Abstract
Siderophores are best known as small iron binding molecules that facilitate microbial iron transport. In our previous study we identified a siderophore-like molecule in mammalian cells and found that its biogenesis is evolutionarily conserved. A member of the short chain dehydrogenase family of reductases, 3-OH butyrate dehydrogenase (BDH2) catalyzes a rate-limiting step in the biogenesis of the mammalian siderophore. We have shown that depletion of the mammalian siderophore by inhibiting expression of bdh2 results in abnormal accumulation of cellular iron and mitochondrial iron deficiency. These observations suggest that the mammalian siderophore is a critical regulator of cellular iron homeostasis and facilitates mitochondrial iron import. By utilizing bioinformatics, we identified an iron-responsive element (IRE; a stem-loop structure that regulates genes expression post-transcriptionally upon binding to iron regulatory proteins or IRPs) in the 3′-untranslated region (3′-UTR) of the human BDH2 (hBDH2) gene. In cultured cells as well as in patient samples we now demonstrate that the IRE confers iron-dependent regulation on hBDH2 and binds IRPs in RNA electrophoretic mobility shift assays. In addition, we show that the hBDH2 IRE associates with IRPs in cells and that abrogation of IRPs by RNAi eliminates the iron-dependent regulation of hBDH2 mRNA. The key physiologic implication is that iron-mediated post-transcriptional regulation of hBDH2 controls mitochondrial iron homeostasis in human cells. These observations provide a new and an unanticipated mechanism by which iron regulates its intracellular trafficking.
Keywords: Mammalian siderophore, IRE-IRP regulation, hemochromatosis
Introduction
Iron (Fe) is indispensable for almost all living organisms with the exception of Lactobacilli and Borrelia, which utilize manganese and cobalt for redox reactions (1). The acquisition and transport of Fe is a challenge to all organisms because of its low solubility and high toxicity. To overcome these problems, organisms such as bacteria and fungi synthesize “siderophores” or iron-specific chelating agents. The role of these compounds is to scavenge iron from the environment (1, 2). In contrast, mammalian cells acquire iron from carrier proteins and the newly acquired iron then enters into a cytosolic “labile” iron pool from which it is distributed among various cellular compartments. This pool of iron is postulated to associate with proteins and low-molecular weight compounds such as siderophores and is a key factor in the cell iron-sensing machinery (3, 4, 5, 6).
We have previously identified a mammalian siderophore and found that it is chemically similar to the E. coli Enterobactin – the classical bacterial siderophore (7, 8). Interestingly, the mammalian siderophore is biosynthesized by an evolutionarily conserved pathway and BDH2 (3-hydroxy butyrate dehydrogenase) – a homologue of bacterial EntA catalyzes a rate-limiting step. In this regard, inhibition of bdh2 expression, which subsequently leads to siderophore depletion, alters intracellular iron homeostasis (8). In addition, siderophore depleted mammalian cells, zebrafish, and yeast fail to synthesize heme – an iron-dependent mitochondrial process (8). These results also suggest that siderophore is a regulatory entity but it is not clear if the expression of mammalian siderophore itself is regulated by iron.
Intracellular iron regulates expression of genes involved in cellular iron metabolism, energy metabolism, cell cycle control and oxygen sensing by altering the interaction of iron regulatory proteins (IRPs) with RNA motifs called iron-responsive elements (IREs). These are stem loop structures present in the untranslated regions of a variety of genes that regulate iron uptake, storage, utilization and transport, as well as other cellular processes such as cell cycle, oxygen sensing and energy metabolism (9 – 12). Genes containing an IRE in the 5′-UTR are translationally inhibited when bound by IRPs, whereas mRNAs containing IREs in 3′-UTR are stabilized when bound by IRPs. Intracellular iron negatively regulates interaction of IRE-IRP complexes, by inhibiting association of IRP with IRE. Hence, intracellular iron promotes the translation of messages containing a 5′ IRE, for example ferritin mRNA. Conversely, low intracellular iron levels have the opposite effect – promoting protein synthesis by stabilizing 3′ IRE-containing mRNAs such as transferrin receptor 1 (TfR1) mRNA (9, 11, 13, 14).
IREs are evolutionary conserved hairpin structures of 25–30 nucleotides (15). A typical IRE stem consists of variable sequences that form base pairs of moderate stability (ΔG ≈− 7 kcal/mol), and folds into an α-helix that is slightly distorted by the presence of a small bulge in the middle (an unpaired C residue or an asymmetric UGC/C bulge/loop). The loop contains a conserved 5′-CAGUGH-3′ sequence (H denotes A, C or underlined C and G residues form a base pair. The TfR1 mRNA contains multiple IREs within its long 3′ UTR, while the mRNAs encoding ferritin-H and ferritin-L contain a single IRE in their 5′ UTRs.
Mice lacking IRPs die in utero, thus highlighting the critical importance of the IRE-IRP network (reviewed in refs. 10 and 16). Moreover, mutations in IREs are implicated in human iron disorders (17, 18). Recent studies have also identified non-canonical IREs, which deviate from the above-mentioned sequence, yet confer iron-dependent regulation and in addition they bind IRPs both in vitro and in vivo (12, 19 – 25). In addition, base substitutions in the pseudotriloop region or in the stem structure of a canonical IRE element also confers iron-dependent regulation (25 – 27). For example, human ferritin-H IRE bearing mutations in the 6-nucleotide apical loop sequence that results in conversion of CAGUG(A/C/U) to CAGGG(A/C/U), retains its affinity to IRPs (25).
Mutations in the HighFe (HFE) gene, which encodes a cell surface, atypical major histocompatibility complex I molecule, cause the most common type of hereditary hemochromatosis (HH). The specific mutation that leads to amino acid substitution (C282Y), affects the ability of HFE to interact with its chaperone that facilitate its intracellular processing and transport to the surface (28).
Mitochondria are the major site of iron utilization and imported iron is utilized for the synthesis of heme, iron/sulfur clusters (ISCs; refs. 29, 30). Mitochondrial iron import and heme export are coordinated as alterations in these two processes manifest in abnormal mitochondrial iron homeostasis. Alterations in mitochondrial iron homeostasis lead to changes in cellular iron metabolism, suggesting communication between these two compartments (30).
Given the importance of the mammalian siderophore in intracellular iron homeostasis and heme biogenesis (8), it is important to define the mechanisms that regulate the expression of the mammalian siderophore. We have identified a putative IRE in the 3′-UTR of hBDH2 gene, whose product catalyzes a rate-limiting step in the biosynthesis of the mammalian siderophore. Our studies also demonstrate that this element confers iron-dependent regulation on the expression of hBDH2 and that it associates with IRPs both in vitro and in cells. The iron-dependent regulation of siderophore expression is restricted to hominidae family members. Taken together, our studies show siderophore regulation controls mitochondrial iron import.
Materials and Methods
Cell lines, culture conditions, treatments and transfections
Cells were cultured in Dulbecco’s modified Eagle’s medium (293T; MIMCD), Minimal essential medium (HeLa), Iscove’s modified DMEM (K562), DMEM:F12 (MCF-10 A), Williams E medium (human and primate liver cells) or Medium 200 (HUVEC) supplemented with 10% fetal bovine serum (FBS) or 5% horse serum, 2 mM L-glutamine, 100 units of Penicillin, and 100 μg Streptomycin (Invitrogen). MCF-10 A culture medium contained additional supplements such as insulin, EGFα, cholera toxin B, and hydrocortisone (Sigma). Cells were treated with 100 μM Desferrioxamine (DFO; Calbiochem), or 100 μM Ferric Ammonium Citrate (FAC; Sigma), or 100 μM Hemin (Sigma) for indicated periods of time. Cells were transfected with Lipofectamine (Invitrogen) or X-tremeGENE HP DNA transfection reagent (Roche) as per the manufacturer’s recommended procedure.
