Abstract
The β-branched C3 unit in leinamycin biosynthesis is installed by a set of four proteins, LnmFKLM. In vitro biochemical investigation confirmed that LnmK is a bifunctional acyltransferase/decarboxylase (AT/DC) that catalyzes first self-acylation using methylmalonyl-CoA as a substrate and subsequently trans-acylation of the methylmalonyl group to the phosphopantetheinyl group of the LnmL acyl carrier protein [Liu, T., Huang, Y., and Shen, B. (2009), J. Am. Chem. Soc. 131, 6900-6901]. LnmK shows no sequence homology to proteins of known function, representing a new family of AT/DC enzymes. Here we report the X-ray structure of LnmK. LnmK is homodimer with each of the monomers adopting a double-hot-dog fold. Co-crystallization of LnmK with methylmalonyl-CoA revealed an active site tunnel terminated by residues from the dimer interface. In contrast to canonical AT and ketosynthase enzymes that employ Ser or Cys as an active site residue, none of these residues are found in the vicinity of the LnmK active site. Instead, three tyrosines were identified, one of which, Tyr62, was established, by site-directed mutagenesis, to be the most likely active site residue for the AT activity of LnmK. LnmK represents the first AT enzyme that employs a Tyr as an active site residue and the first member of double-hot-dog fold enzymes that displays an AT activity known to date. The LnmK structure sets the stage to probe the DC activity of LnmK through site-directed mutagenesis. These findings highlight natural product biosynthetic machinery as a rich source of novel enzyme activities, mechanisms, and structures.
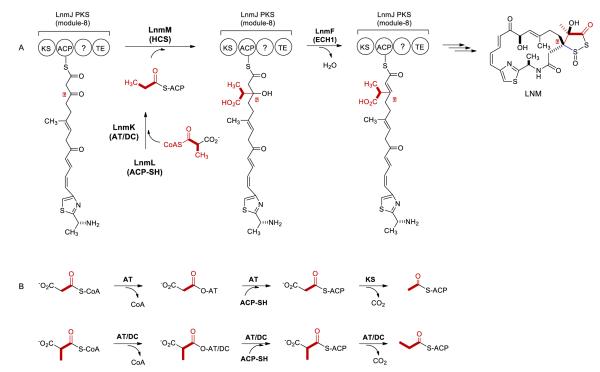
Leinamycin (LNM), a potent antitumor antibiotic, is structurally characterized by an unusual 1,3-dioxo-1,2-dithiolane moiety that is spiro-fused to an 18-membered macrolactam ring of hybrid peptide-polyketide origin (Figure 1A) (1–5). This molecular architecture has not been found in any other natural product to date. We have cloned, sequenced, and characterized the lnm biosynthetic gene cluster from Streptomyces atroolivaceus S-140 (3–7). In vivo and in vitro characterizations of the LNM biosynthetic machinery unveiled that (i) the LNM backbone is assembled from the amino acid and short carboxylic acid precursors by a hybrid nonribosomal peptide synthetase (NRPS)-acyltransferase-less type I polyketide synthase (PKS) (3–5) and (ii) the β-branched C3 unit, which is a part of the unique five-membered 1,3-dioxo-1,2-dithiolane moiety, is installed by a novel pathway for β-alkylation in polyketide biosynthesis (Figure 1A) (8, 9).
Figure 1.
(A) Proposed LNM biosynthetic pathway featuring LnmKLM-catalyzed β-alkylation of the LnmJ PKS module-8 ACP-tethered intermediate in S. atroolivaceus S-140. (B) Two parallel but distinct pathways for the acyl-S-ACP substrate involved in β-alkylation in polyketide biosynthesis, i.e., acetyl-S-ACP from malonyl-CoA by a dedicated ACP, AT, and KS enzymes and propionyl-S-ACP, from methylmalonyl-CoA by a dedicated ACP and a bifunctional AT/DC enzyme
β-Alkylations contribute to the vast structural diversity displayed by polyketide natural products. The β-alkyl branches are installed by hydroxymethylglutaryl-CoA synthases (HCSs) (9–16). HCS catalyzes condensation of an acyl-S-carrier protein (ACP) with the β-keto group of the growing ACP-tethered polyketide intermediate to afford a β-hydroxyacyl-S-ACP intermediate. The latter undergoes further dehydration, decarboxylation, or both by one or both enoyl-CoA hydratases (ECH1 and ECH2) to afford a β-alkylated intermediate in polyketide biosynthesis (Figure 1A).
Two distinct pathways have been discovered for acyl-S-ACPs, i.e., acetyl-S-ACP or propionyl-S-ACP, the substrates of HCSs for β-alkylation in polyketide biosynthesis (Figure 1B) (8–16). For acetyl-S-ACP, it is derived from malonyl-CoA by a dedicated set of three proteins, an ACP, an acyltransferase (AT), and a ketosynthase homologue (KS), and this pathway has been extensively studied in the biosynthesis of bacillaene, curacin, and myxovirescin (for the alkyl branch at C-12) (10–16). For propionyl-S-ACP, it is derived from methylmalonyl-CoA by a dedicated set of two proteins, an ACP and a bifunctional acyltransferase/decarboxylase (AT/DC), and this pathway has been experimentally confirmed in the biosynthesis of LNM and predicted for the biosynthesis of myxovirescin (for the alkyl branch at C-16) (8). Additional homologues of LnmK have also been identified from recent genome sequencing efforts, all of which, however, were annotated as proteins of unknown function (Figure S1).
Previously, we have demonstrated that the β-branched C3 unit in LNM biosynthesis is installed by a set of four proteins, LnmL, an ACP, LnmK, a bifunctional AT/DC, LnmM, a HCS, and LnmF, an ECH1 (8, 9). Inactivation of lnmK, lnmL, or lnmM, respectively, in S. atroolivaceus S-140 all abolished LNM production; the resultant mutant strains instead accumulated an identical set of shunt metabolites, all of which lack the β-branched C3 unit (9). In vitro biochemical investigation confirmed that LnmK is a bifunctional AT/DC: it is first self-acylated using methylmalonyl-CoA, the methylmalonyl group is then transferred from LnmK to the phosphopantetheinyl group of LnmL, and finally the resultant methylmalonyl-S-LnmL is decarboxylated to afford propionyl-S-LnmL (Figure 1B) (8). LnmM-catalyzed condensation between propionyl-S-LnmL and the β-keto group of the growing polyketide intermediate, tethered to the ACP domain of LnmJ PKS-module 8, followed by LnmF-catalyzed dehydration, completes the installation of the β-branch C3 unit to LNM (Figure 1A) (9). While homologues of LnmF, LnmL, and LnmM are known, and their functions have been well characterized from several biosynthetic pathways of β-branched polyketides (10–16), LnmK shows no sequence homology to proteins of known function, representing a new family of AT/DC enzymes (8, 9).
