Abstract
Genome-wide DNA demethylation, including the erasure of genome imprints, in primordial germ cells (PGCs) is a critical first step to creating a totipotent epigenome in the germ line. We show here that, contrary to the prevailing model emphasizing active DNA demethylation, imprint erasure in mouse PGCs occurs in a manner largely consistent with replication-coupled passive DNA demethylation: PGCs erase imprints during their rapid cycling with little de novo or maintenance DNA methylation potential and no apparent major chromatin alterations. Our findings necessitate the re-evaluation of and provide novel insights into the mechanism of genome-wide DNA demethylation in PGCs.
Keywords: cell cycle, chromatin dynamics, genome imprints, primordial germ cells, replication-coupled DNA demethylation
Introduction
Primordial germ cells (PGCs) in mice, the precursors for both the spermatozoa and the oocytes, have long been known to undergo genome-wide epigenetic reprogramming (Surani et al, 2007; Sasaki and Matsui, 2008; Saitou et al, 2012). This includes, most notably, genome-wide DNA demethylation (Popp et al, 2010; Guibert et al, 2012), which leads to the erasure of parental imprints (Hajkova et al, 2002; Lee et al, 2002). Consequently, male and female PGCs acquire an equivalent naïve epigenome at around embryonic day (E) 13.5. They subsequently establish new sex-specific epigenetic modifications along with their progression into spermatogenic or oogenic pathways. Genome-wide epigenetic reprogramming in PGCs has therefore been considered essential as the first step for the creation of the totipotent epigenome in the germ cell lineage. However, despite its importance, the underlying mechanism for genome-wide DNA demethylation in PGCs remains elusive.
DNA demethylation in animals can occur through either an active or a passive mechanism (Saitou et al, 2012). The active mechanism involves enzymes that directly modify 5-methylcytosine (5mC). Recent studies have revealed several potential pathways for active DNA demethylation. One such pathway involves AID (activation-induced cytidine deaminase) and APOBEC1 (apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1), which can deaminate 5mC into thymine (T), and the resultant T/G (guanine) mismatch can be repaired by the base-excision repair (BER) pathway (Morgan et al, 2004). Another pathway involves TET (ten-eleven translocations) proteins (TET1, 2, 3), which oxidize 5mC into 5-hydroxymethylcytosine (5hmC) (Kriaucionis and Heintz, 2009; Tahiliani et al, 2009). The resulting 5hmC can either be deaminated by AID or APOBEC1 and repaired by the BER pathway (Guo et al, 2011), or further oxidized into 5-formylcytosine (5fC) and then into 5-carboxylcytosine (5caC), which is removed and repaired by the BER pathway (He et al, 2011; Ito et al, 2011). In contrast, the passive mechanism involves replication-coupled dilution of 5mC in the absence or under the prevention of maintenance DNA methyltransferase (DNA methyltransferase 1: DNMT1) activity.
There has been a prevailing view that the genome-wide DNA demethylation in PGCs occurs primarily via an active mechanism (Feng et al, 2010). This view is based on (1) the kinetics analysis of the DNA demethylation of the differentially methylated regions (DMRs) of several imprinted genes (Hajkova et al, 2002), (2) the apparently higher genome-wide DNA methylation in Aid (also known as Aicda)-deficient PGCs at E13.5 (Popp et al, 2010), and (3) the activation of the BER pathway and subsequent dramatic chromatin changes, including loss of linker histone H1 and loss of several histone modifications such as histone H3 lysine 27 tri-methylation (H3K27me3), perhaps through histone replacement involving NAP1/NAP1l1 (nucleosome assembly protein 1/nucleosome assembly protein 1 like 1) and HIRA (histone cell-cycle regulation homologue A) at the G2 phase of the cell cycle, which is specifically observed in PGCs at around E11.5 (Hajkova et al, 2008, 2010). However, all these observations include some ambiguity from a quantitative point of view (Saitou et al, 2012) (Results and Discussion), and definitive evidence for the involvement of an active mechanism in the genome-wide DNA demethylation in PGCs is still lacking.
In order to provide a better resolution of the mechanism of genome-wide DNA demethylation in PGCs, we examined the expression and subcellular localization of key genes/proteins for DNA methylation/demethylation in PGCs, and re-visited the proliferation rate of PGCs, and the kinetics of imprint erasure and chromatin dynamics in PGCs. Strikingly, in contrast to the prevailing view, our data indicate that imprint erasure occurs in PGCs in a manner largely consistent with replication-coupled passive demethylation, necessitating the re-evaluation of the mechanism of genome-wide DNA demethylation in PGCs.
Results
Expression of genes involved in DNA methylation/demethylation in PGCs from E9.5 to E13.5
We have shown previously that PGCs repress genes encoding de novo DNA methyltransferases, Dnmt3a/3b, upon their specification (Yabuta et al, 2006; Kurimoto et al, 2008) and show low DNMT3A/3B at least until E12.5 (Seki et al, 2005). We also found that although PGCs express Dnmt1, they repress Uhrf1 (ubiquitin-like with PHD and ring finger domain 1), which encodes an essential factor to recruit DNMT1 to replication foci where maintenance DNA methylation takes place (Bostick et al, 2007; Sharif et al, 2007), at least at the mRNA level upon their specification (Kurimoto et al, 2008).
We set out to obtain a more comprehensive view of the expression of genes involved in DNA methylation/demethylation during PGC development. We mated female mice (BDF1 background) with males bearing the Stella (also known as Dppa3/Pgc7)–EGFP transgenes (C57BL/6 background) (Payer et al, 2006; Seki et al, 2007), isolated Stella–EGFP-positive PGCs from embryos from E10.5 to E13.5 by fluorescence activated cell sorting (FACS), and purified mRNAs from these cells for microarray analysis (Materials and methods). We also analysed the gene expression data in the epiblasts at E5.75, in PGCs at E9.5, and, as a reference, during the in vitro PGC specification from embryonic stem cells (ESCs) (Hayashi et al, 2011).
