Abstract
Uridylyltransferase/Uridylyl-Removing Enzyme (UTase/UR) catalyzes uridylylation of PII and deuridylylation of PII-UMP, with both activities regulated by glutamine. In a reconstituted UTase/UR-PII cycle containing wild-type UTase/UR, the steady-state modification of PII varied from nearly complete modification to nearly complete demodification as glutamine was varied, whether PII was saturating or unsaturating. But when a his-tagged version of UTase/UR was used, robustness to variations in PII concentration was lost, and the range of PII modification states in response to glutamine became smaller as PII concentration increased. The presence of the his-tag on UTase/UR did not alter PII substrate inhibition of the UT activity and had little effect on the level of the UT activity, but resulted in a slight defect in the UR activity. Importantly, at high PII concentration, glutamine inhibition of the UT activity was incomplete. We hypothesized that PII binding to the UR active site in the HD domain was responsible for PII substrate inhibition of the UT activity and, in the his-tagged enzyme, also diminished glutamine inhibition of the UT activity. Consistent with this, three different UTase/UR proteins with HD domain alterations lacked substrate inhibition of the UT activity by PII; in one case the HD alteration eliminated glutamine regulation of the UT activity, while for the other two proteins, alterations of the HD domain partially compensated for the effect of the his-tag in restoring glutamine regulation of the UT activity. We conclude that very strong inhibition of the UT activity was required for the UTase/UR-PII cycle to display robustness to the PII concentration, that in the wild-type enzyme PII brings about substrate inhibition of the UT activity by binding to the HD domain of the enzyme, and that addition of an N-terminal His-tag resulted in an altered enzyme with subtle changes in the interactions between domains such that PII binding to the HD domain interfered with glutamine regulation of the UT domain.
Keywords: Robustness, covalent modification cycle, substrate inhibition, signal transduction
Biological signal transduction systems must produce an accurate output signal in response to an input stimulation in the heterogeneous and stochastic environment of the cell, where variations are routinely experienced in the concentrations of proteins and small molecules that comprise the system as well as in the concentrations of the proteins and small molecules external to the system. In addition to fluctuations in concentration, the enzymatic activities of the proteins of a system may experience fluctuations due to genetic mutations, regulatory covalent modifications, or alternative cellular localizations. Robustness is becoming recognized as an important property of biological systems and in particular biological signal transduction systems; robustness is defined as the property that allows a system to maintain its functions in the face of external and internal perturbations (1, 2). The property of robustness always pertains to specific parameters of the system, and system function may be highly robust to changes in certain parameters while remaining fragile to changes in other parameters. For a signaling system, physiologically-important parameters that affect the process of producing an output signal in response to stimulation are the concentrations of proteins and small molecules that comprise the system, and the activities and regulatory properties of the proteins. Some cellular signaling systems are experimentally demonstrated to display robustness to some or all of these parameters (3, 4), and theoretical work argues that such robustness may be a general property of cellular signaling systems, and in particular the many systems that must function over a broad range of conditions (5). However, some systems may be specifically evolved to limit robustness to variation of a parameter, to allow that parameter to control shifting the system between regulatory regimes. The opposite of a robust system is a fine-tuned one, where the output of the system in response to stimulation depends upon the values of the parameters of the system. Of course, "robustness" and "fine-tuning" are human concepts, and in nature we expect systems to have intermediate properties. For example, system function may tolerate variations of parameters within some limits, but be unable to tolerate extreme values such as null.
A common motif of signal transduction systems is the covalent modification cycle, in which the substrate protein of the cycle is subjected to reversible covalent modification that controls its activities. The enzymes that catalyze the modification and demodification of this substrate protein, referred to as the converter enzymes of the cycle, produce an output signal (the level of the modified substrate protein) in response to a stimulation that regulates one or both converter enzymes. Given the ubiquitous occurrence of covalent modification cycles in nature and their importance in central physiological processes, considerable effort has been focused on theoretical and experimental studies of the signal processing properties of such systems, including studies of signal amplification (6), noise filtering (7), and factors affecting sensitivity (8–10). In this paper, we will demonstrate how a kinetic parameter of the converter enzymes, specifically the effectiveness of the inhibition of one of the converter enzymes, eliminated the robustness of a reconstituted covalent modification cycle towards the concentration of its substrate protein. Furthermore, we were able to document the regulatory catastrophe as the cycle substrate protein concentration was increased beyond the range where effective signaling occurred, provisionally providing a diagnostic phenotype for the loss of robustness.
The PII-UTase/UR covalent modification cycle is part of two bi-cyclic cascade systems in E. coli, that participate in the regulation of nitrogen assimilation (reviewed in ref. 11). The PII-UTase/UR-ATase-GS cascade controls the activity of glutamine synthetase (GS) by reversible covalent adenylylation, while the PII-UTase/UR-NRII-NRI cascade controls the phosphorylation state of the enhancer-binding transcription factor NRI (NtrC), and by so doing, regulates the initiation of transcription of nitrogen-regulated genes. In both cascades, the role of the PII-UTase/UR cycle is to communicate the intracellular concentration of glutamine, sensed by UTase/UR, via changes in the uridylylation state of PII. Prior studies of the PII-UTase/UR cycle have revealed the kinetic mechanisms of the UTase and UR activities, elucidated specificity and inhibition constants, established the kinetic mechanism for inhibition by glutamine, and localized the UTase, UR, and glutamine-binding activities to specific domains of the protein (12, 13). Our initial observation of the loss of robustness due to alteration of a kinetic parameter of a converter enzyme was quite fortuitous. We examined a his-tagged version of the bifunctional UTase/UR enzyme, and were surprised to observe that it had a dramatically different behavior than did the untagged protein: the his-tagged converter enzyme only functioned effectively in a reconstituted covalent modification cycle when the PII substrate protein of the cycle was at very low concentration, but not when PII was at high concentration. Comparison of the his-tagged and the wild-type UTase/UR then revealed which kinetic parameter was responsible for robustness of the system to changes in the concentration of its substrate protein. Further studies using his-tagged enzymes with additional alterations then allowed us to present and test an hypothesis for how the activities of the wild-type enzyme were regulated by the stimulatory effector and how this regulation was defective in the his-tagged enzyme.
MATERIALS AND METHODS
Purified Proteins
The preparations of PII, wild-type UTase/UR, and the his-tagged UTase/UR prepared from strain UQ5516 described previously were used (13–15). A second his-tagged but otherwise wild-type UTase/UR enzyme preparation was obtained (from strain SA1) by metal chelate chromatography as described (13), followed by fractionation on a 300 mL Biogel A1.5 M gel filtration column equilibrated in 50 mM Tris-Cl, pH 7.5, 0.1 % EDTA, 10% (v/v) glycerol. The purified enzyme was dialyzed into storage buffer that was the same as the chromatography buffer, except 50% (v/v) glycerol. For clarity, we will distinguish the two different preparations of the his-tagged UTase/UR by referring to the strain from which the enzyme was prepared (UQ5516 or SA1). In Fig S1, the appearance after SDS-PAGE of the wild-type UTase/UR and each of the his-tagged but otherwise wild-type enzymes is shown. Each of these enzymes is approximately 90% pure as judged by visual inspection of the gels. Importantly, the purified enzymes do not appear to be contaminated with ATPases or other activities that interfere with the UT or UR assays (see below). The his-tagged enzymes with alterations in the HD domain (HD-AA [H514A and D515A, from strain UQ5628], HD-QN [H514Q D515N, from strain UQ5629], and D-HD [Δ-A510-D531, from strain UQ5627] were also described previously (13).
Construction and purification of the D107N altered form of UTase/UR
EcoRI and NdeI restriction sites were introduced upstream and a BamHI site downstream of the glnD gene by PCR of the plasmid pDOP (15), using the upstream primer CCCGAATTCATATGAATACCC TTCCAGAACAGTAC and downstream primer GGAATTCGGATCCCTGACGTACCGCCG CTGGTGGCCA. The amplified glnD gene was cloned into pSelect (Promega), forming pSelect-glnD, and this plasmid was mutagenized with the following oligonucleotide: GACGTCAATTTACTGATTTTAAGCCG. After mutagenesis, the ClaI/NsiI fragment of the mutagenized gene was swapped for the corresponding wild-type fragment in plasmid pglnD9 (15), and then the whole of the mutated glnD gene was cloned as an NdeI/EcoRI fragment into NdeI/EcoRI-cleaved pJLA503 (16), resulting in pDOP-D107N. The altered UTase/UR-D107N protein was purified using the same methods that were used for the wild-type UTase/UR (15).
Reconstituted UTase/UR-PII Monocycle
The steady state levels of PII uridylylation at various glutamine concentrations were measured as described previously (14). Briefly, reaction conditions were 100 mM Tris-Cl, pH 7.5, 25 mM MgCl2, 100 mM KCl, 0.3 mg/mL bovine serum albumin, 1 mM DTT, 0.5 mM ATP, 0.2 mM α-ketoglutarate, 0.5 mM [α-32P]-UTP, and with PII and UTase/UR as indicated. Components except ATP and UTP were combined and pre-warmed at 30 °C for 2 min, and reactions were started by addition of a pre-warmed mixture containing the ATP and UTP. Samples were removed at various times and spotted onto Whatman P81 phosphocellulose filters, which were washed in 5% TCA, dried, and counted by liquid scintillation spectroscopy. Where indicated, AMP-PNP was used in place of ATP. For determination of steady state values, long time courses (generally 90 min) were used, and steady states were estimated by averaging values at the latter time samples (generally four samples removed between 30 and 90 min of incubation), when the level of PII uridylylation had achieved a constant value. The steady states observed in this work were quite stable. This indicated that the purified proteins were not contaminated by cellular ATPase activity, as depletion of ATP from the reaction mixtures would result in changing levels of PII uridylylation (23).
Measurement of the UT activity
The initial rate of PII uridylylation was measured as before (14), conditions were 100 mM Tris-Cl, pH 7.5, 25 mM MgCl2, 100 mM KCl, 0.3 mg/mL bovine serum albumin, 0.5 mM ATP or AMP-PNP, as indicated, UMP as indicated, and 0.5 mM α-[32P]-UTP. Reaction mixtures lacking ATP (or AMP-PNP) and UTP were incubated for 2 min at 30 °C, and the uridylylation reactions were started by addition of a pre-warmed mixture of the ATP (or AMP-PNP) and UTP. Samples were removed at various times and spotted onto Whatman P81 phosphocellulose filters, which were washed in 5% TCA, dried, and counted by liquid scintillation spectroscopy. The UT catalytic rate was observed to be almost independent of the enzyme concentration, when the enzyme was varied from 0.01 µM to 1.0 µM, as long as PII was saturating (Fig S2); all of the experiments in this paper were performed within this range of enzyme concentrations. The assay is accurate because the product can be made highly radioactive and is easily meaured when only a tiny amount of the substrate has been converted, allowing good estimation of initial rates.