Bioinformatics approach to identify putative IRE in hBDH2 mRNA
We utilized SIREs (searching for IREs) program available at http://ccbg.imppc.org/sires/index.html to predict iron-response elements in hBDH2 mRNA (31). Multispecies alignment was performed using CLUSTALW program.
Plasmids and molecular cloning strategy
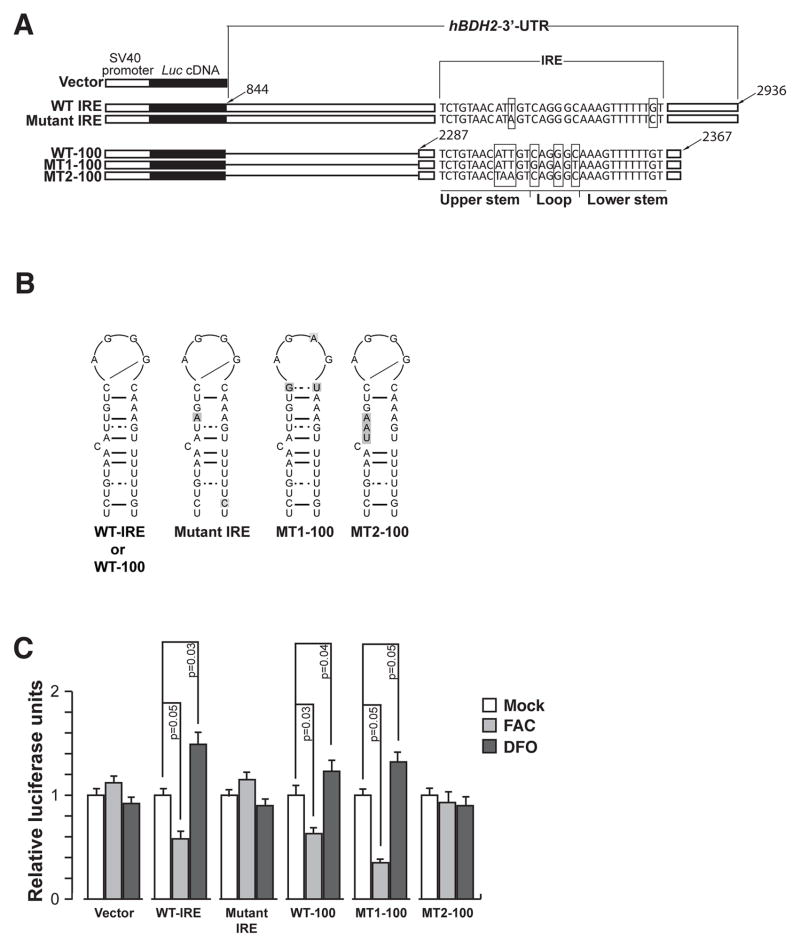
For iron-dependent regulation studies of hBDH2 3′-UTR, a Luciferase reporter containing the hBDH2 3′-UTR was constructed as indicated below. A PCR generated ~2 kb fragment containing hBDH2 3′-UTR was inserted downstream of the Luciferase gene (luc2) in pGL4.13 vector (Promega) to generate the Luc - hBDH2 wt IRE vector. A SV40 early enhancer/promoter drives the expression of the luc2 gene. We also derived a mutant with nucleotide substitutions within the IRE region of hBDH2 3′-UTR (Luc - hBDH2 mut IRE, also see Fig. 5 B) by site-directed mutagenesis. Additionally, we also generated a series of Luciferase reporter plasmids containing either a 100bp oligomer encompassing the wt hBDH2 IRE (Luc - WT 100 hBDH2 IRE) or with nucleotide substitutions in the pseudotriloop region (to convert to a consensus IRE loop sequence; Luc-MT-1 100 hBDH2 IRE) or in the stem structure of the hBDH2 IRE (Luc-MT-2 100 hBDH2 IRE; also see Fig. 5 B) into pGL4.13 vector. The IRE sequences were inserted downstream of the luc2 gene as described above. Finally, we also derived Luciferase reporter plasmids bearing wt hBDH2 IRE (WT-100 5′-LUC) or with nucleotide substitutions in the pseudotriloop region (to convert to a consensus IRE loop sequence; MT1-100 5′-LUC) upstream of the luc2 gene for iron-dependent translational control analysis (Fig. S3 A). All reporter plasmids were sequence verified.
Figure 5. hBDH2 2 IRE and 3′ UTR confer iron-dependent regulation on to a heterologous gene.
(A) Schematic of the Luciferase reporter plasmids. (B) Secondary structures of wt or mutant hBDH2 IREs. Mutant nucleotides are shaded. (C) MCF-10 A cells were transfected with Luciferase reporter plasmids shown in (A) and subsequently treated with 100 μM each of FAC or DFO. Sixteen hours later, cells were assayed for Luciferase activity. Data presented are arbitrary firefly Luciferase light units that have been normalized using the Renilla Luciferase measurements to account for transfection efficiency. All experiments were performed in triplicate, and error bars represent the SD.
Luciferase assays
MCF-10 A cells at 50 to 60% confluency cultured in a 6-well plate were transiently transfected with the indicated Luc-IRE reporter plasmids along with a control Renilla Luciferase plasmid (pGL4.74). An empty vector (pGL4.13) served as a negative control. Following the removal of DNA complexes cells were replated in a 24-well plate and treated for 16 hours with 100 μM solutions of Desferrioxamine (DFO, Calbiochem), Hemin (Sigma), or FAC (Sigma). Firefly and Renilla luciferase activities of lysates were assayed using a Dual Luciferase Assay kit from Promega as per the suggested procedure. All Luciferase measurements were normalized to the Renilla Luciferase expression in order to correct for differences in transfection efficiency.
RNA isolation and gene expression analysis
Total RNA was isolated from naïve or treated cells using Trizol method (Invitrogen). DNase I (Promega) treated RNA was then reverse transcribed using Superscript III RT from Invitrogen as per manufacturer’s recommendations. The resulting cDNAs were subjected to real time PCR analysis using SYBR Green master mix (Promega) following the manufacturer’s recommendations. The fold-change was calculated using ΔΔCT method.
Electrophoretic mobility shift assay (EMSA)
Recombinant His-tagged IRPs (His-IRP-1 and His-IRP-2) were purified from E. coli as described in ref. 21. A 100 bp region containing the hBDH2 wt IRE or with nucleotide substitutions in the pseudotriloop or in the stem structure was (Fig. 5 B) inserted into the pcDNA3.1+ vector (Invitrogen). Similarly, a 60 bp region encompassing the TfR1 B IRE was subcloned from pUC18-TfR B-GH (ref. 32) into the pCDNA3.1+ vector. Radiolabeled wt or mutant hBDH2 IREs, and TfR1 B IRE probes were synthesized by in vitro transcription using T7 polymerase (New England Bio-labs). Briefly, linearized DNA was transcribed in a 20 μl reaction with 1 U of polymerase at 37°C for 1 hr in the presence of 100 μCi of 32P-GTP (Perkin-Elmer) and 2.5 mM each of ATP, CTP, and UTP (Roche). The labeled cRNA was purified in a Sephadex G-50 column (Roche) following the manufacturer’s instructions. Labeled RNA probes had a specific activity of 1–4 × 1010 cpm/μg of RNA. The purified labeled probes were stored at −80°C until further use.
