Abstract
Among the deepest-rooting clades in the human mitochondrial DNA (mtDNA) phylogeny are the haplogroups defined as L0d and L0k, which are found primarily in southern Africa. These lineages are typically present at high frequency in the so-called Khoisan populations of hunter-gatherers and herders who speak non-Bantu languages, and the early divergence of these lineages led to the hypothesis of ancient genetic substructure in Africa. Here we update the phylogeny of the basal haplogroups L0d and L0k with 500 full mtDNA genome sequences from 45 southern African Khoisan and Bantu-speaking populations. We find previously unreported subhaplogroups and greatly extend the amount of variation and time-depth of most of the known subhaplogroups. Our major finding is the definition of two ancient sublineages of L0k (L0k1b and L0k2) that are present almost exclusively in Bantu-speaking populations from Zambia; the presence of such relic haplogroups in Bantu speakers is most probably due to contact with ancestral pre-Bantu populations that harbored different lineages than those found in extant Khoisan. We suggest that although these populations went extinct after the immigration of the Bantu-speaking populations, some traces of their haplogroup composition survived through incorporation into the gene pool of the immigrants. Our findings thus provide evidence for deep genetic substructure in southern Africa prior to the Bantu expansion that is not represented in extant Khoisan populations.
Main Text
Sub-Saharan Africa harbors the deepest-rooting lineages of human mitochondrial DNA (mtDNA), in agreement with an African origin of modern humans supported by both fossil and genetic evidence.1–4 Several studies concurred in placing the root of the mtDNA phylogeny in the southern half of the continent,5–7 and two deep-rooting clades of this phylogeny—haplogroups L0d and L0k—have been unanimously associated with so-called Khoisan populations.6–9 The generic term “Khoisan” covers hunter-gatherer and pastoralist populations of southern Africa who speak non-Bantu indigenous languages and share some linguistic features (one of the most characteristic being the heavy use of click consonants in their languages); however, these similarities might be the effect of contact.10 Haplogroups L0d and L0k are present nearly exclusively in Khoisan populations and neighboring Bantu-speaking populations that have been in documented close contact with them;11–14 the only known exceptions are sporadic occurrences of haplogroup L0d in East Africa (e.g., in the Sandawe from Tanzania)7 and in an individual from Yemen6 as well as an individual from Kuwait6 who belongs to haplogroup L0k. Specialists recognize three independent language families among Khoisan, namely Tuu, Kx’a, and Khoe-Kwadi,15–17 which are spoken by a large number of different ethnolinguistic groups comprising both foragers and pastoralists. The forager populations of the central Kalahari, who speak languages belonging to the Tuu and Kx’a families, are assumed to be the descendants of autochthonous Late Stone Age populations, whereas the Khoe-Kwadi languages may have been brought to the area by pastoralist populations around 2,000 years ago.18–20 The populations speaking Bantu languages, in contrast, are known for their expansion over almost half the African continent and are associated with the concomitant spread of the Bantu language family, an agricultural lifestyle, and iron technology.3,21,22 Archeological data suggest that they may have reached southern Africa not earlier than 2,000–1,200 years ago,3,23,24 where they met populations who were probably ancestral to current Khoisan populations.
The most recent comprehensive study that focused on the deepest-rooting lineages of the mtDNA phylogeny was undertaken by Behar et al.,6 who analyzed a total of 624 full mtDNA sequences belonging to haplogroup L∗(xM,N). Although this was the first substantial collection of complete mtDNA genome sequences from Africa, some limitations arose from the inclusion of a large number of sequences from diverse published sources that were not always of high quality; furthermore, for some sequences the source population or the country of origin was not clearly specified. Nevertheless, the sequences considered in that study still represent the vast majority of the haplogroup L∗(xM,N) data set included in the most recent version of Phylotree (Build 15, September 201225), a comprehensive database of mtDNA genome sequences that is periodically updated when more data become available. It thus represents the most accessible resource for studying mtDNA variation and is a widely used reference for mtDNA nomenclature.26
Behar et al.6 focused particularly on the root of the phylogeny, i.e., the age and variability of the Khoisan-specific haplogroups L0d and L0k, with the aim of investigating the most likely model of origin and isolation of Khoisan populations. With their data they were able to suggest a time frame for the dispersal of the main lineages and the split of Khoisan and other modern humans, which they dated not later than 90 thousand years ago (kya); furthermore, they suggested that the early human settlement of Africa was matrilineally structured. These hypotheses are relevant for the interpretation of early human demography and evolution; however, their results were substantially limited by the fact that only one ethnolinguistically undefined “Khoisan” sample of 38 individuals was included, thereby missing the potentially immense variability of the different ethnolinguistic populations subsumed under the generalized label Khoisan. In addition, only 30 sequences from haplogroup L0d and 7 from L0k were included, representing only a small and probably incomplete fraction of the overall variation in these haplogroups.
