Abstract
The Ndc80 complex is the key microtubule-binding element of the kinetochore. In contrast to the well-characterized interaction of Ndc80-Nuf2 heads with microtubules, little is known about how the Spc24-25 heterodimer connects to centromeric chromatin. Here, we present molecular details of Spc24-25 in complex with the histone-fold protein Cnn1/CENP-T illustrating how this connection ultimately links microtubules to chromosomes. The conserved Ndc80 receptor motif of Cnn1 is bound as an α helix in a hydrophobic cleft at the interface between Spc24 and Spc25. Point mutations that disrupt the Ndc80–Cnn1 interaction also abrogate binding to the Mtw1 complex and are lethal in yeast. We identify a Cnn1-related motif in the Dsn1 subunit of the Mtw1 complex, necessary for Ndc80 binding and essential for yeast growth. Replacing this region with the Cnn1 peptide restores viability demonstrating functionality of the Ndc80-binding module in different molecular contexts. Finally, phosphorylation of the Cnn1 N-terminus coordinates the binding of the two competing Ndc80 interaction partners. Together, our data provide structural insights into the modular binding mechanism of the Ndc80 complex to its centromere recruiters.
Keywords: centromere, chromosome segregation, histone-fold, kinetochore, Ndc80/Hec1
Introduction
The ability to faithfully distribute the genetic material between emerging daughter cells is a fundamental prerequisite for all development. The accuracy of segregating chromosomes in eukaryotes is remarkable: in budding yeast, the loss rate for a given chromosome per generation is approximately only 10−5 (Murray et al, 1986). This high-fidelity segregation is achieved through the action of kinetochores, large multi-protein structures that assemble on chromosomal domains termed as centromeres (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). Centromeres are characterized by the presence of specialized nucleosomes in which the canonical histone H3 is replaced by the histone variant CENP-A (Guse et al, 2011). The deposition of CENP-A nucleosomes marks centromeres for the recruitment and assembly of additional polypeptides, collectively referred to as the constitutive centromere-associated network (CCAN) (Foltz et al, 2006; Okada et al, 2006; Hori et al, 2008). A crucial role of these proteins is to function as an assembly platform for the microtubule-binding interface of the kinetochore, the so-called KMN network, consisting of KNL-1, the Mtw1/MIS12 complex and the Ndc80 complex (Lampert and Westermann, 2011). The microtubule-binding interface connects to dynamic plus-ends and is also required for the generation of a spindle assembly checkpoint signal that coordinates kinetochore attachment with the onset of anaphase (Musacchio and Salmon, 2007).
Within the kinetochore architecture that encompasses >60 polypeptides in yeast, the four-protein Ndc80 complex (composed of Ndc80–Nuf2 and Spc24-25 heterodimers) is of central importance for the attachment of kinetochores to microtubules. Kinetochores contain multiple copies of the Ndc80 complex, in budding yeast between 8 and 20 as measured by fluorescence microscopy and depending on the reference used for quantification (Joglekar et al, 2006; Lawrimore et al, 2011). Initial low-resolution EM analysis of recombinant Ndc80 complex showed an elongated dumbbell shape with globular domains separated by a 570-Å long coiled-coil region (Wei et al, 2005). X-ray crystallography of the isolated globular domains or of an artificially shortened complex lacking the coiled coils has demonstrated that the Ndc80–Nuf2 end folds into a dimeric arrangement of calponin-homology domains that contacts the microtubule surface through conserved positively charged residues (Wei et al, 2006a; Ciferri et al, 2008; Alushin et al, 2010). Impairment of these interactions through mutations results in loss of kinetochore-microtubule attachments in cells (Sundin et al, 2011). On the centromere-proximal end of the Ndc80 complex, the Spc24-25 subunits form a globular domain (Wei et al, 2006b). In contrast to the Ndc80-microtubule interaction, however, the interface between the complex and the inner kinetochore has remained structurally uncharacterized. Initial reconstitution experiments employing C. elegans proteins have indicated that the Ndc80 complex associates with the four-protein Mtw1/Mis12 complex (Cheeseman et al, 2006), an interaction that was later confirmed with recombinant human and yeast complexes (Maskell et al, 2010; Petrovic et al, 2010; Hornung et al, 2011). The Mtw1 complex is localized centromere-proximal compared to the Ndc80 complex (Joglekar et al, 2009), in part through interaction with the conserved DNA-binding protein CENP-C (Przewloka et al, 2011; Screpanti et al, 2011), suggesting that Mtw1 can act as a kinetochore receptor for Ndc80. In addition, recent studies have identified the mammalian CCAN component CENP-T, and its yeast homologue Cnn1, as direct interaction partners of the Ndc80 complex (Gascoigne et al, 2011; Schleiffer et al, 2012). CENP-T proteins contain a histone-fold domain (HFD) at the C-terminus used for associating with one or more additional small histone-fold proteins (CENP-W, -S and -X) and they may form a nucleosome-like structure at the centromere (Hori et al, 2008; Nishino et al, 2012). Here, we provide insights into the kinetochore interactions of the Ndc80 complex from the crystal structure of the budding yeast Spc24-25/Cnn1 interface. We prove that the two distinct Ndc80 complex recruiters, Cnn1 and the Mtw1 complex, target the same binding site on Spc24-25 and we identify a region on the Mtw1 complex essential for the Ndc80–Mtw1 interaction. Furthermore, we propose a model in which phosphorylation modulates the affinity of the competitive Ndc80 receptors towards their binding partner.
Results
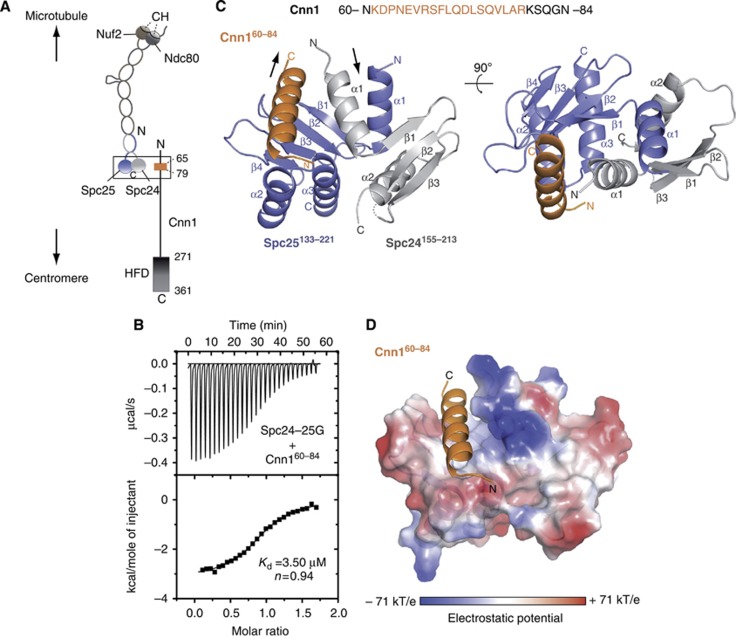
Overall architecture of the Ndc80–Cnn1 interface
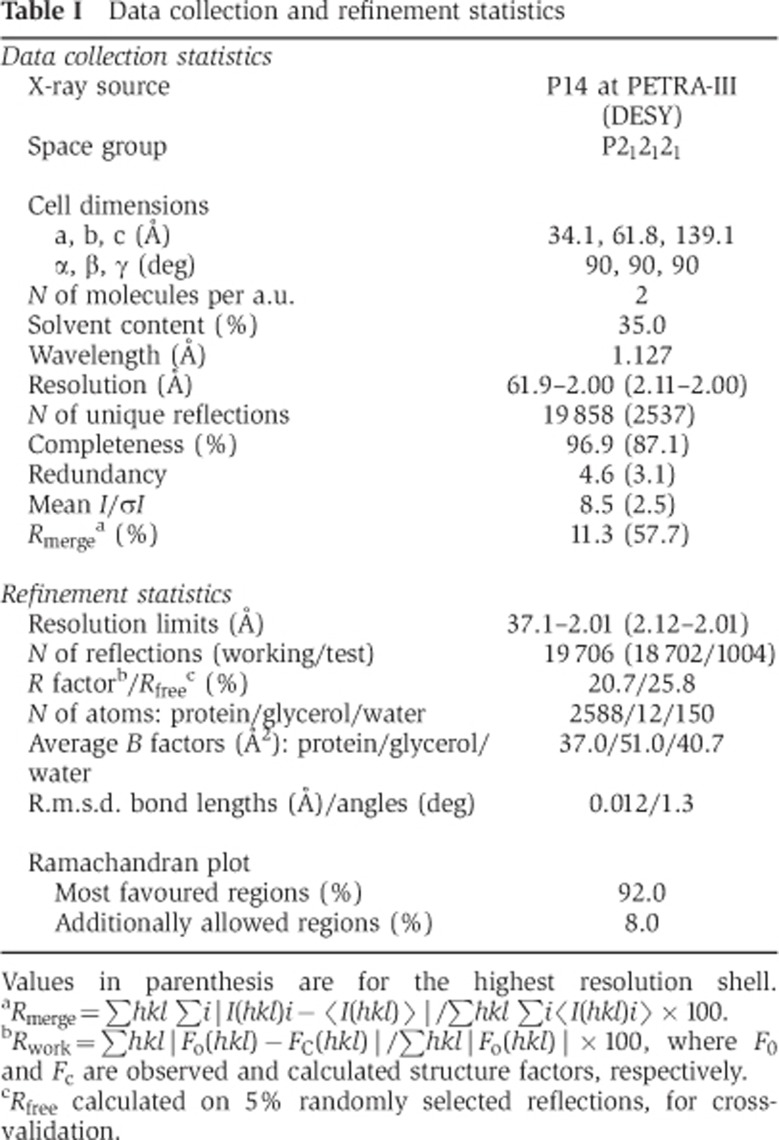
Our previous work has shown that budding yeast Cnn1CENP-T associates with the Spc24-25 heterodimer and we have identified a conserved peptide motif in the N-terminus of Cnn1 (aa 65–79) that is strictly required for this association (Schleiffer et al, 2012; Figure 1A). We first asked whether the globular domain of the Spc24-25 complex, lacking the coiled coil (Spc24155–213–Spc25133–221), is sufficient for binding to Cnn1. In isothermal titration calorimetry (ITC) experiments, a 25aa Cnn1-derived peptide including the N-terminal conserved motif (Cnn160–84) interacted with the Spc24-25 globular domain with an apparent dissociation constant of 3.5 μM (Figure 1B), similar to the affinity for the full-length Spc24-25 heterodimer (Schleiffer et al, 2012). This indicates that the binding site of the Cnn1 N-terminus must be located in the globular domain of Spc24-25. To uncover the structural details of the Cnn1–Ndc80 complex association, Spc24155–213-25133–221 was purified and co-crystallized with Cnn160–84. The high-resolution structure of the complex was solved by molecular replacement, using the crystal structure of the budding yeast Spc24-25 globular domain as search model (Wei et al, 2006), and refined at 2.0 Å resolution to an R factor of 20.7% (Rfree of 25.8%) (Table I). The asymmetric unit of the crystal contains two Spc24155–213-25133–221 heterodimers (B-A and D-C): each Spc subunit shows an α/β tertiary structure composed of an anti-parallel β sheet of three (Spc24) or five (Spc25) β strands flanked by either two (Spc24) or three (Spc25) α helices (Figure 1C). Notably, this characteristic fold resembles the RWD domain (RING finger-, WD-repeat-, and DEAD-like proteins), recently found in several kinetochore-related proteins of budding yeast, such as Csm1, a subunit of the Monopolin complex, the checkpoint protein Mad1 and the Ctf19 and Mcm21 subunits of the kinetochore COMA complex (Corbett et al, 2010; Kim et al, 2012; Schmitzberger and Harrison, 2012). During Spc24-25 refinement, additional continuous electron density was observed in both Spc24-25 globular domains of the asymmetric unit that could be unequivocally modelled and refined as the amino acids 61–79 (in B-A) or 62–79 (in D-C) of Cnn160–84 (Figure 1C; Supplementary Figure S1). The two Spc24155–213-25133–221–Cnn160–84 molecules display a very similar structure (r.m.s.d. 0.54 Å, calculated on 162 Cα pairs over 165) and thus the general features of the complex will be discussed from here on.
