alliance for Lupus Research (K.L.M., T.B.N., L.A.C. and C.O.J.), the Arthritis National Research Foundation Eng Tan Scholar Award (T.B.N.), the Arthritis Foundation (A.M.S., and P.M.G.), the Korea Healthcare Technology R&D Project, Ministry for Health and Welfare, Republic of Korea (A111218-11-GM01; S.C.B.), and Korean R&D Program of MKE/KEIT (10035615; Y.W.S.). Additional funding awarded from the Swedish Research Council, Swedish Association Against Rheumatism and the King Gustaf Vth 80th Jubilee Foundation and the Fundación Instituto de Salud Carlos III PS0900129 partially funded through European FEDER funds and the Consejería de Salud de Andalucía PI-0012, the Wenner Gren Foundation for support (C.G.), the Wellcome Trust (T.J.V.), Arthritis Research UK (T.J.V.), CTSA Grant Number I ULI RR025014-02 (A.M.S.) from the National Center for Research Resources (NCRR), Kirkland Scholar Award (L.A.C.), Wake Forest University Health Sciences Center for Public Health Genomics (C.D.L., M.C.C., M.C.M., and P.S.R.) and the Federico Wilhelm Agricola Foundation Research grant (B.A.P.E.).
Footnotes
Competing interests: None
REFERENCES
- 1.Harley IT, Kaufman KM, Langefeld CD, et al. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009 May;10(5):285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Curr Opin Rheumatol. 2010 Sep;22(5):478–482. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser R, Criswell LA. Genetics research in systemic lupus erythematosus for clinicians: methodology, progress, and controversies. Curr Opin Rheumatol. 2010 Mar;22(2):119–125. doi: 10.1097/BOR.0b013e3283361943. [DOI] [PubMed] [Google Scholar]
- 4.Moser KL, Kelly JA, Lessard CJ, et al. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009 Jul;10(5):373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010 Dec;6(12):683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawalha AH, Webb R, Han S, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS One. 2008;3(3):e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob CO, Zhu J, Armstrong DL, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2009 Apr 14;106(15):6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacob CO, Reiff A, Armstrong DL, et al. Identification of novel susceptibility genes in childhood-onset systemic lupus erythematosus using a uniquely designed candidate gene pathway platform. Arthritis Rheum. 2007 Dec;56(12):4164–4173. doi: 10.1002/art.23060. [DOI] [PubMed] [Google Scholar]
- 9.Suarez-Gestal M, Calaza M, Endreffy E, et al. Replication of recently identified systemic lupus erythematosus genetic associations: a case-control study. Arthritis Res Ther. 2009;11(3):R69. doi: 10.1186/ar2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb R, Wren JD, Jeffries M, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009 Apr;60(4):1076–1084. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartupee J, Li X, Hamilton T. Interleukin 1alpha-induced NFkappaB activation and chemokine mRNA stabilization diverge at IRAK1. J Biol Chem. 2008 Jun 6;283(23):15689–15693. doi: 10.1074/jbc.M801346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinarello CA. The IL-1 family and inflammatory diseases. Clin Exp Rheumatol. 2002 Sep-Oct;20(5) Suppl 27:S1–S13. [PubMed] [Google Scholar]
- 13.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006 Nov;6(11):823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nan X, Ng HH, Johnson CA, Laherty CD, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998 May 28;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 15.Lorincz MC, Schubeler D, Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol Cell Biol. 2001 Dec;21(23):7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawalha AH. Xq28 and lupus: IRAK1 or MECP2? Proc Natl Acad Sci U S A. 2009 Jun 16;106(24):E62. doi: 10.1073/pnas.0904068106. author reply E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009 Nov;41(11):1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adrianto I, Wen F, Templeton A, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011 Mar;43(3):253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lessard CJ, Adrianto I, Kelly JA, et al. Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am J Hum Genet. 2011 Jan 7;88(1):83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Wu H, Khosravi M, et al. Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet. 2011 May;7(5):e1002079. doi: 10.1371/journal.pgen.1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997 Sep;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Hoggart CJ, Parra EJ, Shriver MD, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003 Jun;72(6):1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoggart CJ, Shriver MD, Kittles RA, et al. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004 May;74(5):965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKeigue PM. Prospects for admixture mapping of complex traits. Am J Hum Genet. 2005 Jan;76(1):1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009 Jun;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Liu G, Tsuruta Y, Gao Z, et al. Variant IL-1 receptor-associated kinase-1 mediates increased NF-kappa B activity. J Immunol. 2007 Sep 15;179(6):4125–4134. doi: 10.4049/jimmunol.179.6.4125. [DOI] [PubMed] [Google Scholar]
- 30.Arcaroli J, Silva E, Maloney JP, et al. Variant IRAK-1 haplotype is associated with increased nuclear factor-kappaB activation and worse outcomes in sepsis. Am J Respir Crit Care Med. 2006 Jun 15;173(12):1335–1341. doi: 10.1164/rccm.200603-341OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieude P, Bouaziz M, Guedj M, et al. Evidence of the contribution of the X chromosome to systemic sclerosis susceptibility: association with the functional IRAK1 196Phe/532Ser haplotype. Arthritis Rheum. 2011 Dec;63(12):3979–3987. doi: 10.1002/art.30640. [DOI] [PubMed] [Google Scholar]
- 32.Lech M, Kantner C, Kulkarni OP, Ryu M, Vlasova E, Heesemann J, et al. Interleukin-1 receptor-associated kinase-M suppresses systemic lupus erythematosus. Ann Rheum Dis. 2011 Dec;70(12):2207–2217. doi: 10.1136/ard.2011.155515. [DOI] [PubMed] [Google Scholar]
- 33.Samaco RC, Neul JL. Complexities of Rett syndrome and MeCP2. J Neurosci. 2011 Jun 1;31(22):7951–7959. doi: 10.1523/JNEUROSCI.0169-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fehrmann RS, Jansen RC, Veldink JH, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011 Aug;7(8):e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broides A, Ault BH, Arthus MF, et al. Severe combined immunodeficiency associated with nephrogenic diabetes insipidus and a deletion in the Xq28 region. Clin Immunol. 2006 Aug;120(2):147–155. doi: 10.1016/j.clim.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Puig N, Fersht AR. Characterization of the native and fibrillar conformation of the human Nalpha-acetyltransferase ARD1. Protein Sci. 2006 Aug;15(8):1968–1976. doi: 10.1110/ps.062264006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnesen T, Thompson PR, Varhaug JE, et al. The protein acetyltransferase ARD1: a novel cancer drug target? Curr Cancer Drug Targets. 2008 Nov;8(7):545–553. doi: 10.2174/156800908786241113. [DOI] [PubMed] [Google Scholar]
- 38.Rope AF, Wang K, Evjenth R, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011 Jul 15;89(1):28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi S, Ogasawara H, Takahashi K, et al. Identification of a domain conferring nucleotide binding to the N-acetyl-d-glucosamine 2-epimerase (Renin binding protein) J Biochem. 2002 Apr;131(4):605–610. doi: 10.1093/oxfordjournals.jbchem.a003140. [DOI] [PubMed] [Google Scholar]
- 40.Teplitsky V, Shoenfeld Y, Tanay A. The renin-angiotensin system in lupus: physiology, genes and practice, in animals and humans. Lupus. 2006;15(6):319–325. doi: 10.1191/0961203306lu2306rr. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.