Abstract
Crystal (Cry) proteins are globally used in agriculture as proteinaceous insecticides. They have also been recently recognized to have great potential as anthelmintic agents in targeting parasitic roundworms (e.g. hookworms). The most extensively characterized of the anthelmintic Cry proteins is Cry5B. We report here the 2.3 Å resolution structure of the proteolytically activated form of Cry5B. This structure, which is the first for a nematicidal Cry protein, shows the familiar three-domain arrangement seen in insecticidal Cry proteins. However, domain II is unusual in that it more closely resembles a banana lectin than it does other Cry proteins. This result is consistent with the fact that the receptor for Cry5B consists of a set of invertebrate-specific glycans (attached to lipids), and also suggests that domain II is important for receptor binding. We found that not only is galactose an efficient competitor for binding between Cry5B and glycolipids, but so too is N-acetylgalactosamine (GalNAc). GalNAc is one of the core arthroseries tetrasaccharides of the Cry5B receptor, and galactose an antennary sugar that emanates from this core. These and prior data suggest that the minimal binding determinant for Cry5B consists of a core GalNAc and two antennary galactoses. Lastly, the protoxin form of Cry5B was found to bind nematode glycolipids with equal specificity as activated Cry5B, but with lower affinity. This suggests that the initial binding of Cry5B protoxin to glycolipids can be stabilized at the nematode cell surface by proteolysis. These results lay the groundwork for the design of effective Cry5B-based anthelmintics.
Keywords: Crystal proteins, Bacillus thuringiensis, Nematicidal, Anthelmintic, Lectin
Introduction
Crystal (Cry) proteins are widely used on a global scale as proteinaceous insecticides. These proteins target caterpillars, beetles, mosquitoes, and black flies, but have no effects on higher animals; they also lack the harmful side effects that small molecule pesticides often have on higher animals (1–5). Cry proteins are produced by the Gram-positive soil bacterium Bacillus thuringiensis (Bt), and over 200 varieties of such proteins are known (with sequence identities spanning <20% to >90%) (2). These proteins are produced as crystalline inclusions in sporulating Bt, and following ingestion by invertebrates, are solubilized in the invertebrate gut as monomeric protoxins. Monomeric protoxins are then activated by host proteases, which remove N-terminal and, in almost all cases, C-terminal regions of the protoxins to yield ~60–70 kDa activated Cry proteins. Activated Cry proteins bind specific receptors (2) on midgut epithelial cells and insert into the plasma membrane of these cells to form cytotoxic pores. In some cases, more than one receptor has been identified, and receptor binding has been noted to occur sequentially (6). Receptor binding has also been noted to promote oligomerization of activated Cry proteins, which appears to enhance subsequent pore formation (7).
The structures of seven insecticidal Cry proteins (Cry1Aa, Cry2Aa, Cry3Aa, Cry3Bb, Cry4Aa, Cry4Ba, and Cry8Ea1) have been determined and are available in the Protein Data Bank (8–14). These proteins have a characteristic compact three-domain architecture. Domain I is an all α-helical bundle responsible for pore formation (5). Evidence suggests that the pore is initiated by the insertion of an α-helical hairpin into the membrane, which is then followed by the insertion of the other helices (15). Domains II and III are rich in β-sheets and resemble lectins of the β-prism fold and jellyroll topology families, respectively (16). Domains II and III have been implicated in receptor binding (6, 17–22), and domain III also modulates pore activity (23).
Cry proteins are active not only against insects but also nematodes, including those that infest crops or are parasites of animals (24–29). The most extensively characterized nematicidal Cry protein is Cry5B, which has been shown to be a potent in vivo anthelmintic, providing therapeutic effects against the hookworm parasite Ancylostoma ceylanicum in hamsters and the intestinal roundworm parasite Heligmosomoides bakeri in mice (24, 25). The potential use of Cry5B as an anthelmintic has immense implications as intestinal roundworms infect ~2.3 billion people worldwide, and there is an urgent need for new and better anthelmintics (30–32). The receptor for Cry5B consists of a set of invertebrate-specific glycans that are attached to lipids on the surface of intestinal epithelial cells (33, 34). These glycans are composed of an arthroseries tetrasaccharide core (GalNAcβ1-4GlcNAcβ1-3Manβ1-4Glc) decorated with antennary glycans (Gal, Glc, Fuc, and 2-O-Me-Fuc), or single sugars (Fig. S1) (33, 35).
Due to the importance of Cry5B as an anthelmintic and as a roundworm-active Cry protein (as opposed to an insect-active Cry protein), we undertook to characterize the structure of Cry5B and characterize its glycan-binding properties in further detail. We found that Cry5B is the most structurally divergent Cry protein described to date, with domain II being especially divergent. Domain II most closely resembles a banana lectin, suggesting that this domain is important for Cry5B glycolipid receptor binding. We found that GalNAc acts as an effective competitor for binding between Cry5B and nematode glycolipids, which in combination with prior results, suggests that the minimal binding determinant for Cry5B consists of a core GalNAc and two antennary galactoses. Lastly, we found that the protoxin form of Cry5B binds nematode glycolipids with the same specificity as activated Cry5B, but with weaker affinity. This suggests that the binding of Cry5B protoxin to the nematode cell surface may be strengthened by subsequent proteolysis. Our results provide a foundation for the design of effective anthelmintics based on Cry5B.
Experimental Procedures
Expression, purification and crystallization of Cry5B protein
Cry5B protoxin was purified from the crystal protein-deficient B. thuringiensis (Bt) strain HD1 that had been transformed with a plasmid encoding cry5B (36). Cry5B protoxin was purified as previously described from spore crystal lysates (24) and stored as a precipitate in water at −80 °C. For experimental manipulations, aliquots of Cry5B protoxin were solubilized in 20 mM HEPES, pH 8.0 at a final concentration of 5 mg/mL. The protein concentration was determined using a calculated ε280 of 162,510 M−1cm−1.
For expression in E. coli, residues 1–772 of cry5B were cloned with an N-terminal His- tag into vector pQE9, and expressed in E. coli M15. Bacteria were grown at 37 °C to mid-log phase (OD600 0.6–0.8), and expression of Cry5B(1-772) was induced with 0.15 mM isopropyl β-D-1-thiogalactopyranoside. After induction, bacteria were grown at 25 °C for 10 hours, and then harvested by centrifugation (6328 × g, 20 min, RT). Bacteria were lysed by sonication in phosphate buffered saline (PBS), and the lysate was centrifuged (17,418 × g, 20 min, 4 °C). The supernatant, which contained Cry5B(1-772), was applied to a Ni2+-nitrilotriacetic acid (NTA) agarose column, the column was washed with 3 column volumes of NiC buffer (0.5 M NaCl, 20 mM HEPES, pH 8.0), and Cry5B(1-772) was eluted from the column with NiC buffer supplemented with 0.5 M imidazole. Cry5B(1-772) was dialyzed in 50 mM NaCl, 20 mM HEPES, pH 8.0 and concentrated by ultrafiltration to 5 mg/mL. The concentration of Cry5B(1-772) was determined using a calculated ε280 of 108,525 M−1 cm−1. For phase determination, selenomethionine (SeMet) was biosynthetically incorporated into Cry5B(1-772) as previously described (37), and SeMet-labeled Cry5B(1-772) was expressed and purified as above.