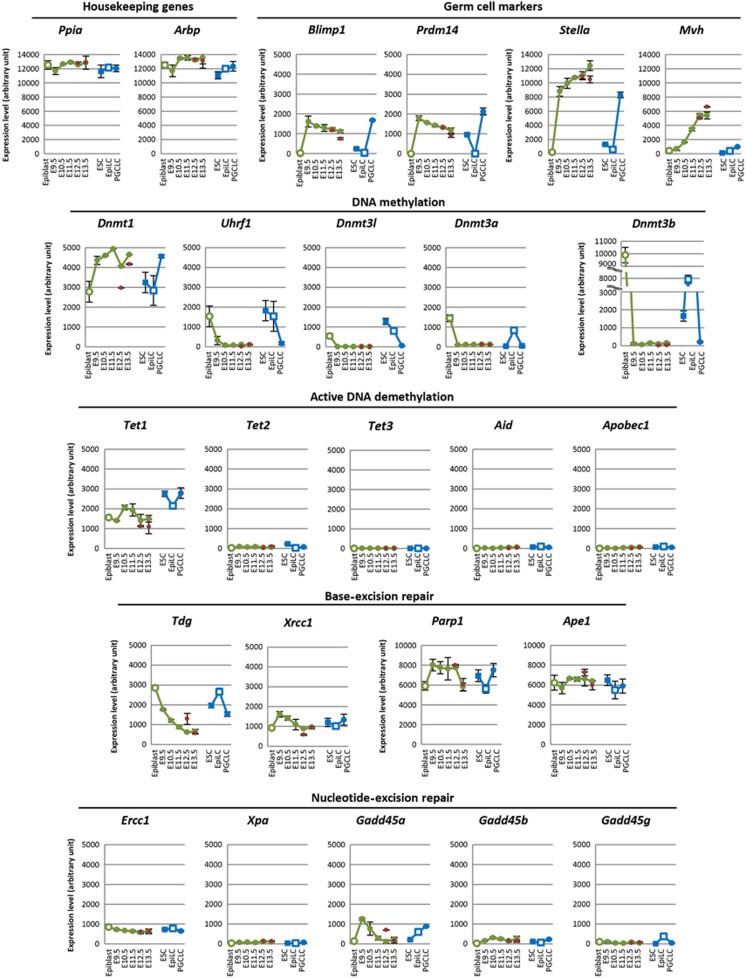
To validate the microarray analysis, we first looked at the expression of genes specific to PGCs: Blimp1 (also known as Prdm1) and Prdm14 were constantly detected in PGCs from E9.5 to E13.5, with a slight decrease in their expression at E13.5, Stella showed very high expression in PGCs from E9.5 to E13.5, and Mvh (mouse vasa homologue, also known as Ddx4) exhibited progressive upregulation from E9.5 to E13.5 (Figure 1). These data are consistent with previous findings (Tanaka et al, 2000; Chang et al, 2002; Saitou et al, 2002; Sato et al, 2002; Yamaji et al, 2008), corroborating the accuracy of our microarray analysis. We previously estimated that the expression levels of Blimp1 and Stella in PGCs were ∼100 and 1000 copies per cell, respectively (Yabuta et al, 2006; Kurimoto et al, 2008). Therefore, their arbitrary expression levels in the microarray analysis, roughly 1000 and 10 000, can be considered to correspond to 100 and 1000 copies per cell, respectively (Figure 1).
Figure 1.
Expression of key genes for DNA methylation/demethylation during PGC development in vivo and in vitro. Expression levels of housekeeping genes, genes specific to PGCs, and key genes involved/potentially implicated in DNA methylation/demethylation during PGC development in vivo (green plots: E5.75 epiblasts, E9.5, E10.5, and E11.5 PGCs of mixed sexes, and E12.5 and E13.5 male PGCs; red plots: E12.5 and E13.5 female PGCs) and in vitro (blue plots: male ESCs, male epiblast-like cells (EpiLCs) at day 2 of induction, and male PGC-like cells (PGCLCs) at day 6 of induction) as determined by microarray analysis. We used E9.5–E11.5 PGCs of mixed sexes because gene expression of PGCs is indistinguishable until E11.5 in both sexes (Jameson et al, 2012). Expression data of E5.75 epiblasts, E9.5 PGCs, ESCs, day 2 male EpiLCs and day 6 male PGCLCs were obtained from our previous publication (Hayashi et al, 2011) (GEO database accession number GSE30056). The vertical axis represents arbitrary expression levels determined by microarray analysis. The average values with s.d.’s (two independent experiments) are indicated.
We next looked at the expression of Dnmts and Uhrf1. We found that Dnmt1 shows strong expression (roughly 300–400 copies per cell) in PGCs from E9.5 to E13.5, whereas Uhrf1 is acutely downregulated in PGCs and shows consistently low expression (around or <10 copies per cell) from E9.5 to E13.5 (Figure 1). Dnmt3a, 3b, and 3l were also downregulated and were present in consistently low quantities (around or <10 copies per cell) in PGCs from E9.5 to E13.5 (Figure 1). Combined with our previous findings (Seki et al, 2005; Yabuta et al, 2006; Kurimoto et al, 2008), these data demonstrate that the key genes implicated in maintenance and de novo DNA methylation are transcriptionally repressed in PGCs from their specification up until E13.5.
We next looked at the expression of genes implicated in active DNA demethylation. Tet1 showed consistent expression in PGCs from E9.5 to E13.5 (around 100 copies per cell), whereas the expression of Tet2 was consistently low (around or <10 copies per cell) and Tet3 was undetectable (Figure 1). Aid and Apobec1 were also undetectable or expressed at very low levels (around or <10 copies per cell) in PGCs from E9.5 to E13.5 (Figure 1). Thymine DNA glycosylase (Tdg), which can remove T from T/G mismatches and also 5caC from 5caC/G pairs for BER (Gallinari and Jiricny, 1996; He et al, 2011), showed relatively high expression in early PGCs (∼around 100–200 copies per cell at E9.5) and was slightly downregulated thereafter (<100 copies per cell at E12.5 and E13.5) (Figure 1). Genes involved in BER (Xrcc1: X-ray repair complementing defective repair in Chinese hamster cells 1; Parp1: poly (ADP-ribose) polymerase family, member 1; Ape1: apurinic/apyrimidinic endonuclease 1) were constantly expressed in PGCs as well as in ESCs and epiblasts, but genes involved in nucleotide-excision repair (NER) (Ercc1: excision repair cross-complementing rodent repair deficiency, complementation group 1; Xpa: xeroderma pigmentosum, complementation group A; Gadd45a: growth arrest and DNA-damage-inducible 45 α; Gadd45b: growth arrest and DNA-damage-inducible 45 β; Gadd45g: growth arrest and DNA-damage-inducible 45 γ) were not (Figure 1). All the genes examined exhibited the levels of expression expected based on observation of the in vivo PGCs in the in vitro PGC specification system (Figure 1). These findings provide quantitative evidence in support of the idea that PGCs consistently possess little maintenance and de novo DNA methylation potential after their specification, and that, among the proposed pathways for active DNA demethylation, it may only be the TET1-TDG pathway that can operate in PGCs.
