Abstract
Pseudomonas aeruginosa possesses an unusual pathway for 5′-methylthioadenosine (MTA) metabolism involving deamination to 5′-methylthioinosine (MTI) followed by N-ribosyl phosphorolysis to hypoxanthine and 5-methylthio-α-D-ribose 1-phosphate. The specific MTI phosphorylase of P. aeruginosa has been reported (Guan, R., Ho, M. C., Almo, S. C. and Schramm, V. L. (2011) Biochemistry 50, 1247–1254) and here we characterize MTA deaminase from P. aeruginosa (PaMTADA). Genomic analysis indicated the PA3170 locus to be a candidate for MTA deaminase (MTADA). Protein encoded by PA3170 was expressed and shown to deaminate MTA with 40-fold greater catalytic efficiency for MTA than for adenosine. The kcat/Km value of 1.6 × 107 M−1s−1 for MTA is the highest catalytic efficiency known for an MTA deaminase. 5′-Methylthiocoformycin (MTCF) is a 4.8 pM transition state analogue for PaMTADA but causes no significant inhibition of human adenosine deaminase or MTA phosphorylase. MTCF is permeable to P. aeruginosa and exhibits an IC50 of 3 nM on cellular PaMTADA activity. PaMTADA is the only activity in P. aeruginosa extracts to act on MTA. MTA and 5-methylthio-α-D-ribose are involved in quorum sensing pathways, thus PaMTADA is a potential target for quorum sensing. The crystal structure of PaMTADA in complex with MTCF shows the transition state mimic 8-R-hydroxyl group in contact with a catalytic site Zn2+, the 5′-methylthio group in a hydrophobic pocket and the transition state mimic of the diazepine ring in contact with a catalytic site Glu.
Keywords: 5′-methylthioadenosine, 5′-methylthioinosine, quorum sensing, transition state analogue, MTA deaminase, 5′-methylthiocoformycin, tight-binding inhibitor, Pseudomonas
Pseudomonas aeruginosa is a Gram-negative bacterium and a major opportunistic human pathogen, accounting for approximately 15% of all hospital infections.1 Immunocompromised patients and patients with comorbid illnesses are especially susceptible to the infection.1,2 P. aeruginosa has multiple antimicrobial resistance mechanisms making infections difficult to treat.3 High morbidity and mortality rates have been reported in P. aeruginosa infections, especially for late-onset ventilator associated pneumonia.4,5 In P. aeruginosa, the production of virulence factors and biofilm formation are regulated by quorum sensing (QS) systems.6 QS involves bacterial cell-to-cell communication by small molecules. QS allows bacteria populations to adjust behavior in response to environmental conditions.4 Communication in QS relies on signaling molecules including the N-acyl-homoserine lactones (AHLs) found in P. aeruginosa and most other Gram-negative bacteria. AHLs are synthesized as the bacterial cell density increases. When the concentrations of AHLs reach a critical threshold, the signal molecules bind to specific receptors and regulate target genes expression. A major QS system in P. aeruginosa includes las and rhl, which use 3-oxo-C12-homoserine lactone and C4-homoserine lactone as signaling molecules, respectively. QS signaling is correlated with the virulence of P. aeruginosa infections. Deletion of single or multiple QS genes in P. aeruginosa reduced virulence in several mouse models.5 The presence of QS signaling molecules and expression of QS-responsive genes in P. aeruginosa have been detected in sputum samples of cystic fibrosis patients. And most recently, production of QS-dependent virulence factors of P. aeruginosa have been linked to the development of ventilator-associated pneumonia.6 Since inhibition of QS biosynthetic pathways does not affect cell growth, blocking QS synthesis has been proposed as a strategy to attenuate the virulence of bacterial infections without causing drug resistance.7
AHL synthase catalyzes the production of AHL using S-adenosylmethionine (SAM) and acylated-acyl carrier protein as precursors. The reaction produces 5′-methylthioadenosine (MTA) as a product. MTA is also an important product from polyamine biosynthesis and is recycled by a SAM salvage pathway.8 In most bacteria, MTA is degraded by 5′-methylthioadenosine nucleosidase (MTAN) to adenine and 5-methylthio-α-D-ribose. Inhibition of E. coli and V. cholerae MTANs with transition state analogue inhibitors or by gene deletion, disrupts quorum sensing, and reduces biofilm formation, supporting MTAN as a target for QS in most Gram negative bacteria.9 Mammals do not express an MTAN, nor do they have QS pathways, giving species specificity to this target.
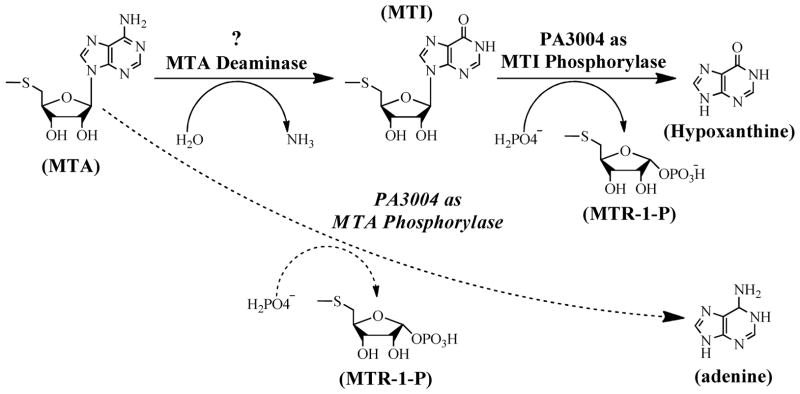
In eukaryotes and archaea, MTA degradation is catalyzed by 5′-methylthioadenosine phosphorylase (MTAP) which converts MTA and phosphate to adenine and 5-methylthio-α-D-ribose 1-phosphate.10 P. aeruginosa was originally thought to be a bacterial anomaly, possessing an MTAP (PA3004 gene) instead of MTAN. We recently characterized the PA3004-encoded protein and found it to prefer methylthioinosine (MTI) as substrate.11 It remains the only known example of a specific MTI phosphorylase (MTIP). The discovery of MTIP suggested that MTA must be deaminated in P. aeruginosa. We examined MTA catabolism in P. aeruginosa using [8-14C]MTA. A MTA→MTI→hypoxanthine pathway was established and no significant MTAP or MTAN activity was observed.11 These results established a functional PaMTIP in cells and extracts and implicated the existence of an MTA deaminase (MTADA) to convert MTA to MTI (Fig. 1). If MTADA is directly and solely responsible for MTA degradation in P. aeruginosa, inhibiton of PaMTADA would be functionally similar to that of MTAN in other bacterial species, causing MTA product inhibition of AHL synthase and disruption of quorum sensing in P. aeruginosa.12 This pathway is unprecedented in bacteria, but Plasmodium species also possess a similar two-step pathway of MTA degradation. In the case of Plasmodium species, both the purine nucleoside phosphorylase and the adenosine deaminase (ADA) are broad-specificity enzymes, capable of functioning as MTIP and MTADA, respectively. However, inosine and adenosine are preferred substrates and MTI and MTA are secondary substrates.13,14
Figure 1.