Measurement of the UR activity
PII-[32P]-UMP was prepared as described previously (14); briefly this involved extended incubation of PII with UTase/UR in the absence of glutamine followed by brief heating at 60 °C to inactivate the UTase/UR. The initial rate of deuridylylation of PII-UMP was examined at 30 °C as before (14), in reaction mixtures that contained 100 mM Tris-Cl, pH 7.5, 25 mM MgCl2, 100 mM KCl, 0.3 mg/mL bovine serum albumin, 1 mM α-ketoglutarate or as indicated, 0.5 mM ATP or AMP-PNP, as indicated, and with glutamine and PII-UMP as indicated. Reaction mixtures were incubated in the absence of PII-UMP for 2 min, and initiated by addition of pre-warmed PII-UMP. Samples were removed at various times and spotted onto Whatman P81 phosphocellulose filters, which were washed in 5% TCA, dried, and counted by liquid scintillation spectroscopy.
The catalytic rates measured in the standard UR assay were observed to depend upon the enzyme concentration; the higher the enzyme concentration, the lower the apparent UR kcat in the assay (Fig S2). Furthermore, the assay was variable from day to day, as it was very difficult to provide the substrate PII-UMP at identical concentration and modification state. Because of this, meaningful comparisons could only be made in experiments where UR rates were measured side-by-side at the same enzyme concentration, as we did in this study. This deficiency in the assay is likely due to underestimation of the inital reaction rates; the assay is based upon watching the labeled substrate become unlabeled and consequently a significant fraction of the substrate is converted in the assay.
In order to attempt to obtain more accurate initial rates in the UR assay, we used a thin-layer chromatography method to separate PII-UMP from UMP. This procedure allowed us to measure the product of the UR reaction (UMP) as opposed to simply measuring dissapearance of the PII-UMP substrate in the standard UR assay method. For the TLC-based assay method, reaction conditions were as in the standard UR assay, and 4 µL samples were removed at various times and immediately mixed with 1 µM of 0.5 mM EDTA to stop the reaction. After all samples had been collected, 1 µL aliquots were spotted onto Cellulose PEI thin layer chromatography plates (J. T. Baker, Inc), and plates were developed using 0.2M KPi, pH 8.0, as the solvent. [Prior to spotting the samples, the positions where samples would be spotted were marked lightly with a pencil, and each plate was chromatographed in water, dried briefly in air, and spotted with 1 µL of a mixture of UTP, UDP, and UMP (30 mM each) which served as a carrier and to indicate the position of UMP.] After development, plates were dried in air, visualized under hand-held UV light, and the positions of the UMP spots was marked with a pencil. A typical chromatogram that had been marked with a pencil to indicate the nucleotide spots and then subjected to autoradiography is shown in Fig S3. The origin (containing PII-UMP) and the UMP spots were cut from the plates, and the slices were counted by liquid scintillation counting. The fraction of counts appearing in the UMP spot relative to the origin spot was used to calculate the concentration of UMP. This assay method had the advantage that the product of the UR reaction was measured directly, and therefore it was possible to determine initial rates under conditions where only a small fraction of the initial substrate had been converted. Higher initial rates of catalysis were observed using this method than in the standard assay procedure, which we attribute to better estimation of the initial reaction velocities (Fig S2). This TLC-based assay method also showed a dependence of UR catalytic rate on enzyme concentration; while similar kcat were obtained at 0.01 µM enzyme and 0.1 µM enzyme, much lower kcat were obtained than were expected when the enzyme was 1 µM (Fig S2C). Consequently, this UR assay method was also only suitable for side-by-side comparisons of different enzyme samples at the identical enzyme concentration.
RESULTS
The UTase/UR-PII covalent modification cycle is robust to changes in the PII concentration
Using purified proteins, we studied the responses of the UTase/UR-PII cycle to lutamine in systems that contained different concentrations of PII and UTase/UR. We show elsewhere that good glutamine-signaling properties were obtained when PII was used at 100 µM, the highest concentration we were able to provide, and that fairly low enzyme concentrations did not prevent effective signaling, although when the enzyme was low relative to PII, the reactions had to be incubated for a very long time to obtain the steady state level of PII uridylylation (17). We will also show elsewhere that variation of PII and enzyme concentration alters the sensitivity of responses and the midpoint of responses (17); such effects are well known in theory (8–10, 18). For the purposes of the current study, we focus on a fairly narrow range of PII concentrations where the wild-type system displayed excellent responsiveness to glutamine (Fig 1A). PII is a homotrimeric protein that can be reversibly modified at a unique site (Y51) on each subunit, such that its modification state can range from zero to three modifications/trimer. When PII was at 36 µM and UTase/UR was at 1.2 µM, PII modification state went from 2.99 uridylyl groups in the absence of glutamine to 0.26 uridylyl groups/trimer in the presence of 10 mM glutamine (Fig 1A); therefore, the range of modification states sampled during this transition was ~ 2.73 out of a possible range of 3. This range was highly reproducible; in four additional repeats of the experiment, the range of PII modification states varied from 2.69–2.73. Since PII is a homotrimer and the UTase/UR is a monomer, at this ratio of substrate to enzyme there were 90 PII modification sites for every converter enzyme active site. The range of PII modification states was only slightly diminished when PII was at the low concentration of 0.5 µM and the UTase/UR was present at 0.02 µM, in which case the range of uridylylation states was ~2.59 (Fig 1A). At this ratio of substrate to enzyme, there were 75 substrate sites per converter enzyme active site. In four repeats of the experiment at these conditions, and in one additional experiment where PII was at 0.5 µM and the UTase/UR was present at 0.05 µM, the range of uridylylation states was again found to be quite consistent, varying between 2.53 and 2.61. The two conditions noted so far were chosen to ensure that both ultrasensitive and hyperbolic regimes were sampled (Fig 1A), and the variation of conditions discernibly shifted the midpoint of the response, as expected (17). These differences notwithstanding, a wide range of steady-state uridylylation states was consistently obtained in response to changes in the glutamine concentration, regardless of whether the PII concentration was high or low.
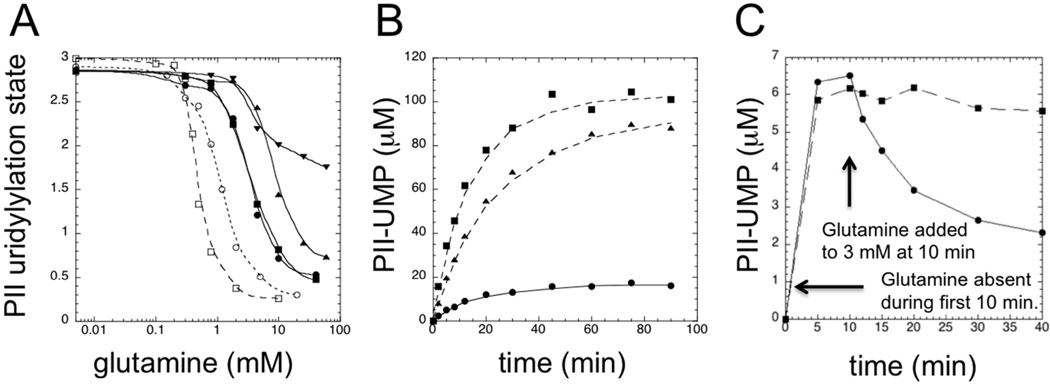
Figure 1. A his-tagged version of the UTase/UR was defective in steady-state glutamine signaling in reconstituted PII-UTase/UR covalent modification cycles.
A. Steady state glutamine responses of reconstituted cycles. Experiments were conducted as in Materials and Methods, and the steady-state levels of PII modification at various glutamine concentrations are shown, stated in terms of modified subunits/tetramer. Symbols: □, PII at 36 µM and wt UTase/UR at 1.2 µM; ○, PII at 0.5 µM and wt UTase/UR at 0.02 µM; ▼, PII at 36 µM and his-tagged UTase/UR (UQ5516) at 1.2 µM; ▲, PII at 3 µM and his-tagged UTase/UR (UQ5516) at 0.1 µM; ■, PII at 0.5 µM and his-tagged UTase/UR (UQ5516) at 0.017 µM; ●, PII at 0.2 µM and his-tagged UTase/UR (UQ5516) at 0.0067 µM. B. Approach to the steady state in reconstituted systems containing 10 mM glutamine. All systems contained 36 µM PII and 1.2 µM enzyme. Symbols: ●, wt UTase/UR; ▲, his-tagged UTase/UR (UQ5516); ■, his-tagged UTase/UR (SA1). C. Response of reconstituted covalent modification cycles to the addition of glutamine. Systems contained 3 µM PII and 0.2 µM enzyme. For the first 10 min, systems were incubated in the absence of glutamine, after which glutamine was added to a final concentration of 3 mM. Symbols: ●, wt UTase/UR; ■, his-tagged UTase/UR (SA1).
A his-tagged version of the UTase/UR displayed an altered response to glutamine in reconstituted monocycles
We examined two different preparations of a his-tagged version of UTase/UR that were produced by cloning the glnD structural gene into the common expression vector pET15b (13). The two preparations of the enzyme were made in our two laboratories, using slightly different procedures (MATERIALS AND METHODS). For our studies, we conducted experiments shown here with both enzyme preparations, which behaved the same and are referred to by the strain that served as the source of the enzyme: UQ5516 (Wisconsin) and SA1 (Michigan). The altered form of the enzyme resulting from expression from pET15b contains the sequence : met-gly-ser-ser-his-his-his-his-his-his-ser-ser-gly-leu-val-pro-arg-gly-ser-his added to the N-terminus of the protein. The his-tagged UTase/UR (UQ5516), when used at 1.2 µM, provided a very shallow response to glutamine when PII was at 36 µM, and the midpoint of the response was shifted to significantly higher glutamine concentration, relative to the results obtained with the wild-type enzyme under the same conditions (Fig1A). In this experiment, a narrow range of uridylylation states (~ 1.1) signaled glutamine concentration, and this narrow range was biased towards high uridylylation states. Using the his-tagged UTase/UR UQ5516 enzyme preparation, we also examined three other conditions where the ratio of PII modification sites to enzyme active sites was also 90:1; specifically, systems where enzyme was at 0.1 µM and where PII was at 3 µM, where the enzyme was 0.017 µM and PII was 0.5 µM, and where enzyme was at 0.0067 µM and PII was at 0.2 µM (Fig 1A). In the system where PII was at 3 µM, a wider range of uridylylation states signaled glutamine concentration (~ 2.1) than when PII was at 36 µM, and when PII was at 0.5 µM and 0.2 µM a still wider range of uridylylation states signaled the glutamine concentration (~ 2.4), almost equal to the range obtained when the wild-type UTase/UR (lacking his-tag) was used (Fig 1A). These results suggested that the absolute PII concentration was the important parameter controlling the range of uridylylation states obtained, and not the ratio of PII to the enzyme (which was fixed at 90:1). In addition to affecting the range of uridylylation states obtained in response to glutamine variation, the PII concentration also controlled the midpoint of the glutamine response (Fig 1A).