We here report analyses of 500 mtDNA genome sequences belonging to haplogroups L0d and L0k, of which 15 have already been published in Barbieri et al.,14 leading to a more than 10-fold increase in the available complete mtDNA genome sequences from southern Africa (Phylotree ver. 1525). With this rich data set, we aim to elucidate the phylogenetic relationships, the patterns of diversity, and the distribution of these relatively understudied haplogroups that represent some of the deepest-rooting lineages in the maternal phylogeny of modern humans. The broader data set from which the subset of L0d and L0k sequences was chosen consists of mtDNA genome sequences generated from saliva samples collected in Botswana, Namibia, Zambia, and Angola after prior approval by the relevant institutional review boards and with the consent of the donors after the aims of the study had been explained to them with the help of local translators, where necessary. Details of the samples have been described elsewhere.11,27,28 The sequence data set analyzed here comprises 45 ethnolinguistic groups, who speak Khoisan languages belonging to all three accepted language families as well as different Bantu languages; individuals were assigned to populations on the basis of the ethnic affiliation of their maternal grandmother (Table S1 available online).
Libraries enriched for mtDNA29,30 were sequenced on the Illumina GAIIx platform, resulting in an average 400-fold coverage. Sequences were manually checked with BioEdit and read alignments were screened with ma31 to exclude alignment errors and confirm indels. The two poly-C regions (np 303–315, 16,183–16,194) were excluded from the analysis. To minimize the impact of missing data, we applied imputation and resolved unknown positions by comparison to at least two otherwise identical haplotypes in the data set. Before imputation, 74 sequences included positions with missing data; after imputation, only 26 sequences still had missing positions. In the final alignment, 32 positions were left with an unknown nucleotide call (26 of which corresponded to polymorphic sites) and were excluded from the analyses (see Table S2 for a list of the excluded positions). Basic haplogroups were defined with the web tool Haplogrep.26 Mutations that did not fit the overall phylogeny were checked manually in the read alignments to exclude the possibility of erroneous base calls. Although we took into account published data on the frequency of haplogroups L0d and L0k, only the 500 sequences that were generated with the same technology and from individuals for whom we know the place of sampling and ethnicity were included in the phylogenetic analyses. We did not include previously published sequences, because they do not add substantial information to our analysis and often pose problems because of missing positions6 or missing ethnolinguistic information. The only exceptions are the L0k2 sequence from Yemen6 and the six L0d3 sequences from South Africa, Kuwait,6 and Tanzania,5 which we included to clarify the structure in L0k and L0d3 discussed below.
First, we compared the frequency and distribution of haplogroups L0d and L0k in our data set and in the available literature (where in most cases haplogroups were assigned based on partial mtDNA sequence variation and/or RFLP typing; cf. Table S1 and Figure S1 for details) and plotted the frequencies of each haplogroup (Figures 1A and 1B) with the software Surfer ver. 10.4.799 (Golden Software). The maps show a concentration of both L0d and L0k in the southern part of the continent, with L0d present in high frequency in populations from South Africa, Namibia, and Botswana, and sporadically (<5%) in some populations of Zambia, Mozambique, and Angola, as well as in the Sandawe from Tanzania. The highest frequencies (90%–100%) are found in Khoisan foragers of central Botswana, as well as in South African populations with Khoisan ancestry.32,33 In general, other studies did not distinguish between L0d and L0k as typical “Khoisan” lineages; and yet, interestingly, the distribution of haplogroup L0k is far more restricted than that of L0d, with a maximum frequency of 33% in the !Xuun foragers of Namibia; it is also found in frequencies >10% in several populations of foragers in Botswana and Namibia who speak languages belonging to all three Khoisan linguistic families (see Table S1), as well as in the Bantu-speaking Fwe from southwestern Zambia.
Figure 1.
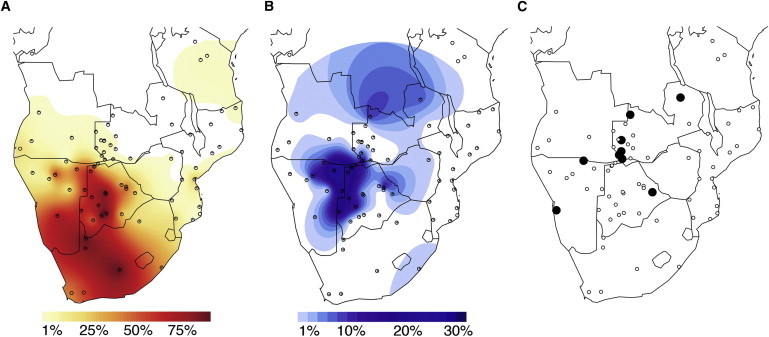
Surfer Maps Displaying the Spatial Distribution of Haplogroup Frequencies
Dots indicate sample locations.
(A) Haplogroup L0d.
(B) Haplogroup L0k. Note that the scale in (B) is different from that in (A).
(C) Presence of haplogroups L0k1b and L0k2 in southern Africa (large black dots). The actual sampling location of one Topnaar Nama individual with haplogroup L0k1b is shown here; in (A) and (B) this individual was included with the general Nama population sample.