MTA degradation in Pseudomonas aeruginosa. The dashed line indicates the previous (and incorrect) annotation of MTA phosphorylase activity for PA3004 (in italics). The PA3004 protein is now identified as a MTI phosphorylase and the conversion of MTA to MTI requires the existence of MTA deaminase.11
Recently, the first specific MTA deaminase has been reported in Thermotoga martima.15 The TmMTADA can deaminate MTA, S-adenosylhomocysteine and adenosine but prefers MTA. TmMTADA was identified by using structure-based docking with high-energy forms of potential substrates and the activity validated by enzymatic assays with purified protein. A crystal structure of TmMTADA in complex with S-inosylhomocysteine, the product of SAH deamination, was determined in the same study, revealing the key residues for binding substrates in the active site.15 These findings on TmMTADA guided our search for PaMTADA.
Here we report the PA3170 encoded protein to be a MTA deaminase of P. aeruginosa PAO1. The substrate specificity was characterized and we identified several powerful transition state analogue inhibitors. The inhibition of cellular PaMTADA activity was investigated using the transition state analogue inhibitor, 5′-methylthiocoformycin (MTCF), an inhibitor interacting with picomolar affinity. The crystal structure of PaMTADA in complex with MTCF defines the catalytic site interactions responsible for transition state stabilization in 6-aminopurine deaminase reactions.
MATERIALS AND METHODS
Chemicals
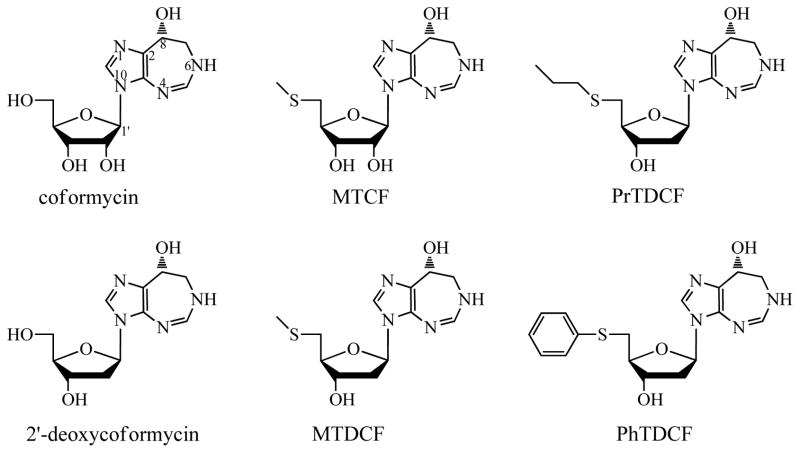
Coformycin (CF), 5′-methylthiocoformycin (MTCF), 2′-deoxycoformycin (DCF), 5′-methylthio-2′-deoxycoformycin (MTDCF), 5′-propylthio-2′-deoxycoformycin (PrTDCF) and 5′-phenylthio-2′-deoxycoformycin (PhTDCF) were synthesized by methods reported earlier (Fig. 2).16 [8-14C]MTA was synthesized as described previously.17 All other chemicals and reagents were obtained from Sigma or Fisher Scientific, and were of reagent grade.
Figure 2.
Inhibitors of PaMTADA. Coformycin and 2′-deoxycoformycin are transition state analogues of adenosine deaminase. MTCF and other 5′-functionalized-2′-deoxycoformycins are transition state analogues of MTA deaminases.16
Plasmid construction
A synthetic gene was designed from the predicated protein sequence of gene PA3170 in Pseudomonas Genome Database.18 Gene PA3170 belongs to Pseudomonas aeruginosa PAO1 and encodes a conserved hypothetical protein. The synthetic gene was purchased from DNA2.0 Inc. in a pJexpress414 expression vector. The encoded protein has an additional 14 amino acids at the N-terminus which includes a His6 tag.
Enzyme purification and preparation
BL21-CodonPlus(DE3)-RIPL E. coli were transformed with the synthetic plasmid and grown overnight at 37 °C in 100 mL of LB medium with 100 μg/mL ampicillin. The culture was transferred into 1 L of LB/ampicillin medium and growth continued at 37 °C to an O.D.600 of 0.7. Expression was induced for 4 h at 37 °C by addition of 1 mM IPTG. The cells were harvested by centrifugation at 4500 × g for 30 min. The cell pellet was suspended in 20 mL of lysis buffer (50 mM phosphate, pH 8.0, containing 15 mM imidazole and 300 mM NaCl), with addition of 2 tablets of EDTA-free protease inhibitor (from Roche Diagnostics) and 20 mg lysozyme (from chicken egg). Cells were disrupted by two passes through a French pressure cell and centrifuged at 20,000 × g for 30 min. The supernatant was loaded onto a 4 mL column of Ni-NTA Superflow resin equilibrated with 20 mL of lysis buffer. The column was washed with 20 mL of wash buffer (50 mM phosphate, pH 8.0, containing 50 mM imidazole and 300 mM NaCl), and the target protein was eluted with 12 mL of elution buffer (50 mM phosphate, pH 8.0, containing 250 mM imidazole and 300 mM NaCl). Eluted protein was immediately dialyzed against dialysis buffer A (50 mM phosphate, pH 8.0, containing 300 mM NaCl and 10% glycerol) to remove the imidazole, followed by dialysis against dialysis buffer B (50 mM Hepes, pH 7.4, containing 10% glycerol). Dialyzed protein was concentrated to 7.8 mg/ml and was > 95% pure as judged by SDS-PAGE. All of the purification procedures were performed at 4 °C but the Ni-NTA column was run at room temperature (25 °C). Concentrated protein was stored at −80 °C. The extinction coefficient of PaMTADA is 48.93 mM−1cm−1 at 280 nm, as calculated by the ProtParam program from ExPASy and was used to estimate protein concentration (http://ca.expasy.org/seqanalref/).