Loss of UHRF1 and of DNMT1 recruitment to replication foci in PGCs
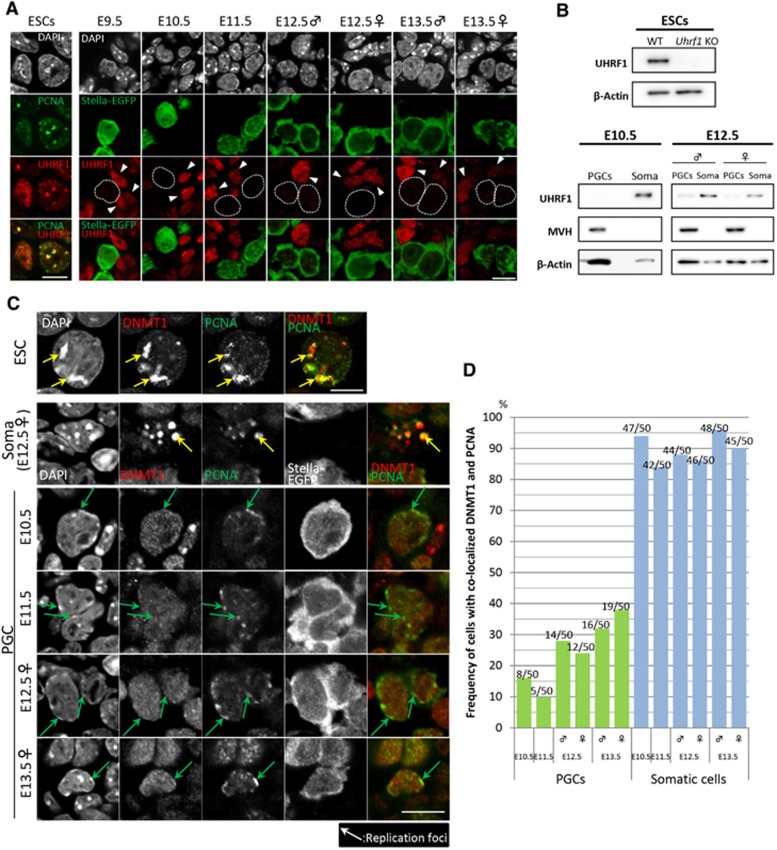
We next set out to examine the expression and localization of UHRF1 and DNMT1 protein in PGCs after E9.5. In particular, we examined whether DNMT1 is recruited to replication foci in PGCs from E10.5 to E13.5. PCNA (proliferating cell nuclear antigen) is a typical marker for the replication foci and co-localizes with DNMT1 at replication foci with characteristic subnuclear patterns along the progression of the S phase of the cell cycle (Leonhardt et al, 2000). Especially at the mid–late S phase, PCNA and DNMT1 co-localize at late replicating heterochromatic foci (including DAPI-positive peri-centromeric heterochromatin) with relatively larger size (O’Keefe et al, 1992; Leonhardt et al, 2000). Importantly, UHRF1 also becomes upregulated at the mid–late S phase and localizes at these sites (Miura et al, 2001). In Uhrf1-knockout ESCs, DNMT1 is not successfully targeted to replication foci, and as a result, these cells show genome-wide DNA demethylation (Bostick et al, 2007; Sharif et al, 2007).
We first stained ESCs for UHRF and PCNA. Consistent with a previous report (Sharif et al, 2007), the results showed that UHRF1 exhibited a broad distribution in the nuclei of cells outside the S phase, whereas it showed specific and conspicuous localization at PCNA-positive replication foci in cells at the mid–late S phase (Figure 2A). Western blot analysis confirmed that there was robust expression of UHRF1 in wild-type ESCs and no expression of UHRF1 in the Uhrf1-knockout ESCs (Figure 2B) (Sharif et al, 2007). We then examined the expression of UHRF1 in PGCs. Consistent with the microarray analysis, immunofluorescence analysis showed that Stella–EGFP-positive PGCs migrating in the hindgut endoderm at E9.5 exhibited no/low expression of UHRF1, whereas the hindgut endoderm showed UHRF1 expression (Figure 2A). We found that PGCs in the incipient genital ridges at E10.5 and E11.5 also exhibited no/low expression of UHRF1, whereas somatic cells in the genital ridges expressed UHRF1 (Figure 2A). PGCs exhibited slight expression of UHRF1 after E12.5 both in males and in females (Figure 2A). We confirmed by western blot analysis that PGCs at E10.5 lack UHRF1, whereas PGCs at E12.5 showed UHRF1 at a low level both in males and in females (Figure 2B). These findings demonstrate that PGCs, especially those between E9.5 and E11.5, possess little, if any, UHRF1 activity.
Figure 2.
UHRF1 expression and DNMT1 localization in PGCs. (A) Immunofluorescence analysis of UHRF1 expression (red) in PGCs (green) after E9.5. As a control, UHRF1 (red) and PCNA (green) expression in ESCs are shown in the left column. PGCs were identified as Stella–EGFP-positive cells. The nuclear architecture of PGCs and somatic cells is shown by DAPI staining (white). The position of the PGC nucleus is delineated by a dotted line and the somatic cells are marked by arrowheads in the red (UHRF1) channel. Bar, 10 μm. (B) Western blot analysis of UHRF1 expression in 5 × 103 wild-type and Uhrf1-knockout ESCs (top) and in 2 × 104 Stella–EGFP-positive PGCs and Stella–EGFP-negative somatic cells at E10.5 and E12.5 (both in males and in females) (bottom). MVH was used as a germ cell marker and β-Actin was used as a control. (C) Localization of DNMT1 and PCNA in PGCs and somatic cells. As a control, localization of DNMT1 and PCNA in ESCs is shown in the top row. PGCs (Stella–EGFP-positive, from E10.5 to E13.5) and somatic cells (E12.5) at the mid–late S phase as judged by PCNA localization and DAPI staining are shown. Green and yellow arrows indicate PCNA-positive replication foci in PGCs and in somatic cells and ESCs, respectively. Bar, 10 μm. (D) Percentage of co-localization of DNMT1 with PCNA in PGCs and somatic cells at the mid–late S phase after E10.5. 50 PGCs and 50 somatic cells were counted.
We next examined whether DNMT1 is recruited to replication foci in PGCs from E10.5 to E13.5. We stained PGCs and gonadal somatic cells for DNMT1 and PCNA. To facilitate the evaluation of co-localization of DNMT1 with PCNA, we selected cells at the mid–late S phase based on the localization of PCNA at DAPI-positive peri-centromeric heterochromatin (major axis >0.4 μm, relative staining intensity more than twice that of the nucleoplasm). As a control, we stained ESCs, which showed that DNMT1 localizes at PCNA-positive replication foci at the mid–late S phase (Figure 2C). As shown in Figures 2C and D, ∼90% of the gonadal somatic cells at the mid–late S phase from E10.5 to E13.5 exhibited localization of DNMT1 at the PCNA-positive replication foci, indicating that, in these cells, maintenance DNA methylation occurs in a replication-coupled manner, at least at late replicating heterochromatic foci (including DAPI-positive peri-centromeric heterochromatin) and most likely in other regions of the genome. In sharp contrast, only ∼10–20% and ∼30% of the PGCs at the mid–late S phase showed localization of DNMT1 at replication foci at E10.5–E11.5 and at E12.5–E13.5, respectively (Figures 2C and D). These findings suggest the possibility that a majority of PGCs fail to undergo/are inefficient for replication-coupled maintenance DNA methylation, at least at late replicating heterochromatic foci and presumably also in other regions of the genome, especially at E10.5 and E11.5, and hence erase their DNA methylation via replication-coupled passive DNA demethylation.
Constant and rapid proliferation of PGCs after E9.5
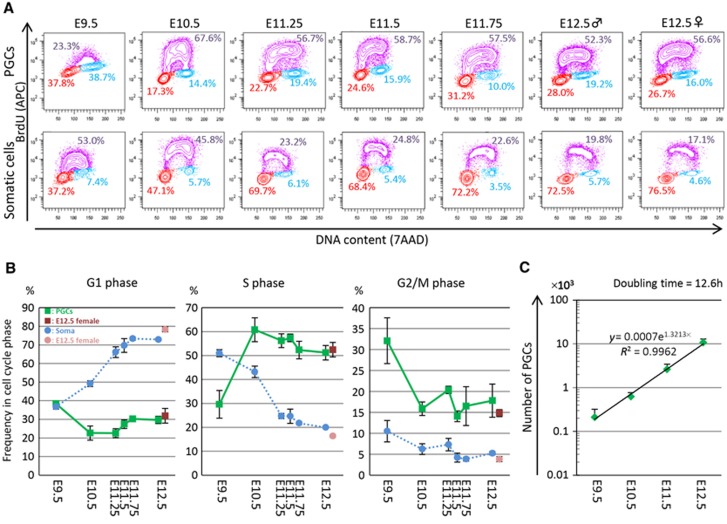
Our previous study showed that a majority (∼60%) of PGCs migrating into the hindgut (E7.75–E8.75) are arrested at the G2 phase of the cell cycle, and when they come out into the mesentery after E9.5, they start rapid proliferation (Seki et al, 2007). To further explore the possibility of replication-coupled passive DNA demethylation in PGCs, we next set out to determine the cell-cycle distribution and proliferation of PGCs after E9.5. We mated females (ICR background) with males bearing the Stella–EGFP transgenes (C57BL/6 background), and injected BrdU into pregnant females at six different time points (E9.5, E10.5, E11.25, E11.5, E11.75, and E12.5). We isolated embryos and stained the dissociated embryonic cells containing PGCs for their BrdU incorporation and DNA contents, which were analysed by FACS.