We next reconstructed a phylogeny of the L0d and L0k mtDNA genome sequences from the most probable tree out of 10 million MCMC chains with BEAST (v1.7.234) and identified the mutations defining different branches by viewing the aligned sequences in BioEdit in comparison to the Reconstructed Sapiens Reference Sequence (RSRS35). The node branches were dated with the mutation rate of 1.26 × 10−8 for the coding region only,36 which makes our estimates comparable to those from Behar et al.6 The complete tree of sequences showing mutations that characterize the major branches is available in the Supplemental Data (Figure S2); further discussion of some of these mutations is found in Table S3. Figure 2 summarizes the tree topology and the TMRCA of lineages, with confidence intervals indicated for the major nodes.
Figure 2.
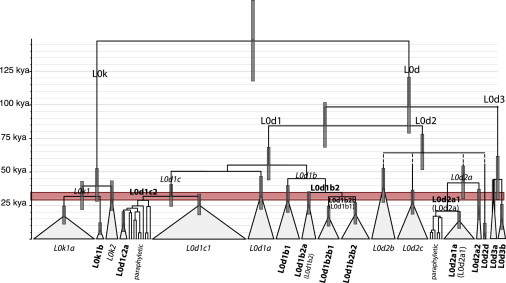
Simplified Tree Topology for the Major Lineages of L0d and L0k, Based on Coding Region Sequences and with Time Scale Indicated
Previously undetected branches are labeled in bold font; when a previously reported branch is renamed, the old label is given in brackets. Confidence intervals for the TMRCA of the major nodes are indicated by vertical bars. The red shading highlights the time span that was associated with the deterioration of climate in the central Kalahari area.
The tree coalesces 145 kya (95% C.I.: 118–179 kya), corresponding to the time of split between L0d and L0k. From the topology of the tree, different sublineages can be distinguished for both the L0d and L0k haplogroups. For L0d, three main branches (L0d3, L0d1, and L0d2) separate around 95 kya (95% C.I.: 79–121 kya), whereas L0k splits into L0k1 and L0k2 approximately 40 kya (95% C.I.: 28–53 kya). The first branch of L0d is the uncommon L0d3, which is found in a population with South African Khoisan ancestry (Karretjie People) at 13% and in a Coloured population at 10%,32 as well as being attested in one undefined Khoi and one individual from Kuwait6 and three Sandawe and one Burunge from Tanzania (our identification, based on sequences from Gonder et al.5). In our data set, it is found in only five individuals (two Nama and one Hai||om, who speak Khoe languages, and two Kgalagadi, who speak a Bantu language). As can be seen from the tree (Figure 2), L0d3 splits into two branches (L0d3a and L0d3b) 45 kya (95% C.I.: 30–61 kya), with eight mutations defining L0d3b (Figure S2 and Table S3). Interestingly, this split reflects geographic substructure: L0d3a is restricted to East Africa and the Middle East, being found in the individuals from Kuwait and Tanzania, and L0d3b is restricted to southern Africa, being found in the five individuals of our data set plus the Khoi sequence published by Behar et al.6
L0d1 is the most common subhaplogroup: it is present in all Khoisan populations, all Bantu-speaking populations of our data set from Botswana and Namibia, and a few individuals from Bantu-speaking populations of Zambia and Angola. It coalesces approximately 55 kya (95% C.I.: 44–68 kya) and comprises two branches, of which the first includes haplogroups L0d1a and L0d1c. L0d1a is a monophyletic clade; however, two sites, namely T199C and C16266A, previously assumed to define this clade, pose problems for reconstructing the history of mutations (see Table S3 for details).
In L0d1c, substantial variation emerges from our expanded data set that pushes the coalescence date back to 32 kya (95% C.I.: 24–41 kya), 10 ky older than previously estimated.6 A low posterior probability is associated with the first nodes; these are represented by paraphyletic clades that are characterized by a large number of private mutations. In addition to the paraphyletic clades, L0d1c contains two monophyletic clades. The first is the previously attested L0d1c1, which is defined by only two of the mutations previously associated with it (Figure S2 and Table S3). The second monophyletic clade in L0d1c, which we here define as L0d1c2a, is represented by six haplotypes and supported by four mutations (Figure S2).
The second basal branch of L0d1 is subhaplogroup L0d1b, which coalesces approximately 45 kya (95% C.I.: 35–56 kya) and is thus 10 ky older than previously estimated.6 As shown by our data, this is characterized by only one mutation, T3618C, splitting immediately into several subhaplogroups. Because the haplogroup previously labeled L0d1b1 is only the second of three hierarchical splits, the nomenclature is revised as follows: we propose to assign the label L0d1b1 to the first branch, which is characterized by four mutations (Figure S2). This is followed by a branch that we label L0d1b2, which is defined by several of the mutations previously assigned to L0d1b (Figure S2). This splits into L0d1b2a—represented by a monophyletic clade labeled in the latest version of Phylotree (ver. 1525) as L0d1b2—and L0d1b2b, which was previously defined as L0d1b1 and again contains two subclades: L0d1b2b1 and L0d1b2b2 (Figure 2).