The experiments presented in Fig 1A using wild-type and his-tagged UTase/UR were performed on different occasions, and because of the complexity of the experiments it is highly desirable to have side-by-side comparisons performed under identical conditions. In Fig 1B we show the approach to the steady state in side-by-side experiments for systems containing 10 mM glutamine, 36 µM PII, and 1.2 µM enzyme (90:1 ratio of PII modification sites to UT and UR active sites). When the wild-type enzyme was used, a low PII uridylylation state was obtained, whereas when the his-tagged enzymes were used, a high PII uridylylation state was obtained (Fig 1B). The his-tagged UTase/UR prepared from strain SA1 resulted in slightly higher PII uridylylation states than did the his-tagged UTase/UR prepared from strain UQ5516, and this behavior was consistently obtained, as shown in the experiments to follow and numerous additional experiments. Differences of this magnitude were also obtained in activity measurements of wild-type UTase/UR preparations made on different occasions, and probably reflect differences in loss of activity during purification (Fig S4). The high steady-state levels of PII uridylylation obtained with the his-tagged enzymes in the presence of 10 mM glutamine are consistent with the results from Fig 1A.
In another experiment using reconstituted covalent modification cycles, we explored the effect of allowing the systems to reach the steady state in the absence of glutamine, such that PII was highly uridylylated, and then adding glutamine to 3 mM, a concentration expected to result in an intermediate level of PII uridylylation (Fig 1B). For these experiments, the trimeric PII was at 3 µM and the enzyme was at 0.2 µM, such that the ratio of substrate to catalytic sites was 45:1. When the system contained the wild-type UTase/UR, addition of glutamine resulted in an immediate decrease in the level of PII uridylylation and the system approached the steady state characteristic of the final glutamine concentration and conditions (Fig 1C). This experiment was repeated with three independent preparations of the wild-type UTase/UR with similar results (Fig S4B). By contrast, when the system contained the his-tagged UTase/UR (SA1), glutamine addition was essentially without effect and the PII uridylylation state remained high (Fig 1C). This experiment was repeated on another occasion with similar result. Together, the experiments in Fig 1 indicated that the his-tagged enzymes displayed a severe defect in glutamine signaling when used in reconstituted covalent modification cycles, particularly when PII was at the relatively high concentrations of 3 µM or 36 µM.
Additional side-by-side experiments were used to compare the wild-type and his-tagged enzymes in reconstituted covalent modification cycles at different PII and enzyme concentrations, and in the presence or absence of 10 mM glutamine. In Fig 2 we show a series of experiments in which the wild-type enzyme and the his-tagged (UQ5516) enzyme were examined at a fixed ratio of PII subunits to enzyme active sites of 90:1. Although for clarity the data are presented in separate panels for the wild-type enzyme (Fig 2A) and the his-tagged (UQ5516) enzyme (Fig 2B), the experiments were performed side-by-side for both enzymes for each PII concentration; in addition all experiments with the exception of those using 3 µM PII were also performed side-by-side with the SA1 his-tagged preparation, and those data are not shown only because they are quite similar to the data for the hig-tagged enzyme prepared from UQ5516. As shown in Fig 2A, the wild-type system was robust to changes in the PII concentration when the ratio of substrate to enzyme was held constant. By contrast, the system containing the his-tagged enzyme was not robust to PII concentration over the same range (Fig 2B), even though the ratio of substrate to enzyme was held constant. At high PII concentrations, the systems with the his-tagged enzymes were unable to maintain a low level of PII modification in the presence of 10 mM glutamine (shown for UQ5516 in Fig 2B). Another consistent, but less dramatic, result was that for all three enzyme samples (wild type, UQ5516, and SA1), a slightly higher PII uridylylation state was obtained in the presence of glutamine when PII was 0.2 µM, relative to that obtained when PII was at 0.5 µM (shown for wild-type and his-tagged UTase/UR (UQ5516) in Fig 2).
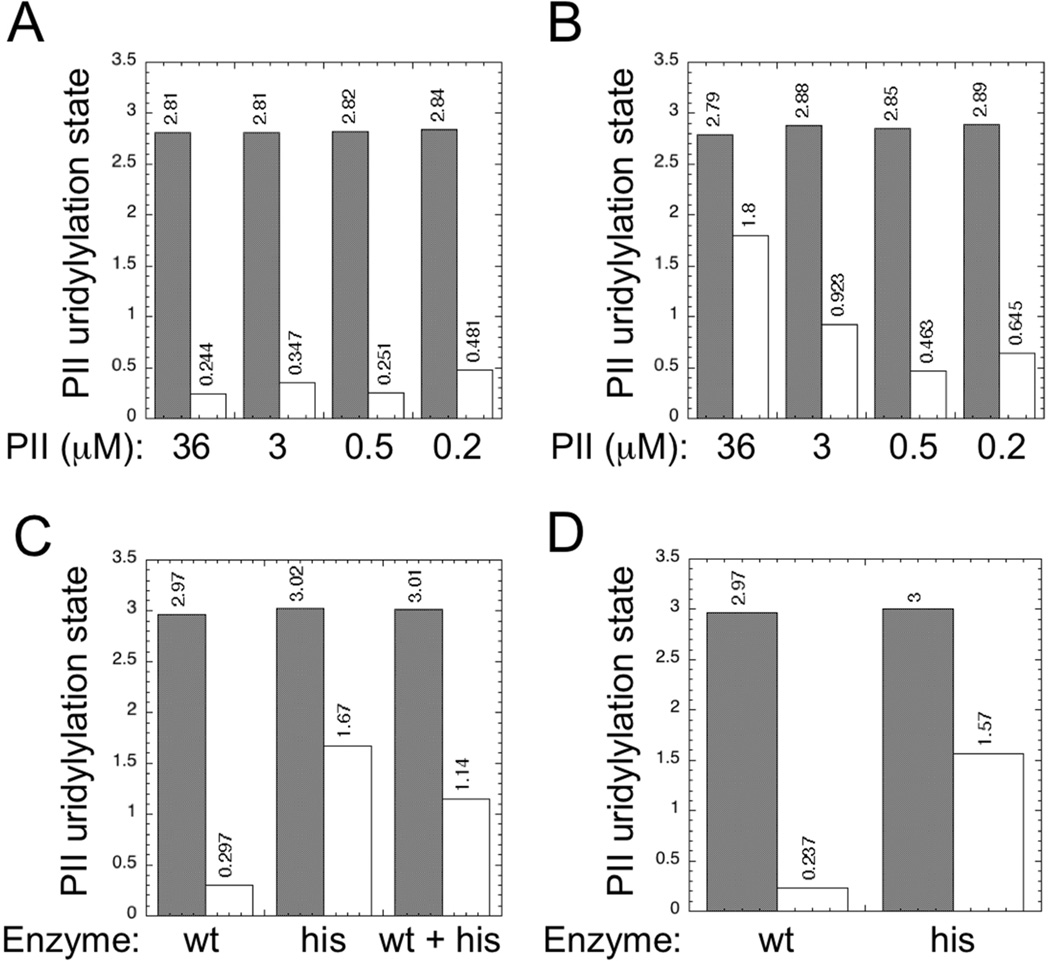
Figure 2. Comparison of steady-state PII modification levels in reconstituted covalent modification cycles containing wild-type or his-tagged UTase/UR.
Results of experiments where glutamine was absent are shown with filled bars; results of experiments where glutamine was at 10 mM are shown with unfilled bars. A. Results of system containing wild-type UTase/UR. B. Results of systems containing his-tagged UQ5516 UTase/UR. Experiments for panels A and B for each concentration of PII were performed side-by-side as described in the text. To maintain a fixed ratio of PII to catalytic sites in the set of experiments, when PII was at 36 µM, UTase/UR was at 1.2 µM; when PII was at 3 µM, UTase/UR was at 0.1 µM; when PII was at 0.5 µM, UTase/UR was at 0.017 µM; and when PII was at 0.2 µM, UTase/UR was at 0.0067 µM. C. Result of combining wild-type and his-tagged enzymes. PII was at 36 µM, and wt and/or his-tagged enzymes were present at 1.2 µM each, as indicated. D. Steady state levels of PII uridylylation in systems where AMP-PNP was present instead of ATP. Experiments were as in panel A and panel B where PII was at 36 µM and UTase/UR was at 1.2 µM, except that ATP was replaced by 0.5 µM AMP-PNP.
In another side-by-side comparison, we examined systems that contained 36 µM PII and 1.2 µM wild-type or his-tagged (UQ5516) enzyme, or a combination of both enzymes at 1.2 µM each, +/− 10 mM glutamine (Fig 2C). Again, the wild-type system displayed an excellent response to glutamine, whereas the system containing the his-tagged (UQ5516) enzyme displayed elevated levels of PII modification at 10 mM glutamine. The system containing both enzymes produced a level of PII-UMP at 10 mM glutamine that was intermediate between the levels obtained with either of the two enzymes. This suggested that neither enzyme preparation contained an activator or inhibitor but rather that the enzyme catalytic rates were balanced against one another.
The results described so far could have been explained by a deficiency in the UR activity of the his-tagged enzyme, by a defect in activation of the UR activity by glutamine, or by a defect in the ability of glutamine to inhibit the UT activity of the enzyme. Such defects could result in elevated PII modification states in the presence of glutamine. However, the mechanism to explain this defect must also account for its dependence on the PII concentration at fixed ratio of PII to enzyme.