Enzymatic assays
Deaminase activity on MTA, adenosine, SAH, and adenine was measured by the absorbance change at 265 nm. The extinction coefficients are 8.1 mM−1cm−1 for MTA and adenosine (13), 8.55 mM−1cm−1 for SAH, and 6.7 mM−1cm−1 for adenine.19,20 Deaminase activity on guanosine was measured at 260 nm with an extinction coefficient of 3.9 mM−1cm−1.21 HsMTAP and EcMTAN activity on MTA was determined by conversion of product adenine to 2,8-dihydroxyadenine using xanthine oxidase (XOD) as the coupling enzyme.22 The extinction coefficient is 15.5 mM−1cm−1 at 305 nm. Reactions of deaminase were carried out at 25 °C in 1 cm cuvettes. Assay mixtures of 1 mL contained 50 mM Hepes, pH 7.4, 100 mM NaCl, 100 μg/mL BSA, variable concentrations of substrate, and appropriate amounts of purified PaMTADA. Reactions were initiated by addition of enzyme and the initial rates were monitored with a CARY 300 UV-Visible spectrophotometer. Control rates (no PaMTADA) were subtracted from initial rates. Kinetic parameters of PaMTADA were obtained by fitting initial rates to the Michaelis-Menten equation using GraFit 5 (Erithacus Software).
Inhibition assays
Inhibition assays for PaMTADA were carried out by adding 0.15 nM PaMTADA into reaction mixtures at 25 °C containing 50 mM Hepes, pH 7.4, 100 mM NaCl, 100 μM MTA, 100 μg/mL BSA, and variable concentrations of inhibitor. Inhibition assays of HsMTAP were carried out at 25 °C by adding 0.8 nM enzyme into reaction mixtures containing 50 mM Hepes, pH 7.4, 100 mM phosphate, pH 7.4, 100 μM MTA, 1 mM DTT, 1 unit of XOD, and variable concentrations of MTCF. Inhibition assays of EcMTAN were carried out at 25 °C by adding 0.15 nM enzyme into reaction mixtures containing 100 mM Hepes, pH 7.4, 100 mM NaCl, pH 7.4, 50 μM MTA, 1 mM DTT, 1 unit of XOD, and variable concentrations of MTCF. Controls having no enzyme and no inhibitor were included in all of the inhibition assays. The inhibition constant was obtained by fitting initial rates with variable inhibitor concentrations to equation (1) using GraFit 5 (Erithacus Software):
| (1) |
where vi is the initial rate in the presence of inhibitor, vo is the initial rate in the absence of inhibitor, Km is the Michaelis constant for MTA, [S] and [I] are MTA and inhibitor concentrations, respectively, and Ki is the inhibition constant. The inhibitor concentration was corrected using equation (2) when it was less than 10 times of the enzyme concentration:23
| (2) |
where [I]′ is the effective inhibitor concentration, [I] is the inhibitor concentration in the reaction mixture, Et is the total enzyme concentration.
Crystallization, data collection, and structure determination of PaMTADA in complex with MTCF
To obtain the PaMTADA:MTCF complex, the PaMTADA was concentrated to 35 mg/ml in 50 mM Hepes, pH 7.5, and 10% glycerol followed by incubation with 1.2 mM MTCF. The PaMTADA:MTCF complex crystallized in 1.26 M sodium phosphate (monobasic) and 0.14 M potassium phosphate (dibasic) at a final pH of 5.6 at 18 °C using hanging drop or sitting drop vapor diffusion method. Crystals were transferred to mother liquor supplemented with 20% glycerol and flash-cooled in liquid N2 prior to data collection. X-ray diffraction data were collected at the X29A beamline of Brookhaven National Laboratory on an ADSC Q315 detector at 100K. Data were processed using HKL2000 program suite and summarized in Table 1.24
Table 1.
Data collection and refinement statistics
| PaMTADA:MTCF complex | |
|---|---|
| PDB code | 4GBD |
| Data collection | |
| Space group | C2 |
| Cell dimension | |
| a, b, c (Å) | 119.8, 120.3, 77.3 |
| α, β, γ (°) | 90.0, 108.0, 90.0 |
| Resolutions (Å) | 50.00–2.00 (2.07–2.00) |
| Rsym (%) | 9.0 (65.6) |
| I / σI | 11.5 (1.7) |
| Completeness (%) | 97.6 (98.3) |
| Redundancy | 3.7 (3.6) |
|
| |
| Refinement | |
| Resolution (Å) | 50.00–2.00 |
| No. unique reflections | 70236 |
| Rwork / Rfree (%) | 19.6/23.5 |
| B-factors (Å2) | |
| Protein | |
| (main chain) | 40.8 |
| (side chain) | 46.2 |
| Water | 44.5 |
| Ligand* | 42.3 |
| No. of Atoms | |
| Protein | 6694 |
| Water | 243 |
| Ligand | 44 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.61 |
|
| |
| Ramachandran analysis | |
| favored region | 96.5% |
| allowed region | 3.1% |
| disallowed region | 0.4% |
| Coordinate Error by Luzzati plot (Å) | 0.24 |
Numbers in parentheses are for the highest-resolution shell. One crystal was used for each data set.
The ligand occupancy is 1.0.
The structure of PaMTADA:MTCF complex was determined by molecular replacement with the program Molrep,22 using the crystal structure of the amidohydrolase family protein OLEI061672_1_465 from Oleispira antarctica (PDB ID: 3LNP), without bound ligand as the search model. Models without inhibitor were iteratively rebuilt in COOT and refined in Refmac5.25,26 Manual inhibitor building was initiated only after the Rfree decreased below 30% and was guided by clear ligand density in Fo – Fc electron density maps contoured at 3σ. Data processing and refinement statistics are summarized in Table 1.