EMSAs were performed as previously described in refs. 21 and 33. Concisely, 2–10 × 104 cpm of radio labeled RNA probes were initially heat denatured prior to incubation with either 10 ng of purified IRPs for 30 min at room temperature (RT) in a reaction volume of 15μl made up with a binding buffer (25 mM Tris-HCl, pH 7.5; 40 mM KCl; 1% Triton-X 100). Subsequently, RNase T1 (1 unit) was added to the reaction for 10 min at RT, followed by heparin (10 μg/μl; Roche) for an additional 10 min at RT. RNA loading dye was added to each sample and the RNA:protein complexes were resolved for 45 min at 100 v in 0.5 x TBE on a 1.5 mm 4% polyacrylamide gel (GIBCO-BRL) using 0.5 x TBE. Gels were vacuum dried and subjected to autoradiography.
Immunopurification of mRNPs
Immunoprecipitation of IRP-RNA complexes was previously described (34). Briefly, 10 μg of anti-IRP-1 or IRP-2 specific antibody was conjugated to prewashed Protein G-Sepharose (PGS, Invitrogen) in the presence of 200 units each RNAsin (Promega) and Protector RNAse inhibitor (Roche) at 4°C for 1 hour with gentle agitation. The beads were then washed with PBS to remove unbound Ab.
Cells were treated with 100 μM each DFO or hemin for 16 hours at 37°C. Cytoplasmic extracts were prepared by adding 250 μl of lysis buffer (20 mM Hepes, pH 7.6, 25 mM KCl, 0.5% NP-40 and 1 mM PMSF) to the 60-mm dish and incubated at 4°C for 30 min. The clarified supernatants were then incubated with PGS beads coated with anti-IRP antibodies or with uncoated PGS beads (as a control) in binding buffer (20 mM Hepes, pH 7.9, 150 mM NaCl, 0.05% Triton X-100 and 200 units of RNAsin) at 4°C for 1 hour with agitation. The beads were washed twice with the binding buffer and twice with the wash buffer (20 mM Hepes, pH7.9, 150 mM NaCl, 1% Triton X-100, and 200 units of RNAsin). The beads were resuspended in 100 μl of RNase free water, and the bound RNA was recovered by phenol-chloroform extraction and ethanol precipitation. The immunoprecipitated RNA was then subjected to quantitative real time PCR as described above.
In a related set of experiments IRP-RNA complexes from 293T cells stably expressing FLAG-IRPs (35) were precipitated using beads coated with anti-FLAG Ab. Precipitated RNA was recovered as described above.
RNAi knockdown of IRP-1 and IRP-2 genes
The shRNAs specific for IRP-1 and IRP-2 were obtained from Susy Torti (Wake Forest University). HeLa or MCF-10 A cells were transfected with IRP-1 or IRP-2 specific shRNA expressing plasmids and selected for Neomycin resistance (400μg/ml). To achieve a complete knockdown of IRP-1 or IRP-2, the shRNA expressing cells were further transfected with siRNA oligomers specific for IRP-1 or IRP-2 or control siRNA oligomers (Dharmacon). The efficiency of the knockdown was assessed by RT-PCR using gene-specific primers or with IRP-1 and IRP-2 specific antibodies (Santa Cruz biotechnology).
Analysis of hBDH2 mRNA stability
The stability of hBDH2 mRNA was analyzed by following the procedure described in ref 36. Briefly, Actinomycin D (5 μg/ml) was added to untreated (mock) cells and to cells that were prior treated with 100 μM each DFO or hemin for 16 hours at 37°C. Cells were collected at various times points and the relative levels of hBDH2 mRNA was quantified by quantitative real time PCR as described above. The hBDH2 and TfR1 mRNA levels were normalized to rlp13a, a ribosomal gene. Data were plotted on a semi logarithmic graph and the first order decay slope was calculated using lineal regression. Half-lives of hBDH2 and TfR1 mRNA were calculated using the equation: t1/2 = ln2/(ln10 × the slope of the regression line). The slope is log10 (N0/N)/t, where N0 is the initial amount of mRNA, N is the amount of mRNA at time t, and t is the elapsed time. Half-lives was calculated from values of three independent experiments.
Assessment of intracellular iron levels
Labile iron pools were measured using the iron-sensitive probes Calcein Green (CALG; Molecular Probes) and rhodamine B-[(1,10-phenanthrolin-5-yl)aminocarbonyl]benzyl ester (RPA) or rhodamine B-[(phenanthren-9-yl)aminocarbonyl]benzyl (RPAC; Axxora), for cytosolic and mitochondrial compartments, respectively. The fluorescent mitochondrial iron indicator RPA contains a rhodamine moiety for mitochondrial targeting and a 1,10-phenanthroline moiety for iron chelation; iron binding quenches the fluorescence of RPA (37). As a control, we used a structurally similar compound, RPAC, which contains the same fluorophore and linker as RPA but lacks iron-chelating properties and thus its fluorescence is not responsive to iron (37).
Histological procedures
BDH2 or TfR1 levels in paraffin embedded human liver samples from Dr. Gretta Jacobs (Case Western Reserve University) were analyzed using validated anti-human BDH2 (http://www.origene.com/antibody/ab_short.mspx?product=TA501293;Origene) or TfR1 (Invitrogen; ref. 38) antibodies as per the manufacturer’s instructions. The nuclei were counterstained with methylene blue. Non-heme iron in human liver tissues was visualized by diaminobenzidine(DAB)-enhanced Perls Prussian blue staining. Non-heme iron stains blue. Hematoxylin and Eosin staining was performed utilizing a kit from Sigma as per the manufacturer’s procedure.
Statistical analysis
Statistical analyses were performed by one-way analysis of variance and Tukey HSD (honestly significant difference) test was performed for multiple comparisons using SAS/STAT software. p values of less than 0.05 were considered statistically significant. Error bars shown in figures represent SD.
Results
Iron-dependent regulation of human bdh2 expression
We have previously shown that 2,5-dihydroxy benzoic acid (2,5-DHBA) is a component of the mammalian siderophore, which is similar to 2,3-DHBA, the Fe-binding moiety of E. coli Enterobactin (7, 8). We also found that BDH2, a homolog of bacterial EntA is responsible for 2,5-DHBA synthesis (8). Depletion of the siderophore from mammalian cells, yeast, and zebrafish embryos caused abnormal accumulation of cytoplasmic Fe and mitochondrial Fe deficiency. Thus, these results suggest that the mammalian siderophore is a critical regulator of cellular iron homeostasis.
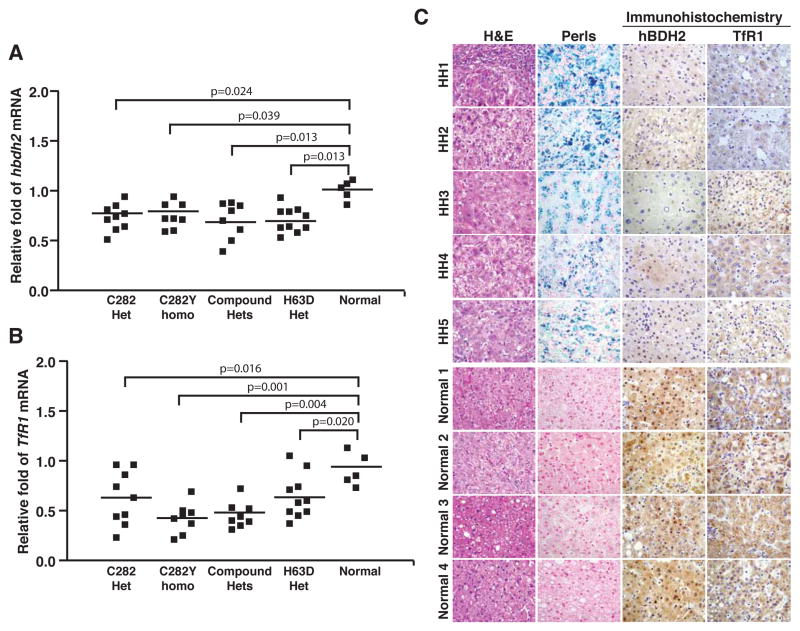
Based on these findings, we hypothesize that the siderophore may be in turn regulated by iron. To test this prediction, we assessed hBDH2 expression in liver samples obtained from hemochromatosis patients. Quantitative real-time PCR analysis with specific primers (Table S1) demonstrated a decrease in hBDH2 expression in liver samples from hemochromatosis patients compared with liver samples from normal controls (Fig. 1 A). Further, the reduction in hBDH2 expression inversely coincides with iron indices as well as the genetic status of the disease (Figure 1 A and Table S2). The mRNA levels of TfR1, a well characterized iron regulated gene was also decreased in these samples (Fig. 1 B). Transcript levels were normalized to rlp13a mRNA, which is not iron regulated.