Figure 1.
Structure determination of the Spc24-25/Cnn1 interface. (A) Schematic representation illustrating the general centromere-microtubule orientation of the Cnn1–Ndc80 complex. Nuf2 and Ndc80 are shown in brown and dark grey, respectively, with the microtubule-binding calponin homology (CH) domains highlighted; Spc24 and Spc25 are shown in light grey and slate blue, respectively. The Cnn1 conserved N-terminal motif (65–79) is displayed in orange and its histone-fold domain (HFD) is highlighted. A rectangle indicates the interface visualized in the crystal structure. (B) Isothermal titration calorimetry performed by titrating the Spc24-25 globular domain (Spc24-25G) with a Cnn1 peptide including the Ndc80-binding motif (Cnn160–84). The Kd was estimated fitting a non-linear curve to the binding isotherms derived from the data. (C) Overall architecture of the Spc24155–213–Spc25133–221–Cnn160–84 crystal structure. Spc24, Spc25, and Cnn1 are represented according to their secondary structure and coloured in light grey, slate blue, and orange, respectively. Arrows indicate the opposite N-terminal/C-terminal orientation of Cnn1 relative to Spc24-25. A dotted line indicates the residue Lys184 of Spc24 not visible in the electron density (molecule B-A-F of the asymmetric unit). (D) Localization of Cnn160–84 in the hydrophobic pocket at the interface between Spc24155–213 and Spc25133–221, shown as electrostatic surface (transparency 20%). Cnn1 secondary structure is displayed in orange.
Table 1. Data collection and refinement statistics.
Values in parenthesis are for the highest resolution shell.
aRmerge=∑hkl ∑i|I(hkl)i−〈I(hkl)〉|/∑hkl ∑i〈I(hkl)i〉 × 100.
bRwork=∑hkl|Fo(hkl)−FC(hkl)|/∑hkl|Fo(hkl)| × 100, where F0 and Fc are observed and calculated structure factors, respectively.
cRfree calculated on 5% randomly selected reflections, for cross-validation.
The bound Cnn160–84 peptide adopts an α-helical structure accommodated in a hydrophobic pocket at the interface between Spc24155–213 and Spc25133–221, forming a three-helix bundle with the two α-helices α1 of both Spc subunits (Figure 1D and C). The majority of the Cnn1-binding interface is constructed by Spc25133–221, while Spc24155–213 only donates helix α1 for complex formation (corresponding interfaces: 597.0 and 240.3 Å2, respectively). The localization of Cnn1, near the beginning of the Ndc80–Nuf2-interacting coiled coils, and its anti-parallel orientation relative to Spc24-25, are not consistent with a linear head-to-tail arrangement of the centromere-binding Cnn1 and the microtubule-binding Ndc80 complex (Figure 1A and C). In the observed configuration the HFD of Cnn1 would indeed project in the direction of the microtubule instead of pointing towards the centromere.
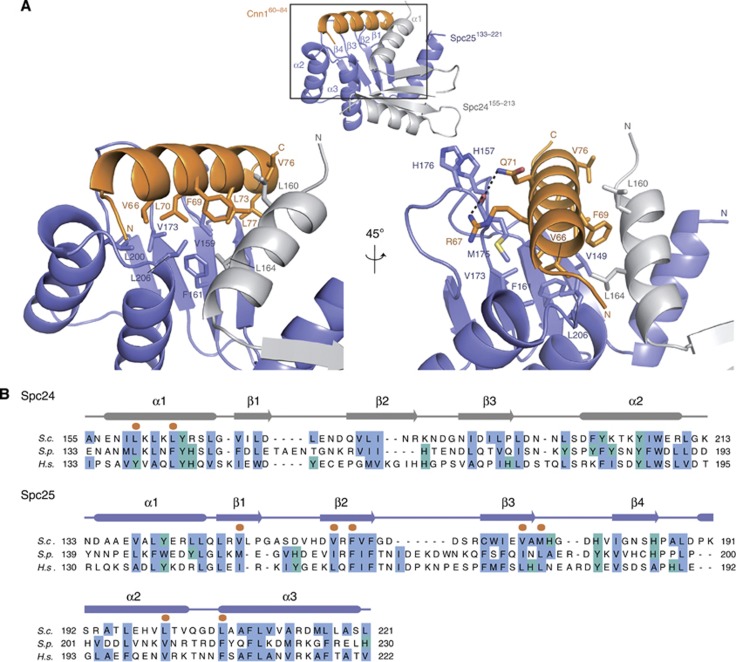
Atomic details and validation of the Ndc80–Cnn1 binding pocket
The hydrophobic side of the Cnn1 helix, in particular residues Phe69, Leu70, and Leu73 that characterize the N-terminal conserved motif of Cnn1/CENP-T proteins, faces the hydrophobic pocket of the Spc24-25 globular domain, establishing extensive van der Waals contacts with several Spc25 residues (Val149, Val159, Phe161, Val173, Leu200, and Leu206) and with Leu160 and Leu164 of the Spc24 helix α1 (Figure 2A). The presence of hydrophobic residues at these sites is conserved among eukaryotic Spc24-25 proteins (Figure 2B; Supplementary Figure S2). The only polar interactions of the Spc24-25/Cnn1 interface consist of two hydrogen bonds formed by the side chains of Arg67 and Gln71 (Cnn1) with the carbonyl oxygen of the Met175 main chain, located in the β3–β4 loop of Spc25. Comparison with the Spc24155–213–Spc25133–221 structure reveals that Cnn1 binding induces a reorientation of the β3–β4 loop (Supplementary Figure S1), thus further exposing the hydrophobic pocket required to host the Cnn1 α helix. In addition, the remodelled orientation of this loop is stabilized by ring-stacking interactions between His176 (Spc25 β3–β4 loop) and His157 in the Spc25 β1–β2 loop, which also reorients markedly from its configuration in the unbound Spc24-25 globular domain (Figure 2A; Supplementary Figure S1). Consistent with this, the average B factors of both loops are still relatively high (32.5–45.0 Å2 for β1–β2 loop and 37.0–61.7 Å2 for β3–β4 loop compared to the average protein B factor of 37.0 Å2), pointing to an inherent flexibility of these active-site bordering loops that seems to be required for an induced-fit Cnn1 binding.
Figure 2.
Details of the Spc24-25–Cnn1 interaction. (A) Close-up view of the Spc24155–213-25133–221–Cnn160–84 interaction network. The secondary structure of Spc24, Spc25, and Cnn1 is displayed in light grey, slate blue, and orange, respectively. The interacting residues are highlighted as sticks, with O in red, N in blue, and S in yellow. The established hydrogen bonds are represented as black dashed lines. (B) Sequence alignment of the Spc24 and Spc25 homologues from Saccharomyces cerevisiae (S.c.), Schizosaccharomyces pombe (S.p.), and Homo sapiens (H.s.). The conservation of non-polar residues is highlighted according to the Clustalx colouring scheme and Cnn160–84-interacting residues are marked with an orange dot. The secondary structure is derived from the X-ray coordinates of Spc24155–213–Spc25133–221–Cnn160–84 using the program STRIDE (Heinig and Frishman, 2004). See Supplementary Figure S5 for alignment of more sequences.