Haplogroup L0d2, which coalesces around 65 kya (95% C.I.: 52–78 kya), is less common than L0d1 and is found at frequencies >10% only in populations from Botswana (mainly Khoisan foragers, but also the Bantu-speaking Tswana and Kgalagadi) and in the pastoralist Nama and forager Hai||om of Namibia (Table S1). With our data the diversity of this part of the tree is substantially increased: the earliest splits appear almost simultaneously, and we are unable to cleanly resolve the phylogeny (with a very low posterior probability for each of the nodes). From these splits arise four monophyletic clades: the previously defined L0d2a, L0d2b, and L0d2c, as well as a previously unreported branch that we here define as L0d2d. Although the clade previously defined as L0d2c is not changed by our data, subhaplogroup L0d2a is much more diverse than previously known, as is also reflected by our TMRCA estimate of approximately 40 ky (95% C.I.: 30–54 kya) versus the previous estimate of 9 ky.6 Some of the mutations previously thought to be characteristic of L0d2a actually define a subclade of L0d2a (Figure S2), which we here call L0d2a1, whereas the branch previously called L0d2a1 is shown to be a subclade of L0d2a1 and is therefore correspondingly labeled L0d2a1a (Figure 2). Two further previously undetected branches emerge from our data: L0d2a2, a sister clade of L0d2a1, and the very divergent subclade of L0d2 mentioned above, which we here define as L0d2d.
L0k separated from L0d approximately 145 kya (95% C.I.: 118–179 kya) and has a TMRCA of approximately 40 ky (95% C.I.: 28–53 kya). The majority of L0k lineages can be unambiguously assigned to the branch previously defined as L0k1;6 however, with our expanded data set we are now able to identify variation within L0k1, which consists of two sister clades: L0k1a (proposed in the latest version of Phylotree based on a sequence from Barbieri et al.14) and L0k1b (defined here), which we find in four individuals of our data set (Figure 2). Haplogroup L0k2 had previously been found in only one ethnolinguistically undefined individual from Yemen;6 in our data set, nine individuals from Bantu-speaking populations of Zambia and northeast Botswana belong to this haplogroup (Table S1).
The branching structure of the mtDNA phylogeny may have been shaped by events of climate change occurring at different periods in southern Africa. Thus, the deep splits in haplogroups L0k, L0d1b2, and L0d1c and the diversification of haplogroups L0d1a, L0d1b1, and L0d2c, which all happened approximately 30–40 kya, might be associated with the deterioration of climate in the central Kalahari area ∼35–27 kya.37 The aridification of this area, which was partly concurrent with a milder and more moist climate in the Eastern Cape,38 would have led to the dispersal of foragers to more suitable environments, with the subsequent separation and isolation of populations leading to the diversification of the mtDNA tree. Conversely, the shallow branches of L0k1a, L0d1c1, and L0d2a1a (Figure S2), which started to diversify 15–10 kya, suggest population expansions that may be associated with the postglacial amelioration of the climate and concomitant environmental diversification.39 Such expansions are also visible in the Bayesian Skyline Plots generated with BEAST34 (Figure S3): thus, L0k shows a signal of expansion at ∼5 kya and L0d1 and L0d2 expand ∼3–4 kya. Archeological evidence suggests an increase in population size beginning approximately 14 kya that peaked ∼4 ky,18 in good accordance with the genetic evidence.
The separation between L0k1a, L0k1b, and L0k2 is particularly evident from a network (Figure 3), where different patterns of diversity characterize the three haplogroups: whereas L0k1a has short branches and shows signals of expansion in its star-like pattern, L0k1b and especially L0k2 are composed of long separate branches and unique haplotypes that might represent the remains of an ancient and richer diversity. Although L0k was previously tentatively associated with a relatively late immigration of pastoralist Khoe populations rather than with central Kalahari foragers,40 our more comprehensive data demonstrate that this haplogroup, together with L0d, is in fact characteristic of the central Khoisan genetic profile, being absent only from South Africa. Seventy sequences are identified as belonging to L0k1a, coming predominantly from the Khoisan populations of Botswana (plus the Khoe-speaking Hai||om of Namibia) that also carry high frequencies of L0d1. In contrast, the distribution of L0k1b and L0k2 is highly restricted, being found only in the northern range of the L0d/L0k distribution, predominantly in Zambia (Figure 1C). Interestingly, and in contrast to L0k1a, L0k1b and L0k2 are found almost exclusively in Bantu-speaking populations (Figure 3; Table S1), who probably acquired it after contact with Khoisan groups; the only exceptions are an individual from Yemen with L0k2 and a Topnaar Nama (speaking a Khoe language) with L0k1b.
Figure 3.