Cry5B protoxin produced in B. thuringiensis and Cry5B(1-772) produced in E. coli were activated by cleavage with elastase (overnight, RT, 200:1 Cry5B:elastase mass ratio) in 20 mM HEPES, pH 8.0. Activated Cry5B was purified by gel filtration chromatography (Superdex 200 26/60) in 200 mM NaCl, 20 mM HEPES, pH 8.0. Activated Cry5B was then dialyzed in 50 mM NaCl, 20 mM HEPES, pH 8.0 and concentrated to 5 mg/mL. The protein concentration of activated Cry5B was determined using a calculated ε280 of 72,825 M−1 cm−1. The site of elastase cleavage was determined by N-terminal sequencing. For this, activated Cry5B was run on SDS-PAGE and blotted onto a polyvinylidene fluoride membrane. The molecular mass of elastase cleaved Cry5B was determined by MALDI-TOF mass spectrometry to be 66,145 Da (calculated 65,815 Da for residues 112–170 and 173–698, the nicked protein that constitutes elastase-cleaved Cry5B).
Crystallization, data collection, structure determination, and refinement
Crystals of elastase-activated Cry5B produced in B. thuringiensis were grown at RT by the vapor diffusion, hanging drop method. One μL of 5 mg/mL activated Cry5B was mixed with 1 μL of 20% PEG 3350, 0.2 M NaCH3COO as the precipitant. Crystals of SeMet-labeled, elastase-activated Cry5B, which had been prepared from Cry5B(1-722) produced in E. coli, were obtained under the same condition. Crystals of activated Cry5B were soaked in the precipitant solution supplemented with 15% glycerol as a cryoprotectant, before being mounted and cooled to 100 K in a N2 stream.
Diffraction intensities from native and SeMet-labeled Cry5B crystals were collected at the Advanced Light Source (ALS, Berkeley, CA) beamline 4.2.2 (Table S1). Integration, scaling, and merging of intensities were carried out using Mosflm and Scala (38). Phases were determined by the single anomalous dispersion (SAD) method from crystals of SeMet-labeled Cry5B using Phenix (39), and were also refined using Phenix. Three selenomethionine positions were identified in the asymmetric unit, which contained a single molecule of elastase-activated Cry5B. The initial electron density map clearly showed an α-helical bundle corresponding to domain I and β-sheets corresponding to domains II and III. Residues 187–326 were built automatically with Phenix, and residues 112–161 (corresponding to the α4 helix in domain I) were manually built using Coot (40), as were domains II and III. The partial model encompassing ~450 residues, which was missing the loops connecting the β strands in domains II and III, was then refined against the higher resolution native data set. The electron density map generated from the native data enabled tracing of the rest of the chain, except for residues 171 and 172, which were removed by elastase and are missing in the model. Simulated annealing with torsion angle dynamics was carried out from 3000 K using the slow-cool protocol of CNS (41). Ten cycles of maximum likelihood restrained refinement was subsequently carried out using REFMAC (38), each cycle being followed by manual rebuilding into σA-weighted 2mFo-DFc and mFo-DFc maps using Coot. Waters were added in the later stages of the refinement using Phenix with default parameters (3σ peak height in mFo-DFc maps), followed by inspection of maps.
Structure validation was performed using Procheck (42) and Molprobity (43). In the final model, 93.1% and 98.8% of residues were in allowed and generously allowed Ramachandran regions, respectively. The final map had correlation coefficients of 0.997 and 0.995 for the main chain and side chains, respectively, as calculated with OVERLAPMAP (38). The Molprobity clash score was 19.4 (61st percentile) and the overall score was 2.57 (55th percentile). The atomic coordinates and structure factors have been deposited with the Protein Data Bank (accession code 4D8M).
Structure figures were produced with the program PyMOL (44). Structure-based sequence alignments were generated using Expresso (45) and displayed using ESPript (46). Calculations of structural superposition and sequence identity were carried out with Coot.
Purification of upper phase glycolipids from nematodes
Caenorhabditis elegans N2 and Pristionchus pacificus PS312 were grown on high growth medium plates seeded with E. coli OP50. Once near starvation, nematodes were harvested from the plates, and pellets of mixed-life stage worms amounting to 0.5 mL were washed three times with water. The pellets were resuspended in three pellet volumes of water and sonicated five times for 2 min each at 11–14 watts, with chilling on ice between sonication steps. Upper phase glycolipids were purified based on the Svennerholm partitioning method (47–49).
Glycolipid overlay assay
The thin-layer chromatography (TLC) assay was carried out as previously described (49). For biotinylation, elastase-activated Cry5B (6 μL of a 2.5 mg/mL solution in 200 mM NaCl, 20 mM HEPES, pH 8.0) was treated with a 3.16-fold molar excess of N-Hydroxysuccinimido biotin in 10 μL total volume of 20 mM HEPES, pH 8.0. After 2 h at RT on a rocker, 90 μL of 20 mM HEPES, pH 8.0 was added. Cry5B protoxin was biotinylated similarly, except that a 22-fold molar excess of N-Hydroxysuccinimido biotin was used.
Developed HPTLC plates were fixed for 60 s in 40 mL hexanes (mixture of isomers, 99% pure, Acros Organics) and 40 mL hexanes containing 0.02 % polyisobutylmethacrylate. After drying for at least 5 min at 45 °C, the plates were blocked with 9.9 mL of blocking buffer (PBS containing 0.5 % bovine serum albumin and 0.02 % Tween). When monosaccharides were added to compete for binding, the HPTLC plates were blocked in 8.9 mL blocking buffer and 1 mL of 1 M monosaccharide. After 30 min on a rocker, 100 μL of labeled Cry5B was added. After a further 2 h on the rocker, the plates were washed with 10 mL blocking buffer for 1 min and then for a second time for 5 min. To visualize Cry5B, the plates were incubated for 1 h with 40 μL avidin DH and biotinylated alkaline phosphatase H (Reagent A and B, Vectastain ABC-AP kit) dissolved in 6 mL blocking buffer. The plates were washed with 10 mL blocking buffer three times for 1 min each, and then incubated in the alkaline phosphatase substrate NBT/BCIP (30 μL of each NBT and BCIP per 5 mL solution, Vector Labs) for 2–4 h. There were slight variations in the sharpness of bands from experiment to experiment, but the result that Gal and GalNAc blocked binding by Cry5B to glycolipids was consistent throughout.