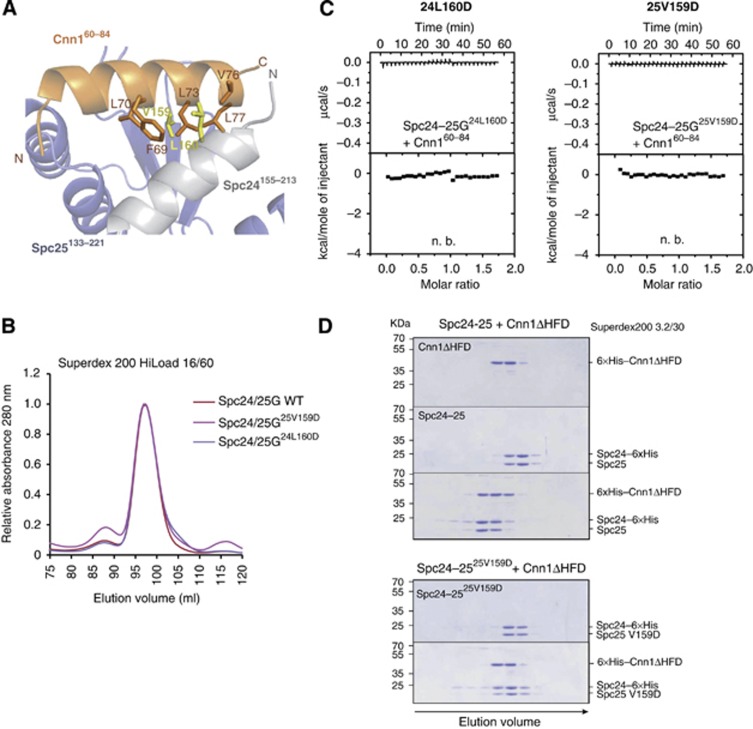
In order to validate the observed interaction between Spc24-25 and Cnn1, the single mutants V159D of Spc25 (25V159D) and L160D of Spc24 (24L160D) were generated to distort the hydrophobic character of the ligand-binding groove, preventing Cnn1 interaction. These residues (highlighted in yellow in Figure 3A) were chosen for their position at the surface of the hydrophobic pocket, so that the structural integrity of the Spc24-25 complex was not affected, as validated by analytical size-exclusion chromatography comparing the elution volumes of the mutants and the wild-type Spc24155–213-Spc25133–221 heterodimers (Figure 3B). In ITC experiments, both single mutations completely abolished Cnn160–84 binding (Figure 3C), confirming our structural model according to which Spc24 and Spc25 form a composite hydrophobic binding pocket required to tether Cnn1. Similar experiments were performed by mutating individually or in combination three conserved residues of Cnn160–84, Phe69, Leu70, and Leu73, and testing the binding of the corresponding peptides to the wild-type Spc24-25 globular domain. All introduced mutations abrogated the interaction (Supplementary Figure S3) thus providing strong evidence that the observed Spc24-25/Cnn1 interface is relevant in solution.
Figure 3.
Validation of the Spc24-25/Cnn1 interface. (A) Role of L160 (Spc24) and V159 (Spc25) in Cnn160–84 binding. The secondary structure of Spc24, Spc25, and Cnn1 is displayed in light grey, slate blue, and orange, respectively (transparency 40%). The residues L160 (Spc24) and V159 (Spc25), mutated to aspartic acid for validating the Spc24155–213–Spc25133–221/Cnn160–84 interface, are shown as sticks and highlighted in yellow. The Cnn1 residues that establish van der Waals contacts with L160 or V159 are displayed as sticks. (B) Elution profiles of size-exclusion chromatography performed with Spc24-25 globular domain (Spc24-25G) wild-type (in red) or Cnn1-binding mutants, Spc24-25G25V159D (in violet) and Spc24-25G24L160D (in magenta). Identical elution profiles indicate a similar complex shape and integrity. (C) Isothermal titration calorimetry performed by titrating Spc24-25G25V159D or Spc24-25G24L160D with Cnn160–84 (n.b.=no binding). (D) Coomassie-stained gels of analytical size-exclusion chromatography using 12 μM Cnn1ΔHFD (1–270), 12 μM full-length Spc24-25 wild-type and their combination (upper part); 12 μM full-length Spc25-25V159D and its combination in equimolar amount with Cnn1ΔHFD (lower part). Complex formation is visualized by a shift to earlier elution volumes of both molecules.
Both mutations 24L160D and 25V159D also compromised the Spc24-25–Cnn1 interaction when testing the full-length Spc24-25 heterodimer with the entire amino-terminal part of Cnn1 missing only the HFD (Cnn1ΔHFD) (Figure 3D). These results confirm that the Spc24-25 hydrophobic pocket observed in the minimal complex of our crystal structure is the main binding site for the Cnn1–Ndc80 complex interaction.
Cnn1 and the Mtw1 complex target the same binding site on Spc24-25
We next analysed the phenotype of yeast strains bearing mutations in the Ndc80–Cnn1 binding pocket in vivo. For this purpose, wild-type Spc25 or Spc25V159D, expressed from its endogenous promoter, was integrated into the URA3 locus of a ΔSpc25/Spc25 diploid strain and the asci dissected after sporulation. In contrast to integration of the wild-type gene, Δspc25 Spc25V159D spores failed to be recovered (Figure 4A) and the same result was observed for the Spc24L160D mutant (Supplementary Figure S4), indicating that altering the Cnn1-binding site on Spc24-25 is lethal. This observation is in contrast with the finding that a Cnn1 deletion is viable in budding yeast (Bock et al, 2012; Schleiffer et al, 2012) leading us to speculate that the 25V159D and 24L160D mutations were affecting not only the kinetochore recruitment of the Ndc80 complex through Cnn1, but also the Ndc80–Mtw1 complex interaction, thereby impairing the binding of the Ndc80 complex with both kinetochore partners. In order to test this hypothesis in vitro, recombinant Ndc80 complex including the 25V159D mutation was produced in bacteria and its binding to the Mtw1 complex was tested by analytical size-exclusion chromatography. For this purpose, we used a version of the Mtw1 complex harbouring a deletion of the first 171 amino acids of Dsn1 (Dsn1ΔN); this deletant is functional in vivo and less prone to degradation in solution (Hornung et al, 2011). Confirming our hypothesis, the Ndc8025V159D complex failed to form a 1:1 interaction with the Mtw1Dsn1ΔN complex when mixed in equimolar concentrations (Figure 4B). The same result was observed for the Ndc8024L160D complex (Supplementary Figure S4), demonstrating that Cnn1 and the Mtw1 complex are not only competitors for binding to the Ndc80 complex (Schleiffer et al, 2012), but that they also target the same Spc24-25 hydrophobic pocket.
Figure 4.
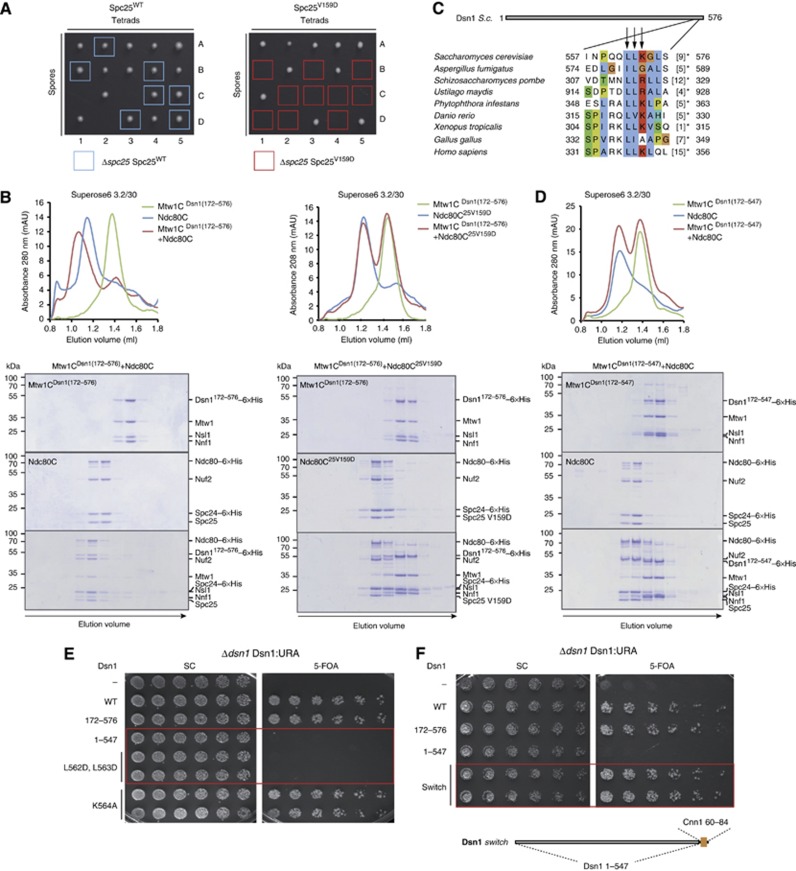
Identification of an essential Ndc80–Mtw1 interface. (A) Asci dissection of Δspc25/Spc25 Spc25WT (left) and Δspc25/Spc25 Spc25V159D (right). The viable Δspc25 Spc25WT spores are highlighted with a light blue square. The Δspc25 Spc25V159D spores, marked with a red rectangle, failed to be recovered, indicating that V159D is a lethal Spc25 mutation. (B) Elution profiles and Coomassie-stained gels of analytical size-exclusion chromatography performed with 4 μM Mtw1Dsn1(172–576) complex (Mtw1CDsn1(172–576)), 4 μM wild-type Ndc80 complex (Ndc80C) and their combination (left); 4 μM Mtw1Dsn1(172–576) complex, 4 μM Ndc80 complex including the Spc25V159D mutant (Ndc80C25V159D) and their combination (right). Complex formation is visualized by a shift to earlier elution volumes of both molecules. (C) Multiple-sequence alignment of a conserved C-terminal motif in Dsn1 proteins. Background colouring of residues is based on the Clustalx colouring scheme. Amino acids omitted before the stop are given in brackets. Amino acids mutated and tested in the plasmid shuffle assays are marked with an arrow. (D) Elution profiles and Coomassie-stained gels of analytical size-exclusion chromatography performed with 4 μM Mtw1CDsn1(172–547), 4 μM wild-type Ndc80C and their combination. The lack of a shift to earlier elution volumes of both molecules indicates abolished complex formation. (E) Plasmid shuffle assays performed transforming Δdsn1 Dsn1:URA strain with Dsn1 wild type (WT), Dsn1172–576 (172–576), Dsn11–547 (1–547) and the mutants Dsn1L562D, L563D (L562D, L563D) and Dsn1K564A (K564A). Single clones were spotted in two-fold dilutions on minimal medium lacking (left panel, SC) or containing (right panel) 5-FOA. Red rectangles highlight the lethality of mutants Dsn11–547 and Dsn1L562D, L563D. (F) Plasmid shuffle assay executed as in (E) testing the functionality of a Dsn1 version where the last C-terminal 29 amino acids were substituted by the Cnn1 residues 60–84 (Dsn1 switch, in schematic representation underneath the panel). The red rectangle encloses the viable Dsn1 switch mutant.