Inhibition of cellular PaMTADA activity
Inhibtion of PaMTADA activity in cell lysates was carried out as follows. P. aeruginosa PAO1 (ATCC number: 15692) was grown at 37 °C to stationary phase in LB medium for 16 h. Cells were collected by centrifugation at 16,100 × g and washed three times with 100 mM phosphate, pH 7.4. Cells were lysed using BugBuster reagent (Novagen). Clarified lysate (47 μL) was incubated with 0–1000 nM of MTCF and [8-14C]MTA (15 μL containing approximately 0.1 μCi 14C) in 100 mM phosphate, pH 7.4, for 20 min, with a total volume of 80 μL. Reaction mixtures were quenched with perchloric acid (1.8 M final concentration) and neutralized with potassium hydroxide. Precipitates were removed by centrifugation and carrier hypoxanthine, adenine, MTI, and MTA were added to the supernatant. Separation of the metabolites was carried out on a C18 Luna HPLC column (Phenomenex) with a gradient of 5 to 52.8% acetonitrile in 20 mM triethylamine acetate, pH 5.2. The UV absorbance at 260 nm was monitored. The retention times were 5.1 min (hypoxanthine), 7.5 min (adenine), 20.4 min (MTI), and 21.9 min (MTA), respectively. Fractions were collected in scintillation vials, dried, reconstituted in 200 μL deionized water prior to addition of 10 mL ULTIMA GOLD LSC-Cocktail scintillation fluid. The cpm of 14C was counted at 20 min per cycle for three cycles using a Tri-Carb 2910TR liquid scintillation analyzer. Cell lysate was replaced by lysis buffer in reaction mixtures in control experiments. Inhibition of cellular PaMTADA was investigated with addition of MTCF to the LB medium instead of addition to the cell lysate. The final concentrations of MTCF used in the LB varied from 0 to 1000 nM. Culture growth in the presence of MTCF used 1% inoculums by volume in all cultures. All other procedures were the same as described above. A third experiment was carried out with addition of 500 nM MTCF in LB medium and addition of 0 or 100 nM MTCF to the cell lysate after the BugBuster lysis. The IC50 for MTCF was obtained by fitting the percentage of degraded MTA to the concentration of MTCF using equation (4) and the GraFit 5 (Erithacus Software):
| (4) |
where y is the percentage of degraded MTA at certain [I] (inhibitor concentration), y0 is the percentage of degraded MTA at zero [I], c is the maximum difference between y and y0, and IC50 is the inhibitor concentration giving half maximal inhibition.
RESULTS AND DISCUSSION
The hunt for PaMTADA
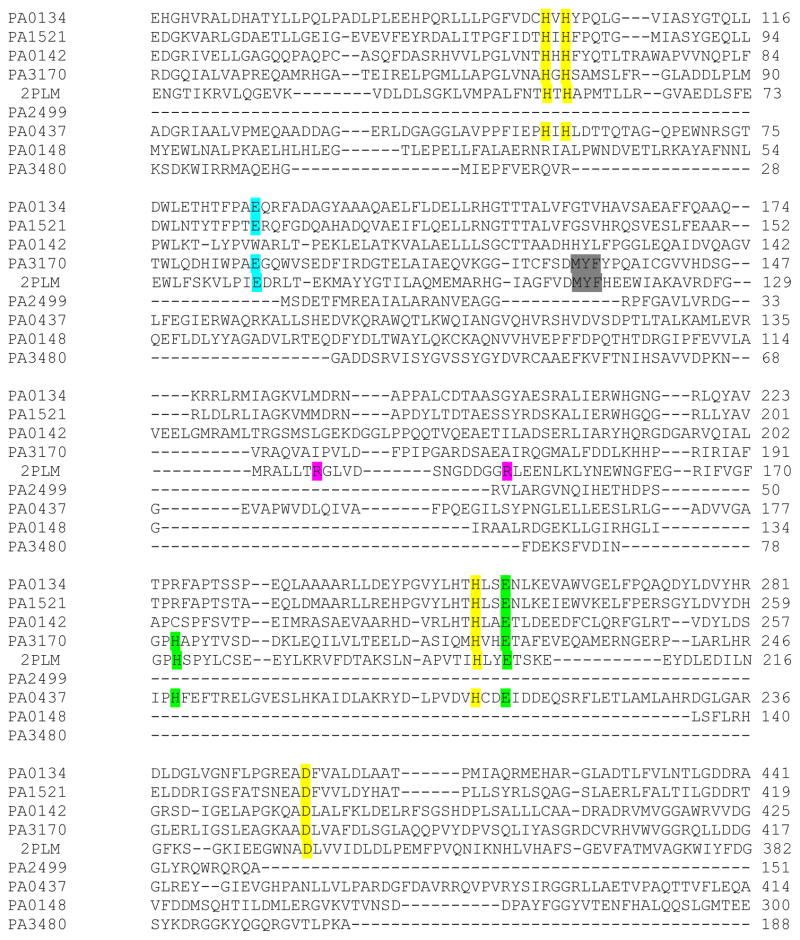
There is no gene annotated as a MTA deaminase in Pseudomonas but our previous discovery of the pathway from MTA to hypoxanthine via MTI indicated the existence of a MTA deaminase in P. aerguinosa.11 The active site of MTA deaminase was expected to contain zinc, purine and methylthioribose binding sites. The Pseudomonas genome database contains several genes annotated as putative deaminases based on zinc binding motif. These included PA0134 (guanine deaminase), PA1521 (guanine deaminase), PA0142 (guanine deaminase), PA0148 (adenosine deaminase), PA2499 (unspecified deaminase), PA3480 (deoxycytidine triphosphate deaminase), PA0437 (cytosine deaminase), and PA3170 (guanine deaminase). We searched all of the corresponding protein sequences against the PDB database. One of the hits was MTA deaminase from Thermotoga maritima (TmMTADA; PDB ID: 1J6P). TmMTADA also deaminates SAH and adenosine but favors MTA as the substrate.15 The crystal structure (PDB ID: 2PLM) of TmMTADA in complex with S-inosylhomocysteine (SIH) revealed catalytic site residues for recognition of ribosyl and homocysteine moieties of SIH. Glu84 interacts with the ribosyl group by forming two hydrogen bonds with 2′ and 3′ hydroxyl group. Met114, Try115, and Phe116 create a hydrophobic pocket surrounding the methythio group of homocysteine. Arg136 and Arg148 are involved in the binding of carboxylate group of homocysteine. Multiple sequence alignments show Glu84, Met114, Try115 and Phe116 to be conserved in PA3170 but Arg136 and Arg148 are not (Fig. 3). His173 and Glu203 of TmMTADA interact with the adenine base and are conserved in PA3170. Our analysis supports assignment of PA3170 as a MTA deaminase with differences in the ability to use SAH as a substrate.
Figure 3.
Sequence alignment of TmMTADA (2PLM) and the putative MTADA proteins of P. aeruginosa PAO1. Based on the structural analysis of 2PLM, the residues interacting with Zn-, ribose-, adenine, and methylthio- groups are highlighted in yellow, cyan, green and gray, respectively. The two arginine residues of 2PLM are responsible for carboxylate binding of SAH and are highlighted in magenta. Sequences without interactions in the active site are not shown.
MTA deaminase activity of PA3170
The recombinant PA3170 protein was purified and tested for substrate specificity (Table 2). The recombinant protein deaminated MTA and adenosine but was inactive with adenine, SAH and guanosine, suggesting a high specificity for adenosyl compounds with a small 5′-substituent. MTA is the most favorable substrate with a kcat of 24.6 s−1 and Km of 1.5 μM (kcat/Km of 1.6 × 107 M−1s−1). The enzyme is less efficient with adenosine. Although the kcat is 17 s−1, the Km of 46 μM is 30 times higher than that for MTA, causing most of the 40-fold lower catalytic efficiency (kcat/Km) on adenosine (3.7 × 105 M−1s−1). The 30-fold lower Km with MTA supports an important role of the 5′-methylthio-group for MTA binding. The substrate specificity reveals PA3170 protein to be a specific MTA deaminase. The catalytic efficiency on MTA (1.6 × 107 M−1s−1) is the highest of known for MTA deaminase reactions.