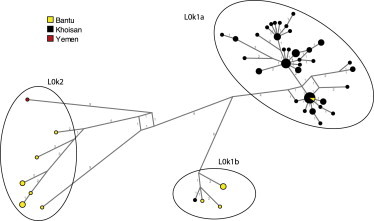
MJ-Network of L0k Based on Full Sequences
The near-exclusive presence of L0k1b and L0k2 haplotypes in Bantu-speaking populations rather than in Khoisan groups requires an explanation. The early separation from L0k1a of L0k2 (almost 40 kya) and L0k1b (around 30 kya) and the absence of recent diversification and branching might in principle suggest a very ancient incorporation into Bantu-speaking populations and subsequent isolation of these relic haplotypes. However, because there is no evidence for people speaking Bantu languages in southern Africa before 2,200 years ago,3 and because L0k is not found in the place of origin of the ancestors of the Bantu-speaking populations in western and central Africa,41,42 the contact between Khoisan and Bantu is unlikely to predate this period.
There are two possible alternative explanations. (1) These L0k1b and L0k2 lineages were incorporated into the Bantu-speaking populations through contact with now-extinct populations whose mtDNA haplogroup composition differed from that found in extant Khoisan groups in that they possessed the divergent L0k types. (2) The ancestors of extant Khoisan populations did possess the divergent L0k types and thus contributed them to Bantu-speaking populations (along with L0d and L0k1a lineages), but the haplogroup composition of the ancestral Khoisan groups was subsequently affected by drift, leading to the loss of L0k1b and L0k2.
We investigated these two alternative scenarios by assessing the probability that L0k1b and L0k2 would be lost from a Khoisan population by drift while being retained in Bantu-speaking populations after incorporation through contact. To do this, we assumed a relatively small effective population size for the Khoisan foragers, who throughout their history have lived in small nomadic bands,18,43 and a 10- to 100-fold higher effective population size for the Bantu-speaking food-producing groups. We simulated the variation in frequency of L0k1b/L0k2 for both the Khoisan and the Bantu virtual populations under three scenarios: (1) assuming Ne = 50 for Khoisan and Ne = 5,000 for Bantu speakers; (2) assuming Ne = 100 for Khoisan and Ne = 1,000 for Bantu speakers; and (3) assuming Ne = 1,000 for Khoisan and Ne = 10,000 for Bantu speakers. All the tests were iterated 10,000 times over 71 generations (about 2,000 years assuming 28 years per generation44), and the final haplogroup composition was checked in a random sample of 30 individuals from each population.
First we evaluated the likelihood of losing L0k1b/L0k2 for a range of initial frequencies of L0k1b/L0k2 in Khoisan (Table 1). The probability of losing L0k1b/L0k2 in the Khoisan is at least 95% for initial frequencies of not more than 3% for Ne = 50, 1.5% for Ne = 100, and 0.3% for Ne = 1,000. We next investigated the minimum amount of unidirectional migration from the Khoisan population necessary to ensure the presence of L0k1b/L0k2 in Bantu-speaking populations in more than 5% of the 10,000 simulated cases (Table 2). To do so, we chose three initial frequencies of L0k1b/L0k2 from Table 1 for Ne = 50, Ne = 100, and Ne = 1,000 that resulted in loss in more than 90% of the simulations, and we created hypothetical ancestral Khoisan populations carrying those frequencies; the rest of the population was assumed to carry other Khoisan haplogroups (i.e., L0d or L0k1a). Finally, we determined the frequency of Khoisan haplogroups other than L0k1b/L0k2 in the Bantu population after 71 generations (Table 2).
Table 1.
Values of the Initial Frequency of L0k1b/L0k2 in the Simulated Khoisan Population with Associated Probabilities of Losing Them after 71 Generations, Based on 10,000 Iterations
|
Initial Frequency of L0k1b/L0k2 in Khoisan |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.05 | 0.03 | 0.02 | 0.015 | 0.01 | 0.006 | 0.005 | 0.004 | 0.003 | 0.002 | |
| Ne = 50 | 93.3 | 96.3 | 96.9 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Ne = 100 | 86 | 91.4 | 94.1 | 97.2 | 97.2 | 100 | 100 | 100 | 100 | 100 |
| Ne = 1,000 | 45 | 63.4 | 74.3 | 80 | 85.8 | 91.9 | 92.9 | 94.3 | 95.4 | 97 |
Three hypothetical cases are considered, with an Ne for Khoisan of 50, 100, and 1,000. Probabilities are expressed in percent.
Table 2.
Migration Rates from Khoisan into Bantu Required to Retain L0k1b/L0k2 in Bantu with a Probability of at Least 5% over 10,000 Iterations, and Corresponding Estimates of the Frequency of Other “Khoisan” Haplogroups Retained in the Bantu
| Khoisan Ne= 50, Bantu Ne= 5,000 | Khoisan Ne= 100, Bantu Ne= 1,000 | Khoisan Ne= 1,000, Bantu Ne= 10,000 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Initial frequency of L0k1b/L0k2 in Khoisan | 0.05 | 0.03 | 0.02 | 0.03 | 0.02 | 0.015 | 0.004 | 0.003 | 0.002 |
| Minimum migration rate | 0.001 | 0.002 | 0.0025 | 0.0015 | 0.003 | 0.012 | 0.01 | 0.023 | –a |
| Frequency of other “Khoisan haplogroups” | 5% | 13% | 18% | 13% | 18% | 57% | 55% | 87% | ND |
The migration rate necessary to incorporate L0k1b/L0k2 into the Bantu-speaking group would be higher than 0.1, so the number of migrants would be larger than the Ne of the Khoisan population.