Here we report the X-ray structure of the bifunctional AT/DC enzyme LnmK. LnmK is homodimer with each of the monomers adopting a double-hot-dog fold (DHDF). Co-crystallization of LnmK with methylmalonyl-CoA revealed an active site tunnel terminated by residues from the dimer partner. In contrast to canonical AT and KS enzymes that employ Ser or Cys as an active site residue, none of these residues are found in the vicinity of the LnmK active site. Instead, three tyrosines were identified, one of which, Tyr62, was established, by site-directed mutagenesis, to be the most likely active site residue for the AT activity of LnmK. LnmK represents the first AT enzyme that employs a Tyr as an active site residue and the first member of DHDF enzymes that displays an AT activity known to date. The LnmK structure sets that stage to probe the DC activity of LnmK through site-directed mutagenesis. These findings highlight natural product biosynthetic machinery as a rich source of novel enzyme activities, mechanisms, and structures.
MATERIALS AND METHODS
Chemicals and Reagents
DL-2-[methyl-14C]malonyl-CoA (American Radiolabeled Chemicals, St. Louis, MO), DL-methylmalonyl-CoA lithium salt (Sigma Aldrich, St. Louis, MO), dNTPs (New England Biolabs, Ipswich, MA), and all other common biochemicals and chemicals were from standard commercial sources. Restriction enzymes and T4 DNA ligase (New England Biolabs), T4 DNA polymerase (Lucigen, Middleton, WI), and Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA) were purchased. The pRSFDuet-1 plasmid and E. coli NovaBlue and BL21(DE3) cells were from Novagen (Madison, WI). Primer synthesis and DNA sequencing were performed at the University of Wisconsin-Madison Biotechnology Center (Madison, WI).
Plasmid Construction
To construct the lnmK expression plasmid pBS3081, two oligos, TEV-1 F and TEV-1 R (see Table S1), were annealed and ligated into the BamHI/XhoI sites of pRSFDuet-1 to afford pBS3080. The oligo insert contains a BsmFI site that allows for ligation independent cloning and encodes a TEV protease site after the His6-tag. pBS3080 was digested with BsmF1 and purified by gel electrophoresis, and the linearized vector was treated with T4 DNA polymerase in the presence of dGTP at 20°C for 30 min followed by heating at 75°C for 20 min to denature the polymerase, affording overhangs with complimentary sequences to clone the PCR amplified lnmK gene for expression. The lnmK gene was amplified by PCR from pBS3050 (8) using the primers LnmK F and LnmK R (Table S1), purified by gel electrophoresis, and similarly treated with T4 DNA polymerase in the presence of dCTP at 20°C for 30 min followed by heating at 75°C. The T4 DNA polymerase treated pBS3080 vector and lnmK fragment were then mixed at room temperature, annealed on ice for 5 min, and transformed into E. coli NovaBlue for ligation independent cloning to construct the lnmK expression plasmid pBS3081. pBS3081 was finally isolated from E. coli NovaBlue and confirmed by DNA sequencing, in which LnmK is produced as a fusion protein with an N-terminal His6-tag that is cleavable by TEV protease. Upon TEV cleavage, the resultant LnmK lacks the N-terminal methionine and contains a Thr2Ser mutation.
To construct the svp and lnmL co-expression plasmid pBS3084, two oligos, TEV-2 F and TEV-2 R (Table S1), were annealed and ligated into NcoI/BamHI sites of pRSFDuet-1 to afford pBS3082. The svp gene was amplified by PCR from pBS18 (17) with the primers Svp F and Svp R (Table S1) and ligated into the BglII site of pBS3082 to afford pBS3083, which was confirmed by DNA sequencing. The lnmL gene was amplified by PCR from pBS3051 (8) with the primers LnmL F and LnmL R (Table S1), and the resultant PCR product was digested and ligated into the BamHI/HindIII sites of pBS3083 to afford pBS3084, which was similarly confirmed by DNA sequencing. Co-expression of svp and lnmL in pBS3084 ensues the production of LnmL in its holo-form, and LnmL is produced as a fusion protein with an N-terminal His6-tag that is cleavable by the TEV protease. Upon TEV cleavage, the resultant holo-LnmL lacks the N-terminal methionine and contains a Thr2Ser mutation.
Expression plasmids for the lnmK mutants (pBS3085-pBS3099) were constructed by following the QuickChange site-directed mutagenesis method (Stratagene, La Jolla, CA) with the primers summarized in Table S1. Thus, the primer pairs for each of the lnmK mutant constructs were used to amplify lnmK from pBS3081. The resulting PCR products were digested with DpnI, purified by gel electrophoresis, and transformed into E. coli NovaBlue. The lnmK mutant constructs were isolated from E. coli NovaBlue, confirmed by DNA sequencing, and named pBS3085 to pBS3097 (Table S1). In these constructs, all LnmK mutants, i.e., Arg144Gln (pBS3085), Asn216Leu (pBS3086), Asn263Leu (pBS3087), Cys24Ala (pBS3088), Ser28Ala (pBS3089), Ser91Ala (pBS3090), Ser100Ala (pBS3091), Ser174Ala (pBS3092), Ser225Ala (pBS3093), Thr103Ala (pBS3094), Tyr62Phe (pBS3095), Tyr222Phe (pBS3096), and Tyr226Phe (pBS3097), are produced as fusion proteins with an N-His6-tag that is cleavable by the TEV protease. Like LnmK from pBS3081, upon TEV cleavage, the resultant LnmK mutants all lack the N-terminal methionine and contain a Thr2Ser mutation.
Protein Production
Expression plasmids for lnmL (pBS3084), lnmK (pBS3081), and the lnmK mutants (pBS3085-pBS3097) were transformed into E. coli BL21(DE3), and the resultant recombinant strains were grown overnight in 50 mL of LB containing 2 mM MgSO4 and 50 μg/mL kanamycin. A 10 mL aliquot of the overnight culture was used to inoculate 1 L of LB containing 2 mM MgSO4 and 50 μg/mL kanamycin, which was then incubated at 37°C with shaking at 250 rpm. Once the OD600 reached ~0.4 - 0.5, the temperature was reduced to 18°C. After the cultures reached thermal equilibrium, gene expression was induced by the addition of isopropyl β-â-thiogalactopyranoside to 50 μg/mL, with incubation for an additional 16 hours. For selenomethionine (Se-Met) containing LnmK, E. coli BL21(DE3) was transformed with pBS3081 and incubated overnight in 50 mL of LB containing 2 mM MgSO4 and 50 μg/mL kanamycin. A 5 mL aliquot of the overnight culture was introduced into autoinducing medium [50 mM Na2HPO4, 50 mM KH2PO4, 25 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeCl3, 4 μM CaCl2, 2 μM MnCl2, 2μM ZnSO4, 0.4 μM CoCl2, 0.4 μM CuCl2, 0.4 μM NiCl2, 0.4 μM Na2MoO4, 0.4 μM Na2SeO3, 0.4 μM H3BO3, 54 mM glycerol, 2.8 mM glucose, 5.6 mM lactose, 200 mg/L each (Ala, Arg, Asn, Asp, Gln, Glu, Gly, His, Ile, Leu, Lys, Pro, Ser, Thr, Trp, Tyr, Val), 10 mg/L Met, 125 mg/L Se-Met and 100 nM vitamin B12](18), containing 50 μg/mL kanamycin. The resultant culture was incubated at 37°C with shaking at 250 rpm for 4 hours, at which time the temperature was reduced to 18°C, and incubation continued for 20 hours. E. coli cells were harvested by centrifugation at 3500g and 4°C for 20 min.