Table 2.
Substrate Specificity of PaMTADA, PfADA, and Human ADA on Adenosine and MTA.
Deaminase activity on MTA has been reported in malarial ADAs and T. maritime MTADA.13–15 The known MTADA enzymes have catalytic efficiency in the range of 1.4 × 104 M−1s−1 to 1.4 × 105 M−1s−1, which are over 100-fold less than that of PaMTADA. Their catalytic efficiency on adenosine is also low and comparable with the ability of PaMTADA to use adenosine, in the range of 9.2 × 107 M−1s−1 to 8.2 × 104 M−1s−1. Human and bovine ADAs do not utilize MTA as substrate and their kcat/Km values for adenosine are 1.6 × 106 M−1s−1 and 1.1 × 106 M−1s−1, respectively (13, 16). PaMTADA has a similar kcat/Km values on adenosine as other ADAs, suggesting this enzyme has the catalytic capacity to act as both ADA and MTADA in biological conditions.
Our kinetic observations support the role of PaMTADA in converting MTA to MTI. The existence of PaMTADA further validates the two step catabolism of MTA in P. aeruginosa (MTA→MTI→hypoxanthine) that we proposed recently on the basis of identification of MTI phosphorylase and the catabolism of 14C-labeled MTA.11
Picomolar Inhibitors of PaMTADA
Coformycin (CF) and 2′-deoxycoformycin (DCF) are natural product transition state analogue inhibitors of adenosine deaminases with picomolar affinity.27 Their 8-R-hydroxyl group mimics the attacking hydroxyl group at the transition state and the 7-membered diazepine ring is protonated at N6, which mimics the N1 protonation proposed to occur with adenosine or MTA at the transition state.28 The molecular electrostatic potential surfaces of the coformycins closely resemble the geometry and charge distribution of the transition states of adenosine deaminases from human, bovine, and Plasmodim falciparum. MTCF and MTDCF possess the transition state features of coformycin and the unique substrate specificity determinants of this enzyme for the 5′-methylthio group.16 MTCF and MTDCF were originally developed as transition-state analogue inhibitors of PfADA, involved in both adenosine and MTA deamination and a potential target for purine salvage in malaria.13 MTCF and MTDCF inhibit PfADA with equlibirium dissociation constants of 400 pM and 700 pM, respectively, but they have no inhibitory effect on human ADA. Inhibition of human ADA is known to cause central nervous system dysfunction and the genetic deficiency of human ADA causes severe immune deficiency disorders.29–31
Six coformycin-based transition state analogue inhibitors (Fig. 2) were tested with PaMTADA and gave Ki values ranging from 4.8 pM to 90 nM (Table 3). Coformycin inhibits PaMTADA with a Ki value of 90 nM. MTCF, however, exhibits more potent inhibition with a 4.8 pM dissociation constant. Thus, MTCF binds to PaMTADA 18,800 times better than CF, and 312,500 times better than the substrate MTA as judged by Km/Ki. The 2′-hydroxyl group has a small effect on the affinity of CF and MTCF. 2′-Deoxycoformycin and MTDCF have Ki values of 37 nM and 8 pM, respectively. Coformycins are transition state analogues for adenosine deaminases, while MTCF is specific for MTA deaminase. PaMTADA has 30 times higher affinity and 40 times higher catalytic efficiency for MTA than for adenosine, which contributes to the more potent inhibition of MTCF than CF. However, the difference in inhibitor affinity is 18,800 times, which cannot be solely attributed to the difference in substrate specificity of the enzymes. The MTCF appears to more precisely capture the transition state features of PaMTADA-catalyzed reaction. Since the transition state features on the purine base are similar in CF and MTCF, the 5′-methylthio group is likely to play a critical role in organizing the substrate and enzyme to an efficient geometry resembling the transition state. However, a detailed transition state structure for PaMTADA has not yet been established. 5′-Propylthiol-2′-deoxycoformycin (PrTDCF) binds PaMTADA 8 times weaker than MTDCF with a Ki value of 67 pM. Similarly, PhTDCF binds 16 times weaker with a Ki value of 130 pM. These results suggest that PaMTADA can accommodate a larger hydrophobic group at the 5′-position of the inhibitor, but prefers the methylthio group.
Table 3.
Summary of Ki values for PaMTADA, PfADA, and human ADA
| Inhibitors | PaMTADA | PfADA a | Human ADA a |
|---|---|---|---|
| Ki (nM) b | Ki (nM) b | Ki (nM) b | |
| Coformycin (CF) | 90 ± 10 | 0.08 ± 0.02 c | 0.11 ± 0.02 c |
| DCF | 37 ± 1 | 0.038 ± 0.009 c | 0.026 ± 0.005 c |
| MTCF | 0.0048 ± 0.0005 | 0.4 ± 0.1 c | > 10000 |
| MTDCF | 0.0080 ± 0.0004 | 0.7 ± 0.2 c | > 10000 |
| PrTDCF | 0.067 ± 0.005 | 12 ± 1 | > 10000 |
| PhTDCF | 0.130 ± 0.009 | 60 ± 10 | > 10000 |
The Ki values of PfADA and human ADA are from Tyler et al.16
Ki is an equilibrium dissociation constant. Unlike the action of coformycins with human and P. falciparum ADAs, no slow-onset inhibition was observed for PaMTADA.
The Ki* values from slow-onset inhibition.
Inhibitor specificity of PaMTADA can be compared to that of PfADA since both enzymes have adenosine and MTA deaminase activities. MTCF, MTDCF, PrTDCF, and PhTDCF bind PfADA with respective Ki values of 400 pM, 700 pM, 12 nM, and 60 nM, representing 5, 9, 150, and 750 times weaker binding affinity than that of CF for the same enzyme.16 2′-Deoxycoformycin binds slightly tighter than CF with a Ki of 38 pM. The preference of PfADA for binding of CF and DCF relative to MTCF and other 5′-functionalized-2′-deoxycoformycins are the opposite of that found for PaMTADA. This establishes the distinct substrate specificity preferences for adenosine and MTA with these enzymes. PfADA prefers adenosine and PaMTADA prefers MTA. In addition, MTCF and 5′-functionalized-2′-deoxycoformycins bind PaMTADA more tightly than PfADA, suggested by the > 80-fold lower Ki, which may be attributed to the 160 times higher catalytic efficiency of PaMTADA on MTA than that of PfADA. Binding of transition state analogues is proportional to catalytic rate enhancement, and the behavior of PaMTADA provides another example of this phenomenon. The preference of MTCF and 5′-functionalized-2′-deoxycoformycins as transition state analogue inhibitors of PaMTADA emphasizes substrate specificity as an essential factor in inhibitor design in addition to transition state features.