As shown in Figures 3A and B, Stella–EGFP-positive PGCs at E9.5 incorporated relatively little BrdU and ∼38, ∼30, and ∼32% of these cells were in the G1, S, and G2/M phases, respectively. In contrast, PGCs at E10.5 incorporated a high amount of BrdU and ∼23, ∼61, and ∼16% of these cells were in the G1, S, and G2/M phases. After E10.5, PGCs constantly incorporated a high amount of BrdU and ∼55% of these cells were in the S phase at least until E12.5 (Figures 3A and B). We did not observe a clear G2 arrest of PGCs around E11.5 (Figures 3A and B). Combined with our previous analysis (Seki et al, 2007), these findings indicate that some of the PGCs at E9.5 are still in the hindgut endoderm and are arrested at the G2 phase; and thereafter, the PGCs exhibit rapid proliferation at a relatively constant rate at least until E12.5. The Stella–EGFP-negative somatic cells, especially those of the genital ridges and mesonephros after E10.5, showed little BrdU incorporation and hence slow proliferation (Figures 3A and B).
Figure 3.
Cell-cycle state and proliferation of PGCs. (A) Representative plots for the cell-cycle state of PGCs (top) and somatic cells (bottom) after E9.5 as analysed by FACS. The vertical axis represents BrdU incorporation and the horizontal axis represents DNA content. Cells in S, G2/M, and G1 phase are shown in purple, blue, and red, respectively, along with the percentage of each. Representative data from three independent experiments are shown. (B) Plots showing the percentage of PGCs (green; E12.5 female PGCs are shown in red) and somatic cells (blue; E12.5 female somatic cells are shown in pink) after E9.5 in the G1, S, and G2/M phase. The average values from three independent experiments are shown with s.d.’s. (C) Plots showing PGC proliferation. The average values from three independent experiments are shown with s.d.’s. The regression line indicates that the doubling time of PGCs from E9.5 to E12.5 is ∼12.6 h.
Based on the number of Stella–EGFP-positive PGCs analysed on FACS, we calculated the number of PGCs per embryo from E9.5 to E12.5 (see Materials and methods). Although the calculated number of PGCs per embryo at each stage varied to a certain extent (Table I), which was also the case in previous studies (Tam and Snow, 1981; Seki et al, 2007), the average number of PGCs at each stage showed a constant exponential increase (Figure 3C), indicating that PGCs proliferate at a constant rate from E9.5 to E12.5. From the slope of the regression line of the cell number plots, the doubling time of PGCs after E9.5 was calculated as ∼12.6 h, indicating that PGCs divide twice per day from E9.5 to E12.5.
Table 1. Number of PGCs from E9.5 to E12.5.
| Experimental number | Number of PGCs per embryo |
|||
|---|---|---|---|---|
| E9.5 | E10.5 | E11.5 | E12.5 male | |
| #1 | 360 | 762 | 2027 | 8938 |
| #2 | 170 | 410 | 3813 | 13 965 |
| #3 | 100 | 688 | 1966 | 9030 |
| Average | 210 | 620 | 2602 | 10 644 |
Kinetics of imprint erasure in PGCs
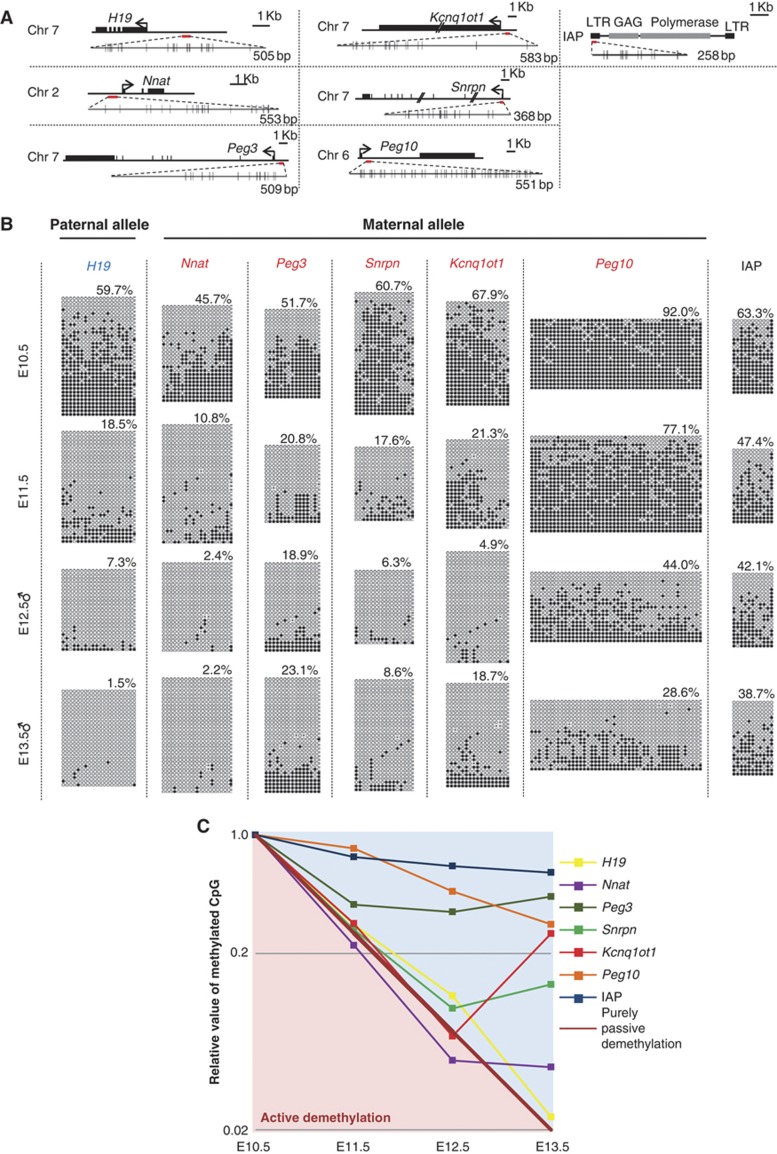
Our analysis has so far shown that during the relevant period for the genome-wide DNA demethylation, PGCs possess little maintenance and de novo DNA methylation potential, and divide more rapidly than previously thought (twice a day; doubling time, ∼12.6 h). We next determined the precise kinetics of the demethylation of the DMRs of imprinted genes by bisulphite sequence analysis. In this analysis, we consider it critical to discriminate the parental alleles, since the original methylation level in a cell is 50% and the bisulphite sequence analysis involves, prior to sequencing, many cycles of PCR amplification and random selection of the amplified clones, which can create ∼10–20% biases/fluctuations relatively easily and lead to misinterpretation of the results. For the discrimination of the parental alleles by single-nucleotide polymorphisms, we crossed C57BL/6 Stella–EGFP females with JF1 males (Koide et al, 1998), isolated PGCs from E10.5 to E13.5 (male PGCs for E12.5 and E13.5), and examined the methylation of the gametic DMRs of one paternally methylated imprinted gene (H19) and five maternally methylated imprinted genes (Nnat (also known as Peg5), Peg10, Snrpn, Peg3, and Kcnq1ot1 (also known as Lit1)) in these cells (Lee et al, 2002; Kobayashi et al, 2006; Tomizawa et al, 2011).