Identification of a Cnn1-related Ndc80 receptor motif in the four-protein Mtw1 complex
In case the Cnn1 N-terminus and the Mtw1 complex are bound similarly to the Spc24-25 globular domain, being accommodated in the same hydrophobic pocket, then a Cnn1-related interaction motif for Ndc80 may be present in any of the subunits of the Mtw1 complex. To uncover such a motif, we built a hidden-Markov model (HMM) from fungal sequences spanning the CENP-T N-terminal motif and searched against homologues of the Mtw1 complex subunits Mtw1, Nnf1, Nsl1, and Dsn1. Although the results were statistically non-significant, eight of the nine hits with a positive score (E values, 0.32–5.4) mapped to the extreme carboxy-terminal region of Dsn1 proteins. An analysis of the conservation pattern and secondary structure predictions from all Mtw1 complex-family members shows that especially in Dsn1 proteins this C-terminal region is separated from the central conserved domain by a polar linker region of variable length. In a focused search for conserved motifs within this region using MEME (Bailey et al, 2009), we identified an 11-amino acid long sequence that overlapped with the hits from the HMM search (S. cerevisiae Dsn1 aa 557–568) (Figure 4C).
As an initial test for the involvement of the Dsn1 carboxy-terminal region in Ndc80 binding, a stop codon was introduced at position 548 of Dsn1172–576 and the interaction between the Mtw1Dsn1(172–547) complex and the Ndc80 complex was analysed by analytical size-exclusion chromatography. Strikingly, the deletion of the 29 terminal residues of Dsn1, although not compromising Mtw1 complex assembly, completely disrupted the binding to the Ndc80 complex (Figure 4D). We next analysed the functional role of the Dsn1 C-terminal part in vivo using a plasmid shuffle assay. To this end, we transformed a haploid Δdsn1 strain, kept viable by expression of wild-type Dsn1 from a URA-based centromeric plasmid, with the Dsn11–547 truncation mutant lacking the last 29 amino acids. In contrast to full-length wild-type Dsn1 and the amino-terminal truncation Dsn1172–576, Dsn11–547 was not able to support growth on 5-FOA plates (Figure 4E). Thus, the Dsn1 C-terminus has an essential function in vivo. In order to further dissect the carboxy-terminal region of Dsn1, point mutations of the amino acids conserved in different Dsn1 proteins were introduced, in particular L562D-L563D or K564A (Figure 4C), and tested in the plasmid shuffle assay. While mutation of K564 did not affect viability, the double mutant Dsn1L562D–L563D was not able to complement the loss of Dsn1, indicating that these conserved hydrophobic amino acids are essential for Dsn1 function. Finally, we tested if the two binding motifs in the Ndc80 recruiters interact similarly with the Spc24-25 hydrophobic pocket by asking whether they were exchangeable. For this purpose, we built a version of Dsn1 in which the last 29 amino acids were substituted with the Cnn160-84 peptide (Dsn1 switch) and performed a Δdsn1 rescue experiment using the plasmid shuffle assay. Expression of Dsn1 switch was able to support viability in the absence of wild-type Dsn1 (Figure 4F). These results strongly indicate that the mode of Dsn1 binding to Spc24-25 is structurally similar to the Cnn1–Spc24-25 interaction and that the two motifs are functionally identical.
A secondary motif in the N-tail is required for Cnn1 function in vivo
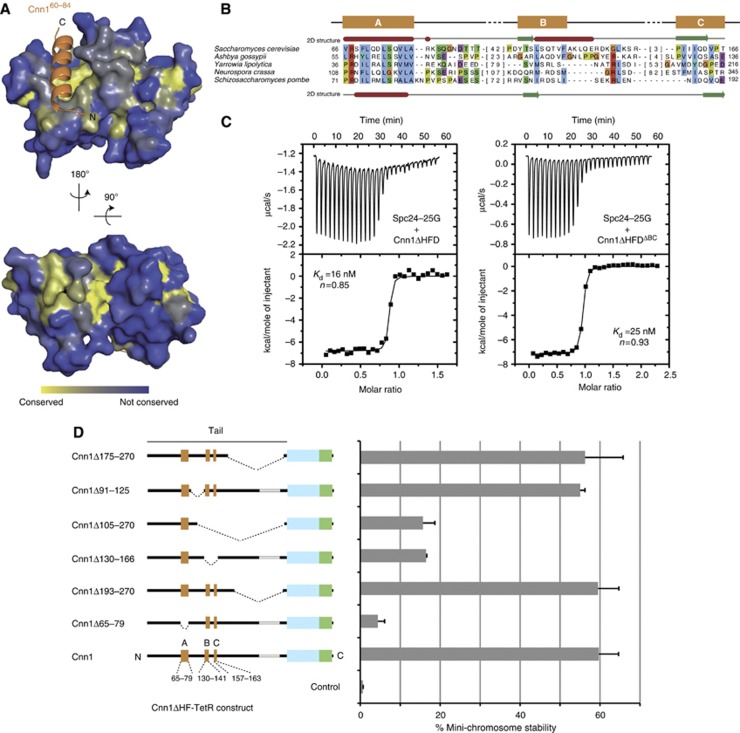
The Cnn1-derived peptide displayed only moderate affinity towards the Spc24-25 domain (Kd=3.5 μM). However, the larger Cnn1 fragment missing only the HFD (Cnn1ΔHFD) interacted with Spc24-25G with a Kd of 16 nM (Figure 5C, left panel), more similar to the nanomolar-range affinities observed between Ndc80 and Mtw1 complexes in vitro (Petrovic et al, 2010). This finding, together with the unexpected binding orientation of the Cnn1 α helix and the presence of a conserved groove located on the opposite side of the Spc24-25 globular domain (Figure 5A), prompted us to ask whether additional parts of the Cnn1 N-terminus might be involved in Spc24-25 binding, thus wrapping around the globular domain and increasing the overall strength of the interaction. A sequence alignment exclusively of fungal CENP-T proteins (Supplementary Figure S5) showed that apart from the main conserved motif, two additional short segments, in budding yeast located ∼40 amino acids carboxy-terminal of the main binding site (Cnn1 aa 130–166), show significant conservation among species (Figure 5B). In order to test if these regions (B and C) were responsible for the increased affinity between Cnn1ΔHFD and Spc24-25G, we performed an ITC experiment using a version of Cnn1ΔHFD deleted of the amino acids 130–166 (Cnn1ΔHFDΔBC). Removing ‘BC’ only had a minor effect on the Kd of the interaction (Figure 5C), suggesting that the amino acids 130–166 are not taking part in the Cnn1/Spc24-25G binding interface, but that other segments, upstream or downstream the Cnn1 main binding motif, could be responsible for establishing additional contacts with the Spc24-25 globular domain and thus increase the overall affinity.
Figure 5.
Functional dissection of the Cnn1 N-tail. (A) Conservation of Spc24-25 plotted onto the structure of the Spc24-25 globular domain. Highly conserved surfaces are coloured in yellow and less conserved surfaces in blue. Note the presence of conserved residues in the Cnn1 binding pocket and on the opposite site of the globular domain. (B) Multiple sequence alignment of an N-terminal region in Cnn1 homologues. S. cerevisiae (above) and S. pombe (below) secondary structure predictions are derived from the Jpred 3 server (α helices in red and β strands in green) (Cole et al, 2008). Background colouring of the residues is based on the Clustalx colouring scheme. The number of omitted residues is indicated in brackets. See Supplementary Figure S5 for alignment of more sequences. (C) Isothermal titration calorimetry performed by titrating the Spc24-25 globular domain (Spc24-25G) with Cnn1 deleted of the histone-fold domain (Cnn1ΔHFD) on the left, or additionally deleted of residues 130–166 (Cnn1ΔHFDΔBC) on the right. Note the very similar Kd values, estimated by fitting a non-linear curve to the derived binding isotherms. (D) Stability of acentric URA3 mini-chromosomes segregated through artificial recruitment of Cnn1ΔHF-TetR. The respective Cnn1 construct is indicated in the left panel (dotted lines indicate the position of the deletion), the corresponding mini-chromosome stability on the right. Error bars denote s.e.m. (n=3).