Figure 1.
Human BDH2 mRNA and protein are down regulated in liver from hemochromatosis patients. (A) Total RNA from normal or hemochromatosis liver samples were subjected to quantitative real time PCR using primers specific for hBDH2 and mRNA levels were normalized to rlp13a, a ribosomal gene. (B) TfR1 mRNA levels were analyzed to assess the impact of iron overloading, an iron-regulated mRNA. (C) Hemochromatosis (HH1–5) or normal liver sections were immunostained with anti-BDH2 Ab (Origene) and Perls Prussian blue staining to detect iron. Non heme iron stains blue. As a positive control, liver sections were also stained with anti-TfR1 Ab. Hematoxylin-Eosin (H&E) staining was performed to demonstrate the liver architecture.
We next asked if a decrease in hBDH2 expression is also reflected at the protein level. To address this question, we analyzed hBDH2 expression by immunohistochemistry in frozen liver sections from hemochromatosis patients and from controls (Table S3). Iron overloading in hemochromatosis samples (HH1 -HH5) was confirmed by Perls prussian blue staining (Fig. 1 C). As expected, TfR1 protein levels are decreased in these samples (Fig. 1 C). In accordance with the real-time PCR data, immunohistochemical staining using an anti-hBDH2 antibody also demonstrated less expression of hBDH2 in liver cells from hemochromatosis patients (Fig. 1 C). Finally, in support of these findings, microarray analysis of liver tissues derived from hemochromatosis patients also demonstrated a decrease in hBDH2 expression (Table S4 and ref. 39). In summary, these results demonstrate that hBDH2 expression is iron regulated.
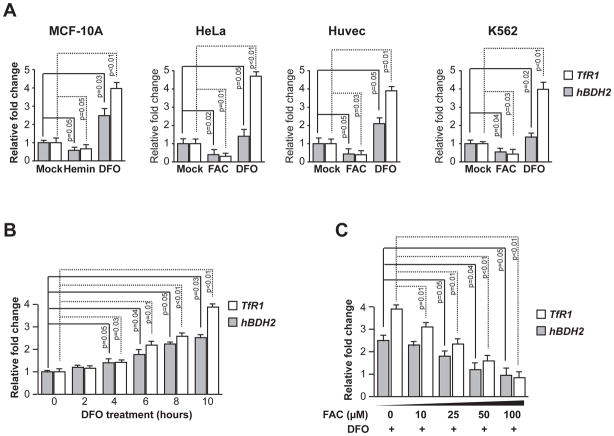
To study the generality of iron-dependent regulation of hBDH2, we next assessed its mRNA levels under iron-replete or iron-deficient conditions in cultured human cells. Specifically, we utilized adherent non-hematopoietic, immortalized cells (MCF-10 A or HUVEC) or transformed cells (HeLa) and non-adherent hematopoietic cells (K562). Cells were either treated with 100 μM DFO or 100 μM each hemin or FAC as iron sources to achieve iron-deficient or iron-replete conditions, respectively. HBDH2 and TfR1 mRNA levels were quantified by qRT-PCR using gene-specific primers (Table S1). As expected, TfR1 mRNA expression increases upon addition of DFO and decreases following iron supplementation in above-mentioned cultured human cells (Fig. 2 A). Similarly, hBDH2 mRNA levels are increased under iron-deficient conditions and decreased under iron-replete conditions in all cells (Fig. 2 A). Transcript levels were normalized to actin mRNA, which is not iron regulated (Fig. 2 A). In addition, immunoblot analysis with a BDH2 specific antibody further confirmed iron dependent regulation of hBDH2 expression in MCF-10 A cells (Fig. S1). A time course analysis indicated hBDH2 mRNA levels accumulated progressively in DFO treated MCF-10 A cells (Fig. 2 B). If hBDH2 mRNA levels were indeed stabilized by iron-delete conditions, then iron supplementation should reverse this process. To test this prediction, we added graded doses of FAC to DFO treated MCF-10 A cells and find that iron supplementation blunted the DFO-induced stabilization of hBDH2 mRNA (Fig. 2 C). Taken together these studies show that iron regulates the expression of hBDH2 mRNA in cultured human cells as well as in patient liver.
Figure 2. Iron-deficiency leads to increased hBDH2 mRNA levels.
(A) Human cell lines were treated with hemin, FAC, DFO or mock-treated for 16 hours and TfR1 and hBDH2 mRNAs were quantified by real time PCR analysis 16 hours post treatment. (B) MCF-10A cells were treated with DFO for the indicated times and hBDH2 and TfR1 mRNAs were quantified as described above. (C) Iron supplementation reverses DFO-induced increase in hBDH2 and TfR1 mRNAs. MCF-10 A cells were treated with 100 μM of DFO along with increasing amounts of FAC and relative levels of hBDH2 mRNA were quantified 10 hours post treatment as described above. For A – C, value on y-axis was set at 1.0 for the mRNA levels in untreated cells. The relative mRNA levels in each sample were normalized to actin mRNA. Results shown are the average of three independent experiments with error bars depicting SD.
Identification of an IRE motif in hBDH2 mRNA
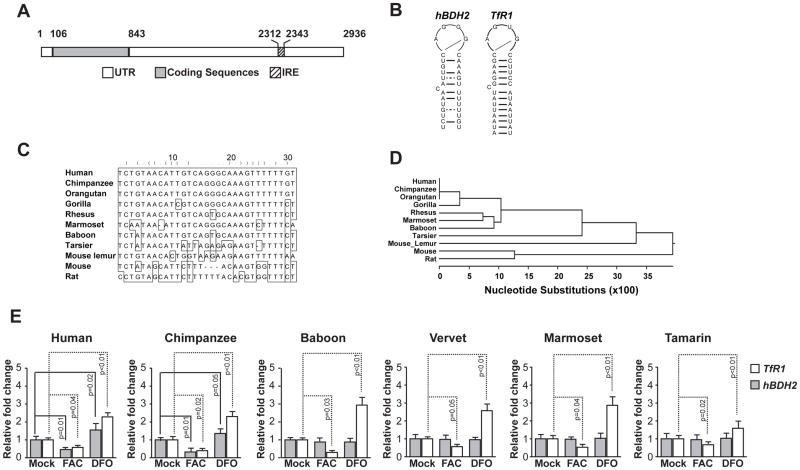
Intracellular iron regulates gene expression post-transcriptionally by IRE-IRP network. To determine whether hBDH2 mRNA contains an IRE-like element, we utilized the SIREs program, which identified a single, IRE-like motif in the 3′-UTR of hBDH2 mRNA (Figs. 3 A, B & C). The IRE sequence of hBDH2 mRNA shares several common features with the previously identified 3′-UTR IRE sequence of the TfR1 gene (Fig. 3 B; ref. 15). For instance, it contains a 6-nucleotide apical loop sequence, CAGGGC, which shares similarities to the canonical IRE sequence of CAGWGH (‘W’ stands for A or U and ‘H’ for A, C, or U). The first C could form a base pair with the fifth G to form an AGG pseudotriloop structure, which may be recognized by the IRPs (25 -27). The IRE of hBDH2 mRNA also contains an upper stem of 4 paired nucleotides and is separated from the lower stem by a single unpaired cytosine (Figs. 3 B & C). A similar motif was found in bdh2 homologues in chimpanzee and orangutan but not in other simian primates (Fig. 3 C & D). Since only members of the hominidae family contain an IRE-like sequence in the 3′-UTR of bdh2, we suggest that iron-dependent regulation of bdh2 is a recent evolutionary acquisition. Thus, IRE-like sequences in the 3′-UTR of bdh2 mRNA is specific to members of the hominidae family, highlighting the adaptation of these species to impose additional mechanisms to regulate the expression of the siderophore.