Results
Overall structure of Cry5B
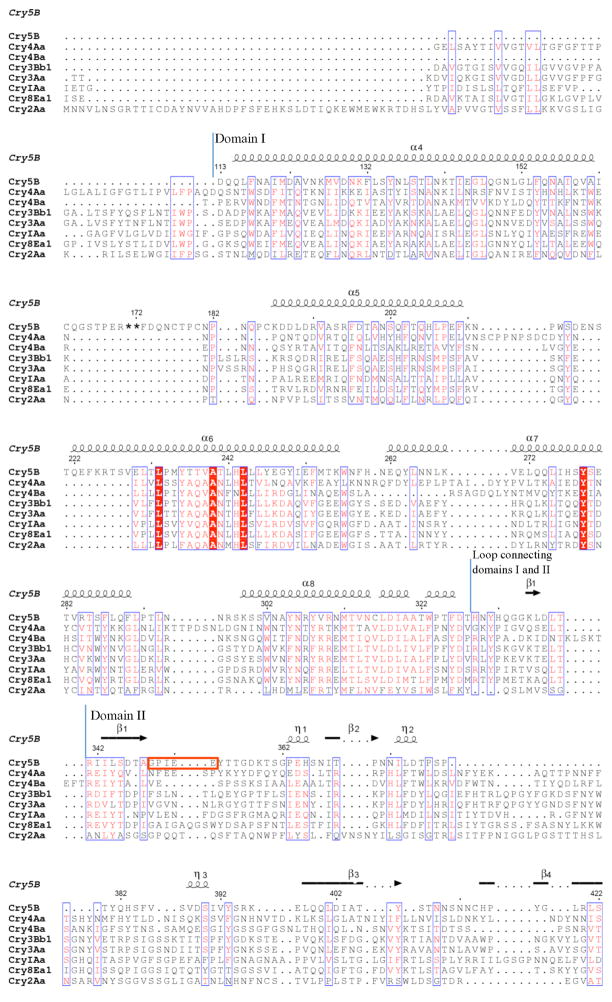
The structure of elastase-activated Cry5B (residues 112–698) was determined by single anomalous diffraction (SAD) and refined to 2.3 Å resolution limit (Table S1). The electron density map was unambiguous and the entirety of the protein chain was clearly defined, except for residues 171 and 172, which are missing in the final model. Elastase-activated Cry5B is nicked at these residues, and as such is composed of two polypeptide chains (residues 112–170 and 173–698) connected by disulfide bonds. This conclusion was verified by N-terminal sequencing and MALDI-TOF mass spectrometry (data not shown). Although the nematicidal Cry5B protein shares only ~16–19% sequence identity with insecticidal crystal proteins, Cry5B has a three-domain structure that resembles that of insecticidal crystal proteins (Fig. 1). The insecticidal crystal proteins of known structure (i.e., Cry1Aa, Cry2Aa, Cry3Aa, Cry3Bb1, Cry4Aa, Cry4Ba, Cry8Ea1) are highly similar to one another. Their Cα positions vary by 1.0–2.1 Å root-mean-square deviation (rmsd) in pairwise comparisons, except for the most dissimilar member of this group, which is Cry2Aa (pairwise rmsd of 2.5–2.8 Å) (10). By comparison, Cry5B has an average rmsd of ~3 Å in pairwise comparisons with the insecticidal crystal proteins. This indicates that Cry5B is the most structurally divergent crystal protein described to date, in accordance with its differing host tropism.
Figure 1. Structure of Cry5B.
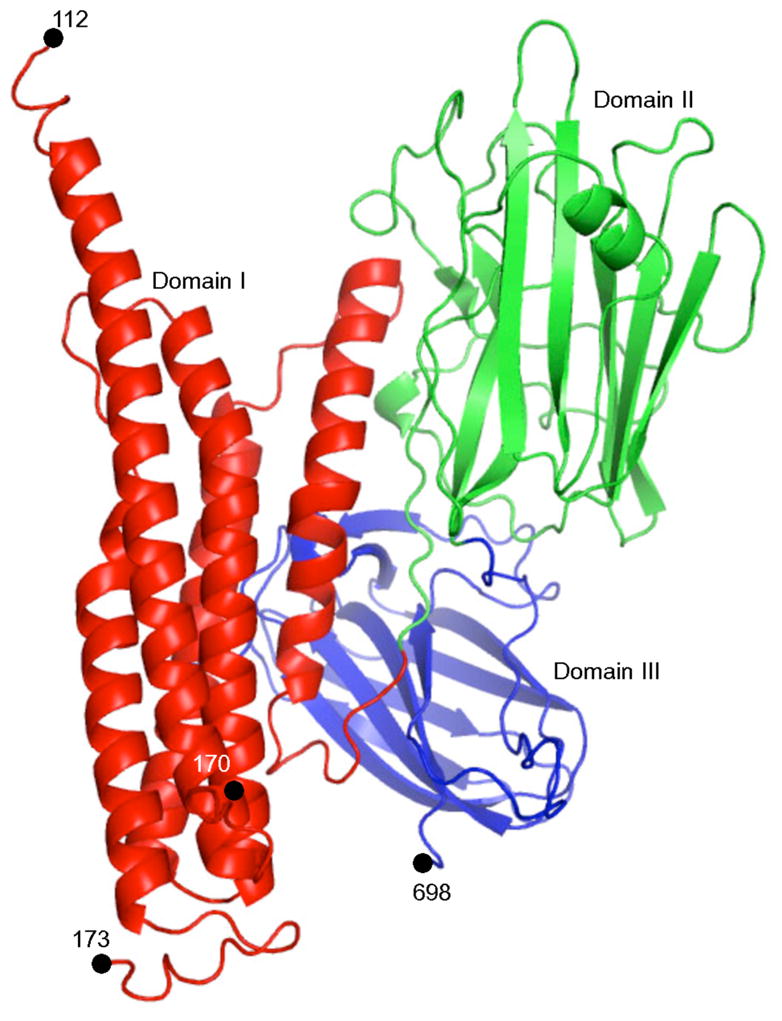
Cry5B is shown in a ribbon diagram. Domain I (red) is a five α-helix bundle, domain II (green) a β-prism domain, and domain III (blue) a β-sandwich domain.
Domain I
Domain I is implicated in pore-formation (8), and in accord with its central mechanistic role, domain I is the most structurally conserved of the three domains of Cry5B. Cry5B domain I (residues 112–328), which is composed of a five α-helix bundle, is superimposable on domain I of the insecticidal Cry proteins with an average rmsd of 1.97 Å (153 Cα positions on average) (Fig. 2a), despite this domain sharing only 22% average sequence identity (Fig. 3). The five helices of activated Cry5B match the five helices of activated Cry4B, in agreement with the fact that these two proteins are cleaved at equivalent locations. In comparison, the protoxin Cry2Aa is uncleaved and therefore extends much farther in the N-terminal direction than does Cry5B; thus the five helices of Cry5B match the last five helices (α4–α8) of Cry2Aa. Similarly, some of the activated Cry proteins (i.e., Cry1Aa, Cry3Aa, Cry3Bb and Cry4Aa) are cleaved at positions N-terminal to activated Cry5B, and here again the five Cry5B helices match the last five helices (α3–α7) of these activated proteins.
Figure 2. Domains of Cry5B.