The two activities of the UTase/UR enzyme are of disproportionate strength; the UT activity has a kcat of ~ 144/min in the absence of glutamine, while the UR activity only displays a kcat of ~ 6/min in the presence of 10 mM glutamine, when measured in our standard assay (12). Furthermore, the UR activity has a basal kcat of ~ 2/min in the absence of glutamine, such that it is only regulated about 3-fold by glutamine (12). By contrast, the UT activity is strongly inhibited by glutamine (12); we show later in this paper that under the conditions of the experiments performed here we obtained about 100-fold inhibition. In the course of our work, we observed that the UR activity of UTase/UR was increased when ATP in the reaction mixtures was replaced by AMP-PNP (Fig S5). (The adenylylate nucleotide in the reaction mixtures is a ligand of PII (19), and is required for PII interaction with UTase/UR). Also, inhibition of the UT activity by glutamine was normal when AMP-PNP replaced ATP in the reaction mixtures (Fig S6). Therefore, by replacing ATP with AMP-PNP, we could modestly elevate the UR activity, while retaining good regulation of the UT activity of the systems. We examined reconstituted UTase/UR-PII monocycles containing AMP-PNP in place of ATP, again using PII at 36 µM and enzyme at 1.2 µM so that the substrate/enzyme ratio was again 90:1 (Fig 2D). Under these conditions, we obtained results that were quite similar to those obtained when the systems contained ATP; that is, the system containing the wild-type enzyme displayed good regulation by glutamine, whereas the system containing the his-tagged (UQ5516) enzyme had elevated levels of PII modification at 10 mM glutamine (Fig 2D). The his-tagged enzyme prepared from strain SA1 was also examined side-by-side in this experiment, and the results were essentially the same as for the enzyme purified from strain UQ5516 (not shown). These results showed that a modest increase in the UR activity had little effect in systems containing either the wild-type or his-tagged enzymes, and provided a clue that a major factor in controlling PII modification state was the inhibition of the powerful UT activity by glutamine.
A combination of his-tagged UTase/UR and an altered form of the enzyme displaying only UR activity effectively regulated PII uridylylation state in response to glutamine
Our conclusion from the experiment shown in Fig 2D, above, was that a modest increase in the UR activity (caused by inclusion of AMP-PNP in the reaction mixtures) was not sufficient to allow changes in the glutamine concentration to bring about broad changes in the steady-state levels of PII modification when the enzyme bore a his-tag. We next examined whether a larger increase in the level of UR activity in the reaction mixtures might allow effective glutamine signaling. To test this, we used an altered form of UTase/UR containing the D107N alteration, constructed and purified as described in MATERIALS AND METHODS. This altered enzyme is similar to a previously described altered versions of the UTase/UR containing different substitutions of D107 (13), except that it does not contain a his-tag. The D107 residue is one of the two critical Mg2+-chelating aspartate residues of the conserved NT domain, and the purified D107N enzyme did not display measurable UT activity. We will characterize the UR activity of this protein in later sections of this report; as we will show, this activity and its regulation by glutamine was nearly the same as for the wild-type enzyme.
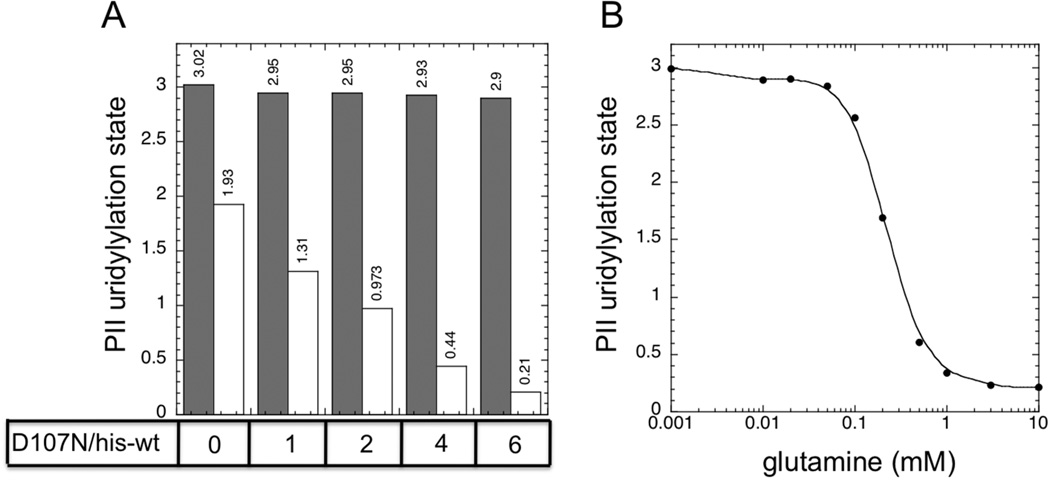
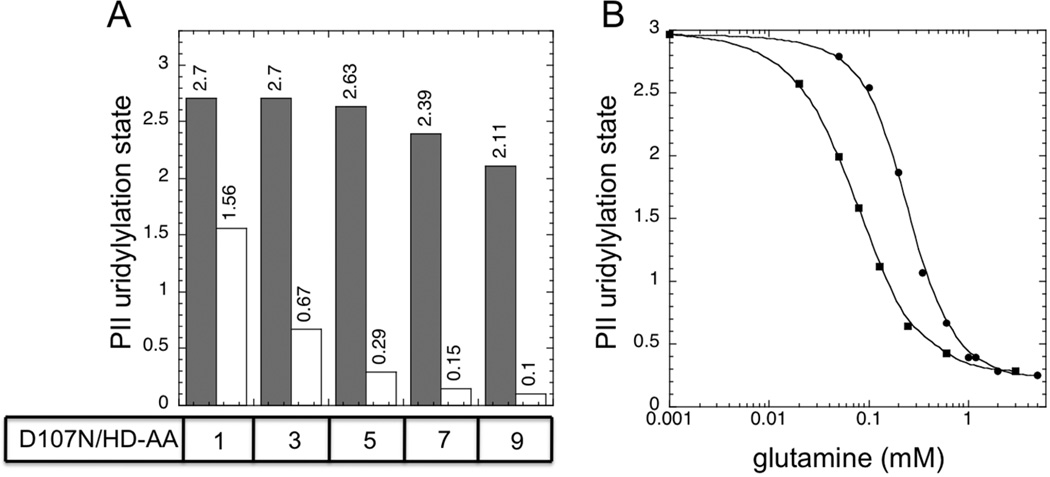
We examined the uridylylation state of PII, present at the high concentration of 36 µM, in the presence and absence of 10 mM glutamine, when the his-tagged UTase/UR (UQ5516) was present at 0.5 µM and the D107N altered enzyme was present at various concentrations (Fig 3A). When the his-tagged (UQ5516) enzyme was the only enzyme present, PII uridylylation state only varied over a narrow range, consistent with the results shown in Fig 1A and Fig 2B. But, the combination of the his-tagged UTase/UR and the altered D107N enzyme resulted in more effective glutamine signaling (Fig 3A). When the D107N enzyme was present at 3 µM in combination with the his-tagged UTase/UR at 0.5 µM (such that the ratio D107N/his-tagged enzyme was 6:1), PII uridylylation state varied from 2.9 in the absence of glutamine to 0.21 in the presence of 10 mM glutamine, for a range of ~ 2.7, as typically seen with the wild-type (un-tagged) UTase/UR (Fig 1A). When the steady state responses to a wide range of glutamine concentrations was examined under these conditions, very effective glutamine signaling was observed (Fig 3B), reminiscent of the glutamine signaling by the wild-type (un-tagged) enzyme under similar conditions (Fig 1A). Thus, in practice, the signaling defect of the his-tagged UTase/UR enzyme could be compensated for simply by addition of the monofunctional D107N enzyme that provided additional UR activity (Fig 3B).
Figure 3. A combination of his-tagged UTase/UR and an altered form of the enzyme displaying only UR activity effectively regulated PII uridylylation state in response to glutamine.
A. Steady state PII uridylylation state in the absence of glutamine (filled bars) and in the presence of 10 mM glutamine (unfilled bars). PII was at 36 µM, his-tagged UTase/UR (UQ5516) was at 0.5 µM, and the altered D107N UTase/UR was present at 0 µM, 0.5 µM, 1 µM, 2 µM or 3 µM to provide the indicated ratios of enzymes. B. Glutamine signaling by a reconstituted UTase/UR-PII cycle containing a combination of his-tagged UTase/UR (UQ5516) (0.5 µM) and the altered D107N UTase/UR (3 µM). PII was present at 36 µM.
UT activity of the His-tagged enzymes
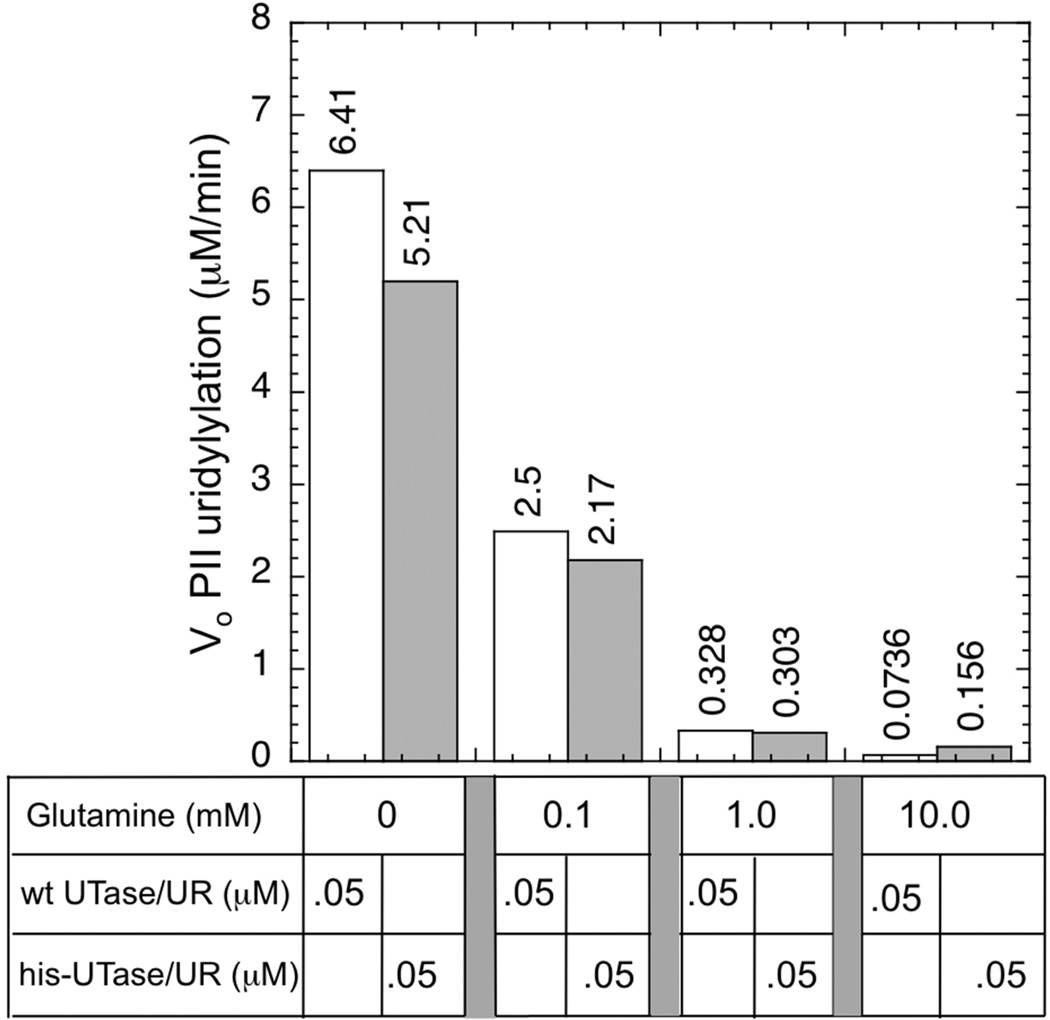
A series of preliminary experiments indicated that both preparations of his-tagged but otherwise wild-type UTase/UR had normal levels of UT activity, which was regulated by glutamine, and which displayed similar Km for PII and substrate inhibition by PII as the wild-type enzyme (Fig S7A, [15]). The his-tagged enzyme preparations displayed a slightly higher inhibition constant for glutamine (0.14 mM) than did the wild-type enzyme (0.06 mM), as shown for the wild-type and UQ5516 enzyme preparations in Fig S7B. Notably, inhibition of the UT activity his-tagged enzyme was incomplete at 10 mM gln, whereas the wild-type enzyme was almost completely inhibited at this glutamine concentration (Fig S7B).