MTCF and other 5′-functionalized-2′-deoxycoformycins have no inhibitory effects on human ADA, suggesting an approach to target P. aerguinosa with minimal side effects to human hosts. MTCF was examined to test if it might be metabolized by other enzymes related to MTA metabolism, namely by human MTA phosphorylase (MTAP) and E. coli MTA nucleosidase (MTAN). No significant degradation of MTCF was observed for either of these enzymes (kobs < 0.001 s−1).
MTCF was tested for its ability to inhibit human MTAP and E. coli MTAN. MTCF binds to human MTAP 625,000 times weaker than to PaMTADA, with a Ki of 3 μM. The Ki of E. coli MTAN is > 5 μM. These results demonstrate the high specificity of MTCF for MTADA activity with minimal interactions with related enzymes.
Inhibition of cellular PaMTADA activity
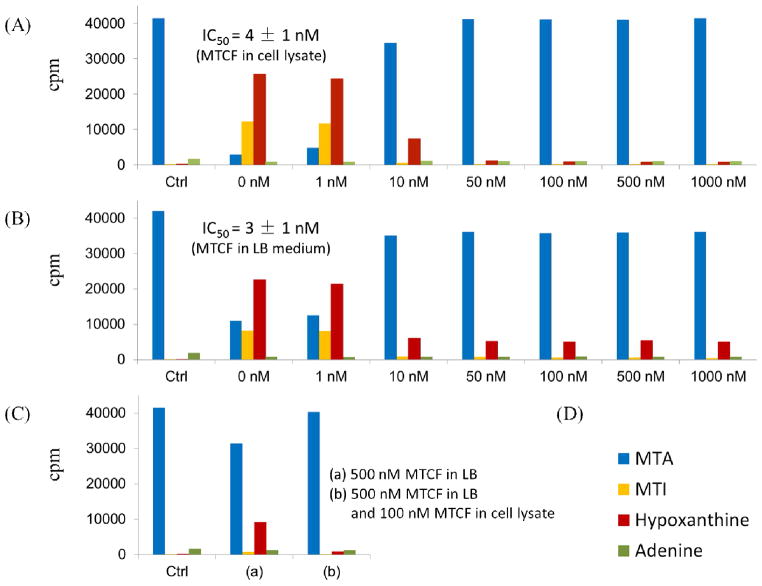
MTCF exhibits tight binding to PaMTADA in enzymatic assays with a 4.8 pM dissociation constant. Its inhibition of PaMTADA was tested in intact P. aerguinosa cells and cell lysate. The inhibition was evaluated by the decrease of MTA degradation. MTA degradation was monitored by tracking the decrease of 14C-label in [8-14C]MTA or its increase in hypoxanthine, adenine, and MTI. Varied concentrations of MTCF were added to P. aeruginosa cell lysates (Fig. 4A). In the absence of MTCF, conversion of [8-14C]MTA to downstream metabolites was nearly complete, indicating that PaMTADA and PaMTIP are functional under these experimental conditions. The degradation of MTA was completely blocked at 50 nM inhibitor, indicating that the deamination of MTA is the only pathway for MTA catabolism in P. aeruginosa extracts. The IC50 was 4 nM, demonstrating the inhibitory potency of MTCF in whole cell lysates.
Figure 4.
Cellular PaMTADA activity and inhibition by MTCF. (A) Effect of MTCF on MTA metabolism in P. aeruginosa cell lysate. (B) Effect of MTCF in P. aeruginosa cell cultures (grown in LB medium). (C) Effect of MTCF in P. aeruginosa cell lysate (grown in LB medium containing MTCF). (D) Color code for the metabolites. The activity of PaMTADA was monitored by the degradation of [8-14C]MTA. Related 14C-metabolites were purified using HPLC and quantitated by scintillation counting. IC50 values were calculated using concentrations of MTCF and the percentage of degraded [8-14C]MTA.
The cellular permeability and in vivo inhibition of PaMTADA by MTCF was examined by the effect of the inhibitor added to LB medium during cell growth (Fig. 4B). We monitored the growth of P. aeruginosa PAO1 with or without MTCF (up to 1 μM) for 36 hours at 37 °C. There was no effect of MTCF on cell growth based on OD600 values. Cells were harvested, wash free of exogenous inhibitor and PaMTADA activity was determined by [8-14C]MTA metabolism in cell extracts. MTA metabolism was clearly inhibited at the PaMTADA step by the growth of cells in the presence of MTCF in LB medium. Maximal inhibition was achieved at a concentration of 10 nM MTCF. However, a small, residual PaMTADA activity was observed independent of the MTCF concentration during growth, even at 1 μM. Two hypotheses were considered for this activity. First, growth on MTCF may induce a deaminase activity resistant to MTCF. Second, the residual activity might arise from diffusional release of the inhibitor during the dilution (and incubation) of a small volume of cell extract into the larger volume of lysis buffer and other incubation buffer. To test these hypotheses, cells were cultured as above in LB medium containing 500 nM MTCF. After preparation of cell extracts at the start of the MTA degradation experiment, either 0 or 100 nM MTCF was added and the degradation of the [8-14C]MTA was monitored (Fig. 4C). If the residual PaMTADA activity arises from a resistant deaminase, the degradation of [8-14C]MTA would be unchanged. If the residual MTA metabolism is due to diffusional loss of inhibitor during extract work-up and incubation, addition of inhibitor in cell lysate would completely quench the residual activity. All PaMTADA activity was inhibited in the experiment. Thus, the residual activity of PaMTADA, clearly present before addition of MTCF to the cell lystate, is a result of slow diffusional loss of inhibitor from the enzyme in cell lysate. Correcting for this residual activity, the IC50 of PaMTADA in LB medium was then calculated to be 3 nM, similar to the 4 nM IC50 of MTCF in cell lysate. These results indicate that MTCF is permeable to P. aeruginosa cells. The cellular PaMTADA concentration can also be estimated in the range of 10 to 50 nM as this concentration of MTCF is required to titrate extracts to zero catalytic activity.