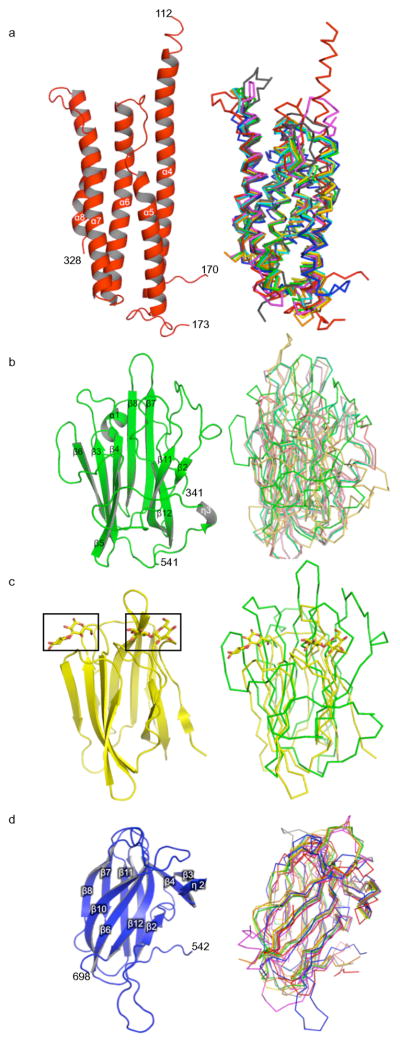
a. Left, domain I in ribbon representation. Right, superposition of Cry5B domain I (red) with domain I from Cry1 Aa (green), Cry2Aa (blue), Cry3Aa (yellow), Cry3Bb (orange), Cry4Aa (grey), Cry4Ba (magenta), and Cry8Ea1 (cyan), all in chain representation.
b. Left, domain II in ribbon representation, and right, superposition with domain II from other Cry proteins, as in panel a.
c. Left, Banana lectin in ribbon representation with bound glycans (yellow) boxed. Right, superposition of banana lectin (yellow), including glycans (laminaribiose), with Cry5B domain 2 (green), in chain representation.
d. Left, domain III in ribbon representation, and right, superposition with domain III from other Cry proteins, as in panel a.
Figure 3. Sequence alignment.
Structure-based sequence alignment of Cry5B and other structurally characterized Cry proteins was carried out with Expresso (Di Tommaso et al., 2011), and annotated with the secondary structure of Cry5B, indicated at top, with ESPript (Gouet et al., 2003). Horizontal red boxes indicate the GXXXE and GG sequences in domain II.
The α6 helix of Cry5B is the central helix in the five-helix bundle and is surrounded by the four other helices. This central α-helix is the most conserved of the α-helices in domain I, containing three residues (Leu233, Ala240, and Leu244) that are absolutely conserved among the structurally characterized Cry proteins (Fig. 3). The central helix along with the preceding helix has been suggested to act as a ‘helical hairpin’ that inserts into the plasma membrane of host midgut epithelial cells and initiates the formation of a cytotoxic pore (15, 50). Mutagenesis studies have demonstrated the crucial role of this central α-helix role in toxicity (2, 51–53).
There are some small differences between Cry5B and the other Cry proteins in domain I. First, the α4 helix of Cry5B at ~45 residues is unusually long for Cry proteins. Second, Cry5B has two disulfide bridges (Cys163-Cys180, Cys177-Cys186) connecting the long loop between α4 and α5, whereas other crystal proteins lack these disulfides, or as in the case of Cry4Aa, have only one disulfide at this location. The long α4–α5 loop contains the nick generated by elastase, which results in the loss of residues 171 and 172. Residues 170 and 173 apparently shift position after the nicking, as deduced from the fact that these residues are too far apart for the missing residues to span the intervening space.
Domain II
Domain II (residues 341–541) consists of a β-prism, as found in other Cry proteins, as well as in the jacalin-related superfamily of lectins (54) (Fig. 2b). β-prism domains generally consist of three four-stranded β-sheets, each with a Greek key topology. These sheets are all parallel to an approximate three-fold axis of symmetry and form the sides of a prism. In Cry5B, one of the β-sheets has four long strands (β5β4β3β6), whereas the other two have a mixture of two long and two short strands. These latter two are composed of β1β12β11β2, with β1 and β2 being short, and β9β8β7β10, with β9 and β10 being short; this latter sheet has an a-helix intervening between β9 and β10.
Domain II is the most structurally divergent of the three domains of Cry5B, with an average rmsd of 3.1 Å (122 average Cα, 8% average sequence identity) as compared to domain II of the insecticidal Cry proteins. A superposition of these domains makes this point clearly (Fig. 2b). In fact, a structural homology search revealed that Cry5B domain II is most similar not to a Cry protein domain II but to a banana lectin (2.7 Å rmsd, 114 Cα, Z-score 9.6, 14% sequence identity) (55) (Fig. 2c). Notably, the structure of the banana lectin (BanLec) has been determined with two different glycans bound, laminaribiose (Glcβ1-3Glc) and Xyl-β1,3-Man-α-O-Methyl (55). Two separate glycan binding sites were observed, both formed by loops at the tips of the β-sheets. The equivalent of the first site in Cry5B consists of the β1β2 and β11β12 loops (emanating from the β1β12β11β2 sheet), and the β3β4 and β5β6 loops (emanating from the β5β4β3β6 sheet) for the second site (Fig. 3). The closer relationship of Cry5B domain II with BanLec as compared to a crystal protein suggests that this domain has a primary role in glycan binding (17).
Domain III
Domain III (residues 542–698) is a β-sandwich with a jellyroll topology, composed of two four-stranded antiparallel β-sheets (Fig. 2d). The β10β6β12β2 sheet packs against domain II, whereas the β5β11β7β8 sheet faces solvent. The β1 and β9 strands are short and part of the β5β11β7β8 and β10β6β12β2 sheets, respectively. Two additional β-strands (β3 and β4) and two short 310-helices lie rougly perpendicular to the β-sandwich and connect β2 to β5. In contrast to domain II, domain III is structurally most similar to domain III of other Cry proteins (2.2 Å average rmsd, 127 average Cα, 20% average sequence identity). As with other Cry proteins, domain III also has structural similarity to carbohydrate binding modules, such as the one in β-agarase (2.4 Å rmsd, 121 Cα, 15% sequence identity, Z-score 13.0). The structure of β-agarase bound to a hexasaccharide is known (56), and the glycan-binding site in β-agarase is equivalent in Cry5B to the solvent-exposed face of the β5β11β7β8 sheet (Fig. 2d).