Protein Purification
E. coli cell pellets, carrying various expression constructs, were resuspended in lysis buffer (1 μg/ml DNAse, 300 mM NaCl, 10 mM imidiazole, 5% glycerol, and 50 mM Tris-HCl, pH 8.0), sonicated (30 × 2 s on ice), and clarified by centrifugation at 50000g and 4°C for 30 min. Supernatant was applied to a 5 mL Ni-Sepharose 6 Fast Flow column (GE Healthcare, Uppsala, Sweden) and washed with lysis buffer using an Äkta FPLC (Pharmacia Amersham Biotech, Uppsala, Sweden). Wash buffer (300 mM NaCl, 20 mM imidiazole, 50 mM Tris-HCl, pH 8.0) was used to further remove contaminants, and proteins were eluted with wash buffer containing 250 mM imidiazole. Proteins were immediately diluted from ~5 mL to 15 mL buffer A (50 mM Tris-HCl, pH 8.0, 10 mM NaCl) plus 1 mM dithiothreitol, 0.1 mM EDTA, and 50 μg/mL TEV protease bearing a C-terminal His6-tag and incubated overnight. The TEV cleaved samples were further diluted to 50 mL with buffer A and passed over a Ni-Sepharose column to remove the TEV protease, and the flowthrough was then loaded onto a MonoQ 10/80 column (Pharmacia). A linear gradient over 12 column volumes from 10 % buffer B to 50 % buffer B (50 mM Tris-HCl, pH 8.0, 1.0 M NaCl) was used to elute the proteins. Purity of protein from the fractions was analyzed by SDS-PAGE. Pure fractions were pooled, concentrated, buffer exchanged into 10 mM Tris-HCl, pH 8.0, 10 mM NaCl, frozen in small aliquots with liquid nitrogen, and stored at −80°C. For the Ser, Thr, Cys, Arg, and Asn mutants of LnmK from pBS3085-pBS3097, the final step of MonoQ 10/80 column chromatography was omitted after the TEV protease removal step. Se-Met LnmK (from pBS3081) was not cleaved by TEV before MonoQ chromatography.
Size Exclusion Chromatography
LnmK was diluted to 1 absorbance unit at 280 nm (~25 μM) in 50 mM Tris-HCl, pH 8.0, 50 mM NaCl, and 100 μl were loaded onto a Sepharose 6 10/300 GL column (GE lifesciences). A second sample with LnmK at 2 absorbance units (~50 μM) was mixed with 10 mM methylmalonyl-CoA and loaded onto the same column to examine the effect of methylmalonyl-CoA on the quaternary structure of LnmK. The column was eluted with 50 mM Tris-HCl, pH 8.0, 50 mM NaCl, at the flow rate of 1.0 mL/min and calibrated with molecular weight (MW) standards of thyroglobulin (660 kDa), ferritin (450 kDa), bovine serum albumin (66 kDa), and ribonuclease A (14 kDa) (Figure S2).
Enzymatic Assays of LnmK
LnmK-catalyzed loading of the methylmalonyl group from methylmalonyl-CoA to holo-LnmL, via a self-acylated intermediate, was assayed by following previously published procedures (8). A typical self-acylation reaction of 50 μL contained 100 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM TCEP, and 20 μM DL-2-[methyl-14C]malonyl-CoA, and the assay was initiated by the addition of 4 μM LnmK or one of the LnmK mutants. For trans-acylation, the same reactions were used but included additional 15 μM holo-LnmL. Reactions were incubated for varying time at 25°C and quenched by the addition of 450 μL of acetone. Proteins were precipitated by centrifugation at 13,000 rpm and 4°C for 10 min. Acetone was removed, and the pellet was allowed to air dry for 10 min. The pellets were resuspended in sample buffer and subjected to SDS-PAGE on 4 - 15% gradient gels (Bio-Rad Laboratories, Hercules, CA). The gels were visualized by Coomassie blue staining and phosphorimaging (LE phosphor screen, Amersham Pharmacia, Molecular Dynamics Division, Piscataway, NJ).
Crystallization
LnmK was screened against 384 crystallization conditions in 200 nL sitting drops at 20°C, set up with a Mosquito (TTPlabtech, Melbourne, United Kingdom) to find initial conditions. Se-Met LnmK (45 mg/mL in 10 mM NaCl and 10 mM Tris-HCl, pH 8.0) plus 2.5 mM methylmalonyl-CoA were screened by hanging drop method over 0.5 mL wells containing 12 - 18% glycerol, 1.3 - 1.6 M (NH4)2SO4, 0.1 M Tris-HCl, pH 7.0 - 8.5, 4 μL drops (1:1, protein:well), which produced crystals in all conditions. LnmK (20 mg/mL in 100 mM NaCl and 10 mM Tris-HCl, pH 8.0) plus 10 mM methylmalonyl-CoA were co-crystallized from 20 - 23% MEPEG 2000, 0.25-0.35 M (NH4)2SO4 and 0.1 M Bis-tris propane-HCl, pH 7.0 in 4 μL drops (1:1, protein:well). LnmK (Tyr62Phe) was co-crystallized with methylmalonyl-CoA in conditions identical to LnmK-methylmalonyl-CoA co-crystallization.
Data Collection, Phasing, Refinement, and Structural Determination
Crystals were frozen directly out of the drops with liquid nitrogen. X-ray diffraction data for Se-Met LnmK was collected at the Advanced Photon Source LS-CAT Beamline 21-ID-D at a wavelength of 0.97941 Å for anomalous data sets and 1.127 Å for native sets. Beamline 21-ID-G at a wavelength of 0.97857 Å was used for LnmK-methylmalonyl-CoA and LnmK (Tyr62Phe)-methylmalonyl-CoA co-crystals. Diffraction intensities were integrated, reduced, and scaled using HKL2000 (19), and data collection and refinement statistics are summarized in Table 1. Experimental phasing of Se-Met LnmK data set was performed using Phenix (20) against a number of space groups in the P622 point group. Refinement was carried out using Refmac (21) in the CCP4i package (22) with automated model building performed by ARP/wARP (23) and manual model building with Coot (24). The refined structure of Se-Met LnmK served as the starting point for molecular replacement with co-crystal structures of LnmK and LnmK (Tyr62Phe) with methylmalonyl-CoA.