Overall, the results of these analyses indicate that there is a high probability of loss of L0k1b/L0k2 lineages in ancestral Khoisan populations if their initial frequency was not more than 1.5% (for Ne = 100, Table 1). With this initial frequency, a Bantu-speaking population with Ne = 1,000 could have retained the L0k1b/L0k2 lineages with a migration rate of 0.012 (Table 2). However, with this migration rate we would expect to find other Khoisan haplogroups (L0d or L0k1a) at a frequency of at least 57% in extant Bantu-speaking populations—and yet the frequency of L0d/L0k1a haplogroups in the Bantu-speaking populations with L0k1b/L0k2 haplotypes is significantly lower in all cases (chi-square test p values < 0.05 for all Bantu-speaking populations). The scenario based on an Ne of 1,000 for Khoisan (Tables 1 and 2) appears even more unlikely. Here the maximum frequency of L0k1b/L0k2 in the ancestral Khoisan population that could be lost by drift with a probability >95% is 0.3% (Table 1); a migration rate of 0.023 is needed in order to retain L0k1b/L0k2 haplogroups in the Bantu-speaking group, which would in turn lead to the incorporation of at least 87% other Khoisan haplogroups (Table 2). The only scenario that would lead to an incorporation of L0d/L0k1a in the Bantu-speaking immigrants compatible with the observed values are that of a Khoisan population of size 50 in contact with a Bantu-speaking population of size 5,000. In this case, if the initial frequency of L0k1b/L0k2 in the Khoisan group was 3%, it could have been incorporated into the Bantu-speaking population with a migration rate of 0.002 and subsequently been lost by drift in the Khoisan group. With such a migration rate, one would expect to find 13% other Khoisan haplogroups in the Bantu speakers, a value compatible with what is found in the Bantu-speaking populations carrying the divergent L0k lineages (Table S1). However, this scenario is based on an implausibly small Ne for the ancestral Khoisan population—because even though these foraging groups live in small bands, the bands are in contact with each other and exchange marriage partners.43 This ethnographic evidence in favor of a larger effective population size in Khoisan is supported by Bayesian Skyline plots for individual Khoisan populations, which show consistent population sizes of at least 1,000 (data not shown).
Overall, the results of this analysis indicate that it is very unlikely that the highly divergent L0k1b/L0k2 lineages were incorporated into the Bantu-speaking populations via gene flow from a population that was ancestral to a Khoisan population in our sample but subsequently lost from the Khoisan population via drift. Instead, these results support the hypothesis that the ancestors of the Bantu-speaking populations carrying the divergent L0k lineages (who now live mainly in Zambia) experienced gene flow from a pre-Bantu population that is nowadays extinct. Alternatively, it is possible that descendants from this pre-Bantu population do exist but have not yet been included in population genetic studies; however, our extensive sampling of populations from Botswana, Namibia, and West Zambia (which includes representatives of nearly all known Khoisan groups) makes it highly unlikely that this pre-Bantu Khoisan population has not yet been sampled. Our data thus indicate the existence of considerable genetic substructure in southern Africa prior to the Bantu expansion (cf. Barbieri et al.14) that is not represented in Khoisan groups today. Unfortunately, individuals from the relevant geographic areas have not yet been included in studies of autosomal DNA variation, making it impossible to assess the overall impact of this substructure on modern genetic diversity in southern Africa. However, from existing Y chromosomal data it appears that the admixture between the pre-Bantu autochthonous groups and the Bantu-speaking immigrants was restricted to the maternal line: the Y chromosome haplogroups found in the Zambian populations included here are not distinct from other sub-Saharan African groups.27 These findings highlight the importance of investigating in more detail other relic haplogroups in more regions of sub-Saharan Africa that might testify to a wider genetic variation in the cradle of modern humans.
In conclusion, with this extensive data set of L0d and L0k sequences, we considerably increase our knowledge of the variation in these basal haplogroups. Our results concerning the geographic and genetic structure within haplogroups L0d and L0k reveal interesting patterns. Whereas L0d1 is common to all the Khoisan populations of our data set and in published sources,6,7,32,33 L0d2 and L0k show a restricted distribution. The presence of divergent L0k haplotypes in populations speaking Bantu languages and their absence from Khoisan populations indicates that it will be possible to learn more about the prehistoric distribution of southern African pre-Bantu peoples by studying Bantu-speaking populations. Several promising areas of southern Africa have yet to be sampled in detail, most notably Zimbabwe, Malawi, and parts of South Africa, Zambia, and Angola; with the retrieval of genetic data from populations located in these areas, we should be able to gain a more complete picture of the genetic variation in southern Africa and better understand the ancient genetic structure.