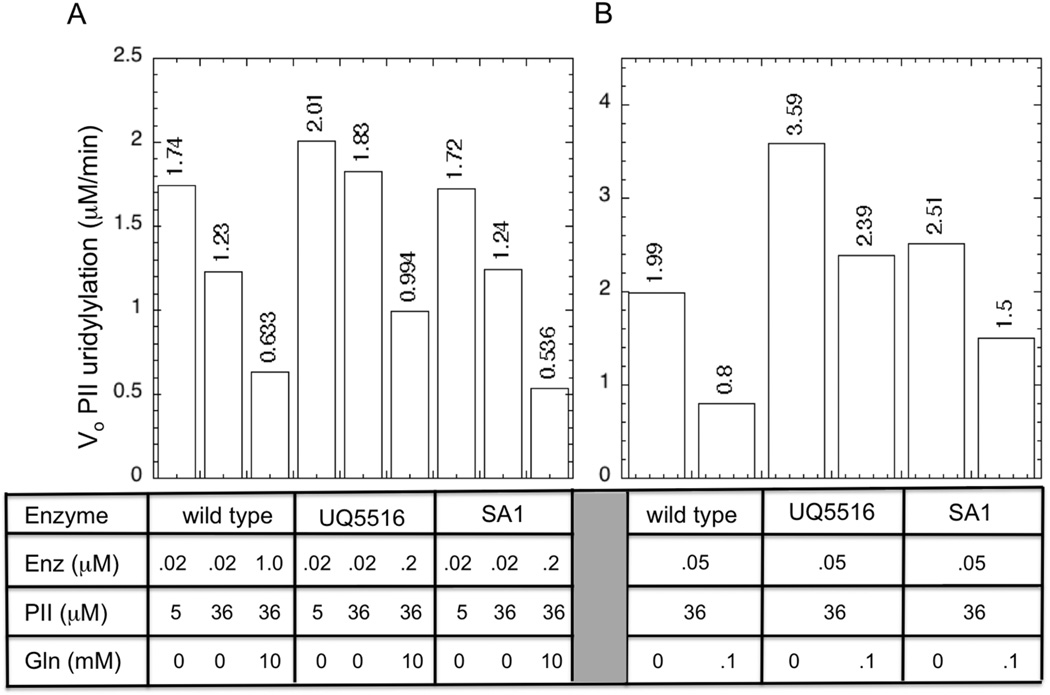
In Fig 4, we show side-by-side comparisons of the wild-type and his-tagged enzymes under various conditions to highlight the similarities and differences of these enzymes. For all three enzyme samples, we measured the initial rate of PII uridylylation (UT activity) in the presence and absence of 10 mM glutamine with PII at 36 µM. Since the UT activity is inhibited by glutamine, the experiments conducted in the presence of glutamine utilized elevated levels of enzyme to obtain easily measurable catalytic rates, as shown in Fig 4. This procedure was used because we verified that the kcat of the UT reaction was largely independent of the enzyme concentration, as long as the PII substrate was saturating (Fig S2). We also measured the UT activity when PII was at 5 µM and glutamine was absent to allow comparison with the rates obtained at 36 µM PII and an assessment of PII substrate inhibition (Fig 4).
Figure 4. Initial rate of PII uridylylation (UT activity) and its regulation by glutamine and UMP.
Measurement of the initial rate of the PII uridylylation was as described in Materials and Methods.
When PII was at 36 µM, the UT activity of the wild-type UTase/UR was regulated ~ 97.2-fold by glutamine (Fig 4). The enzyme displayed substrate inhibition by PII in the absence of glutamine, as the catalytic rate was faster with 5 µM PII than it was with 36 µM PII. The estimate of 97.2-fold regulation from Fig 4 was as follows: when PII was 36 µM, an enzyme concentration of 0.02 µM provided a rate of 1.23 µM/min in the absence of glutamine (corresponding to a kcat of 61.5/min) and an enzyme concentration of 1 µM provided a rate of 0.663 µM/min in the presence of 10 mM glutamine (corresponding to a kcat of 0.633). Thus, glutamine lowered the kcat 97.2-fold.
In contrast to the wild-type enzyme, the his-tagged UTase/UR preparations (SA1 and UQ5516) were only regulated 18-fold and 23-fold, respectively, by 10 mM glutamine (Fig 4A). Thus, when PII was 36 µM, the enzyme at 0.2 µM provided about half the activity when glutamine was present than did the enzyme at 0.02 µM in the absence of glutamine (Fig 4A). These results show that the his-tagged enzyme preparations had a significant defect in glutamine inhibition of the UT activity. In the absence of glutamine, the UT activity of the his-tagged enzymes were similar to that of the wild-type, but in the presence of glutamine the UT activity of the his-tagged enzymes was dramatically higher (Fig 4A).
To allow accurate assessment of the glutamine regulation of these enzyme samples at fixed levels of the enzyme, we examined the effect of glutamine at 0.1 mM on the initial rate of uridylylation when PII was at 36 µM and enzyme was 0.05 µM (Fig 4B). As shown, glutamine regulation of the wild-type enzyme was sharper than that obtained with the his-tagged enzyme preparation; inhibition of the wild-type activity was 59.8%, while the UQ5516 preparation was inhibited 40.2% and the SA1 enzyme preparation was inhibited 33.6% (Fig 4B). Together, the results of Fig 4 show that the his-tagged enzymes had a significant defect in glutamine regulation of the UT activity when PII was 36 µM.
In another set of side-by-side experiments, the glutamine regulation of the UT activity of the wild-type and his-tagged (UQ5516) enzymes were examined in experiments where PII was at 5 µM and the enzyme was at 0.05 µM (Fig 5). Under these conditions the difference in glutamine regulation of the enzymes was discernable, but was less dramatic; the wild-type enzyme was regulated 87-fold by 10 mM glutamine, while the his-tagged UQ5516 enzyme preparation was regulated 33 -fold by 10 mM glutamine (Fig 5). Similar subtle differences were observed in regulation by lower concentrations of glutamine under these conditions (Fig 5). For example, when glutamine was at 0.1 mM, the wild-type enzyme was inhibited 61% while the his-tagged UQ5516 enzyme was inhibited 58.3% (Fig 5). This side-by-side comparison was repeated on another occasion; the wild-type enzyme was inhibited 59.9% by 0.1 mM glutamine while the his-tagged UQ5516 enzyme was inhibited 57.4 % by 0.1 mM glutamine. We conclude that when PII was at 5 µM, glutamine regulation of the wild-type and his-tagged enzymes was nearly the same. By comparison, there was an obvious distinction in the glutamine regulation of the wild-type and his-tagged enzymes when PII was at 36 µM (Fig 4).
Figure 5. Initial rate of PII uridylylation and its regulation by glutamine and UMP.
Initial rate of PII uridylylation was measured as described in Materials and Methods, with PII at 5 µM. The his-tagged enzyme used in this experiment was the UQ5516 preparation.