Table 1.
Data Collection and Refinement Statistics
| Se-Met LnmK | LnmK-CoA | Tyr62Phe-CoA | |
|---|---|---|---|
| Crystallographic Data | |||
| space group | P6122 | P6122 | P6122 |
| cell dimensions (Å) | A=b=60.2 c=311.2 | a=b=60.5 c=311.0 | a=b=59.7 c=311.1 |
| α=β=90° γ=120° | α=β=90° γ=120° | α=β=90° γ=120 | |
| resolution (Å) | 50.0-2.25 | 30.0-1.76 | 30.0-1.77 |
| observations | 292,008 | 323,621 | 301,960 |
| unique reflections | 16,907 | 34,431 | 32,885 |
| completeness %a | 98.8 (98.8) | 97.7 (72.1) | 98.5 (99.8) |
| I/σ(I)a | 28.7 (30.1) | 16.0 (1.9) | 14.7 (4.8) |
| Rmerge%a,b | 0.074 (0.105) | 0.094 (0.673) | 0.107 (0.570) |
| Refinement | |||
| resolution | 28.1-2.25 | 26.1-1.76 | 29.7-1.77 |
| reflections | 15,889 | 32,402 | 30,964 |
| protein atoms | 2,359 | 2,388 | 2,335 |
| waters | 114 | 99 | 160 |
| overall B-factor (Å2) | 23.1 | 34.7 | 27.4 |
| B-factor protein (Å2) | 22.6 | 34.5 | 27.0 |
| B-factor solvent (Å2) | 23.7 | 37.5 | 31.6 |
| R-factorc | 0.202 | 0.219 | 0.227 |
| R-free | 0.254 | 0.249 | 0.275 |
| RMS bonds (Å) | 0.01 | 0.03 | 0.02 |
| RMS angles (deg) | 1.23 | 2.21 | 1.98 |
| Ramachandran plot % | |||
| most favored | 92.3 | 90.7 | 90.6 |
| allowed | 7.7 | 9.3 | 9.4 |
| disallowed | 0 | 0 | 0 |
Values in parentheses indicate statistics for the highest resolution shell.
Rmerge = Σ∣I – <I>∣/Σ<I>, where I is the observed intensity and <I> is the average of intensities obtained from multiple observations of symmetry-related reflections.
R-factor = Σ∥Fo∣ – ∣Fc∥/Σ∣Fo∣, where Fo and Fc are the observed and calculated structure amplitudes, respectively.
RESULTS
Crystallization and Structure Determination of LnmK
Hanging drop experiments of Se-Met LnmK produced hexagonal crystals that appeared in a day and reached full size (~200 μm in length) after about four days. Crystals diffracted maximally to ~1.7 Å in the hexagonal crystal system with cell dimensions of a = b = 60.2 Å, c = 311.2 Å, α = β = 90°, γ = 120°. Due to the very long c cell dimension, very close spots were produced that overlapped at low resolution and appropriate measures were taken to maximize data (25). To obtain low resolution data for phasing, a second data set was only collected to 2.25 Å. Scaling the high and low-resolution data sets resulted in very large Rmerge values, thus higher resolution data was omitted and only the lower resolution data set was used during phasing and refinement.
P6122 space group had the best phasing statistics and produced easily interpretable maps from Phenix AutoSol using the SAD data, indicating it was the correct space group (Table 1). Phenix AutoSol generated ~220 amino acids in five non-continuous strands from the experimentally phased maps. The remainder of the structure was modeled using Coot. The final model consists of residues 11-309, 68 waters, five sulfates, one glycerol, and one Tris molecule. Despite the addition of 2.5 mM methylmalonyl-CoA to the crystallization drops, there is no clear electron density suggesting the presence of CoA or an acylated amino acid residue in the solved Se-Met LnmK structure. The final model refines to an R-factor of 0.209 (Rfree 0.270). There are no residues in the disallowed region of the Ramachandran plot.
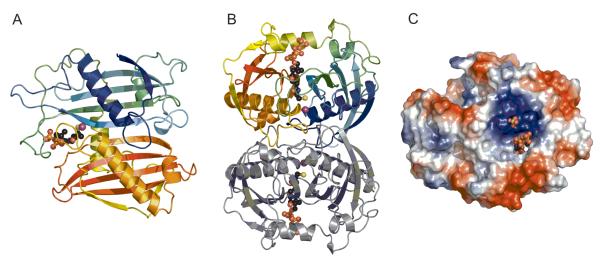
LnmK and LnmK (Tyr62Phe) were co-crystallized in the presence of methylmalonyl-CoA, which hydrolyzed resulting in LnmK-CoA and LnmK (Tyr62Phe)-CoA structures, which share the P6122 spacegroup. Molecular replacement was straightforward using the Se-Met LnmK structure after removing side chains and waters as a starting point for rigid body refinement. Both structures had reasonable data collection and refinement statistics (Table 1). The LnmK-CoA structure is very similar (RMSD 0.3 Å) to the Se-Met LnmK structure except for the presence of the phosphopantetheine moiety of CoA and a chloride ion in the putative active site (Figures 2 and 3). The phosphopantetheine moiety had extra density hanging off the sulfur atom that could not be interpreted biologically and was left unmodeled. The LnmK (Tyr62Phe)-CoA structure has a phosphopantetheine moiety bound in the active site that lacks the extra density off the sulfur atom (Figure 3A) and clearly lacks the Tyr62 hydroxyl group (Figure 3B).
Figure 2.
The X-ray structural of LnmK. (A) Overall fold of LnmK, showing one monomer colored from N-terminal in blue to C-terminal in red, the bound phosphopantetheine moiety in black ball-and-stick model, and the bound chloride as a purple sphere. (B) Biologically relevant homodimer of LnmK with one monomer in color and one monomer in gray. (C) Electrostatic surface of LnmK revealing a basic patch surrounding the substrate binding tunnel to the active site with the phosphopantetheine moiety and a bound sulfate shown in ball-and-stick model.
Figure 3.
The active site of LnmK. (A) Sigma weighted Fo-Fc omit map at 3 σ in green mesh depicting the phosphopantetheine moiety in the co-crystal structure of LnmK (Tyr62Phe) with methylmalonyl-CoA. (B) Tyrosine modeled into the co-crystal structure of LnmK (Tyr62Phe) with methylmalonyl-CoA showing the 2Fo-Fc map in blue at 1 σ and the Fo-Fc map at −3 σ in red. (C) Stereoview showing the amino acid side chains lining the active site tunnel from the co-crystal structure of LnmK with methylmalonyl-CoA. The bound chloride is shown as a purple sphere. Residues emanating from the crystallographic dimer are shown in gray.