Interactions of Cry5B with glycolipids
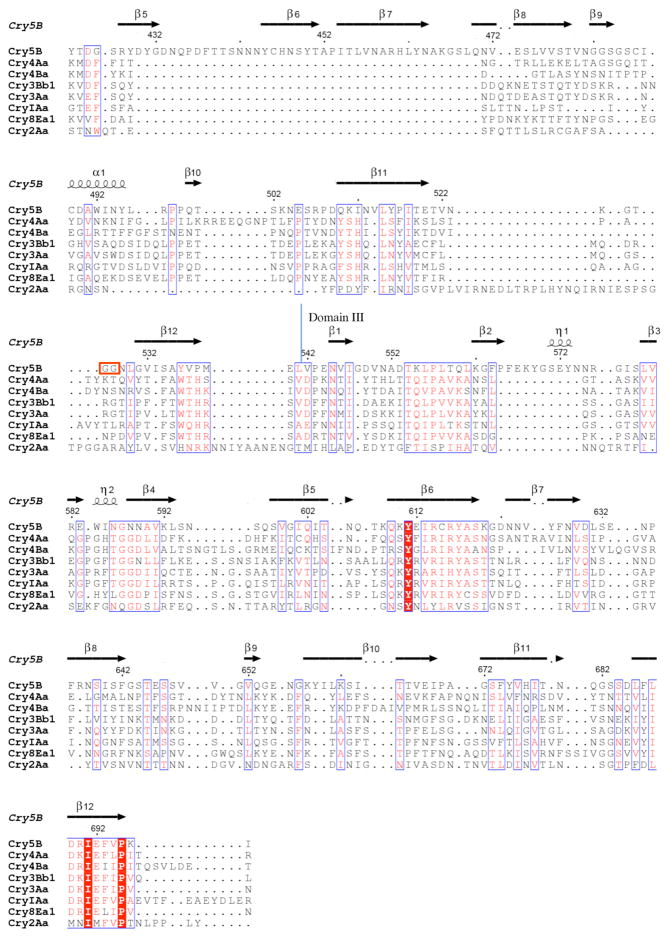
Cry5B has been shown to bind several C. elegans glycolipid species (35), all containing a core arthroseries tetrasaccharide (GalNAcβ1-4GlcNAcβ1-3Manβ1-4Glc) and antennary glycans (Gal, Glc, Fuc, and 2-O-Me-Fuc) (Fig. S1). Significantly, interaction of Cry5B with these glycolipids is effectively blocked by one of the antennary glycans, galactose (35), suggesting that galactose serves at least in part as a Cry5B-binding determinant. To further characterize the interaction of Cry5B with glycolipids, we purified polar glycolipids from P. pacificus, a model nematode that is sensitive to Cry5B (29, 57), along with those from C. elegans. The polar glycolipids from these nematodes were separated by thin-layer chromatography (TLC) and visualized by orcinol staining (Figs. 4a and 4g). This staining showed that P. pacificus has a different set of glycolipids than does C. elegans. Next, Cry5B protoxin and elastase-activated Cry5B, which had both been biotinylated, were overlaid on the TLC plates and visualized. As shown previously, activated Cry5B bound to a number of the C. elegans glycolipids, including the B, C, E, and F species (Figs. 4b and S1), and these interactions were inhibited specifically by galactose (Fig. 4c) (35). For P. pacificus, a double band near the bottom of the TLC plate bound activated Cry5B, and this binding was also inhibited by galactose, demonstrating its specificity (Figs. 4b and 4c). The same pattern of interaction was seen with Cry5B protoxin (Figs. 4h and 4i), but these interactions were much weaker than for activated Cry5B. This was deduced from the fact that galactose completely eliminated glycolipid interactions in the case of Cry5B protoxin but only diminished them in the case of activated Cry5B (c.f. Figs. 4c and 4i). These results indicate that proteolytic activation is not required for the interaction of Cry5B with its glycolipid receptors, and does not change the specificity of such interactions. Activation does, however, increase the affinity of these interactions.
Figure 4. Binding of activated Cry5B and Cry5B protoxin to upper phase glycolipids.
Upper phase glycolipids of C. elegans (C.e.) and P. pacificus (P.p.) were resolved by thin layer chromatography and stained with orcinol (a and g), and overlaid with elastase-activated Cry5B (b–f) or Cry5B protoxin (h–l) in the presence of 100 mM galactose (Gal, c, i), 100 mM glucose (Glc, d, j), 100 mM N-acetylglucosamine (GlcNAc, e, k), or 100 mM N-acetylgalactosamine (GalNAc, f, 1). The glycolipid band species are indicated to the left of panels b and h. This is a representative set from several experiments carried out.
We next asked which other monosaccharides competed with C. elegans and P. pacificus glycolipid binding by Cry5B to get further insight into the binding of Cry5B with its glycolipid receptors. We found that neither glucose nor N-acetylglucosamine (GlcNAc) affected interaction to the same extent as galactose, although there was perhaps a slight diminution with glucose (Figs. 4d, 4e, 4j, 4k). Glucose is at the base of the arthroseries tetrasaccharide core (i.e., attached to ceramide) and is also one of the antennary sugars, and GlcNAc is part of the arthroseries tetrasaccharide core (Fig. S1). Notably, we found that the addition of N-acetylgalactosamine (GalNAc) substantially diminished binding of both Cry5B protoxin and activated Cry5B to the glycolipids (Figs. 4f and 4l). GalNAc is part of the arthroseries tetrasaccharide core and the sugar from which the antennary glycans emanate. These results provide evidence that Cry5B not only interacts with galactose but GalNAc as well.
Discussion
We report here the first structure of a nematicidal Cry protein, and find it to be the most structurally divergent of the Cry proteins characterized to date. The previously characterized Cry proteins, which are all insecticidal, are highly similar in structure to one another (8, 9, 11–14). Of this insecticidal group, Cry2Aa is the most dissimilar (10), which may be related t the fact that it is toxic against both Lepidoptera and Diptera, while the other insecticidal Cry proteins act against only a single order, either Lepidoptera (Cry1Aa) (9), Diptera (Cry4Aa, Cry4Ba) (12, 13), or Coleoptera (Cry3Aa, Cry3Bb, Cry8Ea1) (8, 11, 14). Cry5B is structurally even more divergent than Cry2Aa, with the largest divergence between Cry5B and the insecticidal Cry proteins occurring in domain II. This domain has been implicated in receptor specificity in other Cry proteins (17, 19–22), and in Cry5B this activity would fit its divergent organismal specificity. In contrast, domain I is highly conserved between Cry5B and the insecticidal Cry proteins, which likely reflects its conserved role in pore formation (8). Domain III of Cry5B also bears a close relationship to insecticidal Cry proteins, although not as great as domain I but greater than domain II. This may reflect a role of domain III in modulating both receptor specificity and pore activity, as has been seen in other Cry proteins (6, 18, 21, 23).
The structure of Cry5B was modeled several years ago based on the structure of Cry1Aa (58). The modeled structure, however, is dissimilar in detail from the experimental one reported here, with an overall rmsd of 2.8 Å (401 Cα) between the two; the greatest differences occur in domain II (rmsd 3.4 Å, 131 Cα). This is not surprising given our finding that Cry5B is the structurally most divergent Cry protein characterized to date, and that domain II is the most dissimilar of all the domains.