UR activity of enzymes
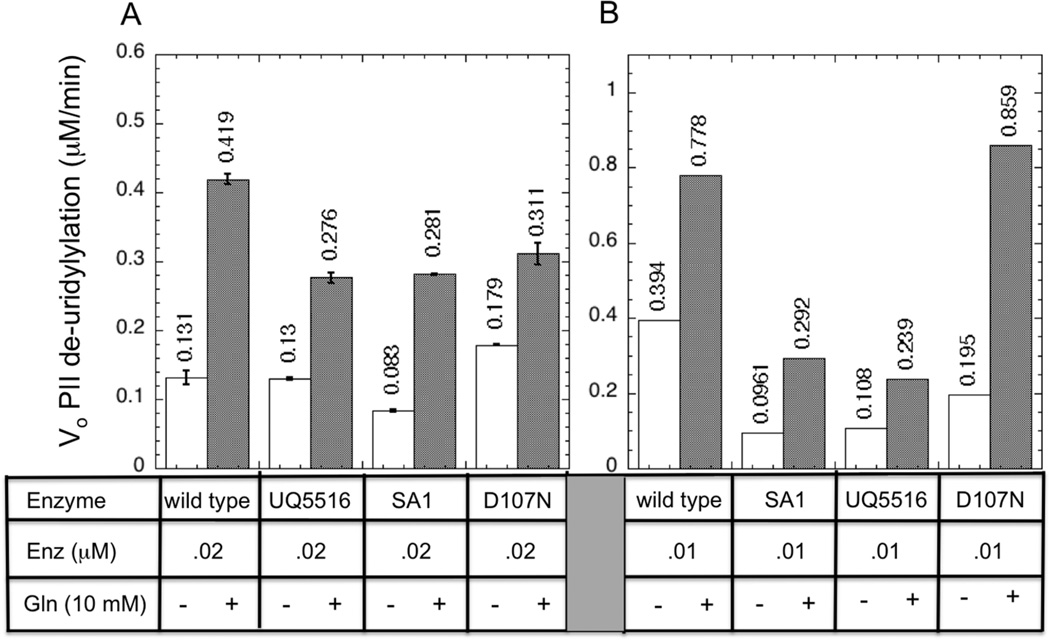
A series of preliminary experiments using the standard UR assay (MATERIALS AND METHODS) indicated that the his-tagged enzymes displayed a modest defect in catalyzing the de-uridylylation of PII-UMP, relative to the wild-type enzyme. In Fig 6A we present side-by-side comparisons of the enzymes, in which the initial rate of PII-UMP deuridylylation was measured in reaction mixtures that contained ATP as adenylylate nucleotide, and contained 4 µM PII-UMP and 0.2 µM enzyme. Under these conditions, the wild-type enzyme displayed a basal UR activity, that was stimulated 3.2-fold by 10 mM glutamine, as expected (19). The UR activity is weak, and the stimulated rate for the wild-type enzyme in this experiment corresponded to ~ 2.1/min. By comparison, the his-tagged enzymes displayed a similar basal UR rate, and 2–3 -fold stimulation by 10 mM glutamine (Fig 6). The D107N altered enzyme displayed a slightly higher basal UR activity than did the wild-type UTase/UR, and this rate was stimulated less than 2-fold by glutamine, such that the stimulated UR rate was slightly lower than that obtained with the wild-type enzyme (Fig 6A). Based on the data in Fig 6A, it appeared that the his-tagged enzymes had only a modest defect in the UR activity.
Figure 6. UR activity of wild-type UTase/UR, his-tagged UTase/UR, and altered D107-N UTase/UR, and its regulation by glutamine in systems containing ATP.
A. Measurements using the standard assay protocol. Initial rates of PII-UMP de-uridylylation were determined as described in Materials and Methods, in experiments where uridylylated PII subunits were initially present at 4 µM. Error bars indicate the standard deviation for duplicate trials, which were performed on the next day. B. Measurements using the TLC-based assay protocol. Initial rates of PII-UMP de-uridylylation were determined as described in Materials and Methods, in experiments where uridylylated PII subunits were initially present at 10.33 µM.
Because of concerns about the accuracy of the standard UR assay, we developed an alternative assay procedure incorporating a thin-layer chromatographic separation of the reaction product (UMP), as described in MATERIALS AND METHODS. Although this new assay procedure is labor-intensive, we believe it allows more accurate estimations of the initial reaction rates, particularly when higher levels of activity are being measured. As shown in Fig 6B, the his-tagged enzymes displayed a significant (~ 3-fold) defect in the level of the UR activity, while the D107N enzyme was again quite similar to the wild-type enzyme. Also, higher UR activities were obtained using the TLC-based assay method as compared to the standard assay method (ca Fig 6A and 6B).
As noted above, the UR activity is stimulated when the ATP in the reaction mixtures is replaced with AMP-PNP. We compared the UR activity of the wild-type and his-tagged (SA1) enzymes in the presence of AMP-PNP in side-by-side experiments that were repeated; the his-tagged enzyme displayed a 5-fold lower level of the basal UR activity and a 2.2-fold lower level of the glutamine-activated UR activity in comparison to the wild-type enzyme (Fig S8).
An hypothesis to explain PII substrate inhibition of the UT activity, glutamine regulation of the UT and UR activities, and the effect of adding a his-tag to the UTase/UR on robustness to PII concentration
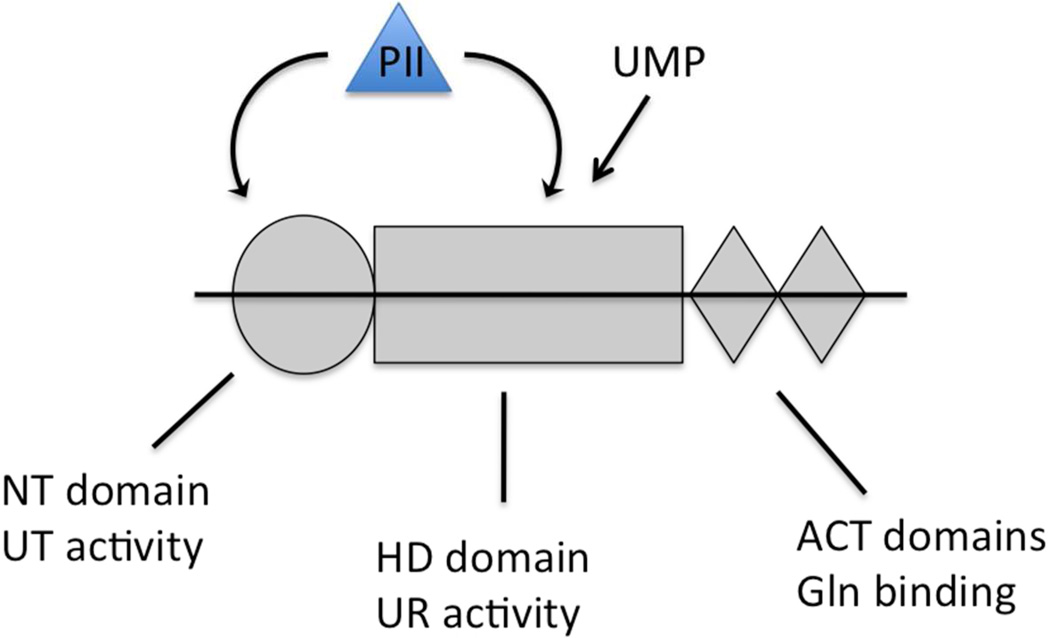
The UTase/UR is a monomeric protein, consisting of 4 functional domains (depicted schematically in Fig 7; [13]). By analogy with other proteins containing ACT domains, it is likely that glutamine binding to the tandem ACT domains at the C-terminal end of the protein is responsible for inhibition of the UT activity and stimulation of the UR activity. Alterations of the ACT domains block glutamine regulation of the UT and UR activities (13). Because the UT activity is tightly regulated by glutamine, we hypothesize that the HD domain is not only responsible for catalysis of the UR activity, but also is responsible for controlling the UT activity in response to glutamine binding. That is, the HD domain has a signal transduction function. The only other possible mechanisms for regulation of the UT activity by glutamine binding to the ACT domains would be for the ACT domains to contact the N-terminal UT domain directly, such as if the protein had an overall curvature or formed an oligomer in which the ACT domains of one subunit were in contact with the NT domain of the opposing subunit. Both of these possibilities seem less likely than if the signal of glutamine binding is passed to the NT (UT) domain by the UR domain. Earlier studies with the related ATase enzyme that reversibly modifies GS showed that in that protein the two catalytic domains regulated each other and had both catalytic and signal transducing functions (19, 20), as we hypothesize for the UTase/UR.
Figure 7. Schematic depiction of the domain arrangement of the UTase/UR and the sites from which UMP and PII exert their inhibitory effects.
The N-terminal NT domain of UTase/UR is depicted as a grey circle, the central HD domain of UTase/UR is depicted as a grey rectangle, and the tandem C-terminal ACT domains of UTase/UR are depicted as grey diamonds. PII, depicted as a blue triangle binds to the NT domain at the site where it is uridylylated (substrate site) and binds to the HD domain at the site where it is formed from PII-UMP (product site). UMP is also a product of the UR activity of the HD domain.
Since PII is a product of the UR reaction catalyzed by the central HD domain and is the substrate of the UT activity of the NT domain, there are two binding sites on the UTase/UR for PII (Fig 7). We hypothesize that PII exerts substrate inhibition of the UT activity upon binding to its site in the HD domain. We also hypothesize that the unusual properties of the his-tagged UTase/UR resulted from an abnormal interaction between the N-terminal nucleotidyltransferase (NT) domain and the central HD domain. Because of this abnormal domain arrangement, the binding of PII to the HD domain (the substrate inhibition site) interferes with the transmission of the glutamine signal from the ACT domains to the NT domain and diminishes the inhibition of the UT activity by glutamine.
A small deletion within the HD domain eliminated glutamine regulation of the UT activity and also eliminated substrate inhibition of the UT activity by PII
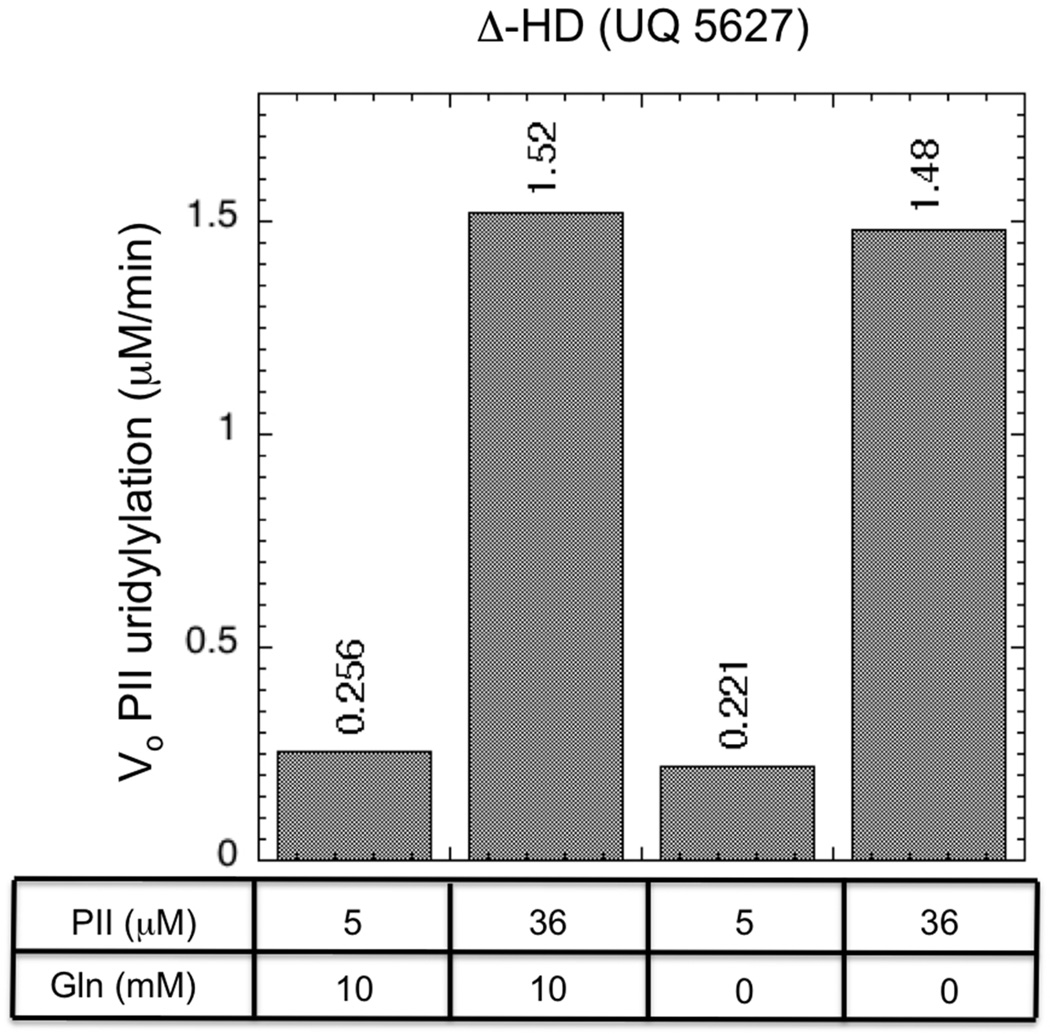
If the PII-mediated substrate inhibition of the UT activity was due to PII binding to the UR active site within the HD domain of UTase/UR, then mutations that alter the UR active site are predicted to eliminate the substrate inhibition. The Δ-HD protein (purified from strain UQ5627, 12) is a his-tagged protein in which 22 residues have been removed by a deletion within the HD domain (Δ-A510-D531), and this protein lacks UR activity (13). This protein displayed fairly weak UT activity, and this activity was not regulated by glutamine (13, Fig 8). Consistent with our hypothesis, the UT activity of the Δ-HD protein did not display substrate inhibition by PII. We interpret the absence of glutamine regulation of the UT activity resulting from the small deletion within the HD domain as reflecting a loss of the signal-transduction function of the HD domain.