The paternally methylated gene and the maternally methylated genes essentially lack DNA methylation of the DMRs on the maternal and paternal alleles, respectively, at all stages examined (Supplementary Figure S1), indicating successful discrimination of the parental alleles in our experiments. Our analysis revealed that, first, many DMRs analysed (those of H19, Nnat, Snrpn, Peg3, and Kcnq1ot1) (Figure 4A) exhibited a substantial erasure of imprints (∼30–∼50% erasure) as early as E10.5 (Figure 4B), indicating that in most imprinted genes, erasure of imprints commences prior to the PGCs’ entry into the genital ridges, most likely in migrating PGCs. Second, the demethylation rates of all the DMRs examined did not exceed those expected from the replication-coupled passive demethylation, considering that the doubling time of PGCs after E9.5 was ∼12.6 h (Figures 4B and C). Third, the demethylation rates of the DMRs examined were heterogeneous and the demethylation of the DMR of one maternally imprinted gene, Peg10, was especially slow and occurred in a similar fashion to that of intracisternal A particles (IAPs) (Figures 4B and C).
Figure 4.
Imprint erasure in PGCs. (A) The location and size of the DMRs (red bars) of one paternally (H19) and five maternally (Peg3, Nnat, Peg10, Snrpn, Kcnq1ot1) imprinted genes analysed in this study are shown. The location and size of the region analysed for the methylation of IAP are also shown (a red bar). The horizontal black bars represent genomic sequences and the black boxes on the bars represent exons of the genes. The arrows indicate the transcription start sites. The DMRs or a portion of the LTR of IAP analysed (red bars) are shown as enlarged bars with the locations of CpG dinucleotides (small horizontal bars). (B) The methylation states in PGCs from E10.5 to E13.5 (male PGCs at E12.5 and E13.5) of the DMRs of the methylated alleles of one paternally (H19) and five maternally (Peg3, Nnat, Peg10, Snrpn, Kcnq1ot1) imprinted genes and of the promoter/long terminal repeat (LTR) of IAPs (a mass population of essentially all IAPs) determined by bisulphite sequence. White and black circles represent un-methylated and methylated cytosine, respectively. The per cent methylation for each DMR/promoter at each stage is indicated. (C) Kinetics of demethylation of the DMRs/promoter analysed. The vertical axis represents the relative methylation rate of each DMR/promoter against the methylation rate of each DMR/promoter at E10.5 shown in a logarithmic scale. The horizontal axis represents the developmental stage. The demethylation kinetics in PGCs expected from purely passive demethylation with no maintenance or de novo methylation when the doubling time of PGCs is ∼12.6 h is shown by a diagonal bold brown line. Note that the demethylation rates of the DMRs/promoter analysed are slower than or close to that expected from the purely passive demethylation.
These findings indicate that the erasure of gametic imprints initiates in migrating PGCs and proceeds at a rate that can be explained largely by replication-coupled passive demethylation, with gene-dependent partial resistances against demethylation.
No apparent major chromatin changes in PGCs at around E11.5
Previous studies have documented that PGCs at around E11.5 are arrested at the G2 phase and undergo dynamic changes in the chromatin state, including nuclear enlargement, loss of chromocentres (intensively DAPI-stained foci), loss of linker histone H1, and loss of many histone modifications, including H3K27me3 (Hajkova et al, 2008, 2010). These changes have been proposed to reflect the genome-wide histone replacement involving NAP1 and HIRA following the genome-wide DNA repair for the genome-wide active DNA demethylation in PGCs (Hajkova et al, 2008, 2010). Since we did not detect the G2 arrest of PGCs at around E11.5 (Figure 3) and our data support the possibility of passive DNA demethylation in PGCs, we next examined whether the reported dynamic changes of the chromatin state could be observed in PGCs in the present study.
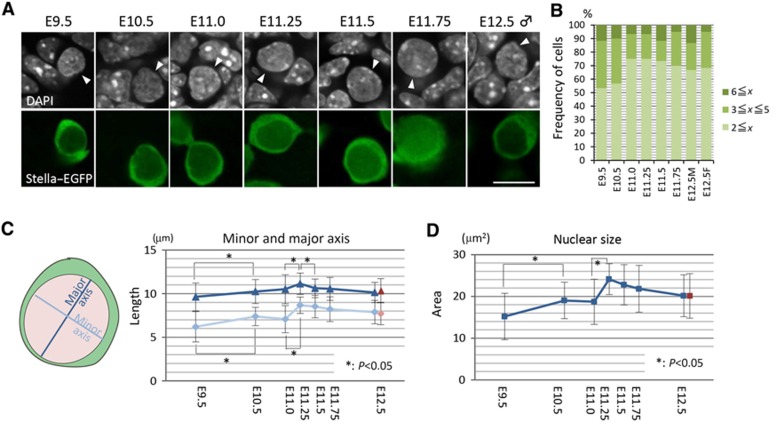
First, we looked at the chromocentres in PGCs. From E9.5 to E12.5, PGCs consistently showed relatively weak DAPI staining throughout their nuclei, with less eminent chromocentres: The chromocentres in PGCs were difficult to observe and smaller in size than those in somatic cells at all the stages examined (Figure 5A and see also Figures 6A and C). We counted the number of detectable chromocentres (major axis >0.4 μm, relative staining intensity more than twice that of the nucleoplasm) in PGCs, but we did not find a specific change of morphology/distribution of chromocentres in PGCs at around E11.5 (Figure 5B). We estimated the nuclear size of PGCs by measuring the length of the major and minor axis of PGCs in confocal sections from E9.5 to E12.5. This analysis showed that the nuclear size of PGCs increases progressively from E9.5 to E11.25 and thereafter decreases slightly (Figures 5C and D).
Figure 5.
Chromocentres and nuclear size of PGCs after E9.5. (A) Nuclear architecture shown by DAPI staining (white, top) of PGCs (white arrowheads, top) (Stella–EGFP-positive, green, bottom) and somatic cells after E9.5. Bar, 10 μm. (B) Number of DAPI-positive chromocentres (major axis >0.4 μm, relative staining intensity more than twice than that of the nucleoplasm) in PGCs after E9.5. (C) Length (μm) of the major (blue; E12.5 female PGCs are shown in red) and minor (pale blue; E12.5 female PGCs are shown in pink) nuclear axis of PGCs after E9.5. At each stage, >50 cells were measured, and the average values with s.d.’s are indicated. (D) Nuclear sizes (μm2) of PGCs after E9.5, calculated from the data in (C). The average values (blue; E12.5 female PGCs are shown in red) with s.d.’s are indicated.