We then wondered whether the conserved region ‘BC’ might contribute to Cnn1 function in vivo. As a cnn1 deletion is viable in budding yeast the assessment of individual Cnn1 mutations in a wild-type background is difficult (Schleiffer et al, 2012). To probe the role of Cnn1 mutations in vivo, we therefore used a plasmid segregation assay in which Cnn1, artificially tethered via the TetR-tetO system, drives the segregation of an acentric mini-chromosome in budding yeast (Kiermaier et al, 2009; Schleiffer et al, 2012). This experiment therefore serves as a quantitative reporter assay for Cnn1 function in vivo. We dissected the function of the Cnn1 N-tail with a panel of deletion mutants (Figure 5D): as demonstrated before, elimination of the conserved motif visualized in the crystal structure (segment ‘A’, residues 65–79) nearly completely abrogated Cnn1-mediated plasmid segregation (Schleiffer et al, 2012), demonstrating that the plasmid-stabilizing activity depended on the ability to interact with the Ndc80 complex. In contrast, large truncations between the binding motif and the histone-fold (delta 193–270 or delta 175–270) were tolerated and had no effect on plasmid segregation. Interestingly, the deletion of residues 130–166, which removes the conserved segments ‘BC’, severely impaired plasmid stability, although not to the same degree as removal of segment ‘A’. By contrast, a substantial portion of the polypeptide chain between segments ‘A’ and ‘BC’ could be eliminated (delta 91–125) without compromising Cnn1 function in this assay. We conclude that the ‘BC’ secondary motif, located carboxy-terminal of the main conserved interaction site, although not directly involved in binding to the Ndc80 complex, makes important contributions to Cnn1 function in vivo.
Phospho-regulation of the Cnn1 N-tail by Cdk1 and Mps1 kinases
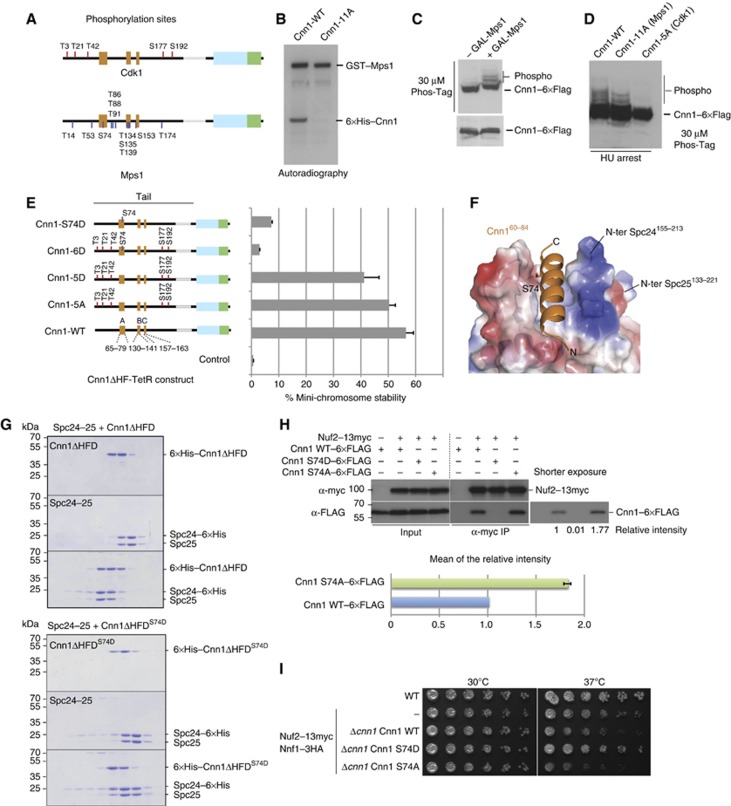
Cnn1 is a phospho-protein in vivo and has been recognized as a target of the mitotic kinases Cdc28 and Mps1 (Loog and Morgan, 2005; Breitkreutz et al, 2011; Bock et al, 2012). We confirmed phosphorylation of Cnn1 by Cdk1 and Mps1 kinases and mapped phosphorylation sites by mass spectrometry following in vitro phosphorylation (Figure 6A; Supplementary Table 3). The target residues for both kinases were located in the N-terminal tail. Interestingly, while the five minimal Cdk1 consensus sites (Thr3, Thr21, Thr42, Ser177, and Ser192) were distributed throughout the N-terminal domain, a number of the mapped Mps1 target sites were located within the conserved regions, most notably Ser74 in the Ndc80-binding element ‘A’ and the cluster Thr134, Ser135, and Thr139 in the functionally essential element ‘B’. Mutation of these sites to alanine eliminated Mps1-dependent Cnn1 phosphorylation in vitro (Figure 6B). Analysis of the Cnn1 migration pattern on Phos-Tag gels supported the notion that Cnn1 is an Mps1 substrate in vivo: upon overexpression of Mps1 from a GAL promoter, an increase in the number of slowly migrating Cnn1 isoforms could be detected by western blot (Figure 6C). Cnn1 phosphorylation over the cell cycle was maximal in S phase, which became apparent in hydroxyurea-arrested cells. Eliminating Mps1 phosphorylation sites with the 11A mutation reduced some of the slowly migrating bands, while mutating the Cdk1 sites (Cnn1-5A) nearly completely abolished the phosphorylation as judged by Phos-Tag western blot (Figure 6D).
Figure 6.
Phospho-regulation of the Cnn1 N-tail by Cdk1 and Mps1 kinases. (A) Position of Cdk1 and Mps1 phosphorylation sites in Cnn1 as obtained from in vitro phosphorylation and mass spectrometry. See Supplementary Table 3 for a detailed description. (B) Mutation of the mapped Mps1 phosphorylation sites eliminates Mps1-dependent phosphorylation of Cnn1 in vitro. Autoradiography showing kinase reactions with recombinant wild-type and 11A-mutant Cnn1ΔHF phosphorylated by GST-Mps1. (C) Overexpression of Mps1 increases phospho-isoforms of Cnn1 in vivo. Phos-Tag western blot of Cnn1–6 × Flag in the absence or presence of Mps1 overexpression. (D) Alanine mutations of the mapped Cdk1 and Mps1 sites reduce or abolish Cnn1 phospho-isoforms as judged by Phos-Tag western blot. (E) Effects of phospho-eliminating or phospho-mimicking mutations on Cnn1-mediated mini-chromosome segregation. The Cnn1 construct is indicated in the left panel, the corresponding plasmid stability on the right. Error bars denote s.e.m. n=3. (F) Location of Ser74 in the Spc24-25–Cnn1 structure. Note that Ser74 is located in the Cnn1 binding pocket. Electrostatic potential coding: blue, −71 kT/e; red, +71 kT/e. (G) Coomassie-stained gels of analytical size-exclusion chromatography performed with 12 μM Spc24-25, 12 μM Cnn1ΔHFD, and their combination (above); 12 μM Spc24-25, 12 μM Cnn1ΔHFDS74D, and their combination (below). The abolished complex formation caused by S74D mutation is indicated by the lack of a shift to earlier elution volumes of both molecules. (H) Co-immunoprecipitation experiments from log-phase cell extracts visualizing the interaction between Cnn1–6 × Flag wild-type and S74D or S74A mutants with Nuf2–13 × myc (Ndc80 complex). The intensity of the bands relative to the wild-type Cnn1 is reported below the western blot and quantified for the S74A mutant compared to the wild type in the lower panel. Error bar denotes s.e.m. (n=3). (I) Spot assays performed by plating four-fold dilutions of the indicated strains at different temperatures on YPD plates.
We analysed the effects of phospho-inhibiting or phospho-mimicking mutations on Cnn1 function in the plasmid segregation assay. In the context of the Cnn1ΔHF-TetR construct, eliminating Cdk1 phosphorylation did not strongly affect mini-chromosome segregation (Figure 6E). This is in contrast to similar targeting experiments with human CENP-T in which the ability of recruiting the Ndc80 complex was abolished upon elimination of Cdk1 phosphorylation (Gascoigne et al, 2011). Mutations mimicking Cdk1 phosphorylation of Cnn1 (Cnn1-5D) decreased the efficiency of plasmid segregation slightly. Interestingly, however, mimicking phosphorylation of the Mps1 target residue Ser74 had a strong inhibitory effect on Cnn1-mediated plasmid segregation either in the context of the Cdk1 mutations (6D mutant) or alone (S74D mutant) (Figure 6E). This result could be explained by the particular position of Ser74 in the Spc24155–213-25133–221–Cnn160–84 structure. The side chain of this residue is located at the interaction interface between Spc24-25 and Cnn1, with a buried surface area of 59.6 Å2, undergoing van der Waals contacts with several residues (Figure 6F). Therefore, the addition of a bulky phosphate group to this serine residue should cause steric and electrostatic problems, impairing complex formation and leading to the plasmid instability observed in the plasmid segregation assay.