Figure 8. UT activity of the his-tagged Δ-HD (UQ5627) protein.
Initial rates of PII uridylylation were determined as in Materials and Methods.
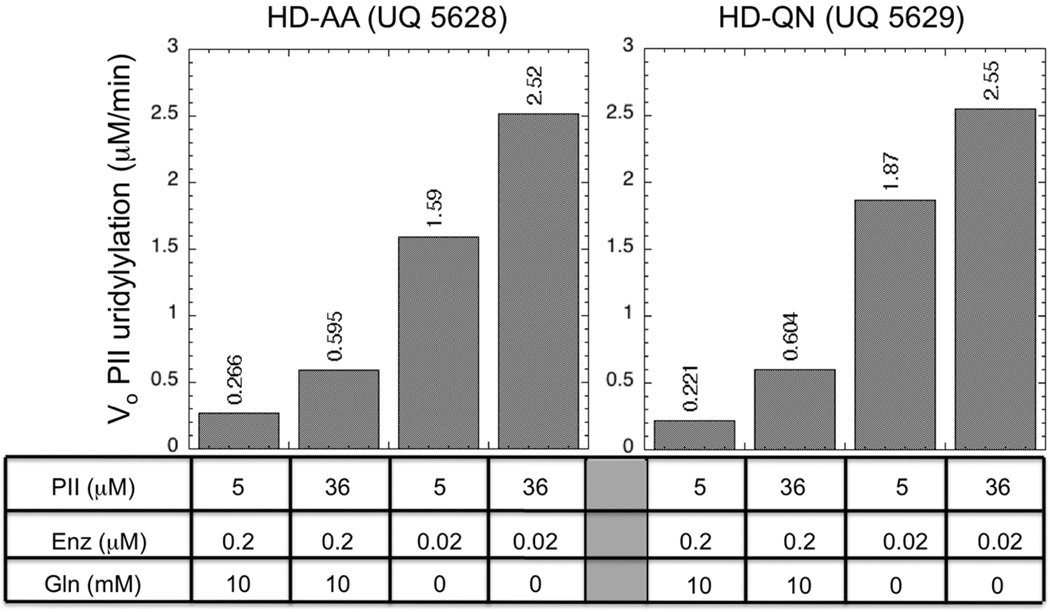
Amino acid substitutions at the catalytic residues within the HD domain eliminated substrate inhibition of the UT activity by PII, and partially restored glutamine regulation of the UT activity
The HD-AA (UQ5628) and HD-QN (UQ5629) proteins are his-tagged enzymes that contain amino acid substitutions at the conserved H and D residues of the HD domain (13). These proteins lack UR activity, but displayed considerable UT activity, which was regulated by glutamine (13, see below). We observed that these proteins displayed better regulation of the UT activity by glutamine than did the his-tagged but otherwise wild-type versions of the UTase/UR (Fig 9). When PII was at 36 µM and UMP was absent, each of these proteins displayed 42-fold regulation of the UT activity by glutamine (Fig 9). That is, when glutamine was absent and the enzyme was at 0.02 µM, the activity was more than 4-fold higher than when glutamine was present and the enzyme was at 0.2 µM. When PII was at 5 µM, the HD-AA protein displayed 59-fold regulation by glutamine and the HD-QN protein displayed 85-fold regulation by glutamine. In additional experiments, we also observed that these two his-tagged proteins with HD domain alterations displayed better regulation than did the his-tagged but otherwise wild-type UTase/UR when glutamine was present at the low concentration of 0.1 mM. Thus, remarkably, alterations within the HD domain partially compensated for the presence of the his-tag in allowing strong glutamine control of the UT activity. Neither the HD-AA nor the HD-QN proteins displayed substrate inhibition of the UT activity by PII (Fig 9), which is consistent with our hypothesis.
Figure 9. UT activity of the his-tagged HD-AA (UQ5628) and HD-QN (UQ5629) proteins.
Initial rates of PII uridylylation were determined as in Materials and Methods.
A reconstituted UTase/UR-PII monocycle comprised of monofunctional UTase and UR enzymes
Since the HD-AA protein lacked UR activity, we used this protein along with the D107N monofunctional UR enzyme to produce a reconstituted UTase/UR-PII cycle comprised of two monofunctional enzymes. The altered HD-AA protein lacked substrate inhibition by PII and its UT activity was well-regulated by glutamine even when PII was at high concentration, we therefore expected that a monocycle containing the monofunctional HD-AA and D107N enzymes should be robust to PII concentration. To examine this, we held PII fixed at 36 µM and examined the effect of combining the HD-AA enzyme at 0.8 µM with various concentrations of the D107N enzyme. The steady-state uridylylation of PII was assessed in systems lacking glutamine or containing 10 mM glutamine, to discern the range of uridylylation states that could be obtained in response to glutamine (Fig 10). As shown, the largest range of uridylylation states in response to glutamine signaling was obtained in this experiment when the ratio of D107N/1HD-AA was 5. Below this ratio, a narrow range of uridylylation states was obtained, biased towards higher uridylylation states, while at a higher ratio of enzymes, a narrow range of uridylylation states was obtained that was biased towards lower uridylylation states (Fig 10A). We therefore explored using a modest excess of the D107N enzyme, relative to the HD-AA enzyme in reconstituted monocycles, and in additional experiments found that a 4:1 ratio of the two enzymes gave the optimal responsiveness to glutamine. The results of using a 4:1 ratio of D107N to HD-AA in a reconstituted UTase/UR-PII monocycle are shown in Fig 10B. PII was used at 36 µM, and unlike the situation when the his-tagged bifunctional UTase/UR was used, a wide range of PII uridylylation states (~ 2.68/3.0) signaled changes in the glutamine concentration (Fig 10B). Thus, the reconstituted system comprised of monofunctional enzymes (Fig 10B) was not defective in signaling when PII was at 36 µM, in contrast to the results obtained with the his-tagged bifunctional enzyme (Fig 1A). In the same experiment, we also examined the performance of a reconstituted UTase/UR-PII cycle containing the D107N and his-tagged but otherwise wild-type UTase/UR (UQ5516), using a 4:1 ratio of D107N to his-tagged UTase/UR (UQ5516). As shown in Fig 10B, a wide range of uridylylation states was also obtained (2.65/3.0) in this system, but other features of the glutamine response differed in the two reconstituted cycles (Fig 10B). The steepness of the response was greater in the system containing the his-tagged UTase/UR (UQ5516) than in the system containing the his-tagged HD-AA altered UTase/UR, and the midpoint of the glutamine response was at a higher glutamine concentration. Since the his-tagged but otherwise wild-type UTase/UR demonstrated PII substrate inhibition of the UT activity, while the HD-AA enzyme did not demonstrate substrate PII inhibition of the UT activity, these results may point to a role of the PII substrate inhibition in determining the sensitivity of the glutamine response of the cycle (16).
Figure 10. Reconstituted UTase/UR-PII cycles containing mixtures of the monofunctional altered HD-AA UTase/UR and D107N UTase/UR.
A. Steady state PII uridylylation state in the absence of glutamine (filled bars) and in the presence of 10 mM glutamine (unfilled bars). PII was at 36 µM, his-tagged altered HD-AA UTase/UR was at 0.8 µM, and the altered D107N UTase/UR was present at 0.8 µM, 2.4 µM, 4 µM, 5.6 µM or 7.2 µM to provide the indicated ratios of enzymes. B. Glutamine signaling by reconstituted UTase/UR-PII cycles containing combination of enzymes. PII was present at 36 µM in all reactions. ■, reactions contained 0.5 µM his-tagged HD-AA UTase/UR and 2 µM D107N UTase/UR. ●, reactions contained 0.5 µM his-tagged UTase/UR (UQ5516) and 2 µM D107N UTase/UR.
DISCUSSION
The addition of various "tags" to proteins to allow the use of common affinity chromatography steps in their purification has greatly advanced the analysis of numerous enzymes, but this procedure is not without risk since the tagged form of the enzyme may display altered properties. Here, we show that the addition of an N-terminal his-tag to the UTase/UR resulted in an altered enzyme that was defective in glutamine signaling when used in reconstituted covalent modification cycles. Specifically, the his-tagged enzyme displayed a defect in glutamine signaling when PII was at high concentration, but displayed only a modest defect in glutamine signaling when PII was at low concentration. That is, the robustness of the covalent modification cycle to changes in PII concentration was lost as a result of adding a his-tag to the enzyme. When PII was at high concentration, a limited range of PII uridylylation states were obtained as glutamine was varied, and these were biased towards high states of uridylylation. This defect was readily evident in side-by-side experiments where the ratio of substrate to enzyme was held constant (Fig 2); thus, it became clear that the PII concentration was the important parameter to which robustness was lost. The fortuitous observation of a defect in robustness of the system to PII concentration allowed us to study the phenomenon and examine the underlying biochemical mechanism.