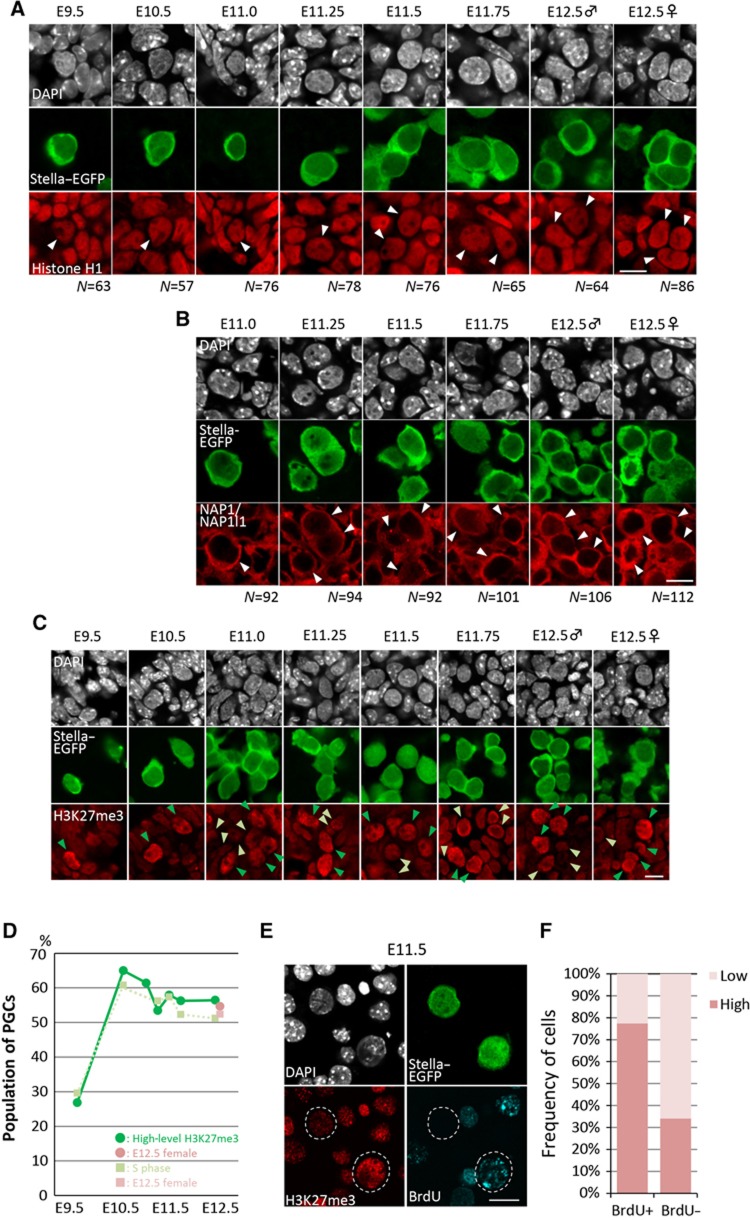
Figure 6.
Chromatin dynamics in PGCs after E9.5. (A) Representative images for the expression of linker histone H1 (red) in PGCs (Stella–EGFP-positive, green) and somatic cells after E9.5. The nuclear architecture of PGCs and somatic cells is shown by DAPI staining (white). The nuclei of PGCs are indicated by white arrowheads in the red channel. The number (N) of PGCs examined is indicated at the bottom. Bar, 10 μm. (B) Representative images for the expression of NAP1/NAP1l1 (red) in PGCs (Stella–EGFP-positive, green) and somatic cells from E11.0 to E12.5. The nuclear architecture of PGCs and somatic cells is shown by DAPI staining (white). Typical PGCs are indicated by white arrowheads in the red channel. The number (N) of PGCs examined is indicated at the bottom. Bar, 10 μm. (C) Representative images for the H3K27me3 state (red) in PGCs (Stella–EGFP-positive, green) and somatic cells after E9.5. The nuclear architecture of PGCs and somatic cells is shown by DAPI staining (white). Green and yellow-green arrowheads indicate PGCs with high and low H3K27me3, respectively. Bar, 10 μm. (D) Percentage of H3K27me3-high PGCs from E9.5 to E12.5 (green; E12.5 female PGCs are shown in pink), plotted with the percentage of PGCs in the S phase (light green; E12.5 female PGCs are shown in pale pink). (E) Representative images for the H3K27me3 state (red) and BrdU incorporation (blue) in PGCs (Stella–EGFP-positive, green) and somatic cells at E11.5. The nuclear architecture of PGCs and somatic cells is shown by DAPI staining (white). PGCs are delineated by dotted lines in red and blue channels. Bar, 10 μm. (F) Percentage of H3K27me3-high (red) or low (pink) PGCs among the BrdU-positive and -negative PGCs.
Second, we examined the expression and localization of the linker histone H1. We consistently detected H1 in PGCs, as well as in somatic cells, from E9.5 to E12.5 at a relatively uniform intensity throughout the period examined (Figure 6A). Although we carefully looked at the expression of H1 in PGCs at around E11.5 (at E11.25, E11.5, and E11.75), we did not find PGCs (N>50 for each stage) showing loss of H1 expression (Figure 6A). Similarly, we consistently detected H3 in PGCs from E9.5 to E12.5 (Supplementary Figure S2). We then went on to examine the expression and localization of NAP1/NAP1l1, a histone chaperone for H1, H2A, and H2B (Zlatanova et al, 2007). As shown in Figure 6B, from E11.0 to E12.5, we consistently detected NAP1/NAP1l1 in PGCs, as well as in somatic cells, predominantly in their cytoplasm. Although we observed some cells that contained relatively higher levels of NAP1/NAP1l1 in their nuclei, these cells had much higher levels of NAP1/NAP1l1 in their cytoplasms; in addition, these cells were not particularly specific around E11.5, but rather were observed throughout the period we examined (Figure 6B).
Third, we examined the H3K27me3 state in PGCs. Consistent with our previous studies, PGCs generally exhibited higher levels of H3K27me3 compared with their somatic neighbours, such as hindgut endoderm and gonadal somatic cells, from E9.5 to E12.5 (Figure 6C) (Seki et al, 2005, 2007). We did, however, detect heterogeneity of H3K27me3 levels in PGCs (Figure 6C, see green and white arrowheads). We quantified the H3K27me3 level in each PGC and classified the PGCs into those bearing high or low H3K27me3 (Materials and methods). As shown in Figure 6D, from E10.5 to E12.5, ∼60% of PGCs showed high H3K27me3 levels and the rest of them exhibited weak H3K27me3. However, we did not observe an abrupt, synchronous loss of H3K27me3 in PGCs at any stage examined.