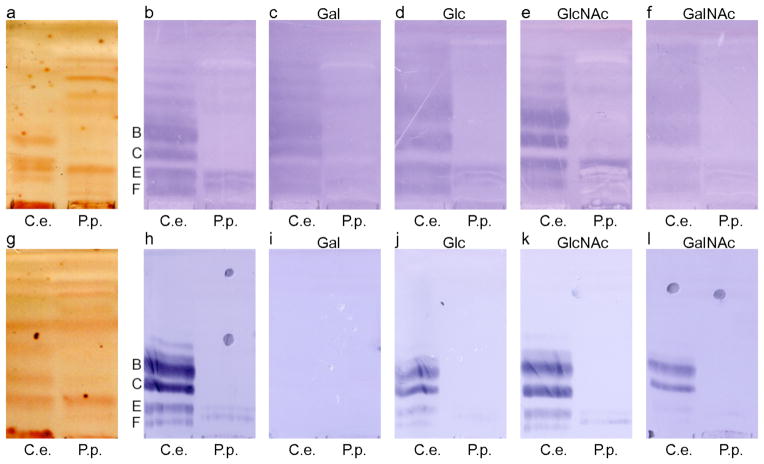
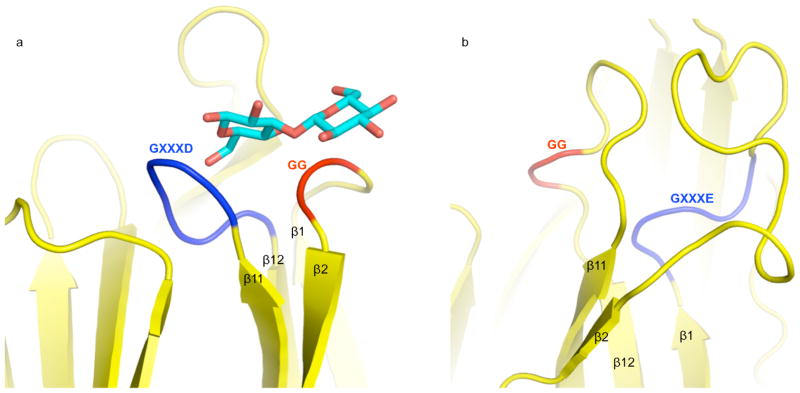
We found that Cry5B domain II is most similar in structure not to another Cry protein, but instead to the lectin BanLec (55). BanLec is a member of the mannose-specific jacalin-related superfamily of lectins, and binds both glucose and mannose. The structure of BanLec with two different bound glycans is known, with each of these glycans being observed to bind at two separate sites (55). Site 1 is conserved in all lectins, and in BanLec is formed by the β1β2 loop, which contains a GG sequence, and the β11β12 loop, which contains a GXXXD sequence (Fig. 5). Site 2 is specific to BanLec and some related lectins, and is formed by the β3β4 loop, which contains a GXXXD sequence, and the β5β6 loop, with contains a GG sequence. In BanLec, most of the hydrogen bonds to the glycans are through main chain amides, while the Asp of the GXXXD sequence makes the only side chain hydrogen bonds. The equivalent of site 1 potentially exists in Cry5B. The β11β12 loop in Cry5B contains a GG sequence, and the β1β2 loop has a GXXXE sequence, closely approximating the GXXXD sequence (Figs. 3 and 5). These loops are considerably longer than in BanLec and may accommodate longer saccharide chain lengths. The equivalent of site 2 does not appear to exist in Cry5B, as the β3β4 and β5β6 loops lack GG and GXXXD sequences. The conservation of the BanLec glycan-binding motifs in Cry5B domain II provides further evidence that domain II is likely to be responsible for recognizing nematode glycolipids.
Figure 5. Glycan-binding motifs.
a. The β1β2 loop of BanLec containing the GG motif (red) and the β11β12 loop containing the GXXD motif (blue) in ribbon representation. The structure of bound laminaribiose is shown in bonds representation with carbons in cyan and oxygens in red.
b. The β1β2 loop of Cry5B domain II containing the GXXE motif (blue), and the β11β12 loop containing the GG motif (red) in ribbon representation.
In contrast, domain III of Cry1Ac has been implicated in binding GalNAc moieties that decorate the surface of its putative receptor aminopeptidase N (59, 60). The GalNAc-binding site in Cry1Ac was suggested by mutational evidence to reside on the solvent-exposed face of the β-sandwich fold in domain III (equivalent to the solvent-exposed surface of the β5β11β7β8 sheet in Cry5B). This supposition was based on modeling, as no structure of Cry1Ac is available, despite the fact that Cry1Ac was reported to be crystallized a number of years ago (61). The putative GalNAc-binding site of Cry1Ac may be unique, in that the Cry1Ac residues identified to be involved in binding GalNAc are dissimilar from other Cry toxins, including the highly related Cry1Aa and the more distant Cry5B (59). Additionally, the functional significance of the interaction between Cry1Ac and GalNAc is questionable, as disruption of this interaction fails to reduce toxicity (59).
It is worth noting that other Cry proteins besides Cry5B are able to bind glycolipids. This includes some insecticidal Cry proteins. Members of the Lepidoptera-specific Cry1A family bind the glycolipids of Manduca sexta and Plutella xylostella (34, 62). This raises the question of why the BanLec-like domain II of Cry5B does not occur in Cry1Aa as well. One possibility is that the interaction of Cry proteins with roundworm and insect glycolipids are quantitatively different, e.g., that Cry5B has developed a more dominant reliance on glycans as receptors. This possibility is in accord with the observation that C. elegans lacking glycolipid receptors is resistant to as much as 1 mg/mL Cry5B (63). Alternatively, there may be a different, as yet unknown, carbohydrate receptor for Cry5B that is present in worms but not insects.
We also found that GalNAc, a part of the arthroseries tetrasaccharide core, is a competitor for interaction between Cry5B and its glycolipid receptors. The competition experiment required high monosaccharide concentration (i.e., 100 mM), indicative of the much higher affinity of Cry5B for its intact glycolipid receptor as compared to simple sugars. This is also consistent with the observation that Cry5B is effective in vivo as an anthelmintic (24, 25), demonstrating that dietary sugars and glycans do not inhibit it from interacting with its receptor. Along with our identification of GalNAc as a binding determinant, prior results provide further definition. These results have shown that Cry5B binds C. elegans glycolipid band E but not band D (Fig. S1) (34). Significantly, band E has two antennary galactoses attached to the tetrasaccharide core, while band D has only one antennary galactose attached to the tetrasaccharide core. Thus, this prior observation combined with our current results suggest that the minimal binding determinant for Cry5B consists of GalNAc from the arthroseries core decorated with at least two antennary galactoses.
Lastly, we found that the protoxin and activated forms of Cry5B bind nematode glycolipids with similar specificities but markedly differing affinities. The protoxin form has weaker receptor affinity, suggesting that the receptor-binding site is partially occluded by the N- or C-terminal regions, or both, that are present in the protoxin but absent in the activated protein. Furthermore, these results suggest that an initially weak binding event between the protoxin and glycolipid receptors could be stabilized by subsequent proteolysis of the protoxin on the nematode cell surface.
In summary, this first structure of a nematicidal Cry protein coupled with biochemical characterization of its interactions with nematode glycolipids have laid the groundwork for detailed dissection of Cry5B function, with the ultimate aim of devising an effective anthelmintic to combat parasitic worms.
Supplementary Material
Acknowledgments
Funding Sources
This works was supported by NSF grant MCB-0517718 (RVA and PG) and NIH grant R01 AI056189 (RVA).
We thank the staff at beamline 4.2.2 for help in data collection, and Melanie Miller for technical assistance.