The structure of PaMTADA and MTCF interaction
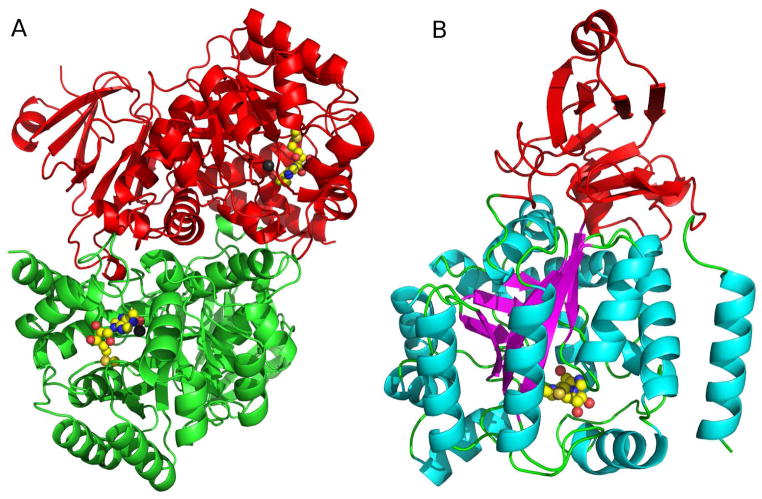
The crystal structure of PaMTADA in complex with MTCF was determined to 2.0 A resolution. PaMTADA forms a homodimer with two zinc ions and two phosphate ions located at the dimer interface (Fig. 5a). The N-terminal 14 amino acids that include the His6 tag, are disordered and are distant from the active site. The protein folds into two domains. The core of the larger domain consists of an (α/β)8 TIM barrel, whereas the smaller domain, including the first 67 amino acid residues and the residues from 361 to 419, is organized into a β sandwich (Fig. 5b).
Figure 5.
The structure of PaMTADA. (A) The homodimeric PaMTADA is shown with one subunit in green and the other in red. The active-site Zn ions and MTCF are drawn as gray spheres and space filling model, respectively. (B) Two distinct domains are present in the PaMTADA monomer. The small domain of PaMTADA is colored in red. The helix, sheet, and loop structures of large domain are colored in cyan, magenta, and green, respectively. MTCF is drawn as a space filling model to show the position of the active site.
The 8-(R)-hydroxydiazepine moiety of MTCF mimics some features of the transition state for N6-deamination of the adenine base. The 8-(R)-hydroxy group at a sp3-bonded center of MTCF mimics the nucleophilic water adding to the sp2 C6 center of the purine ring to create a sp3 transition state. The structure of PaMTADA resembles that of the Plasmodium vivax ADA:MTCF complex reported earlier, where, the N1, N4 and N6 of diazepine ring form hydrogen bonds with surrounding residues or a water molecule (Figs. 6,7).14 The N1 forms hydrogen bonds to the amide of Gly310 and the side chain of Ser313 via a water molecule. The N4 and N6 form hydrogen bonds to side chains of His194 and Glu224, respectively. Both 2′ and 3′ hydroxyl groups of ribose moiety form hydrogen bonds with the side chain of Glu101, consistent with other structures and the predicted sequence analysis (Fig. 6). The 5′-methylthio group resides in a hydrophobic pocket surrounded by Met132, Phe134, Pro155, Leu157, Pro193 and His194 (Fig. 6), explaining the binding advantage afforded by the 5′-methylthio group in MTCF compared to the 5′-hydroxyl group in CF. Transition state features of MTCF include the protonation at N6 and the (R)-hydroxyl with sp3 geometry at C8. The protonated N6 mimics the transition state and forms a hydrogen bond with Glu224 and (R)-hydroxyl group forms an ionic bond with the Zn ion, a mimic of the Zn-OH− nucleophile at the transition state of the normal aromatic nucleophilic substitution reaction. These interactions from the transition state features provide significant binding energy to the inhibitor and thus contribute to the 4.8 pM affinity of MTCF.
Figure 6.
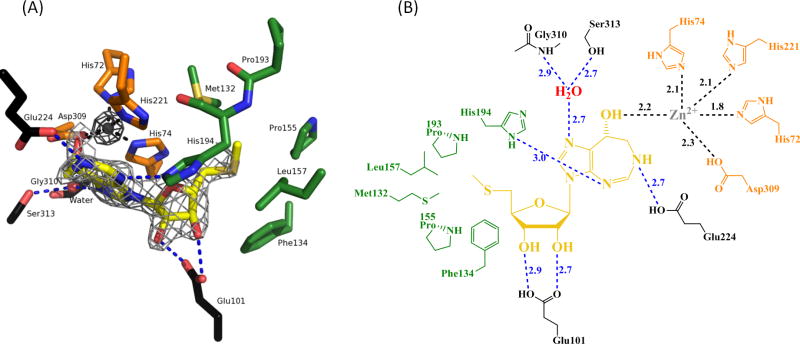
The catalytic site of PaMTADA containing MTCF, Zn ion, and the adjacent amino acid residues. (A) MTCF is in yellow. The Zn ion and water are drawn as gray and red spheres, respectively. The hydrogen bonds between MTCF and surrounding environment are shown as blue dashed lines. The Zn-chelating contacts are shown as black dashed lines. The adjacent amino acid residues chelating Zn, the residues involved in the 5′-hydrophobic interactions of MTCF and other residues hydrogen bonding to MTCF are colored in orange, green and black, respectively. The Zn-omit Fo-Fc density map is shown as a dark gray mesh at a contour level of 20.0 σ. The MTCF-omit Fo-Fc density map is shown as a light gray mesh at a contour level of 5.0 σ. (B) 2D Distance map of the MTADA active site. Color codes are consistent with Fig. 6A. The hydrogen bonds and ionic interactions in the active site are shown as dashed lines. Distances are in angstroms.
Implications for Quorum Sensing
Six transition state analogue inhibitors have been identified for PaMTADA with picomolar dissociation constants. MTCF is the most potent inhibitor with a 4.8 pM Ki in in vitro assays and an IC50 of 3 nM in in vivo studies. It is specific to the MTA deminase activity and has no significant inhibitory effects on human ADA and human MTAP. MTCF is thus a suitable candidate for blocking PaMTADA activity. In P. aeruginosa, MTA degradation follows a unique two-step pathway of MTA → MTI → hypoxanthine, where MTADA is the only enzyme responsible for the first step. Most bacteria utilize MTA nucleosidase for MTA degradation, catalyzing the hydrolysis of MTA to adenine. Inhibition of PaMTADA is expected to increase cellular MTA level and block quorum sensing of P. aeruginosa, similar to the effects of MTAN inhibition in other bacteria. This study assigns the identity of the PA3170 protein as an unusual and specific MTADA and confirms the two-step pathway of MTA metabolism in P. aeruginosa. Transition state analogue inhibitors are identified for PaMTADA with powerful activity and cellular permeability to provide new tools to disrupt QS in P. aeruginosa and other organisms with this unusual pathway.