We assumed that the heterogeneity of H3K27me3 levels in PGCs reflects their cell-cycle state, because the H3K27me3 level has been shown to fluctuate during the cell cycle, being highest at the S phase (Aoto et al, 2008), and the rate of the H3K27me3-high PGCs seems proportional to that of PGCs in the S phase (Figures 1B and 6D). To validate this idea, we mated females (ICR background) with males bearing the Stella–EGFP transgenes (C57BL/6 background), injected BrdU into pregnant females at E11.5, isolated embryos, and stained the dissociated embryonic cells containing PGCs to measure their levels of BrdU incorporation and H3K27me3. This analysis revealed that a majority (>75%) of BrdU-positive PGCs showed higher H3K27me3 levels, whereas a majority of BrdU-negative PGCs exhibited weaker H3K27me3 levels (Figures 6E and F), demonstrating that the heterogeneity of H3K27me3 levels in PGCs indeed reflects their cell-cycle state. Collectively, these findings support the view that PGCs do not abruptly alter their genome-wide chromatin state at around E11.5, but rather show an essentially constant chromatin state after E9.5, although they appear to change their chromatin state in a cell-cycle-dependent manner.
Discussion
There has been a prevailing notion that PGCs erase their genome-wide DNA methylation, including genome imprints, primarily via an active mechanism (Feng et al, 2010). This idea has been based on several independent observations (Hajkova et al, 2002, 2008, 2010; Popp et al, 2010), but some of these results appear to involve quantitative ambiguity and, in our opinion, require further rigorous validation (Saitou et al, 2012). We have shown here that during the critical period of genome-wide DNA demethylation, PGCs consistently bear little de novo or maintenance DNA methylation potential, exhibit rapid cycling, erase genome imprints with varying rates that, however, do not exceed those expected from the replication-coupled passive demethylation, and show no apparent major chromatin alterations. These findings support the idea that, contrary to the prevailing view, PGCs may erase genome imprints, and possibly their genome-wide DNA methylation, mainly through a replication-coupled passive mechanism.
Our data demonstrate that PGCs divide at a doubling time of ∼12.6 h after E9.5, potentially diluting their DNA methylation level by three-quarters per day. PGCs have long been considered to divide constantly at a doubling time of 16 h after E8.5 (Tam and Snow, 1981). However, it is worth noting that, in our previous study, a majority of migrating PGCs from E7.75 to E8.75 were arrested at the G2 phase and PGCs initiated a rapid proliferation after around E9.5 (Seki et al, 2007). In the study by Tam and Snow (1981), PGCs were counted as alkaline phosphatase (AP)-positive cells in tissue sections and the total number of PGCs per embryo was estimated based on a mathematical formula. Since the AP activity is not specific to PGCs (MacGregor et al, 1995), it might be possible that the number of PGCs was slightly overestimated at each stage, leading to an underestimation of the doubling rate of PGCs. Consistent with this assumption, the PGC number at each stage estimated by the previous study was larger than our count, especially at the earlier stages (E9.5 and E10.5). Although a precise determination of the doubling time of PGCs is difficult due to the heterogeneity of embryonic development/the PGC number per embryo, our finding shows that PGCs proliferate more rapidly than previously thought. It is of note that Tam and Snow (1981) also reported that PGCs bear potential to exhibit compensatory rapid proliferation (dividing more than three times between E9.5 and E10.5) after exposure to mitomycin C.
Our data also demonstrate that the genome imprints are substantially erased in PGCs as early as E10.5, indicating that the erasure of imprints is initiated in migrating PGCs. This in turn indicates that the erasure of imprints is not triggered by a signal or signals from embryonic gonads but presumably by a unique property of PGCs. This finding is consistent with a previous study (Lee et al, 2002), but may contradict the finding that the erasure of imprints occurs specifically between E11.5 and E12.5, which was based on the kinetics of imprint erasure determined without the discrimination of the parental alleles (Hajkova et al, 2002). We speculate that the low level of DNMT potential in PGCs triggers genome-wide DNA demethylation, including the erasure of imprints, when PGCs begin to proliferate rapidly after approximately E9.5.
We note that the demethylation rates of the imprints are substantially heterogeneous. The mechanism that creates the gene-by-gene difference in the demethylation rate is unknown. We did not find a clear correlation between the rate of imprint erasure and the presence of tandem direct repeats in the DMRs or the size of the DMRs (not shown). Interestingly, however, Peg10, which exhibited the slowest demethylation rate among the genes examined, is a retrotransposon-derived gene recently acquired in mammals (Ono et al, 2001). Among the retrotransposon sequences, IAPs and other elements with signs of recent insertion are known to be more resistant to demethylation in PGCs (Lane et al, 2003; Popp et al, 2010; Guibert et al, 2012). There might therefore be a mechanism that more efficiently targets the DNMTs, even when their absolute quantity is limited, to some specific sequences, such as those of recently inserted transposons, and this may contribute to the creation of the gene-by-gene heterogeneity of the demethylation rate. In good agreement with this idea, it has been shown previously that the maintenance of imprints and of the methylation of IAPs during pre-implantation development, in which genome-wide DNA demethylation is known to occur via the passive mechanism, depends on the presence of a very small amount of DNMT1, which is below the level of detection by immunofluorescence analysis and is insufficient for the maintenance of the methylation of most other gene loci (Gaudet et al, 2004; Hirasawa et al, 2008).
We have shown previously that PGCs reduce genome-wide H3K9me2 and instead increase genome-wide H3K27me3 during their migration period, and then maintain the H3K9me2-low and H3K27me3-high state after migration (Seki et al, 2005, 2007). Contrary to the previous reports (Hajkova et al, 2008, 2010), we did not find major chromatin changes in PGCs at around E11.5, including loss of chromocentres, loss of the linker histone H1, translocation of NAP1, or loss of H3K27me3, which have been suggested to be part of the genome-wide histone exchange following genome-wide BER for genome-wide active DNA demethylation (Hajkova et al, 2008, 2010). The discrepancy between our present findings and those of these earlier studies may have arisen from the different methods used to evaluate the chromatin states of PGCs. We immunostained the whole genital ridges and examined a large number of PGCs with careful evaluation of the staining level and quality within and between the samples (Materials and methods), whereas in the previous studies, immunostaining was applied to dissociated single PGCs attached on a slide, and thus it appeared that a relatively small number of PGCs were evaluated (Hajkova et al, 2008). We consider that the more limited application of staining may have led to a bias in the results.
There is a report showing that the genome-wide DNA methylation level in Aid-deficient PGCs at E13.5 is higher than that in wild-type PGCs, indicating the presence of active DNA demethylation driven by AID for the genome-wide DNA demethylation in PGCs (Popp et al, 2010). However, this study covers only ∼1% of the genome, and the observed difference in methylation level (22 and 20% in Aid-deficient male and female PGCs, respectively, and 16.3 and 7.8% in wild-type male and female PGCs, respectively) may not represent an actual difference in the function of AID in PGCs, but rather some technical variability. This notion would be supported by the result of another genome-wide study, in which no difference in methylation levels between male and female PGCs at E13.5 was detected (Guibert et al, 2012). We showed that Aid and Apobec1 were expressed in PGCs after E9.5 only at a very low level, if at all (<10 copies per cell), making it difficult to assume that these molecules are involved in the genome-wide DNA demethylation. It is also of note that both Aid-deficient male and female mice are fertile (Muramatsu et al, 2000; Popp et al, 2010). Based on the expression data, it might be that the only active DNA demethylation pathway operating in PGCs is the TET1-TDG pathway. However, Tet1-deficient mice are fertile (Dawlaty et al, 2011) and very recently, Tet1-deficient PGCs have been shown to undergo genome-wide DNA demethylation apparently normally (Yamaguchi et al, 2012), suggesting that this pathway may not be dominant in genome-wide DNA demethylation in PGCs.