The Overall Structure of LnmK
LnmK is a double ‘hot dog fold’ protein (DHDF) (Figure 2B) (26, 27). LnmK is comprised of 10 anti-parallel β-sheet strands with a topology of +2, +1, +1, −3, +5, +3, −1, −1, −2. Residues 11-157 and 196-309 make up the N-terminal and C-terminal hot dog folds (HDF), respectively (Figure 2A). Alignment of the individual HDFs produces high internal structural similarity (RMSD 2.3 Å for 95 residues), yet only 7 % internal sequence identity. The individual folds come together to create an extended β-sheet centered on a pseudo two-fold axis. The residues 158-195 make up a loop extending the length of the protein with an intervening α-helix (residues 175-185, α3) that lies against the extended β-sheet opposite the side containing the wrapped α-helices. Basic residues emanating from α3 create a positively charged surface likely involved in electrostatic interactions with the negatively charged CoA and LnmL (Figure 2C). A tunnel through the protein is present between the individual HDFs, buttressed on one side by α3, and closed off on the opposite side by the C-terminal HDF α-helix and residues emanating from other loops. The hole is blocked on one side by a symmetry mate that forms the active site cavity (Figure 2). The large surface area buried between the crystallographic monomer and symmetry mate is indicative of dimerization of LnmK [1745 Å2, PISA calculated (28)], which was confirmed by size exclusion chromatography. LnmK (calculated MW 35 kD) was eluted as a homodimer with an apparent molecular weight of 66 kD, and addition of methylmalonyl-CoA had no effect on the quaternary structure of LnmK (Figure 4 and Figure S2).
Figure 4.

Quaternary structure of LnmK as a homodimer determined by size exclusion chromatography of LnmK at (A) 1 and (B) 2 absorbance unit at 280 nm with 10 mM methylmalonyl-CoA with UV detection at 260 nm (red) and 280 nm (blue).
Comparison of LnmK by a structural similarity search of the PDB with Dali (29) revealed homology to a variety of enzymes with acyl-CoA or acyl-ACP thioesterase, dehydratase, and isomerase activities, but none with confirmed acyltransferase activity. Very few structural homologues have >15% identity at the primary sequence level, with the highest at 21% for a Pseudomonas aeruginosa hypothetical protein (unpublished PDB 1SH8, RMSD 7.9 Å) comprised of a single HDF. One of the most structurally similar proteins is acyl-CoA thioesterase 12 (hTE12, unpublished PDB 3B7K, 12% sequence identity, RMSD 3.9 Å), and the alignment reveals a similar binding of CoA against α3 (Figure S3).
The AT Active Site of LnmK
Although LnmK and the LnmK (Tyr62Phe) mutant were co-crystallized with methylmalonyl-CoA, density in the active site tunnel clearly represents the CoA hydrolysis product (Figure 3). Hydrolysis of substrate is a known side reaction for malonyl-CoA:ACP acetyltransferases (4) and explains the similar activity for LnmK. Electron density for the adenosine portion of methylmalonyl-CoA is also lacking, likely due to a combination of hydrolysis and conformational heterogeneity. In the LnmK (Tyr62Phe)-CoA structure, a sulfate ion is bound near the phosphate moiety of the phosphopantetheine moiety and is within distance to represent the ribose phosphate, even though there is no clear density for the sugar moiety (Figure 3A). The clear density for the phosphopantetheine moiety in the structure points to an active site, with the phosphate group exposed to solvent and the thiol group buried in the core of the protein (Figures 2 and 3). A tunnel through the enzyme surrounds the phosphopantetheine moiety, which is lined by main chain atoms and hydrophobic residues (Val142, Ala64, Leu221, Gly262, Leu153, Leu261, Tyr66) (Figure 5). The phosphopantetheine moiety interacts with the tunnel via a few specific hydrogen bonds: the Asn263 side chain bonds with the β-alanine carbonyl moiety, the backbone carbonyl of Tyr260 bonds with the β-alanine amide, the carbonyl of Phe65 bonds with the cysteamine moiety amide, the Leu153 amide interacts with the pantoate carbonyl via a water, the pantoate hydroxyl moiety is bonded to the Leu261 carbonyl and to the Tyr66 hydroxyl via a water molecule, and the terminal thiol is positioned 3.5Å away from the amide of Phe65 (Figure 5). Phe83 and Pro21 from the symmetry mate form a plug at one end of the tunnel forming an active site cavity (Figure 3C). A chloride ion is bound in the base of the active site by the backbone carbonyl of Phe223 and Asn216 (Figure 3C). There are no serine, threonine or cysteine residues in the cavity to act as a nucleophile for accepting and donating the methylmalonyl unit, as would be predicted from acyltransferases known to date. However, there are three tyrosines (Tyr62, Tyr222 and Tyr226) bearing the hydroxyl groups, serving as alternative but unprecedented candidates for the active site residue of an AT (Figure 3C).
Figure 5.

Flattened map of the LnmK active site tunnel showing key interactions between the bound phosphopantetheine moiety and the amino acid residues lining up the active site tunnel.
Comparison of the Se-Met and native LnmK structures reveals conformational changes for two tyrosines at the entrance of active site tunnel upon CoA/LnmL binding. In the Se-Met structure, Tyr66 blocks the active site tunnel entrance, and, upon rotation away from the tunnel, it would clash with Tyr179, which must move in response. The latter is exactly what was observed in the co-crystal structures of LnmK and LnmK (Tyr62Phe) with methylmalonyl-CoA (Figure S4). Also observed in the Se-Met LnmK structure, one primary hydroxyl of a glycerol molecule is bound in the tunnel in a similar position as the sulfur in the co-crystal structures of LnmK and LnmK (Tyr62Phe) with methylmalonyl-CoA (Figure S5), and this may explain why the phosphopantetheine moiety is not bound in the Se-Met LnmK structure. One of the bound solvent sulfate ions is found around the basic surface near the entrance to the active site tunnel.