Prior studies have shown that the UTase/UR consists of three functional elements: an N-terminal NT domain, that catalyzes the UT activity, a central HD domain that catalyzes the UR activity, and a pair of tandem ACT domains at the C-terminus of the protein that is responsible for glutamine sensation (12). ACT domains are commonly found in a tandem, paired, arrangement, so the two ACT domains may comprise a functional unit responsible for passing the glutamine signal to the other domains. We hypothesize that the domains of the UTase/UR interact with and regulate their neighboring domains, and that the C-terminal glutamine-sensing ACT domains do not directly regulate the N-terminal NT domain but rather, must pass their signal indirectly, through the central HD domain, to control the UT activity of the NT domain (Fig 7). A key to understanding the effect of PII concentration on the enzyme is the observation of PII substrate inhibition of the UT activity (15). Substrate inhibition results from the substrate binding to not only the catalytic site of an enzyme, but also to another site from which inhibition occurs. In the case of the UTase/UR enzyme, substrate inhibition by PII is very likely to reflect PII binding to both the catalytic site in the N-terminal UT domain and to a site in the central HD domain. PII is a product of the UR activity, thus there is certainly a PII binding site in the HD domain. We observed that the UT activity of the his-tagged enzymes was subject to substrate inhibition by PII, thus it seems likely that PII binds normally to the site in the HD domain in these enzymes. We also observed that altered enzymes with either a small deletion within the HD domain or two point mutations within the HD domain lacked substrate inhibition of the UT activity by PII, and that the regulation of the UT activity by glutamine was altered by mutations in the HD domain. These observations are consistent with PII binding to the HD domain as the source of substrate inhibition, and with the HD domain having a role in passing the glutamine signal from the ACT domains to the UT domain.
We hypothesize that the presence of the his-tag resulted in a subtle alteration of the interactions between the N-terminal nucleotidyltransferase (UTase) domain and the central HD (UR) domain. As long as PII was present at low concentration, such that there was little occupancy of the HD domain site from which PII exerts substrate inhibition of the UT activity, glutamine regulation of the UT activity of the his-tagged enzyme was nearly normal (Fig 5). But, at high PII concentration, where PII occupied both the N-terminal catalytic site and the HD domain PII-binding site, glutamine signaling by the his-tagged enzyme was clearly defective (Fig 4). Our hypothesis to explain these observations is that, for the his-tagged enzyme, when PII bound to the HD domain it interfered with the passage of the glutamine signal to the N-terminal NT domain.
We observed that both the his-tagged (but otherwise wild-type) UTase/UR and the his-tagged HD-AA monofunctional (UTase only) enzyme could be combined with the monofunctional (UR-only) D107N altered enzyme to produce reconstituted covalent modification cycles that responded well to glutamine when PII was at the high concentration of 36 µM. Thus, the robustness defect of the his-tagged UTase/UR could be offset simply by an increase in glutamine-regulated UR activity. This shows that the relative levels of the antagonistic activities plays a key role in allowing a wide range of PII modification states in response to glutamine signaling, and is consistent with theoretical studies of a covalent modification cycle (21). We observed a defect of the his-tagged UTase/UR in the regulation of the UT activity by glutamine, that became evident as the PII concentration was increased. But, other types of defects could likely result in the similar reduction in PII modification states in response to glutamine, such as simple defects in either catalytic activity. Furthermore, if an enzyme had a defect in catalysis, this could perhaps be compensated for by increased glutamine regulation of an activity so as to restore the normal relative level of the two antagonistic enzyme activities. That is, the relative levels of the UT and UR activities are likely to determine the range of modification states in response to glutamine, as predicted (21), and a variety of mechanisms may alter or restore the natural relative levels of the activities.
Interestingly, when we compared reconstituted cycles that contained the his-tagged UTase/UR and the HD-AA altered UTase/UR in combination with the D107N altered enzyme, we observed that cycles containing the his-tagged but otherwiswe wild-type enzyme had a steeper response to glutamine than did cycles containing the HD-AA protein (Fig 10B). Although further studies will be necessary, this finding suggests that PII substrate inhibition of the UT activity may play a role in increasing the sensitivity (apparent kinetic order, Hill coefficient) of glutamine responses of the cycle. The ability of substrate inhibition to increase the sensitivity of responses of a covalent modification cycle was demonstrated by Guidi and Goldbeter (16).
One general conclusion from our study is that robustness of the covalent modification cycle to the concentration of its substrate depended critically on the catalytic rates of the antagonistic converter enzymes and their regulation, consistent with theory (21). Depending on the parameters of the system, a fairly modest defect in regulation of an activity can bring about a dramatic change in the steady state responses of the covalent modification cycle and eliminate robustness to the substrate concentration. Another important observation from our study is that a loss of robustness to the concentration of the cycle substrate was manifested by a diminished range of modification states in response to the stimulatory effector. We hypothesize that this may be a common manifestation of the loss of robustness to the substrate concentration.
Supplementary Material
Acknowledgements
We thank Gary Roberts for sharing information and materials, and for helpful comments on the work and the manuscript.
Footnotes
Y. Z was supported by grant GM65891 to Gary Roberts
Supplemental materials may be accessed free of charge online at htp://pubs.acs.org.
Supplemental materials for this paper consists of 8 figures.
Contributor Information
Peng Jiang, Email: pejiang@umich.edu.
Yaoping Zhang, Email: yzhang8@wisc.edu.
M. R. Atkinson, Email: mark1073@me.com.
REFERENCES
- 1.Savageau MA. Parameter sensitivity as a criterion for evaluating and comparing the performance of biochemical systems. Nature. 1971;229:542–544. doi: 10.1038/229542a0. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. Towards a theory of biological robustness. Mol Syst Biol. 2007;3:137. doi: 10.1038/msb4100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 4.Shinar G, Milo R, Martinez MRT, Alon U. Input-output robustness in simple bacterial signaling systems. Proc. Natl. AScad. Sci. USA. 2007;104:19931–19935. doi: 10.1073/pnas.0706792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soyer OS, Pfeiffer T. Evolution under fluctuating environments explains observed robustness in metabolic netwoorks. PLoS Comp. Biol. 2010;6:e10000907. doi: 10.1371/journal.pcbi.1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chock PB, Stadtman ER. Superiority of interconvertable enzyme cascades in metabolic regulation: analysis of multicyclic systems. Proc. Natl. Acad. Sci. USA. 1977;74:2766–2770. doi: 10.1073/pnas.74.7.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Uribe C, Verghese GC, Mirny LA. Operating regimes of signaling cycles: statics, dynamics, and noise filtering. PLoS Comp. Biol. 2007;3:e246. doi: 10.1371/journal.pcbi.0030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardenas ML, Cornish-Bowden A. Characteristics necessary for an interconvertible enzyme cascade to generate a highly sensitive response to an effector. Biochem. J. 1989;257:339–345. doi: 10.1042/bj2570339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldbeter A, Koshland DE. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koshland DE, Jr, Goldbeter A, Stock JB. Amplification and adaptation in regulatory and sensory systems. Science. 1982;217:220–225. doi: 10.1126/science.7089556. [DOI] [PubMed] [Google Scholar]
- 11.Ninfa AJ, Jiang P, Atkinson MR, Peliska JA. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr Top Cell Regul. 2000;36:31–75. doi: 10.1016/s0070-2137(01)80002-9. [DOI] [PubMed] [Google Scholar]
- 12.Jiang P, Peliska JA, Ninfa AJ. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII proteins. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Pohlmann EL, Conrad MC, Roberts GP. Mutagenesis and functional characterization of the four domains of GlnD, a bifunctional nitrogen sensor protein. J. Bacteriol. 2010;192:2711–2721. doi: 10.1128/JB.01674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura AC, Jiang P, Van Wassenhove L, Del Vecchio D, Merajver SD, Ninfa AJ. Signaling properties of a covalent modification cycle are altered by a downstream target. Proc. Natl. Acad. Sci. USA. 2010;107:10032–10037. doi: 10.1073/pnas.0913815107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamberov ES, Atkinson MR, Chandran P, Feng J, Ninfa AJ. Sensory components controlling bacterial nitrogen assimilation. Cell Mol. Biol. Res. 1994;40:175–191. [PubMed] [Google Scholar]
- 16.Schauder B, Blocker H, Frank R, McCarthy JE. Inducible expression vectors incorporating the E. coli atpE translational initiation region. Gene. 1987;52:279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- 17.Jiang P, Ninfa AJ. Characterization of the Reconstituted UTase/UR-PII-NRII-NRI Bicyclic Signal Transduction System that Controls the Transcription of Nitrogen-Regulated (Ntr) Genes in Escherichia coli. 2012 doi: 10.1021/bi300575j. (submitted, accompanying manuscript) [DOI] [PubMed] [Google Scholar]
- 18.Guidi GM, Carlier MF, Goldbeter A. Bistability in the isocitrate dehydrogenase reaction: an experimentally based theoretical study. Biophys J. 1998;74:1229–1240. doi: 10.1016/S0006-3495(98)77837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamberov ES, Atkinson MR, Ninfa AJ. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 20.Jiang P, Mayo AE, Ninfa AJ. Escherichia coli glutamine synthetase adenylyltransferase (ATase, E.C. 2.7.7.49): Kinetic characterization of regulation by PII, PII-UMP, glutamine, and a-ketoglutarate. Biochemistry. 2007;46:4133–4146. doi: 10.1021/bi0620510. [DOI] [PubMed] [Google Scholar]
- 21.Jiang P, Pioszak AA, Ninfa AJ. Structure/function analysis of glutamine synthetase adenylyltransferase (ATase, E.C. 2.7.7.49) of Escherichia coli. Biochemistry. 2007;46:4117–4132. doi: 10.1021/bi0620508. [DOI] [PubMed] [Google Scholar]
- 22.Stadtman ER, Chock PB. Superiority of interconvertable enzyme cascades in metabolic regulation: analysis of monocyclic systems. Proc. Natl. Acad. Sci. USA. 1977;74:2761–2765. doi: 10.1073/pnas.74.7.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.