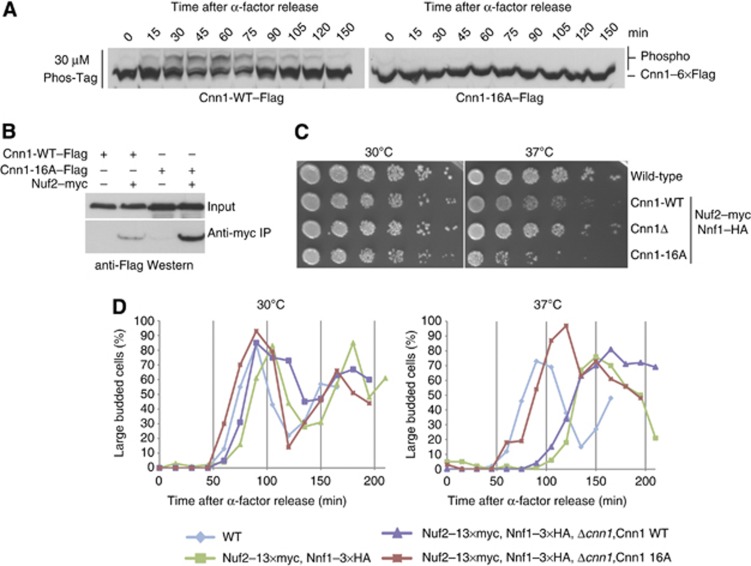
To confirm this hypothesis, we tested the interaction between Spc24-25 full length and the phopsho-mimicking mutant Cnn1ΔHFDS74D in vitro. Mutation of this single amino acid completely abrogated the Cnn1–Spc24-25 interaction, as judged by analytical size-exclusion chromatography (Figure 6G). To further investigate the effect of Ser74 phosphorylation in vivo, we performed immunoprecipitation experiments in log-phase extracts testing both the phoshpo-mimicking Cnn1 S74D and the phospho-inhibiting Cnn1 S74A mutants. We observed a complete abolishment of the Cnn1–Ndc80 interaction in the case of S74D, as expected from the analytical gel filtration experiments, and an increased association for the single mutant S74A (Figure 6H). Thus, Cnn1 Ser74 is involved in the Cnn1–Ndc80 phospho-regulation in vivo. As an effect of these altered interactions, we noticed interesting phenotypes in the strain background used for the co-immunoprecipitation experiments, which contained epitope-tagged Nuf2 (Ndc80 complex) and Nnf1 (Mtw1 complex) subunits. Upon integration of Cnn1-WT, the strain displayed growth defects at 37°C which are rescued by the S74D mutant and slightly increased in the case of S74A (Figure 6I). However, the weak phenotype of the single S74A mutation suggested that additional phosphorylation sites could be involved in regulating the Cnn1–Ndc80 complex interaction. For this reason, we constructed a Cnn1 mutant in which the mapped Cdk1 and Mps1 sites were mutated to alanine (Cnn1-16A). When analysed by Phos-Tag western blot, this mutant abolished the slowly migrating phospho-isoforms over the cell cycle (Figure 7A). In co-immunoprecipitation experiments using log-phase extracts, Cnn1-16A displayed a strongly increased association with the Ndc80 complex, consistent with the notion that multiple phosphorylation sites could serve to modulate the interaction (Figure 7B). Moreover, we noticed that in the Nuf2–13myc Nnf1–3HA strain background, expression of Cnn1-16A caused a growth defect at 37°C. Importantly, this phenotype was the opposite of deleting Cnn1 which improved growth at 37°C (Figure 7C). In order to further characterize the Cnn1-16A phenotype, we followed the cell-cycle progression after α-factor release and observed chromosome segregation using immunofluorescence microscopy. The Nuf2–13myc Nnf1-3HA background strain displayed a pronounced cell-cycle delay at 37°C that was partially restored by the expression of Cnn1-16A, but not Cnn1 wild-type (Figure 7D). When spindles were visualized by microscopy, however, the Nuf2–13myc Nnf1–3HA Cnn1-16A strain showed pleiotropic mitotic defects, such as unequal segregation and short spindles with ‘cut’ DNA (Supplementary Figure S6).
Figure 7.
Characterization of the phospho-inhibiting Cnn1-16A mutant. (A) Phos-Tag western blot showing wild-type or phosphorylation-deficient Cnn1 in the course of the cell cycle after an α-factor release. Note the absence of phospho-isoforms in the Cnn1-16A mutant. (B) Co-immnuoprecipitation experiment testing the association between Nuf2–myc (Ndc80 complex) and Cnn1 wild-type or phosphorylation-deficient Cnn1-16A mutant. (C) Serial dilution assay comparing growth of Cnn1 wild type, Cnn1 deletion, and non-phosphorylatable Cnn1 in a Nuf2−myc, Nnf1−HA, background. Note the temperature sensitivity of the Cnn1-16A mutant. (D) Percentage of large budded cells after α-factor release, counted for 100 cells at each time point at permissive or restrictive temperature (37°C).
These experiments indicate that while deleting Cnn1 does not produce a strong phenotype in wild-type budding yeast, non-regulatable Cnn1 may compete too early with the Mtw1 complex for Ndc80 binding leading to mitotic defects. Thus, the control of the Cnn1–Ndc80 interaction, including the phosphorylation of Ser74 by Mps1, is important to support proper chromosome segregation.
Discussion
Principles of Ndc80 complex kinetochore recruitment
For proper kinetochore function, the linkage of the Ndc80 complex to the inner centromere should fulfill several criteria. First, since multiple Ndc80 complexes must engage with the same microtubule plus-end, it should provide structural flexibility, allowing the complexes to move relative to each other in order to establish a firm connection to the microtubule. Second, the connection must be very secure to resist the pulling forces of the mitotic spindle on the chromosome. This becomes critical in anaphase when maximum forces act on the chromosomes, but error correction mechanisms that would detect unattached chromosomes have been deactivated (Mirchenko and Uhlmann, 2010). Finally, the linkage between the Ndc80 complex and the inner centromere should be regulated in order to control recruitment or differential interactions with one or more kinetochore partners. The Cnn1–Ndc80 complex crystal structure provides the first insights of how the kinetochore fulfills these diverse requirements.
The Ndc80 receptor motif of Cnn1 is located in a largely unstructured N-terminal tail of Cnn1, adopting an α-helical conformation in complex with its conserved cognate binding interface (Figure 8A). The anti-parallel orientation compared to Spc24-25 suggests that other residues could be involved in the binding interface, increasing the affinity to nanomolar range and allowing the histone-fold containing C-terminus to project towards the centromere. In agreement with this hypothesis, the crystal structure of a chicken 35-aa CENP-T peptide in complex with Spc24-25 indicates the same conserved helix as main binding site, but a number of additional amino acids wrapping around the Spc24-25 globular domain (Tatsuo Fukagawa, personal communication). These residues are not as conserved as the Cnn1 helix, but still may contribute to increase the Cnn1–Spc24-25 interaction strength. In this configuration, Cnn1/CENP-T could effectively act as a ‘molecular hook’ and securely tether the Ndc80 complex to the inner kinetochore. This type of configuration shows similar high-affinity binding than large en-bloc interfaces but has the benefit of providing the kinetochore larger conformational flexibility.
Figure 8.
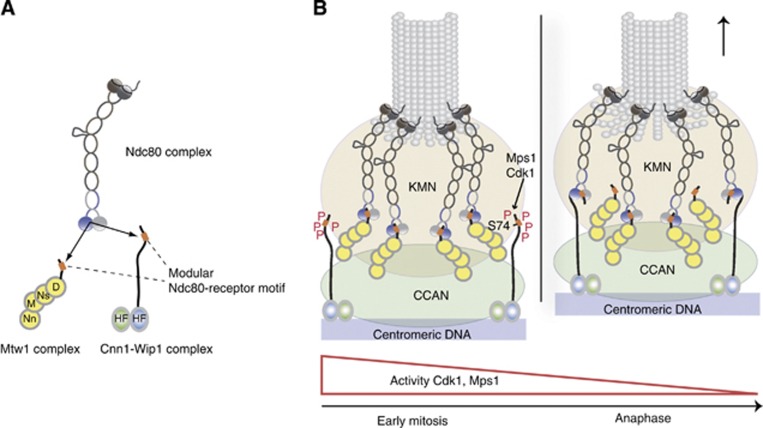
Model for coordinated interaction of the Ndc80 complex with two centromere recruiters. (A) The Mtw1 complex and the histone-fold protein complex Cnn1-Wip1 are mutually exclusive binding partners for the Ndc80 complex, containing structurally related binding motifs (orange boxes) that interact with the same hydrophobic pocket in Spc24-25. (B) Coordination of Ndc80 binding requires phosphorylation of the Cnn1 N-terminus. During early mitosis, where the activity of Cdk1 and Mps1 is high, phosphorylation of the Cnn1 N-terminus decreases the affinity for Ndc80 binding and thus promotes formation of the essential Ndc80–Mtw1 linkage. With decreasing Cdk1 and Mps1 activity over the cell cycle, Cnn1 effectively competes with the Mtw1 complex for Ndc80 binding.
In the attempt to find other Spc24-25 binding sites downstream the main Cnn1 motif, we analysed the two conserved segments ‘B’ and ‘C’. The ITC and the mini-chromosome segregation experiments suggest that, although not implicated in Spc24-25 binding, these segments are required for Cnn1-dependent plasmid segregation. ‘B’ and ‘C’ may establish important contacts with other KMN or CCAN components that are present in the Cnn1-driven artificial kinetochores. Additional experiments will be required in the future to uncover the exact role of these conserved motifs.