Acknowledgments
We are grateful to all the individuals who voluntarily participated in this study and to the governments of Angola, Botswana, Namibia, and Zambia for supporting our research. We thank Hongyang Xu for help with the imputation process, Roland Schröder for lab assistance, and Martin Kircher and Mingkun Li for support in processing raw data and read alignments. This work has been carried out within the EUROCORES Programme EuroBABEL of the European Science Foundation and was supported by funds from the Deutsche Forschungsgemeinschaft and the Max Planck Society. J.R. was partially supported by a grant from Fundação para a Ciência e a Tecnologia (FCT; PTDC/BIA-BDE/68999/2006) and M.V. is supported by STAB VIDA, Investigação e Serviços em Ciências Biológicas, Lda., and by the Portuguese Ministry for Science, Technology and Higher Education through PhD grant SFRH/BDE/51828/2012.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
BioEdit Software, http://www.mbio.ncsu.edu/BioEdit/bioedit.html
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
HaploGrep, http://haplogrep.uibk.ac.at/
Phylotree, http://www.phylotree.org/
Accession Numbers
The GenBank accession numbers for the 485 sequences reported in this paper are KC345764–KC346248.
References
- 1.Campbell M.C., Tishkoff S.A. The evolution of human genetic and phenotypic variation in Africa. Curr. Biol. 2010;20:R166–R173. doi: 10.1016/j.cub.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum M.G.B., Jakobsson M. Deep divergences of human gene trees and models of human origins. Mol. Biol. Evol. 2011;28:889–898. doi: 10.1093/molbev/msq265. [DOI] [PubMed] [Google Scholar]
- 3.Phillipson D.W. Cambridge University Press; Cambridge: 2005. African Archaeology. [Google Scholar]
- 4.Tattersall I. Out of Africa: modern human origins special feature: human origins: out of Africa. Proc. Natl. Acad. Sci. USA. 2009;106:16018–16021. doi: 10.1073/pnas.0903207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonder M.K., Mortensen H.M., Reed F.A., de Sousa A., Tishkoff S.A. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 6.Behar D.M., Villems R., Soodyall H., Blue-Smith J., Pereira L., Metspalu E., Scozzari R., Makkan H., Tzur S., Comas D., Genographic Consortium The dawn of human matrilineal diversity. Am. J. Hum. Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tishkoff S.A., Gonder M.K., Henn B.M., Mortensen H., Knight A., Gignoux C., Fernandopulle N., Lema G., Nyambo T.B., Ramakrishnan U. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol. Biol. Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y.S., Olckers A., Schurr T.G., Kogelnik A.M., Huoponen K., Wallace D.C. mtDNA variation in the South African Kung and Khwe-and their genetic relationships to other African populations. Am. J. Hum. Genet. 2000;66:1362–1383. doi: 10.1086/302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight A., Underhill P.A., Mortensen H.M., Zhivotovsky L.A., Lin A.A., Henn B.M., Louis D., Ruhlen M., Mountain J.L. African Y chromosome and mtDNA divergence provides insight into the history of click languages. Curr. Biol. 2003;13:464–473. doi: 10.1016/s0960-9822(03)00130-1. [DOI] [PubMed] [Google Scholar]
- 10.Güldemann T. The Kalahari basin as an object of areal typology: A first approach. In: Schladt M., editor. Language, Identity and Conceptualization among the Khoisan. Rüdiger Köppe; Köln: 1997. pp. 137–169. [Google Scholar]
- 11.Coelho M., Sequeira F., Luiselli D., Beleza S., Rocha J. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol. Biol. 2009;9:80. doi: 10.1186/1471-2148-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlebusch C.M., Naidoo T., Soodyall H. SNaPshot minisequencing to resolve mitochondrial macro-haplogroups found in Africa. Electrophoresis. 2009;30:3657–3664. doi: 10.1002/elps.200900197. [DOI] [PubMed] [Google Scholar]
- 13.Quintana-Murci L., Harmant C., Quach H., Balanovsky O., Zaporozhchenko V., Bormans C., van Helden P.D., Hoal E.G., Behar D.M. Strong maternal Khoisan contribution to the South African coloured population: a case of gender-biased admixture. Am. J. Hum. Genet. 2010;86:611–620. doi: 10.1016/j.ajhg.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbieri C., Butthof A., Bostoen K., Pakendorf B. Genetic perspectives on the origin of clicks in Bantu languages from southwestern Zambia. Eur. J. Hum. Genet. 2012 doi: 10.1038/ejhg.2012.192. Published online August 29, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heine B., Honken H. The Kx’a family: a new Khoisan genealogy. J. Asian Afr. Stud. 2010;79:5–36. [Google Scholar]
- 16.Güldemann T. Institut für Afrikanistik, Universität Leipzig; Leipzig: 2005. Studies in Tuu (Southern Khoisan). University of Leipzig Papers on Africa, Languages and Literatures 23. [Google Scholar]
- 17.Güldemann, T., and Elderkin, E.D. (2010). On external genealogical relationships of the Khoe family. In Khoisan Languages and Linguistics: Proceedings of the 1st International Symposium January 4–8, 2003, M. Brenzinger and C. König, eds. (Riezlern/Kleinwalsertal. Quellen zur Khoisan-Forschung. Köln: Rüdiger Köppe,), pp. 15–52.