Testing the AT Active Site Residues of LnmK
To test the hypothesis that LnmK employs a tyrosine as the catalytic nucleophile, Tyr62, Tyr222 and Tyr226 were mutated to phenylalanine (Figure 3C) and tested using DL-2-[methyl-14C]malonyl-CoA as a substrate (Figures 1B). LnmK Tyr222Phe and Tyr226Phe mutants behaved as the native enzyme, but Tyr62Phe lost both self- (Figure 6A) and trans-acylation activities (Figures 6B and 6C) implicating Tyr62 as the active site residue with its hydroxyl group acting as the catalytic nucleophile (Figure 6). Asn216 and Asn263 were mutated to leucine, which reduced AT activity (lanes 5 and 6, Figure S6), and this finding is consistent with their predicted roles in anchoring CoA/LnmL within the active site tunnel (Figure 3C). All conserved Ser, Thr, and Cys among LnmK and homologues were identified (Figure S1) and mutated to interrogate their function in AT catalysis. The Cys24Ala, Ser28Ala, Ser100Ala, Thr130Ala, Ser174Ala, and Ser225Ala mutants all retained activity (Figure S6), and the diminished activity of the Ser91Ala mutant resulted from its thermo-instability (lane 12, Figure S6). The Ser91Ala mutant precipitated out upon warming the assay solution to 25°C, and this is consistent with the observation that Ser91 is buried and makes important contacts for LnmK structural integrity. Taken together, these results point to Tyr62 as the active site residue, the hydroxy group of which acts as the nucleophile and forms an acyl-O-Tyr intermediate in AT catalysis (Figure 1B). Similar to known acyltransferases that employ Ser at their active sites, LnmK also hydrolyzes substrate methylmalonyl-CoA (Figure 6A) and product propionyl-S-LnmL (Figure 6B) over extended incubations, which is likely the reason for the absence of methylmalonyl-CoA or an acylated residue in the active site of the crystal structures of LnmK.
Figure 6.
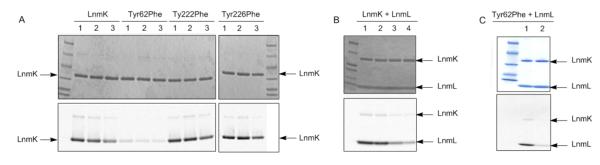
Enzymatic assays and time courses of LnmK and the LnmK Tyr62Phe, Tyr222Phe, and Tyr226Phe mutants for their AT activity with DL-2[methyl-14C]malonyl-CoA as a substrate upon SDS-PAGE (4-15%) (top panels) and autoradiography (bottom panels) analyses. (A) Self-acylation of LnmK in comparison to the three Tyr62Phe, Tyr222Phe, and Tyr226Phe mutants with varying incubation times (lane 1, 10 s; lane 2, 140 s; and lane 3, 1000 s). (B) Trans-acylation to LnmL by LnmK with varying incubation times (lane 1, 0.5 min, lane 2, 5 min; lane 3, 20 min, and lane 4, 60 min). (C) Trans-acylation to LnmL by LnmK in comparison with the Tyr62Phe mutant with 1 min incubation time.
DISCUSSION
LnmK Representing a New Family of AT/DC Enzymes
β-Alkyl branches in polyketide biosynthesis are installed by HCSs, which utilize acetyl-S-ACP and propionyl-S-ACP to furnish the β-methyl and β-ethyl/propionyl branches, respectively (9–16). Two parallel but distinct pathways have been discovered for acetyl-S-ACP and propionyl-S-ACP biosynthesis (Figure 1B). Acetyl-S-ACP from malonyl-CoA is catalyzed by a dedicated set of ACP, AT, and KS (acting as a DC). This pathway has been characterized from several polyketide biosynthetic machinery, and both the AT and KS enzymes show high sequence homology to proteins of known AT and KS activities (8–16). Propionyl-S-ACP from methylmalonyl-CoA by a dedicated ACP (LnmL) and a bifunctional AT/DC (LnmK) was first reported for the LNM biosynthetic machinery (Figure 1A) (8). Although both the AT and DC activity of LnmK were established biochemically (Figure 1B) (8) and homologues of LnmK have been identified from other biosynthetic pathways (Figure S1), LnmK shows no sequence homology to either ATs or DCs known to date, representing a new family of bifunctional AT/DC enzymes.
We now report the X-ray crystal structures of LnmK at 1.7Å resolution (Table 1). LnmK is a homodimer composed of two monomeric DHDF (Figure 2). Each monomer possesses an active site tunnel terminated by residues emanating from the dimer partner, which is revealed through co-crystallization with the substrate methylmalonyl-CoA (Figure 3). The crystal structures set the stage to explore the utility of LnmK in engineering polyketide structural diversity by providing a molecular surface to interrogate the interaction with LnmL and by revealing the putative active site residues to determine LnmL-tethered substrate specificity. Most importantly, these structures reveal surprising deviations from the known chemical mechanisms of either AT and KS enzymes and DHDF enzymes. LnmK represents the first AT that employs a Tyr as an active site residue and the first member of DHDF enzymes that displays an AT activity (Figures 3C and 6). These findings highlight natural product biosynthetic machinery as a rich source of novel enzyme activities, mechanisms, and structures. Mechanistic and structural characterization of novel enzymes for natural product biosynthesis will greatly facilitate efforts to engineer natural product structural diversity for drug discovery and development by combinatorial biosynthesis strategies.
A New Member of the HDF Family
Members of the HDF family often share little sequence homology, but yet can be structurally similar; this leads to a large number of functions within this structure family (26, 27). Nevertheless, AT activity has not been demonstrated by a HDF enzyme to date. The majority of HDFs are acyl-CoA thioesterases and dehydratases involved in primary metabolism. Recently, the activities of this family have grown from the characterizations of natural product biosynthetic machinery, including dehydratase domains with DHDFs from erythromycin (PDB ID 3EL6) (30) and curacin (PDB IDs, 3KG6, 3KG7, 3KG8, 3KG9) (31) type I polyketide synthases, the product template domains with DHDFs from fungal type I polyketide synthase (PDB IDs, 3HRR and 3HRQ) (32, 33), and thioesterases with HDFs from enediyne biosynthetic machinery (PDB IDs, 2XFL, 2XEM, 2W3X) (34–36). LnmK shares a similar overall fold with these enzymes associated with natural product biosynthesis, yet their structures are quite different, with RMSD values in the rage of 4.4 to 4.9 Å for the DHDF enzymes and 2 to 3.2 Å for the single HDF enzymes (Figure S7). The active site entrance of LnmK is located at the meeting point between the individual HDFs (Figure 2), while the active site entrance of the product template domain and dehydratases are located within one of the HDFs (Figure S7). LnmK is structurally more similar to the DHDF acyl-CoA and acyl-ACP thioesterases and dehydratases from primary metabolism with respect to the location of the active site (Figure S7). The dehydratases utilize conserved Asp and His in the active sites for activity, and the DHDF thioesterases use a variety of mechanisms for catalysis, including an Asp, Thr, and Gln catalytic triad to activate a water molecule for nucleophilic attack, or direct attack of an Asp or Glu onto a thioester activated by backbone amides (26, 27). LnmK lacks these typical catalytic residues in the active site making the enzymatic mechanism enigmatic (Figure 3). Although LnmK is structurally very similar to hTE12, an acyl-CoA thioesterase, the binding modes of CoA are very different; whereas the interactions of LnmK with CoA are mainly through the phosphopantetheine moiety and electrostatic interactions with the ribose phosphate, hTE12 binds the adenine moiety (Figure S3). These comparisons serve to highlight the unique structure of LnmK as a DHDF family member that catalyzes unprecedented AT/DC activity and the versatility of this unique structural fold in acquiring and evolving new enzymatic activity.