ABBREVIATIONS
- BanLec
banana lectin
- Bt
Bacillus thuringiensis
- Cry
crystal
- Gal
galactose
- GalNac
N-acetylgalactosamine
- Glc
glucose
- GlcNAc
N-acetylglucosamine
- NTA
Ni2+-nitrilotriacetic acid
- rmsd
root-mean-square deviation
- SAD
single anomalous diffraction
- SeMet
selenomethionine
- TLC
thin-layer chromatography
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information. Supplemental Figure 1 and Supplemental Table 1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Betz FS, Hammond BG, Fuchs RL. Safety and advantages of Bacillus thuringiensis-protected plants to control insect pests. Regul Toxicol Pharmacol. 2000;32:156–173. doi: 10.1006/rtph.2000.1426. [DOI] [PubMed] [Google Scholar]
- 2.de Maagd RA, Bravo A, Crickmore N. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 2001;17:193–199. doi: 10.1016/s0168-9525(01)02237-5. [DOI] [PubMed] [Google Scholar]
- 3.Griffitts JS, Aroian RV. Many roads to resistance: how invertebrates adapt to Bt toxins. Bioessays. 2005;27:614–624. doi: 10.1002/bies.20239. [DOI] [PubMed] [Google Scholar]
- 4.Roh JY, Choi JY, Li MS, Jin BR, Je YH. Bacillus thuringiensis as a specific, safe, and effective tool for insect pest control. J Microbiol Biotechnol. 2007;17:547–559. [PubMed] [Google Scholar]
- 5.Bravo A, Gill SS, Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49:423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkins JL, Lee MK, Valaitis AP, Curtiss A, Dean DH. Bivalent sequential binding model of a Bacillus thuringiensis toxin to gypsy moth aminopeptidase N receptor. J Biol Chem. 2000;275:14423–14431. doi: 10.1074/jbc.275.19.14423. [DOI] [PubMed] [Google Scholar]
- 7.Gomez I, Sanchez J, Miranda R, Bravo A, Soberon M. Cadherin-like receptor binding facilitates proteolytic cleavage of helix alpha-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002;513:242–246. doi: 10.1016/s0014-5793(02)02321-9. [DOI] [PubMed] [Google Scholar]
- 8.Li JD, Carroll J, Ellar DJ. Crystal structure of insecticidal delta-endotoxin from Bacillus thuringiensis at 2.5 A resolution. Nature. 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 9.Grochulski P, Masson L, Borisova S, Pusztai-Carey M, Schwartz JL, Brousseau R, Cygler M. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J Mol Biol. 1995;254:447–464. doi: 10.1006/jmbi.1995.0630. [DOI] [PubMed] [Google Scholar]
- 10.Morse RJ, Yamamoto T, Stroud RM. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure. 2001;9:409–417. doi: 10.1016/s0969-2126(01)00601-3. [DOI] [PubMed] [Google Scholar]
- 11.Galitsky N, Cody V, Wojtczak A, Ghosh D, Luft JR, Pangborn W, English L. Structure of the insecticidal bacterial delta-endotoxin Cry3Bb1 of Bacillus thuringiensis. Acta Crystallogr D Biol Crystallogr. 2001;57:1101–1109. doi: 10.1107/s0907444901008186. [DOI] [PubMed] [Google Scholar]
- 12.Boonserm P, Davis P, Ellar DJ, Li J. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J Mol Biol. 2005;348:363–382. doi: 10.1016/j.jmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Boonserm P, Mo M, Angsuthanasombat C, Lescar J. Structure of the functional form of the mosquito larvicidal Cry4Aa toxin from Bacillus thuringiensis at a 2.8-angstrom resolution. J Bacteriol. 2006;188:3391–3401. doi: 10.1128/JB.188.9.3391-3401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo S, Ye S, Liu Y, Wei L, Xue J, Wu H, Song F, Zhang J, Wu X, Huang D, Rao Z. Crystal structure of Bacillus thuringiensis Cry8Ea1: An insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela. J Struct Biol. 2009;168:259–266. doi: 10.1016/j.jsb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Gazit E, La Rocca P, Sansom MS, Shai Y. The structure and organization within the membrane of the helices composing the pore-forming domain of Bacillus thuringiensis delta-endotoxin are consistent with an “umbrella-like” structure of the pore. Proc Natl Acad Sci U S A. 1998;95:12289–12294. doi: 10.1073/pnas.95.21.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Maagd RA, Bravo A, Berry C, Crickmore N, Schnepf HE. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu Rev Genet. 2003;37:409–433. doi: 10.1146/annurev.genet.37.110801.143042. [DOI] [PubMed] [Google Scholar]
- 17.Gomez I, Dean DH, Bravo A, Soberon M. Molecular basis for Bacillus thuringiensis Cry1Ab toxin specificity: two structural determinants in the Manduca sexta Bt-R1 receptor interact with loops alpha-8 and 2 in domain II of Cy1Ab toxin. Biochemistry. 2003;42:10482–10489. doi: 10.1021/bi034440p. [DOI] [PubMed] [Google Scholar]
- 18.Atsumi S, Mizuno E, Hara H, Nakanishi K, Kitami M, Miura N, Tabunoki H, Watanabe A, Sato R. Location of the Bombyx mori aminopeptidase N type 1 binding site on Bacillus thuringiensis Cry1Aa toxin. Appl Environ Microbiol. 2005;71:3966–3977. doi: 10.1128/AEM.71.7.3966-3977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacheco S, Gomez I, Arenas I, Saab-Rincon G, Rodriguez-Almazan C, Gill SS, Bravo A, Soberon M. Domain II loop 3 of Bacillus thuringiensis Cry1Ab toxin is involved in a “ping pong” binding mechanism with Manduca sexta aminopeptidase-N and cadherin receptors. J Biol Chem. 2009;284:32750–32757. doi: 10.1074/jbc.M109.024968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roh JY, Nair MS, Liu XS, Dean DH. Mutagenic analysis of putative domain II and surface residues in mosquitocidal Bacillus thuringiensis Cry19Aa toxin. FEMS Microbiol Lett. 2009;295:156–163. doi: 10.1111/j.1574-6968.2009.01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez I, Arenas I, Benitez I, Miranda-Rios J, Becerril B, Grande R, Almagro JC, Bravo A, Soberon M. Specific epitopes of domains II and III of Bacillus thuringiensis Cry1Ab toxin involved in the sequential interaction with cadherin and aminopeptidase-N receptors in Manduca sexta. J Biol Chem. 2006;281:34032–34039. doi: 10.1074/jbc.M604721200. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez LE, Perez C, Segovia L, Rodriguez MH, Gill SS, Bravo A, Soberon M. Cry11Aa toxin from Bacillus thuringiensis binds its receptor in Aedes aegypti mosquito larvae through loop alpha-8 of domain II. FEBS Lett. 2005;579:3508–3514. doi: 10.1016/j.febslet.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 23.Masson L, Tabashnik BE, Mazza A, Prefontaine G, Potvin L, Brousseau R, Schwartz JL. Mutagenic analysis of a conserved region of domain III in the Cry1Ac toxin of Bacillus thuringiensis. Appl Environ Microbiol. 2002;68:194–200. doi: 10.1128/AEM.68.1.194-200.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cappello M, Bungiro RD, Harrison LM, Bischof LJ, Griffitts JS, Barrows BD, Aroian RV. A purified Bacillus thuringiensis crystal protein with therapeutic activity against the hookworm parasite Ancylostoma ceylanicum. Proc Natl Acad Sci U S A. 2006;103:15154–15159. doi: 10.1073/pnas.0607002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Georghiou SB, Kelleher AJ, Aroian RV. Bacillus thuringiensis Cry5B protein is highly efficacious as a single-dose therapy against an intestinal roundworm infection in mice. PLoS Negl Trop Dis. 2010;4:e614. doi: 10.1371/journal.pntd.0000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Platzer EG, Bellier A, Aroian RV. Discovery of a highly synergistic anthelmintic combination that shows mutual hypersusceptibility. Proc Natl Acad Sci U S A. 2010;107:5955–5960. doi: 10.1073/pnas.0912327107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XQ, Tan A, Voegtline M, Bekele S, Chen CS, Aroian RV. Expression of Cry5B protein from Bacillus thuringiensis in plant roots confers resistance to root-knot nematode. Biological Control. 2008;47:97–102. [Google Scholar]