It is interesting to compare the Ndc80–Cnn1 interface to the recently reported structure of the RWD domain protein Csm1 in complex with its binding partner Mam1 (Corbett and Harrison, 2012). In this structure, Mam1 shows an extended conformation that drapes across the Csm1 surface, which resembles the binding mode of longer Cnn1 fragments. On the other hand, Mam1 adopts only short helical segments and does not display obvious similarity with the quite long and well-defined Cnn1 helix. Together with the divergent primary sequence of RWD proteins, this suggests that there could be structural plasticity in their interaction with respective binding partners.
Mechanism of KMN network assembly in budding yeast
Our structural characterization of the Ndc80–Cnn1 interface has allowed us to identify a similar interaction domain in the Mtw1 complex. The stable association between these two four-protein complexes, and thus the assembly of the KMN network, depends on a hydrophobic peptide motif located in the carboxy-terminus of the Dsn1 subunit (Figure 8A). This small region behaves similarly to Cnn1 segment ‘A’, as the two motifs can be switched without affecting viability. In human cells, the C-terminal domain of the Nsl1 subunit is required for the interaction between Ndc80 and Mtw1 complexes (Petrovic et al, 2010). The budding yeast Nsl1 carboxy-terminus lacks sequence homology to its human counterpart and the yeast Mtw1 complex may therefore rely critically on the Dsn1 subunit for connecting to Ndc80. However, additional binding motifs within the Mtw1 complex, possibly present in other subunits, could also take part in the interaction with the Ndc80 complex. Interestingly, the hydrophobic region we identified in the Dsn1 subunit (in particular a pair of conserved Leucines, residues S.c. Leu562, Leu563) is present in human Dsn1. It will be interesting to test whether this motif also contributes to KMN assembly in human cells.
Our results show that, although the connection between Mtw1 and Ndc80 complexes is essential in budding yeast, while the Ndc80–Cnn1 interaction is not, both Ndc80-recruitment pathways rely on similar binding modules, located at the terminal regions of the Cnn1 and Dsn1 subunits. The functional differences between these two types of Ndc80 linkages to the inner kinetochore therefore remain to be established. Experiments designed to compare kinetochore-microtubule interactions in the presence or absence of Ndc80–Cnn1 linkages both in vitro and in vivo will be required to address this point.
Differential requirements for Cnn1/CENP-T phospho-regulation between yeast and humans
Cdk1 phosphorylation of the N-terminus of CENP-T has been shown to play an important role in kinetochore assembly in human cells (Gascoigne et al, 2011) and it is required for a high-affinity interaction between chicken CENP-T and Spc24-25 (Tatsuo Fukagawa, personal communication). Our experiments indicate that Cdk1 phosphorylation of the Cnn1 N-terminal tail is not required for the interaction between Ndc80 complex and Cnn1. Supporting this observation, the association between the proteins in budding yeast is predominantly detected in anaphase, when Cdk1 activity is low (Bock et al, 2012; Schleiffer et al, 2012). These differences in phospho-regulation of CENP-T proteins between budding yeast and humans may reflect the fundamental differences in the regulation of kinetochore assembly: in budding yeast, kinetochores stay assembled and attached to microtubules throughout most of the cell cycle, with the exception of a brief period during S phase when centromeric DNA is replicated (Kitamura et al, 2007). By contrast, the microtubule-binding interface of the kinetochore in human cells starts to assemble during prophase and microtubules become available for interaction after nuclear envelope breakdown (Cheeseman and Desai, 2008). Our experiments indicate that phosphorylation by the mitotic kinase Mps1 plays a role in regulating the interaction between the Ndc80 complex and Cnn1, in particular by phosphorylating Ser74 which is located at a key position in the binding interface. The coordination of the Ndc80 interaction with its centromere receptors is critical for the cell: our experiments using point mutants in Dsn1 clearly demonstrate that the Mtw1–Ndc80 linkage is required for viability. Mtw1 and Ndc80 complexes seem to be present in a near stoichiometric ratio at kinetochores and their copy number does not change significantly over the cell cycle (Joglekar et al, 2006). We propose that high Cdk1 and Mps1 activity during S phase and early mitosis promotes the formation of the essential Mtw1–Ndc80 linkage by inhibiting the Cnn1–Ndc80 interaction. With decreasing Cdk1 and Mps1 activity, as cells go through anaphase, dephosphorylation promotes the formation of Cnn1–Ndc80 connections leading to a structural rearrangement of the kinetochore, which may be optimized for chromosome segregation (Figure 8B). The profile of Mps1 activity, which is high during S phase and early mitosis and decreases gradually as yeast cells go through anaphase (Palframan et al, 2006), is consistent with the timing of the Ndc80–Cnn1 association in vivo. Our analysis of phosphorylation-deficient Cnn1 indicates that establishing the interaction between Cnn1 and Ndc80 prematurely can be problematic, because it interferes with the formation of the essential Ndc80–Mtw1 complex linkage. Recent studies indicate substantial anaphase-specific structural alterations in the CENP-A nucleosome present at centromeres (Shivaraju et al, 2012). This finding supports the notion that kinetochore architecture may change dynamically over the cell cycle. We note that the observed phospho-regulation of Cnn1 did not prevent the chimaeric Dsn1 switch protein from supporting yeast growth. The reason may be that Ser74 is not effectively phosphorylated in the context of the hybrid protein because Dsn1 switch is lacking the other Cdk1 and Mps1 target sites, whose phosphorylation could be required for Ser74 modification.
In summary, the structure of the Cnn1–Ndc80 complex interface provides detailed insights into the association between kinetochore components. The identification of a Cnn1-related motif in the Mtw1 complex may point to the presence of short interaction modules as a general mechanism for the establishment of kinetochore architecture. Finally, histone tails are well known for their fundamental roles in the regulation of chromatin. The tail domain of the histone-fold protein Cnn1 provides an interesting example for the usage of such a domain in the context of the kinetochore architecture, revealing what might be an evolutionary old principle to link microtubule-binding factors to chromosomes.
Materials and methods
Protein biochemistry
Mutant versions of recombinant proteins were generated using Quikchange Multi Site-Directed mutagenesis (Agilent Technologies). All versions of His-tagged Spc24-25 full length and globular domain were cloned in cassettes 1 and 2 of the pETduett-1 vector (Novagen) and co-expressed in E. coli. Mtw1Dsn1(172–576) complex, Mtw1Dsn1(172–547) complex, and Ndc80 complex subunits Nuf2 and Ndc80 were cloned and expressed as described in Schleiffer et al (2012) and Lampert et al (2010). For details, see Supplementary experimental procedures.
Crystallization and data collection
Prior to crystallization trials, 0.34 mM Spc24155–213-25133–221 was pre-incubated with 5.8 mM synthetic Cnn1 peptide, comprising residues 60–84 (Cnn160–84). Sitting drop crystallization experiments yielded rectangular crystals that appeared after 3 days of vapour diffusion at 20°C in a 0.2-μl drop containing a 1:1 mixture of protein and precipitant solution (15% PEG6000, 5% glycerol). Prior to flash freezing in liquid nitrogen, a single crystal was shortly soaked in a cryoprotectant solution composed of 60% precipitant solution, 18% glycerol, and 22% gel filtration buffer (10 mM Bis-Tris propane pH 7.6, 250 mM NaCl, and 10 mM DTT). The crystal diffracted to a maximum resolution of 2.0 Å at the P14 beamline of PETRA-III at DESY (Deutsches Elektronen-Synchrotron, Hamburg, Germany). The diffraction data were processed with XDS (Kabsch, 2010) and intensities merged with SCALA of the CCP4 program suite (CCPN, 1994).
Crystal structure determination, refinement, and analysis
The structure was solved by molecular replacement with phaser (McCoy et al, 2007) using the crystal structure of the Spc24155–213-25133–221 (PDB code 2FTX, Wei et al, 2006) as search model. The two molecules in the asymmetric unit (chains B-A and D-C) were refined with CNS (Brunger et al, 1998). Additional electron density appearing in the hydrophobic pocket at the interface between Spc24 and Spc25 of both molecules was manually built-in using Coot (Emsley and Cowtan, 2004) as the Cnn1 peptide containing residues 61–79 (chain F, in B-A) and 62–79 (chain E, in D-C). In later refinement stages, several loops undergoing rearrangements compared to the Spc24155–213-25133–221 structure had to be manually fitted into the Spc24155–213-25133–221–Cnn160–84 electron density. Corresponding loops are composed of Spc24 residues 174–177, 183–186, and 194–199 and of Spc25 residues 152–157, 164–168 and 175–180. The resultant complete and adjusted model was refined with Phenix (Afonine et al, 2012), applying non-crystallographic symmetry (NCS) restraints. Water molecules were added with Phenix and manually checked. The last refinement cycles were performed with Buster (Blanc et al, 2004), using TLS (Translation Libration Screw-motion) and NCS restraints. The program PROCHECK (Laskowski et al, 1996) was used for validating the stereochemistry of the final model as summarized in Table I.
In the final model, all residues of Spc24155–213 and Spc25133–221 are well defined by the electron density, with exception of Lys184 (Spc24, B) and Lys184-Asn185 (Spc24, C), located in the flexible β2–β3 loop (average B values of 69.9–85.1 Å2 compared to the average protein B factor of 37.0 Å2).