Acknowledgments
Funding. This work was supported by NIH research grant GM41916.
Structural data for this study were measured at beamline X29A of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
Abbreviations
- MTA
5′-methylthioadenosine
- MTI
5′-methylthioinosine
- QS
quorum sensing
- AHLs
N-acyl-homoserine lactones
- MTAP
MTA phosphorylase
- MTIP
MTI phosphorylase
- MTAN
MTA nucleosidase
- ADA
adenosine deaminase
- MTADA
MTA deaminase
- SAM
S-adenosylmethionine
- CF
coformycin (IUPAC(2R,3R,4S,5R)-2-((R)-8-hydroxy-7,8-dihydroimidazo[4,5-d][1,3]diazepin-3(6H)-yl)-5-(hydro xymethyl)tetrahydrofuran-3,4-diol)
- DCF
2′-deoxycoformycin
- MTCF
5′-methylthiocoformycin
- MTDCF
5′-methylthio-2′-deoxycoformycin
- PrT
5′-propylthio-
- PhT
5′-phenylthio-
Footnotes
Accession Code. The atomic coordinates and structure factors of PaMTADA in complex with MTCF have been deposited in the Protein Date Bank with an accession code (PDB ID) of 4GBD.
References
- 1.Bodey GP, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 2.Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strateva T, Yordanov D. Pseudomonas aeruginosa - a phenomenon of bacterial resistance. J Med Microbiol. 2009;58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 4.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 5.Rumbaugh KP, Griswold JA, Hamood AN. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2000;2:1721–1731. doi: 10.1016/s1286-4579(00)01327-7. [DOI] [PubMed] [Google Scholar]
- 6.Kohler T, Guanella R, Carlet J, van Delden C. Quorum sensing-dependent virulence during Pseudomonas aeruginosa colonisation and pneumonia in mechanically ventilated patients. Thorax. 2010;65:703–710. doi: 10.1136/thx.2009.133082. [DOI] [PubMed] [Google Scholar]
- 7.Smith RS, Iglewski BH. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J Clin Invest. 2003;112:1460–1465. doi: 10.1172/JCI20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams-Ashman HG, Seidenfeld J, Galletti P. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem Pharmacol. 1982;31:277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho MC, Almo SC, Schramm VL. Transition state analogs of 5′-methylthioadenosine nucleosidase disrupt quorum sensing. Nat Chem Biol. 2009;5:251–257. doi: 10.1038/nchembio.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckoreelall K, Wilson L, Parker WB. Identification and characterization of two adenosine phosphorylase activities in Mycobacterium smegmatis. J Bacteriol. 2011;193:5668–5674. doi: 10.1128/JB.05394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan R, Ho MC, Almo SC, Schramm VL. Methylthioinosine phosphorylase from Pseudomonas aeruginosa. Structure and annotation of a novel enzyme in quorum sensing. Biochemistry. 2011;50:1247–1254. doi: 10.1021/bi101642d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci U S A. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ting LM, Shi W, Lewandowicz A, Singh V, Mwakingwe A, Birck MR, Ringia EAT, Bench G, Madrid DC, Tyler PC, Evans GB, Furneaux RH, Schramm VL, Kim K. Targeting a Novel Plasmodium falciparum Purine Recycling Pathway with Specific Immucillins. J Biol Chem. 2005;280:9547–9554. doi: 10.1074/jbc.M412693200. [DOI] [PubMed] [Google Scholar]
- 14.Ho M-C, Cassera MB, Madrid DC, Ting L-M, Tyler PC, Kim K, Almo SC, Schramm VL. Structural and Metabolic Specificity of Methylthiocoformycin for Malarial Adenosine Deaminases. Biochemistry. 2009;48:9618–9626. doi: 10.1021/bi9012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–U772. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyler PC, Taylor EA, Froehlich RFG, Schramm VL. Synthesis of 5′-Methylthio Coformycins: Specific Inhibitors for Malarial Adenosine Deaminase. J Am Chem Soc. 2007;129:6872–6879. doi: 10.1021/ja0708363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh V, Lee JE, Nunez S, Howell PL, Schramm VL. Transition state structure of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli and its similarity to transition state analogues. Biochemistry. 2005;44:11647–11659. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- 18.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39:D596–600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zappia V, Galletti P, Carteni-Farina M, Servillo L. A coupled spectrophotometric enzyme assay for methyltransferases. Anal Biochem. 1974;58:130–138. doi: 10.1016/0003-2697(74)90449-7. [DOI] [PubMed] [Google Scholar]
- 20.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Zielke CL, Suelter CH. Purine, purine nucleoside, and purine nucleotide aminohydrolases. Enzymes. (3) 1971;4:47–78. [Google Scholar]
- 22.Kung PP, Zehnder LR, Meng JJ, Kupchinsky SW, Skalitzky DJ, Johnson MC, Maegley KA, Ekker A, Kuhn LA, Rose PW, Bloom LA. Design, synthesis, and biological evaluation of novel human 5′-deoxy-5′-methylthioadenosine phosphorylase (MTAP) substrates. Bioorg Med Chem Lett. 2005;15:2829–2833. doi: 10.1016/j.bmcl.2005.03.107. [DOI] [PubMed] [Google Scholar]
- 23.Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 27.Agarwal RP. Inhibitors of adenosine deaminase. Pharmacol Ther. 1982;17:399–429. doi: 10.1016/0163-7258(82)90023-7. [DOI] [PubMed] [Google Scholar]
- 28.Luo M, Singh V, Taylor EA, Schramm VL. Transition-State Variation in Human, Bovine, and Plasmodium falciparum Adenosine Deaminases. J Am Chem Soc. 2007;129:8008–8017. doi: 10.1021/ja072122y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margolis J, Grever MR. Pentostatin (Nipent): a review of potential toxicity and its management. Semin Oncol. 2000;27:9–14. [PubMed] [Google Scholar]
- 30.Johnson SA. Clinical pharmacokinetics of nucleoside analogues: focus on haematological malignancies. Clin Pharmacokinet. 2000;39:5–26. doi: 10.2165/00003088-200039010-00002. [DOI] [PubMed] [Google Scholar]
- 31.Cristalli G, Costanzi S, Lambertucci C, Lupidi G, Vittori S, Volpini R, Camaioni E. Adenosine deaminase: functional implications and different classes of inhibitors. Med Res Rev. 2001;21:105–128. doi: 10.1002/1098-1128(200103)21:2<105::aid-med1002>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]