An AT Mechanism Based on Tyrosine
Canonical (methyl)malonyl-CoA:ACP acyltransferases use a Ser as the catalytic nucleophile (37–40). The active site of these ATs is rigid in nature with all the catalytic residues set in place, such that conformational changes during catalysis are minimal (37–40). The co-crystal structures of LnmK and LnmK (Try62Phe) with methylmalonyl-CoA reveal no Ser in the vicinity of the active site but instead three tyrosines (Tyr62, Tyr222, and Tyr226) as alternative active site residues of an AT (Figure 3C). Site-directed mutagenesis indeed confirmed that Tyr62 is the most likely residue to act as the catalytic nucleophile; the LnmK (Tyr62Phe) mutant completely loses its AT activities in both self-acylation (Figure 6A) and trans-acylation to LnmL (Figures 6B and 6C), whereas the LnmK (Tyr222Phe) and LnmK (Tyr226Phe) mutants retain the same activity as the native LnmK (Figure 6)). The crystal structure of the LnmK (Tyr62Phe) confirms that the mutant is still properly folded and should be catalytically competent if the Tyr62 hydroxyl was not necessary for activity. Unfortunately, LnmK (Tyr62Phe) still hydrolyzed methylmalonyl-CoA, which otherwise would have revealed the binding interactions of the methylmalonyl moiety. In all of the structures the Tyr62 hydroxyl faces away from the active site, such that there would have to be rotation about Cα-Cβ to bring the hydroxyl near the bound thioester (Figure 3C). Evidence for such a conformational change is hinted at by a distorted β-sheet bulge between Tyr62-Leu63 that upon relaxation could drive the repositioning of Tyr62 into the active site. This needed conformational change masks the identity of the catalytic base responsible for activation of the tyrosine. Together the use of a tyrosine as the catalytic nucleophile and the need for conformational changes make the AT mechanism of LnmK unique. These findings are sure to inspire and inform the search for ATs and other natural product biosynthetic machinery that employ noncanonical active sites.
AT and DC Activity from the Same Active Site
The decarboxylation reaction most likely takes place within the phosphopantetheine binding site, as the only pockets of conserved residues lie in this active site and dimerization interface (Figure S1). There are various ways enzymes catalyze the decarboxylation of β-keto acids. Minimally these enzymes must maintain a deprotonated carboxylate, align electron orbital overlap, and stabilize the enolate intermediate. The well-studied acetoacetate decarboxylase uses a lysine to form an enamine with the ketone group which stabilizes the intermediate enolate and an arginine to maintain a deprotonated carboxylate (41). Methylmalonyl-CoA decarboxylase is proposed to use two backbone amides to stabilize the thioester enolate oxygen, while a nearby histidine likely protonates the α-carbon (42). KSs utilize a conserved histidine to stabilize the thioester enolate oxygen (43–45). The lack of acids and bases in the active site of LnmK make it difficult to propose a mechanism. Nevertheless, in the co-crystal structures of LnmK and LnmK (Tyr62Phe) with methylmalonyl-CoA, a chloride is bound between the backbone amide of Phe223 and side chain of Asn216, which may be analogous to the carbonyl-binding mode in methylmalonyl-CoA decarboxylase (Figure 3). The LnmK structure will allow us to probe the DC reaction through targeted mutagenesis of the active site residues and analysis of the resulting acyl group carried by LnmL, in order to understand the complex chemistry carried out by this novel bifunctional AT/DC enzyme.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kyowa Hakko Kogyo Co. Ltd (Tokyo, Japan) for the wild-type S. atroolivaceus S-140 strain and the Advanced Photon Source LS-CAT beamline staff, especially David Smith, for assistance with X-ray data collection. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences (Contract No. DE-AC02-06CH11357), and use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Funding. This work was supported in part by NIH NIGMS Protein Structure Initiative Grant GM094596 (G.N.P. and B.S.) and NIH Grant CA106150 (B.S.).
ABBREVIATIONS
- ACP
acyl carrier protein
- AT
acyltransferase
- DC
decarboxylase
- ECH
enoyl-CoA hydratase
- HCS
hydroxymethylglutaryl-CoA synthase
- KS
ketosynthase
- LNM
leinamycin
- MW
molecular weight
- NRPS
nonribosomal peptide synthetase
- PKS
polyketide synthase
- Se-Met
selenomethionine
Footnotes
Accession codes. The atomic coordinates and structure factors of Se-Met LnmK and LnmK and LnmK (Tyr62Phe) in complex with CoA have been deposited within the Protein Data Bank as entry 4HZN, 4HZO and 4HZP, respectively.
Supporting Information. Oligos and primers used (Table S1), LnmK and its homologues and conservation scores painted on the LnmK structure (Figure S1), calibration with molecular weight standards for size exclusion chromatography of LnmK on Sepharose 6 10/300 GL column (Figure S2), structural alignment of LnmK with hTE12 (Figure S3), conformational change upon substrate binding as revealed by comparison between the structures of Se-Met LnmK and LnmK and LnmK (Tyr62Phe) in complex with phosphopantetheine (Figure S4), structure of Se-Met LnmK with glycerol bound at the active site tunnel as compared with the structures of LnmK and LnmK (Tyr62Phe) in complex with CoA (Figure S5), enzymatic assays of LnmK and selected mutants for their AT activity in transacylation to LnmL with DL-2[methyl-14C]malonyl-CoA as a substrate upon SDS-PAGE and autoradiography analyses (Figure S6), and selected enzymes characterized from natural product biosynthetic machinery that adopt a HDF (Figure S7). This material is available free of charge via the Internet at http://pubs.acs.org.
Notes. The authors declare no competing financial interests.