Figure 3. Structure, sequence, and phylogenic conservation of hBDH2 3′-UTR IRE.
(A) Location of the hBDH2 IRE in the 3′-UTR. IRE. Black boxes indicate exons and the hatched box indicates the location of the IRE. (B) Predicated secondary structures of 3′-UTR IREs of hBDH2 and hTfR1. (C) Multispecies alignment of hBDH2 3′-UTR IRE sequences. (D) Phylogenetic distribution of bdh2 IRE. (E) Iron-dependent regulation of hBDH2 mRNA is restricted to members of the hominidae family. TfR1 and hBDH2 mRNAs were quantified by real time PCR in transformed human and primate liver cell lines 16 hours after treatment with FAC or DFO. The value on y-axis was set at 1.0 for the mRNA levels in untreated cells. The relative mRNA levels in each sample were normalized to eif2b2 (an internal standard for RT-PCR analysis of primate mRNA samples). Positive control includes TfR1, whose expression is affected by iron. Results shown are the average means of three independent experiments with error bars depicting SD.
Iron-dependent regulation of the mammalian siderophore is restricted to hominidae
We assessed iron-dependent regulation of bdh2 mRNA in transformed liver cells derived from various non-human primates and - as a reference - in transformed human liver cells (Fig. 3 E). Liver is appropriate for these studies since bdh2 is abundantly expressed in this organ (8). We found that bdh2 mRNA levels were decreased by hemin and increased by DFO in liver cell lines from chimpanzee and human, but not in baboon, vervet, marmoset and tamarin cells (Fig. 3 E). We evaluated bdh2 mRNA levels in cultured rodent cells and found that iron does not regulate bdh2 mRNA. As expected, TfR1 levels changed based on iron content in all cell types (Fig. 3 E and Fig. S2). Collectively, the results of Figure 3 suggest that the presence of an IRE-like motif determines the iron-dependent regulation of bdh2. These results further confirm the prediction that the IRE motif in bdh2 mRNA is restricted to hominidae family (Fig. 3).
Recruitment of IRPs on to hBDH2 IRE-like motif in vitro
Our data suggest that the IRE-like motif in the hBDH2 mRNA confers iron-dependent regulation. To examine the association between hBDH2 IRE and IRPs in cells, two sets of co-immunoprecipitation experiments were performed.
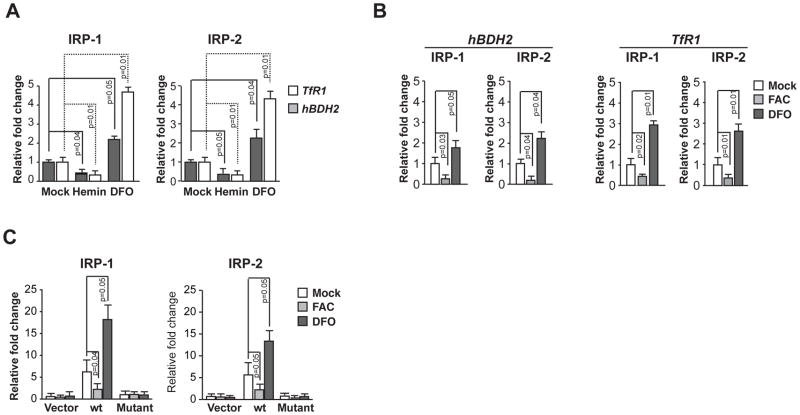
In the first set of experiments, we immunoprecipitated endogenous IRP-1 and IRP-2 complexes from naïve cells or cells treated with DFO or hemin using antibodies specific for IRP-1 and IRP-2. Anti-IRP antibodies effectively immunoprecipitated endogenous hBDH2 mRNA in cytosolic extracts of naïve MCF-10A cells (Fig. 4 A). DFO treatment enhanced immunoprecipitation between IRPs and hBDH2 mRNA, whereas hemin treatment markedly reduced co-immunoprecipitation of hBDH2 mRNA with IRPs (Fig. 4 A). TfR1 also co-immunoprecipitated with IRPs (Fig. 4 A). Together, these data suggest that hBDH2 mRNA associates with IPRs in cells. In complementary experiments, we immunoprecipitated endogenous hBDH2 mRNA from 293T cells stably expressing FLAG IRPs (ref. 35) with an anti-Flag antibody and confirm the association of hBDH2 mRNA with IRPs in cells (Fig. 4 B).
Figure 4. IRP-1 and IRP-2 associate with hBDH2 mRNA.
(A) MCF-10 A cells were initially treated with 100 μM each hemin or DFO for 16 hours. Cytoplasmic extracts of naïve or hemin or DFO treated MCF-10A cells were incubated with anti-IRP-1 Ab or anti-IRP-2 Ab and the RNA-protein complexes were precipitated with protein G Sepharose beads and the mRNAs in the immunoprecipitates were quantified in a real-time PCR analysis. (B) Co-immunoprecipitation of IRPs with endogenous hBDH2 mRNA. 293T cells stably expressing FLAG-IRP-1 or FLAG-IRP-2 were initially treated with 100 μM each FAC or DFO for 16 hours. Cytoplasmic extracts of naïve or FAC or DFO treated 293T cells stably expressing FLAG-IRP-1 or FLAG-IRP-2 were incubated with anti-FLAG Ab coated beads and the mRNAs in immunoprecipitates were quantified in a real-time PCR analysis. For A and B, the value on y-axis was set at 1.0 for the mRNA levels in naïve cells. The relative mRNA levels in each sample were normalized to the input hBDH2 mRNA. Results shown are the average means of three independent experiments with error bars depicting SD. (C) 293T cells stably expressing FLAG-IRPs were transiently transfected with a Luciferase reporter plasmid or Luciferase reporter plasmids containing hBDH2 wt IRE or hBDH2 mutant IRE (also see Figure 5 A). Transfected cells were then treated with 100 μM each hemin or DFO for 16 hours. Cytoplasmic extracts of transfected and treated cells were then incubated with anti-FLAG Ab coated beads and the luc mRNAs in immunoprecipitates were quantified in a real-time PCR analysis. The value on y-axis was set at 1.0 for the mRNA levels of cells transfected naïve Luciferase reporter plasmid. The relative mRNA levels in each sample were normalized to the input firefly luc mRNA. Results shown are the average means of three independent experiments with error bars depicting SD.