- 18.Deacon H.J., Deacon J. Altamira Press; Walnut Creek, CA: 1999. Human Beginnings in South Africa: Uncovering the Secrets of the Stone Age. [Google Scholar]
- 19.Güldemann T., Stoneking M. A historical appraisal of clicks: a linguistic and genetic population perspective. Annu. Rev. Anthropol. 2008;37:93–109. [Google Scholar]
- 20.Güldemann T. A linguist’s view: Khoe-Kwadi speakers as the earliest food-producers of southern Africa. South Afr. Humanit. 2008;20:93–132. [Google Scholar]
- 21.Ehret C. Bantu expansions: Re-envisioning a central problem of early African history. Int. J. Afr. Hist. Stud. 2001;34:5–41. [Google Scholar]
- 22.Pakendorf B., Bostoen K., de Filippo C. Molecular perspectives on the Bantu expansion: a synthesis. Language Dynamics and Change. 2011;1:50–88. [Google Scholar]
- 23.Kinahan J. A History of Namibia: From the Beginning to 1990, M. Wallace. Hurst and Company; London: 2011. From the beginning: the archaeological evidence; pp. 15–43. [Google Scholar]
- 24.Reid A., Sadr K., Hanson-James N. Herding traditions. In: Lane P., Reid A., Segobye A., editors. Ditswa MMung: The Archaeology of Botswana. Pula Press and The Botswana Society; Gaborone: 1998. pp. 81–100. [Google Scholar]
- 25.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 26.Kloss-Brandstätter A., Pacher D., Schönherr S., Weissensteiner H., Binna R., Specht G., Kronenberg F. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 27.de Filippo C., Barbieri C., Whitten M., Mpoloka S.W., Gunnarsdóttir E.D., Bostoen K., Nyambe T., Beyer K., Schreiber H., de Knijff P. Y-chromosomal variation in sub-Saharan Africa: insights into the history of Niger-Congo groups. Mol. Biol. Evol. 2011;28:1255–1269. doi: 10.1093/molbev/msq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickrell J.K., Patterson N., Barbieri C., Berthold F., Gerlach L., Güldemann T., Kure B., Mpoloka S.W., Nakagawa H., Naumann C. The genetic prehistory of southern Africa. Nat. Commun. 2012;3:1143. doi: 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 30.Maricic T., Whitten M., Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briggs A.W., Good J.M., Green R.E., Krause J., Maricic T., Stenzel U., Lalueza-Fox C., Rudan P., Brajkovic D., Kucan Z. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- 32.Schlebusch C.M., de Jongh M., Soodyall H. Different contributions of ancient mitochondrial and Y-chromosomal lineages in ‘Karretjie people’ of the Great Karoo in South Africa. J. Hum. Genet. 2011;56:623–630. doi: 10.1038/jhg.2011.71. [DOI] [PubMed] [Google Scholar]
- 33.Henn B.M., Gignoux C.R., Jobin M., Granka J.M., Macpherson J.M., Kidd J.M., Rodríguez-Botigué L., Ramachandran S., Hon L., Brisbin A. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behar D.M., van Oven M., Rosset S., Metspalu M., Loogväli E.L., Silva N.M., Kivisild T., Torroni A., Villems R. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas D.S.G., Shaw P.A. Late Quaternary environmental change in central southern Africa: new data, synthesis, issues and prospects. Quat. Sci. Rev. 2002;21:783–797. [Google Scholar]
- 38.Lewis C.A. Late Quaternary climatic changes, and associated human responses, during the last 45000 yr in the Eastern and adjoining Western Cape, South Africa. Earth Sci. Rev. 2008;88:167–187. [Google Scholar]
- 39.Mitchell P. Cambridge University Press; Cambridge: 2002. The Archaeology of Southern Africa. [Google Scholar]
- 40.Schlebusch, C.M. (2010). Genetic variation in Khoisan-speaking populations from southern Africa. PhD thesis, University of the Witwatersrand, Johannesburg.
- 41.Veeramah K.R., Connell B.A., Ansari Pour N., Powell A., Plaster C.A., Zeitlyn D., Mendell N.R., Weale M.E., Bradman N., Thomas M.G. Little genetic differentiation as assessed by uniparental markers in the presence of substantial language variation in peoples of the Cross River region of Nigeria. BMC Evol. Biol. 2010;10:92. doi: 10.1186/1471-2148-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batini C., Lopes J., Behar D.M., Calafell F., Jorde L.B., van der Veen L., Quintana-Murci L., Spedini G., Destro-Bisol G., Comas D. Insights into the demographic history of African Pygmies from complete mitochondrial genomes. Mol. Biol. Evol. 2011;28:1099–1110. doi: 10.1093/molbev/msq294. [DOI] [PubMed] [Google Scholar]
- 43.Barnard A. Cambridge University Press; Cambridge, UK: 1992. Hunters and herders of southern Africa: a comparative ethnography of the Khoisan peoples. [Google Scholar]
- 44.Fenner J.N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