The interacting surfaces between protein molecules in the asymmetric unit were analysed with PISA (Krissinel and Henrick, 2007). The program CONTACT (CCP4 suite) was used to analyse the specific interactions between Spc24-25 and the Cnn1 peptide. Superposition of the Cα atoms of Spc24155–213-25133–221–Cnn160–84 and Spc24155–213-25133–221 was performed using Superpose (Krissinel and Henrick, 2004) considering all residues of Spc25133–221. All graphical representations were generated with PyMol (DeLano Scientific, San Carlos, CA, USA, http://www.pymol.org), using the B-A-F complex in the asymmetric unit.
Isothermal titration calorimetry
ITC experiments were performed in 20 mM HEPES (pH 7.5) and 120 mM NaCl at 25°C using a VP-ITC calorimeter (Microcal Inc., Northampton, MA, USA). In all, 51 μM Spc24155–213-25133–221 wild type or mutants in the temperature-controlled sample cell was titrated with 400 μM Cnn160–84 in 29 individual injections of 10 μl. In all, 300 μM Cnn1ΔHFD or 275 μM Cnn1 ΔHFDΔBC was used to titrate 40 μM Spc24155–213-25133–221 or 26 μM Spc24155–213-25133–221, respectively. Thermodynamic values of the interactions were obtained using Origin 7 (OriginLab Corp.) by fitting the binding isotherms with the non-linear least squares method, assuming one set of binding sites.
Interaction studies
Analytical size-exclusion chromatography was performed in 20 mM HEPES (pH 7.5) and 120 mM NaCl, with the addition of 5% glycerol for interaction analysis involving Cnn1ΔHFD. Spc24-25 wild type or mutants and Cnn1ΔHFD were used at 12 μM concentration and 50 μl was loaded on Superdex200 3.2/30. All variants of Mtw1 and Ndc80 complexes were diluted to 4 μM and 50 μl injected on Superose 6 3.2/30. When used in combination, proteins were incubated on ice for 30 min to allow complex formation. Fractions of 100 μl were collected and analysed by SDS–PAGE. In the interaction studies between Mtw1 and Ndc80 complexes, 40 μl of the analysed fractions was precipitated with acetone prior to SDS–PAGE. For co-immunoprecipitation experiments, lysates from log-phase cultures were prepared in 25 mM HEPES pH 8, 150 mM NaCl, 2 mM EDTA, 1 mM DTT and 5% glycerol (Lysis buffer) using a BeadBeater (BioSpec). In all, 3.5 mg of extract was incubated ON in presence of protease and phosphatase inhibitor cocktails (COMPLETE, PhosSTOP, Roche) with 30 μl pure Dynabeads Protein G (Novex) coupled with mouse anti-myc 9E10 Monoclonal antibody (COVANCE). The beads were washed five times with lysis buffer and proteins were eluted by boiling the sample 5 min in SDS–PAGE loading buffer. In all, 30 μg of the initial lysates as input and 5 or 20 μl of eluted proteins were separated by SDS–PAGE and analysed by western blot with mouse anti-FLAG-M2-Peroxidase (HRP) conjugated (SIGMA), mouse anti-myc and secondary anti-mouse-HRP conjugated (Jackson) antibodies. The relative intensities of the bands were quantified using ImageJ.
Sequence analysis
Sequence homologues were collected within the NCBI non-redundant protein database, using NCBI-BLAST (version 2.2.24, E-values <0.01) (Altschul et al, 1997) and aligned with MAFFT (version 6, L-INS-I method) (Katoh and Toh, 2008). Sequence accession numbers are listed in Supplementary Table S2. The alignments were visualized and processed with Jalview (Clustalx colouring scheme) (Waterhouse et al, 2009). Profile HMM searches were performed with HMMER version 2.3.2 (Eddy, 1998). The C-terminal motif in Dsn1 homologues was defined with MEME (one occurrence per sequence, motif width 5–15) (Bailey et al, 2009). Spc24 and Spc25 sequence conservation values were calculated based on the alignments in Supplementary Figure S2 using al2co, with an independent count-based sequencing weighting scheme, the sum-of-pairs measure conservation calculation method and the BLOSUM62 scoring matrix (Pei and Grishin, 2001).
Yeast genetics
Yeast strains are based on the S288C background and were generated by standard procedures. A list of the used yeast strains can be found in Supplementary Table S1. The mini-chromosome segregation assay was performed as described previously (Schleiffer et al, 2012). For the plasmid shuffle assay, the S. cerevisiae strain SWY344 was transformed with pRS315 plasmids (LEU) carrying wild-type or mutated versions of Dsn1. Serial two-fold dilutions of ON cultures were prepared on 96-well plates in minimal medium starting from OD600=0.5. The dilutions were spotted on minimal medium without (SC) or with 5-FOA and grown at 25°C for 3 days. Spot assays for the analysis of Cnn1 phosphorylation mutants were performed by plating on YPD plates four-fold dilutions of ON cultures starting from OD600=0.6.
Immunofluorescence microscopy
Large budded cells were fixed with 5% formaldehyde after an α-factor release to the restrictive temperature (37°C). For microtubule visualization, spheroplasts were incubated ON with anti-alpha-tubulin monoclonal antibody (YOL134, Abcam) followed by secondary anti-rat Rhodamine-conjugated antibody (Abcam). DAPI (4′,6-diamidino-2-phenylindole) was used for DNA staining. Samples were visualized with an Axioplan2 microscope using a × 63/1.4 plan-apochromat Oil DIC objective and a CoolSnap HQ camera. Images were acquired and analysed with MetaMorph.
Accession numbers
The coordinates and the structure factors of the budding yeast Spc24-25–Cnn1 complex have been deposited in the Protein Data Bank with accession code 4GEQ.
Supplementary Material
Acknowledgments
We thank all members of the Westermann lab for discussion. We thank Gleb Bourenkov for help with data collection, Tobias Krojer for help with structure refinement and analysis, Mathias Madalinski for peptide synthesis, and Otto Hudecz for help with presentation of mass spectrometry results. We thank Frank Uhlmann and Mark Winey for yeast strains. Research in the Westermann lab receives funding from the European Research Council under the European Community’s Seventh Framework Programme (SW, FP7/2007–2013)/ERC grant agreement n° 203499, and from the Austrian Science Fund FWF (SW, SFB F34).
Author contributions: FM performed structural, biochemical, and genetic experiments. GL performed genetic and biochemical experiments. AS contributed bioinformatic analysis. AH helped with structure determination and analysis. KM contributed the mass spec analysis. TC guided the structural analysis. SW guided the project and performed genetic and biochemical experiments. FM, TC, and SW wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- 4 CCPN. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50(Pt 5): 760–763 [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68(Pt 4): 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E (2010) The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 467: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(Web Server issue): W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1): 2210–2221 [DOI] [PubMed] [Google Scholar]
- Bock LJ, Pagliuca C, Kobayashi N, Grove RA, Oku Y, Shrestha K, Alfieri C, Golfieri C, Oldani A, Dal Maschio M, Bermejo R, Hazbun TR, Tanaka TU, De Wulf P (2012) Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat Cell Biol 14: 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras AC, Nesvizhskii AI, Tyers M (2011) A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54(Pt 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, Salek M, Rappsilber J, Moores CA, Salmon ED, Musacchio A (2008) Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36(Web Server issue): W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Harrison SC (2012) Molecular architecture of the yeast monopolin complex. Cell Rep 1: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC (2010) The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1): 2126–2132 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, Cleveland DW (2006) The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469 [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145: 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF (2011) In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32(Web Server issue): W500–W502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T (2008) CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052 [DOI] [PubMed] [Google Scholar]
- Hornung P, Maier M, Alushin GM, Lander GC, Nogales E, Westermann S (2011) Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J Mol Biol 405: 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bloom K, Salmon ED (2009) In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol 19: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED (2006) Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol 8: 581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr D66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kiermaier E, Woehrer S, Peng Y, Mechtler K, Westermann S (2009) A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol 11: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Kim S, Sun H, Tomchick DR, Yu H, Luo X (2012) Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci USA 109: 6549–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU (2007) Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev 21: 3319–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1): 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lampert F, Westermann S (2011) A blueprint for kinetochores—new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol 12: 407–412 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED (2011) Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol 195: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog M, Morgan DO (2005) Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434: 104–108 [DOI] [PubMed] [Google Scholar]
- Maskell DP, Hu XW, Singleton MR (2010) Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol 190: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchenko L, Uhlmann F (2010) Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr Biol 20: 1396–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Schultes NP, Szostak JW (1986) Chromosome length controls mitotic chromosome segregation in yeast. Cell 45: 529–536 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T (2012) CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148: 487–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR 3rd, Desai A, Fukagawa T (2006) The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Palframan WJ, Meehl JB, Jaspersen SL, Winey M, Murray AW (2006) Anaphase inactivation of the spindle checkpoint. Science 313: 680–684 [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV (2001) AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics 17: 700–712 [DOI] [PubMed] [Google Scholar]
- Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, Maiolica A, Stark H, Musacchio A (2010) The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol 190: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM (2011) CENP-C is a structural platform for kinetochore assembly. Curr Biol 21: 399–405 [DOI] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A (2009) The life and miracles of kinetochores. EMBO J 28: 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S (2012) CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol 14: 604–613 [DOI] [PubMed] [Google Scholar]
- Schmitzberger F, Harrison SC (2012) RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep 13: 216–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A (2011) Direct binding of cenp-C to the mis12 complex joins the inner and outer kinetochore. Curr Biol 21: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL (2012) Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell 150: 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin LJ, Guimaraes GJ, Deluca JG (2011) The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell 22: 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC (2006a) The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol 14: 54–59 [DOI] [PubMed] [Google Scholar]
- Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC (2006b) Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure 14: 1003–1009 [DOI] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC (2005) Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA 102: 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.