In a second set of experiments, we immunoprecipitated IRP-1 and IRP-2 complexes from cells transfected with a hBDH2 3′-UTR reporter plasmid. In these experiments we utilized 293T cells stably expressing FLAG IRP-1 or FLAG IRP-2 (ref. 35). These cells were transfected with reporter plasmids containing either hBDH2 wt IRE (luc-hBDH2 wt IRE-3′-UTR) or hBDH2 mutant IRE (luc-hBDH2 mutant IRE-3′-UTR). The mutant hBDH2 3′-UTR reporter plasmid contains 2 mutations in the IRE that are known to disrupt IRE-IRP interactions (Fig. 5 A for description of these plasmids). An empty Luciferase reporter plasmid served as a negative control. Ectopically expressed hBDH2 wt IRE 3′-UTR was enriched significantly by immunoprecipitation in both IRP-1 and IRP-2 expressing cells (Fig. 4 C). The mutant reporter plasmid was expressed normally as judged by Luciferase expression (Fig. 5) but failed to associate with either of IRPs (Fig. 4 C). As expected, the empty Luciferase reporter plasmid lacking the hBDH2 3′-UTR did not associate with IRP-1 and IRP-2 (Figure 4 C). Together, these data show that hBDH2 mRNA contains a functional IRE-like motif, which associates with IRPs in cells and is required for iron-dependent regulation of hBDH2 mRNA expression.
hBDH2 3′-UTR IRE is required for iron-dependent regulation
The results presented so far suggest that IRPs interact with the hBDH2 3′-UTR IRE. A Luciferase reporter system was used to directly confirm iron-dependent post-transcriptional regulation by the hBDH2 3′-UTR. HeLa cells were transiently transfected with a Luciferase reporter gene containing the ~2 kb hBDH2 3′-UTR (Fig. 5 A). The control plasmid contained the same promoter but lacked the hBDH2 3′-UTR. Following overnight transfection, cells were then treated with FAC or DFO. Luciferase expression was increased by DFO and decreased by FAC in cells expressing hBDH2 wt IRE 3′-UTR (Fig. 5 C). Mutation of the IRE in the hBDH2 3′-UTR abrogated iron-dependent regulation of Luciferase expression (Fig. 5 C).
To further rigorously test the functional importance of IRE in the iron-dependent regulation of hBDH2, we placed sequences encompassing hBDH2 IRE downstream of Luciferase coding sequences and assessed the Luciferase expression under iron-replete or iron-deficient conditions (Fig. 5 C). DFO addition led to an increase in Luciferase expression in the wt-100 hBDH2 IRE-luc and MT1-100 hBDH2 IRE-luc transfectants compared with MT2-100 hBDH2 IRE-luc transfectants (Fig. 5 C). To additionally confirm these results, we placed sequences encompassing hBDH2 IRE upstream of Luciferase coding sequences and assessed the Luciferase expression under iron-replete or iron-deficient conditions (Fig. S3 A). Again, hBDH2 IRE presence conferred iron-dependent expression of Luciferase activity (Fig. S3 C). Collectively, the results of Figs. 5 & S3 indicate that hBDH2 3′-UTR or hBDH2 IRE confers iron-dependence to a heterologous gene.
Abrogation of IRPs blunts the iron-dependent regulation of hBDH2 mRNA
To further assess the role of the IRPs in the regulation of hBDH2 expression, we abrogated expression of IRP-1 or IRP-2 or both by shRNA and siRNA transfection and determined the expression of hBDH2 under iron-replete or iron-deficient conditions. We initially assessed the efficacy of siRNA transfection by analyzing the expression levels of IRP-1 and IRP-2 by quantitative real time PCR analysis, which demonstrated the near complete knockdown of these two genes (Fig. S4 A). Loss of both IRP-1 and IRP-2 eliminated the iron-dependent regulation of hBDH2 or TfR1 expression as assessed by real time PCR analysis (Fig. S4 B). The effects of IRP reduction were additive, as hBDH2 mRNA expression was greatly reduced in double knockdown cells (Fig. S4 C). Thus, IRPs are required for iron-dependent regulation of hBDH2 expression.
hBDH2 3′-UTR IRE is required for iron-dependent destabilization of hBDH2 mRNA
Binding of IRPs to IREs present in the 3′-UTR prolongs the half-life of mRNA by protecting against endonucleolytic cleavage (ref. 14). IRP-1 is dual function protein that can act as an aconitase under iron-replete conditions or binds IRE when iron levels are low. IRP-2 activity is regulated by proteosomal degradation (reviewed in ref. 13). Thus, iron-replete conditions prevent the binding of IRPs to IREs in the target genes, resulting in degradation of mRNAs with 3′-UTR IREs. Conversely, under iron-deficient conditions IRPs bind and prolong the half-life of mRNAs containing 3′-UTR IREs.
We therefore studied the functional importance of the hBDH2 IRE for mRNA stability. We depleted both IRP-1 and IRP-2 proteins in MCF-10 A cells by RNAi. Data presented in Fig. S5 F confirms the knockdown of IRPs as judged by an immunoblot analysis. Next, we assessed the stability of hBDH2 and TfR1 mRNAs under iron-deficient or iron-replete conditions in control shRNA expressing cells as well as in IRP depleted cells. As expected, iron supplementation destabilized the hBDH2 mRNA, and in contrast, iron scarcity stabilized hBDH2 mRNA levels in control cells (Fig. S5 A & B). Depletion of IRPs decreased the stability of hBDH2 and TfR1 mRNAs (Fig. S5 C & D). Taken together the combined results of Fig. S5 suggest that the iron regulates hBDH2 mRNA stability through IRP proteins.
As noted in Fig. 3, murine bdh2 mRNA does not possess an IRE. To test the prediction that bdh2 mRNA is regulated by iron via IRE-independent mechanisms, we assessed the stability of bdh2 mRNA in a murine pro-B lymphocytic cell line (FL5.12) under iron-deficient or iron-replete conditions. Data presented in Fig. S5 E shows that bdh2 mRNA levels are impervious to iron levels. Thus, the observed hBDH2 regulation is secondary to the IRE (Figs. 1 to 5 and Figs. S3 to S5).
Human BDH2 IRE binds IRPs in vitro
We next examined the binding characteristics of the hBDH2 IRE-like stem-loop to IRP-1 and IRP-2 by RNA electrophoretic mobility shift assays (EMSAs). A radio labeled RNA oligomer encompassing the hBDH2 IRE was incubated with increasing amounts of purified recombinant IRP-1 or IRP-2 or a combination of both IRPs (ref. 21) and the complexes were resolved on non-denaturing polyacrylamide gels and the RNA-protein complexes were visualized by autoradiography. TfR1 B IRE was used as a positive control in these experiments. As expected, TfR1 B IRE binds to both IRPs (Fig. S6 A). The radiolabeled hBDH2 IRE also binds to IRP-1 and IRP-2 proteins (Fig. S6 A). To further confirm the specificity of the binding of IRPs to hBDH2 IRE, we performed EMSA competition assays. Addition of unlabeled wt hBDH2 IRE RNA oligomer efficiently inhibited the binding of IRP-1 or IRP-2 to radiolabeled hBDH2 IRE in a dose-dependent manner (Fig. S6 B). Nucleotide substitutions in the loop region (MT-1; Fig. 5 A), which did not compromise the ability of hBDH2 IRE to confer iron-dependent regulation also abolished the binding of IRPs to radio labeled hBDH2 IRE (Fig. S6 B). However, substitutions at positions in the stem region (MT-2; Fig. 5 A), which blunted the iron responsiveness to hBDH2 IRE is unable to compete (Fig. S6 B). The observation that higher amounts of unlabeled competitor are required to abolish the binding suggests that hBDH2 IRE binds IRPs with lower affinity. As expected, addition of TfR1 IRE efficiently inhibited the binding.