REFERENCES
- 1.Hara M, Asano K, Kawamoto I, Takiguchi T, Katsumata S, Takahashi K, Nakano H. Leinamycin, a new antitumor antibiotic from Streptomyces: producing organism, fermentation and isolation. J. Antibiot. 1989;42:1768–1774. doi: 10.7164/antibiotics.42.1768. [DOI] [PubMed] [Google Scholar]
- 2.Hirayama N, Matsuzawa ES. Molecular structure of a novel antitumor antibiotic leinamycin. Chem. Lett. 1993:1957–1958. [Google Scholar]
- 3.Cheng Y-Q, Tang G-L, Shen B. Identification and localization of the gene cluster encoding biosynthesis of the antitumor macrolactam leinamycin in Streptomyces atroolivaceus S-140. J. Bacteriol. 2002;184:7013–7024. doi: 10.1128/JB.184.24.7013-7024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y, Tang G, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang G, Cheng Y, Shen B. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem. Biol. 2004;11:33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Tang G, Cheng Y, Shen B. Polyketide chain skipping mechanism in the biosynthesis of the hybrid nonribosomal peptide-polyketide antitumor antibiotic leinamycin in Streptomyces atroolivaceus S-140. J. Nat. Prod. 2006;69:387–393. doi: 10.1021/np050467t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang G-L, Cheng Y-Q, Shen B. Chain initiation in the leinamycin-producing hybrid nonribosomal peptide/polyketide synthetase from Streptomyces atroolivaceus S-140. Discrete, monofunctional adenylation enzyme and peptidyl carrier protein that directly load D-alanine. J. Biol. Chem. 2007;282:20273–20282. doi: 10.1074/jbc.M702814200. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, Huang Y, Shen B. Bifunctional acyltransferase/decarboxylase LnmK as the missing link for beta-alkylation in polyketide biosynthesis. J. Am. Chem. Soc. 2009;131:6900–6901. doi: 10.1021/ja9012134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Huang S-X, Ju J, Tang G, Liu T, Shen B. Characterization of the lnmKLM genes unveiling key intermediates for β-alkylation in leinamycin biosynthesis. Org. Lett. 2011;13:498–501. doi: 10.1021/ol102838y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Convergence of isoprene and polyketide biosynthetic machinery: isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu L, Jia J, Liu H, Håkansson K, Gerwick WH, Sherman DH. Metabolic coupling of dehydration and decarboxylation in the curacin A pathway: functional identification of a mechanistically diverse enzyme pair. J. Am. Chem. Soc. 2006;128:9014–9015. doi: 10.1021/ja0626382. [DOI] [PubMed] [Google Scholar]
- 12.Simunovic V, Zapp J, Rachid S, Krug D, Meiser P, Müller R. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. ChemBioChem. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- 13.Calderone CT, Iwig DF, Dorrestein PC, Kelleher NL, Walsh CT. Incorporation of nonmethyl branches by isoprenoid-like logic: multiple beta-alkylation events in the biosynthesis of myxovirescin A1. Chem. Biol. 2007;14:835–846. doi: 10.1016/j.chembiol.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simunovic V, Müller R. 3-hydroxy-3-methylglutaryl-CoA-like synthases direct the formation of methyl and ethyl side groups in the biosynthesis of the antibiotic myxovirescin A. ChemBioChem. 2007;8:497–500. doi: 10.1002/cbic.200700017. [DOI] [PubMed] [Google Scholar]
- 15.Calderone CT. Isoprenoid-like alkylations in polyketide biosynthesis. Nat. Prod. Rep. 2008;25:845–853. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- 16.Buchholz TJ, Rath CM, Lopanik NB, Gardner NP, Håkansson K, Sherman DH. Polyketide β-branching in bryostatin biosynthesis: identification of surrogate acetyl-ACP donors for BryR, an HMG-ACP synthase. Chem. Biol. 2010;17:1092–1100. doi: 10.1016/j.chembiol.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sánchez C, Du L, Edwards DJ, Toney MD, Shen B. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem. Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- 18.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:306–315. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP 4 program suite. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 23.Joosten K, Cohen SX, Emsley P, Mooij W, Lamzin VS, Perrakis A. A knowledge-driven approach for crystallographic protein model completion. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:416–424. doi: 10.1107/S0907444908001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Dauter Z. Carrying out an optimal experiment. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:389–392. doi: 10.1107/S0907444909038578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pidugu LS, Maity K, Ramaswamy K, Surolia N, Suguna K. Analysis of proteins with the “hot dog” fold: prediction of function and identification of catalytic residues of hypothetical proteins. BMC Struct. Biol. 2009;9:37. doi: 10.1186/1472-6807-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dillon SC, Bateman A. The hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinf. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keatinge-Clay A. Crystal structure of the erythromycin polyketide synthase dehydratase. J. Mol. Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akey DL, Razelun JR, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Crystal structures of dehydratase domains from the curacin polyketide biosynthetic pathway. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Image II, Xu W, Tang Y. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J. Biol. Chem. 2010;285:22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill E. a, Kamari-Bidkorpeh O, Tsai S-C, Townsend C. a. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Lanen S. G. Van, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotaka M, Kong R, Qureshi I, Ho QS, Sun H, Liew CW, Goh LP, Cheung P, Mu Y, Lescar J, Liang Z-X. Structure and catalytic mechanism of the thioesterase CalE7 in enediyne biosynthesis. J. Biol. Chem. 2009;284:15739–15749. doi: 10.1074/jbc.M809669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liew CW, Sharff A, Kotaka M, Kong R, Sun H, Qureshi I, Bricogne G, Liang Z-X, Lescar J. Induced-fit upon ligand binding revealed by crystal structures of the hot-dog fold thioesterase in dynemicin biosynthesis. J. Mol. Biol. 2010;404:291–306. doi: 10.1016/j.jmb.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 37.Keatinge-Clay AT, Shelat AA, Savage DF, Tsai S-C, Miercke LJW, O’Connell JD, Khosla C, Stroud RM. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure. 2003;11:147–154. doi: 10.1016/s0969-2126(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 38.Oefner C, Schulz H, D’Arcy A, Dale GE. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:613–618. doi: 10.1107/S0907444906009474. [DOI] [PubMed] [Google Scholar]
- 39.Tang Y, Chen AY, Kim C, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem. Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Y, Kim C, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho M-C, Ménétret J-F, Tsuruta H, Allen KN. The origin of the electrostatic perturbation in acetoacetate decarboxylase. Nature. 2009;459:393–397. doi: 10.1038/nature07938. [DOI] [PubMed] [Google Scholar]
- 42.Benning MM, Haller T, Gerlt JA, Holden HM. New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli. Biochemistry. 2000;39:4630–4639. doi: 10.1021/bi9928896. [DOI] [PubMed] [Google Scholar]
- 43.Pan H, Tsai SC, Meadows ES, Miercke LJW, Keatinge-Clay AT, O’Connell J, Khosla C, Stroud RM. Crystal structure of the priming beta-ketosynthase from the R1128 polyketide biosynthetic pathway. Structure. 2002;10:1559–1568. doi: 10.1016/s0969-2126(02)00889-4. [DOI] [PubMed] [Google Scholar]
- 44.Dreier J, Khosla C. Mechanistic analysis of a type II polyketide synthase. Role of conserved residues in the beta-ketoacyl synthase-chain length factor heterodimer. Biochemistry. 2000;39:2088–2095. doi: 10.1021/bi992121l. [DOI] [PubMed] [Google Scholar]
- 45.Bisang C, Long PF, Cortés J, Westcott J, Crosby J, Matharu AL, Cox RJ, Simpson TJ, Staunton J, Leadlay PF. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature. 1999;401:502–505. doi: 10.1038/46829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.