- 28.Li XQ, Wei JZ, Tan A, Aroian RV. Resistance to root-knot nematode in tomato roots expressing a nematicidal Bacillus thuringiensis crystal protein. Plant Biotechnol J. 2007;5:455–464. doi: 10.1111/j.1467-7652.2007.00257.x. [DOI] [PubMed] [Google Scholar]
- 29.Wei JZ, Hale K, Carta L, Platzer E, Wong C, Fang SC, Aroian RV. Bacillus thuringiensis crystal proteins that target nematodes. Proc Natl Acad Sci U S A. 2003;100:2760–2765. doi: 10.1073/pnas.0538072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albonico M, Allen H, Chitsulo L, Engels D, Gabrielli AF, Savioli L. Controlling soil-transmitted helminthiasis in pre-school-age children through preventive chemotherapy. PLoS Negl Trop Dis. 2008;2:e126. doi: 10.1371/journal.pntd.0000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall A, Hewitt G, Tuffrey V, de Silva N. A review and meta-analysis of the impact of intestinal worms on child growth and nutrition. Matern Child Nutr 4 Suppl. 2008;1:118–236. doi: 10.1111/j.1740-8709.2007.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchuente LA. Control of soil-transmitted helminths in sub-Saharan Africa: Diagnosis, drug efficacy concerns and challenge. Acta Trop. 2011 doi: 10.1016/j.actatropica.2010.07.001. in press. [DOI] [PubMed] [Google Scholar]
- 33.Wiegandt H. Insect glycolipids. Biochim Biophys Acta. 1992;1123:117–126. doi: 10.1016/0005-2760(92)90101-z. [DOI] [PubMed] [Google Scholar]
- 34.Griffitts JS, Haslam SM, Yang T, Garczynski SF, Mulloy B, Morris H, Cremer PS, Dell A, Adang MJ, Aroian RV. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science. 2005;307:922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 35.Griffitts JS, Whitacre JL, Stevens DE, Aroian RV. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science. 2001;293:860–864. doi: 10.1126/science.1062441. [DOI] [PubMed] [Google Scholar]
- 36.Marroquin LD, Elyassnia D, Griffitts JS, Feitelson JS, Aroian RV. Bacillus thuringiensis (Bt) toxin susceptibility and isolation of resistance mutants in the nematode Caenorhabditis elegans. Genetics. 2000;155:1693–1699. doi: 10.1093/genetics/155.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 38.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:263–291. [Google Scholar]
- 43.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeLano WL. The PyMOL User’s Manual 2002 [Google Scholar]
- 45.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svennerholm L, Fredman P. A procedure for the quantitative isolation of brain gangliosides. Biochim Biophys Acta. 1980;617:97–109. doi: 10.1016/0005-2760(80)90227-1. [DOI] [PubMed] [Google Scholar]
- 48.Schnaar RL. Isolation of glycosphingolipids. Methods Enzymol. 1994;230:348–370. doi: 10.1016/0076-6879(94)30024-0. [DOI] [PubMed] [Google Scholar]
- 49.Barrows BD, Griffitts JS, Aroian RV. Caenorhabditis elegans carbohydrates in bacterial toxin resistance. Methods Enzymol. 2006;417:340–358. doi: 10.1016/S0076-6879(06)17021-4. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz JL, Juteau M, Grochulski P, Cygler M, Prefontaine G, Brousseau R, Masson L. Restriction of intramolecular movements within the Cry1Aa toxin molecule of Bacillus thuringiensis through disulfide bond engineering. FEBS Lett. 1997;410:397–402. doi: 10.1016/s0014-5793(97)00626-1. [DOI] [PubMed] [Google Scholar]
- 51.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uawithya P, Tuntitippawan T, Katzenmeier G, Panyim S, Angsuthanasombat C. Effects on larvicidal activity of single proline substitutions in alpha3 or alpha4 of the Bacillus thuringiensis Cry4B toxin. Biochem Mol Biol Int. 1998;44:825–832. doi: 10.1080/15216549800201872. [DOI] [PubMed] [Google Scholar]
- 53.Wu D, Aronson AI. Localized mutagenesis defines regions of the Bacillus thuringiensis delta-endotoxin involved in toxicity and specificity. J Biol Chem. 1992;267:2311–2317. [PubMed] [Google Scholar]
- 54.Raval S, Gowda SB, Singh DD, Chandra NR. A database analysis of jacalin-like lectins: sequence-structure-function relationships. Glycobiology. 2004;14:1247–1263. doi: 10.1093/glycob/cwh140. [DOI] [PubMed] [Google Scholar]
- 55.Meagher JL, Winter HC, Ezell P, Goldstein IJ, Stuckey JA. Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology. 2005;15:1033–1042. doi: 10.1093/glycob/cwi088. [DOI] [PubMed] [Google Scholar]
- 56.Henshaw J, Horne-Bitschy A, van Bueren AL, Money VA, Bolam DN, Czjzek M, Ekborg NA, Weiner RM, Hutcheson SW, Davies GJ, Boraston AB, Gilbert HJ. Family 6 carbohydrate binding modules in beta-agarases display exquisite selectivity for the non-reducing termini of agarose chains. J Biol Chem. 2006;281:17099–17107. doi: 10.1074/jbc.M600702200. [DOI] [PubMed] [Google Scholar]
- 57.Sommer RJ. Pristionchus pacificus. WormBook. 2006:1–8. doi: 10.1895/wormbook.1.102.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia LQ, Zhao XM, Ding XZ, Wang FX, Sun YJ. The theoretical 3D structure of Bacillus thuringiensis Cry5Ba. J Mol Model. 2008;14:843–848. doi: 10.1007/s00894-008-0318-8. [DOI] [PubMed] [Google Scholar]
- 59.Burton SL, Ellar DJ, Li J, Derbyshire DJ. N-acetylgalactosamine on the putative insect receptor aminopeptidase N is recognised by a site on the domain III lectin-like fold of a Bacillus thuringiensis insecticidal toxin. J Mol Biol. 1999;287:1011–1022. doi: 10.1006/jmbi.1999.2649. [DOI] [PubMed] [Google Scholar]
- 60.de Maagd RA, Bakker PL, Masson L, Adang MJ, Sangadala S, Stiekema W, Bosch D. Domain III of the Bacillus thuringiensis delta-endotoxin Cry1Ac is involved in binding to Manduca sexta brush border membranes and to its purified aminopeptidase N. Mol Microbiol. 1999;31:463–471. doi: 10.1046/j.1365-2958.1999.01188.x. [DOI] [PubMed] [Google Scholar]
- 61.Derbyshire DJ, Ellar DJ, Li J. Crystallization of the Bacillus thuringiensis toxin Cry1Ac and its complex with the receptor ligand N-acetyl-D-galactosamine. Acta Crystallogr D Biol Crystallogr. 2001;57:1938–1944. doi: 10.1107/s090744490101040x. [DOI] [PubMed] [Google Scholar]
- 62.Kumaraswami NS, Maruyama T, Kurabe S, Kishimoto T, Mitsui T, Hori H. Lipids of brush border membrane vesicles (BBMV) from Plutella xylostella resistant and susceptible to Cry1Ac delta-endotoxin of Bacillus thuringiensis. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:173–183. doi: 10.1016/s1096-4959(01)00327-x. [DOI] [PubMed] [Google Scholar]
- 63.Griffitts JS, Huffman DL, Whitacre JL, Barrows BD, Marroquin LD, Muller R, Brown JR, Hennet T, Esko JD, Aroian RV. Resistance to a bacterial toxin is mediated by removal of a conserved glycosylation pathway required for toxin-host interactions. J Biol Chem. 2003;278:45594–45602. doi: 10.1074/jbc.M308142200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.