In summary, the in vitro binding experiments suggest that the hBDH2 IRE binds purified recombinant IRPs and is consistent with the association of IRPs to hBDH2 IRE in cells.
Depletion of the siderophore due to increased cytoplasmic iron suppresses iron accumulation in mitochondria
Our data thus far suggest that the hBDH2 IRE confers iron-dependent regulation upon hBDH2. What are the physiological implications of this observation for iron regulation of the siderophore? In our prior study, we showed that depletion of the siderophore led to mitochondrial iron deficiency in yeast, fish and cultured mammalian cells. These studies suggest that siderophore is a major facilitator of mitochondrial iron import (8). Based on these findings, we hypothesize that under iron-replete conditions, down regulation of siderophore via the IRE-IRP network prevents mitochondrial iron overload (Fig. 6 A). We tested this prediction in cultured mammalian cells.
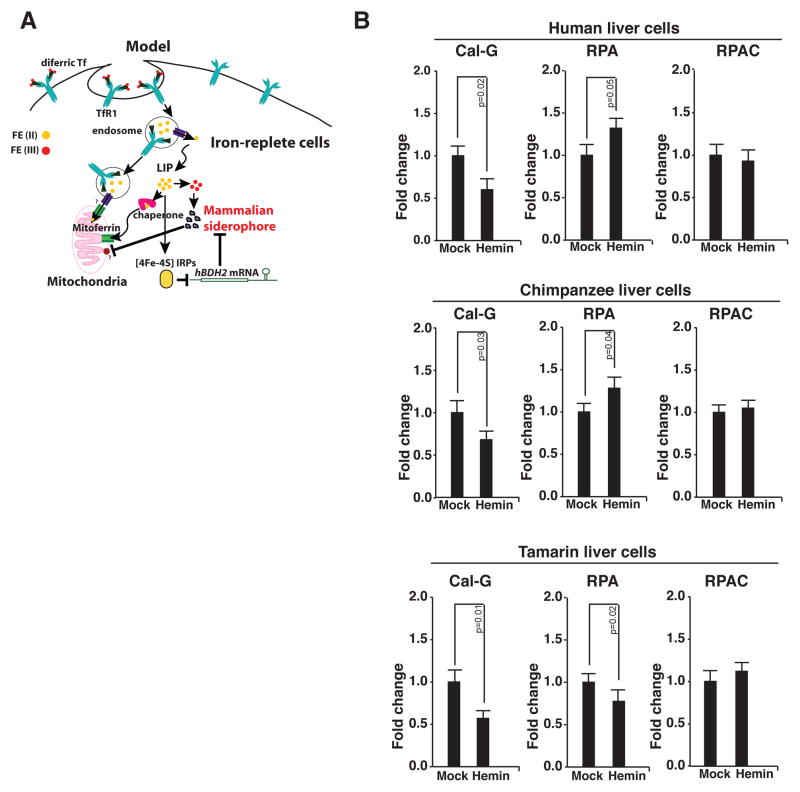
Figure 6. Iron regulation of siderophore controls mitochondrial iron.
(A) Proposed model for iron regulation of siderophore-mediated iron trafficking. In normal human cells a portion of the cytoplasmic free iron pool is composed of the iron-siderophore complex, which is also the form of iron imported into mitochondria. Iron-replete conditions destabilize hBDH2 mRNA leading to reduced siderophore levels. As a consequence, mitochondrial iron levels diminish. (B) Assessment of mitochondrial iron levels in primate cells with or without hBDH2 3′-UTR IRE under replete conditions. RPA or RPAC fluorescence was monitored in control and hemin-treated cells by fluorometry as detailed in methods section. Results shown are the average means of three independent experiments with error bars depicting SD.
We first asked whether suppression of siderophore by increased cytoplasmic iron changes mitochondrial iron content. We tested this idea using a panel of liver cells from great apes some of which lack a functional IRE in the 3′-UTR of bdh2 mRNA (Fig. 3 E). Iron-replete conditions were achieved by treatment with hemin. Cytosolic and mitochondrial iron content was quantified using subcellular iron-specific dyes as outlined in methods section (also see refs 8 & 37). If our hypothesis is correct, mitochondrial iron content should not rise upon an increase in cytoplasmic iron in cells with a functional IRE in the 3′-UTR of bdh2 mRNA. Data presented in Fig. 6 B demonstrate that the mitochondria of liver cells of species with the IRE (human and chimpanzee) are relatively iron deficient despite increased cytoplasmic iron. In contrast, the mitochondrial iron content of cells from a species that does not have the IRE (tamarin) mirrored that of cytoplasmic iron (Fig. 6 B). Thus, the presence of a functional IRE in the hBDH2 mRNA is necessary for regulation mitochondrial iron levels.
Discussion
Most genes involved in the regulation of cellular iron metabolism are in turn regulated by Iron. This group includes the genes involved in cellular iron uptake, storage, and utilization. All are regulated post-transcriptionally via the IRE-IRP network. As indicated by gene knockout studies, this control is vital for the proper maintenance of cellular iron homeostasis (10, 16). It is not known, however, whether such control also regulates expression of genes implicated in intracellular iron trafficking. Mitochondria are the principal consumers of iron yet the precise mechanisms by which iron is imported into the mitochondria remain elusive (22). Iron in the free form is reactive. It therefore must be shielded and chaperoned to the sites of utilization. In our preceding study, we demonstrated that the mammalian siderophore binds and trafficks iron from cytosol into mitochondria. Further, siderophore depletion results in mitochondrial iron deficiency in a variety of model systems (8). Normally, mitochondrial iron import and heme export are in equilibrium and perturbations in this process leads to abnormal mitochondrial iron homeostasis. Since the mammalian siderophore is an important facilitator of mitochondrial iron import, this process must be finely regulated to prevent accidental mitochondrial iron overload. However, the molecular basis by which this regulation is achieved remains to be demonstrated. We hypothesize that the mammalian siderophore is regulated by iron via IRE-IRP network to control the siderophore-dependent iron trafficking into the mitochondria.
Siderophore in human cells is regulated by iron
To examine the above-mentioned hypothesis, in this report, we explored the iron-dependent regulation of the siderophore. In cultured cells and in patient samples, hBDH2 expression is controlled by iron and this regulation is conferred by a single IRE-like motif located in its 3′-UTR. Mutations in the hBDH2 3′-UTR IRE resulted in loss of iron responsiveness. Additionally, in vitro and in vivo experiments found that hBDH2 IRE associates with IRPs and this association is important for the iron-dependent regulation of hBDH2 expression. Finally, hBDH2 IRE also conferred iron responsiveness to a heterologous reporter gene thus, further confirming its functionality.
Human BDH2 belongs to an evolutionarily conserved family of proteins collectively termed short chain dehydrogenases/reductases (SDRs). Interestingly, the presence of IRE is restricted to bdh2 genes of members of the hominidae family, and this finding may suggest that it is a newly acquired regulatory mechanism for iron control of bdh2 genes. Thus, the hBDH2 IRE adds to a growing list of IREs with a restricted phylogenetic distribution. For instance, Cdc14a, succinate dehydrogenase, MRCKα, and AHSP genes although present in all vertebrates, their IREs are restricted to certain species (19 – 21, 24). Thus, the restricted presence of BDH2 IRE to humans and higher order primates further highlight the importance of the bdh2 gene in iron trafficking and cellular iron metabolism and the existence of additional mechanisms necessary to regulate the siderophore-mediated iron trafficking in these species.
Iron-regulated expression of the siderophore controls mitochondrial iron content
What are the physiological implications of the regulation by iron of the expression of mammalian siderophore? Based on gene-ablation studies, we found that the mammalian siderophore facilitates mitochondrial iron import. For instance: 1) siderophore-depleted zebrafish embryos and yeast are heme-deficient; and 2) siderophore suppression results in marked elevation of cytoplasmic iron as a consequence of impaired mitochondrial iron import. Thus, changes in mammalian siderophore levels affect cellular iron metabolism (8). In support of this notion, our studies reveal that under iron-replete conditions, down regulation of siderophore via the IRE-IRP network prevents mitochondrial iron overload. Thus, the presence of a functional IRE in the bdh2 mRNA is necessary for regulation mitochondrial iron levels. In conclusion, based on the previous findings as well as those presented in this report, we hypothesize that iron regulates the siderophore-dependent iron trafficking into the mitochondria.
Supplementary Material
Acknowledgments
We thank Elizabeth Leibold, Matthias Hentze, Kostas Pantopoulos and Susy Torti for providing reagents, Gretta Jacobs for human hemochromatosis liver sections, Joe Willis for normal human liver sections, Sebastian Mueller for providing RNA samples of normal and hemochromatosis liver, Robert Lanford for providing transformed liver cells of human and apes, and Alan Tartakoff for editorial assistance. This work is supported by K01CA113838, R01DK081395, and Case Western Reserve University start up funds to L.R.D and by PS09/00341 from Spanish Health Program (Instituto de Salud Carlos III) and RYC-2008-02352 research contract (Spanish Ministry of Science and Innovation) to M.S. L.R.D. is also a recipient of career developmental awards from March of Dimes and American Society of Hematology.
References
- 1.Guerinot ML. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 2.Nielands JB. Siderophores: structure and functions of microbial iron transport compounds. J Biol Chem. 1995;27:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 3.Kakhlon O, Cabantchik ZI. The labile iron pool: Characterization, measurement, and participation in cellular processes. Free Radical Biol & Med. 2002;33:1037–1046. doi: 10.1016/s0891-5849(02)01006-7. [DOI] [PubMed] [Google Scholar]
- 4.Kruszewski M. Labile iron pool: the main determinant of cellular response to oxidative stress. Mut Res. 2003;531:81–92. doi: 10.1016/j.mrfmmm.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Pol JA. Isolation and characterization of a siderophore-like growth factor from mutants of SV40-transformed cells adapted to picolinic acid. Cell. 1977;14:489–499. doi: 10.1016/0092-8674(78)90235-0. [DOI] [PubMed] [Google Scholar]
- 6.Jones R, Peterson C, Grady R, Cerami A. Low molecular weight iron-binding factor from mammalian tissue that potentiates bacterial growth. J Exp Med. 1980;151:418–428. doi: 10.1084/jem.151.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci USA. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devireddy LR, Hart DO, Goetz DH, Green MR. A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell. 2010;141:1006–1017. doi: 10.1016/j.cell.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenstein RS. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr. 2000;20:627–662. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 10.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: Regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 13.Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binder R, et al. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nature Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 17.Beaumont C. Mutation in the iron responsive element of the L ferritin mRNA in a family with dominant hyperferritinaemia and cataract. Nat Genetics. 1995;11:444–446. doi: 10.1038/ng1295-444. [DOI] [PubMed] [Google Scholar]
- 18.Kato J, et al. A mutation, in the iron-responsive element of H ferritin mRNA, causing autosomal dominant iron overload. Am J Hum Genet. 2001;69:191–197. doi: 10.1086/321261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers JT, et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer’s amyloid precursor protein transcript. J Biol Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- 20.Cmejla R, Petrak J, Cmejlova J. A novel iron-responsive element in the 3′UTR of human MRCKalpha. Biochem Biophys Res Commun. 2006;341:158–166. doi: 10.1016/j.bbrc.2005.12.155. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez M, et al. Iron regulation and the cell cycle. Identification of an iron-responsive element in the 3′-untranslated region of human cell division cycle 14 mRNA by a refined microarray-based screening strategy. J Biol Chem. 2006;281:22865–22874. doi: 10.1074/jbc.M603876200. [DOI] [PubMed] [Google Scholar]
- 22.Meehan HA, Connell GJ. The hairpin loop but not the bulged C of the iron responsive element is essential for high affinity binding to iron regulatory protein-1. J Biol Chem. 2001;276:14791–14796. doi: 10.1074/jbc.M010295200. [DOI] [PubMed] [Google Scholar]
- 23.Sierzputowska-Gracz H, McKenzie RA, Theil LC. The importance of single G in the hairpin loop of the iron responsive element (IRE) in ferritin mRNA for structure: an NMR spectroscopy study. Nucleic Acids Res. 1995;23:146–153. doi: 10.1093/nar/23.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.dos Santos CO, et al. An iron responsive element-like stem-loop regulates a-hemoglobin-stabilizing protein mRNA. J Biol Chem. 2008;283:26956–26964. doi: 10.1074/jbc.M802421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butt J, et al. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. Proc Natl Acad Sci USA. 1996;93:4345–4349. doi: 10.1073/pnas.93.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barton H, Eisenstein RS, Bomford A, Munro H. Determinants of the interaction between the iron-responsive element-binding protein and its binding site in rat L-ferritin mRNA. J Biol Chem. 1990;265:7000–7008. [PubMed] [Google Scholar]
- 27.Henderson BR, Menotti E, Bonnard C, Kuhn LC. Optimal sequence and structure of iron-responsive elements. J Biol Chem. 1994;269:17481–17489. [PubMed] [Google Scholar]
- 28.Feder JN, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 29.Napier I, Ponka P, Richardson DR. Iron trafficking in the mitochondrion: Novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 30.Richardson DR, et al. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci USA. 2010;107:10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campillos M, Cases I, Hentze M, Sanchez M. SIREs: searching for iron-responsive elements. Nucleic Acids Res. 2010;2010:1–8. doi: 10.1093/nar/gkq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casey JL, et al. Iron responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240:924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- 33.Hanson ES, Foot LM, Leibold EA. Hypoxia post-translationally activates iron-regulatory protein 2. J Biol Chem. 1999;274:5047–5052. doi: 10.1074/jbc.274.8.5047. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez M, Galy B, Hentze M, Muckenthaler MU. Identification of target mRNAs of regulatory RNA binding proteins using mRNP immunopurification and microarrays. Nat Protoc. 2007;2:2033–2042. doi: 10.1038/nprot.2007.293. [DOI] [PubMed] [Google Scholar]
- 35.Zumbrennen KB, Wallander ML, Romney SJ, Leibold EA. Cysteine oxidation regulates the RNA-binding activity of iron regulatory protein 2. Mol Cell Biol. 2009;29:2219–2229. doi: 10.1128/MCB.00004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X-L, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNAse-L expression is mediated by the 3′-Untranslated region of its mRNA. J Biol Chem. 2007;282:7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- 37.Rauen U, et al. Assessment of chelatable mitochondrial iron by using mitochondrion-selective fluorescent iron indicators with different iron-binding affinities. Chembiochem. 2007;8:341–352. doi: 10.1002/cbic.200600311. [DOI] [PubMed] [Google Scholar]
- 38.Tampanaru-Sarmeslu, et al. Transferrin and Transferrin receptor in human hypophysis and pituitary adenomas. Am J Pathol. 1998;152:413–422. [PMC free article] [PubMed] [Google Scholar]
- 39.Hagist S, et al. In vitro-targeted gene identification in patients with hepatitis C using a genome-wide microarray technology. Hepatol. 2008;49:378–386. doi: 10.1002/hep.22677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.