In summary, our data show that, in PGCs, the erasure of imprints that occurs as part of genome-wide DNA demethylation may take place primarily through replication-coupled passive demethylation, in the manner of the DNA demethylation in pre-implantation embryos, although we cannot exclude the involvement of an active mechanism. PGCs repress Uhrf1, Dnmt3a, and Dnmt3b upon their specification and at least until E13.5, and this repression of both maintenance and de novo DNA methyltransferase potential may be central for genome-wide DNA demethylation in PGCs. It would be interesting to examine an effect of overexpression in PGCs of key repressed genes on imprint erasure and genome-wide DNA demethylation. Clarification of the mechanism by which these key enzymes are suppressed during PGC specification may be fundamental to understanding the genome-wide DNA demethylation in PGCs.
Materials and methods
Animals
All animals were treated with appropriate care according to the ethical guidelines of Kyoto University. The Stella–EGFP strain was maintained on a largely C57BL/6 background (Payer et al, 2006; Seki et al, 2007). JF1 mice (Koide et al, 1998) were obtained from the Genetics Strains Research Centre at the National Institute of Genetics, Japan. ICR mice and BDF1 mice were purchased from SLC.
Antibodies
The following antibodies were used at the indicated dilution and obtained from the indicated sources: rat anti-GFP (1/250, Nacalai 04404-26); rabbit anti-UHRF1 (1/20, Santa Cruz sc-98817); rabbit anti-DNMT1 (1/20, Santa Cruz sc-20701); mouse anti-PCNA (1/20, Santa Cruz sc-56); mouse anti-H1 (1/200, Abcam ab71594); rabbit anti-H3 (1/200, Abcam ab1791); rabbit anti-H3K27me3 (1/500, Upstate 07449); and rabbit anti-NAP1l1 (1/1000, Abcam ab33076).
The following secondary antibodies from Molecular Probes were used at a 1/250 dilution: Alexa Flour 488 goat anti-rat IgG; Alexa Flour 568 goat anti-rabbit IgG; Alexa Flour 568 goat anti-mouse IgG; and Alexa Flour633 goat anti-mouse IgG.
Cell-cycle analysis and counting of PGC number
ICR females were crossed with Stella–EGFP males to obtain Stella–EGFP transgenic embryos. Noon of the day when the vaginal plugs of mated females were identified was scored as E0.5. Pregnant females were injected intraperitoneally with 1 mg BrdU and embryos were collected after 30 min. The embryonic fragments containing PGCs at E9.5, E10.5, E11.0, E11.25, E11.5, E11.75, and E12.5 were dispersed into single cells by incubation with 0.05% trypsin and 0.5 mM EDTA in PBS at 37°C for 5 min. For the detection of BrdU incorporation, we used an APC-BrdU Flow Kit (BD Biosciences). The dissociated samples were fixed in BD Cytofix/Cytoperm Buffer for 30 min on ice and washed with 1 × BD Perm/Wash Buffer and then incubated in BD Cytoperm Plus Buffer for 10 min on ice. After washing with 1 × BD Perm/Wash Buffer, the samples were re-fixed in BD Cytofix/Cytoperm Buffer for 5 min on ice and washed with 1 × BD Perm/Wash Buffer. The fixed samples were permeabilized in 300 μg/ml DNase solution for 1 h at 37°C, and then incubated in the 1 × BD Perm/Wash Buffer including the APC-conjugated anti-BrdU antibody for 20 min at room temperature. After washing with 1 × BD Perm/Wash Buffer, the DNA of the samples was stained with 7AAD. The stained samples were suspended in the staining solution (3% BSA, 0.09% sodium azide, 1 × PBS) and analysed using a BD FACSAria2 Cell Sorter (BD Biosciences) with FACSDiVa software (BD Biosciences). PGCs were identified as GFP-positive cells. Samples containing a minimum of 5000 PGCs were analysed using a BD FACSAria2 Cell Sorter (BD Biosciences) with FACSDiVa software (BD Biosciences). Three biological replicates were analysed for each sampling point.
Numbers of PGCs were calculated as follows. The number of total living cells in each sample before staining was counted by a hemocytometer. After staining, the living cells were identified as 7AAD-positive cells and the percentage of GFP-positive PGCs among living cells was measured by FACS. We calculated the total PGC number per embryo based on the assumption that the percentage of PGCs in the living cells was the same between before and after the staining.
Immunofluorescence analysis
For immunofluorescence analysis, ICR females were crossed with Stella–EGFP males to obtain Stella–EGFP transgenic embryos. All the experiments were performed at room temperature unless otherwise indicated and the images were captured by confocal microscopy (FV 1000; Olympus).
For the DNMT1, PCNA and UHRF1 immunostaining, the E10.5 embryonic fragments containing PGCs and the E11.5–E13.5 genital ridges were fixed in 4% paraformaldehyde (PFA) in PBS for 30 min on ice. The tissues were then immersed successively in 10 and 30% sucrose in PBS, embedded in OCT compound (Sakura), frozen, and sectioned at a thickness of 10 μm at −20°C. Air-dried sections were washed three times in PBS and treated with 99.8% methanol for 20 min at −20°C. After being washed three times in PBS, the samples were permeabilized with 1% Triton X-100 in PBS, incubated in the blocking solution (3% BSA, 0.1% Tween20, 4 × SSC), and then incubated in the staining solution (1% BSA, 0.1% Tween20, 4 × SSC) containing primary antibodies overnight at 4°C. After washing with 4 × SSC three times, the samples were incubated in the staining solution containing secondary antibodies and 1 ug/ml of DAPI for 1 h. The samples were then washed three times with 4 × SSC and mounted in Vectashield mounting medium (Vector Laboratories) for confocal microscope analysis.
We defined the mid-to-late S phase by the distinctive PCNA staining pattern (Leonhardt et al, 2000). For evaluation of the PCNA-positive replication foci, we defined a focus as follows. (1) The major axis of the focus is >0.4 μm. (2) The signal intensity of the focus is more than twice that of the nucleoplasm (Ma et al, 1998). When there was even one focus in which PCNA and DNMT1 showed co-localization, we evaluated the cell as showing co-localization of DNMT1 with PCNA. We evaluated 50 PGCs in the mid-to-late S phase at each stage.
For the NAP1/NAP1l1 immunostaining, the E11.0–E12.5 genital ridges were fixed in 4% PFA in PBS for 30 min on ice. The tissues were then immersed successively in 10 and 30% sucrose in PBS, embedded in OCT compound (Sakura), frozen, and sectioned at a thickness of 10 μm at −20°C. Air-dried sections were washed three times in PBS and incubated in the blocking solution (0.1% BSA, 0.1% TritonX100, 1 × PBS) for 15 min, and then incubated in the blocking solution (0.1% BSA, 0.1% TritonX100, 1 × PBS) containing primary antibodies overnight at 4°C. After washing with 1 × PBS three times, the samples were incubated in the staining solution containing secondary antibodies and 1 μg/ml of DAPI for 1 h. The samples were then washed three times with 1 × PBS and mounted in Vectashield mounting medium for confocal microscope analysis.
For the H1, H3, and H3K27me3 immunostaining, the E9.5–E10.5 embryonic fragments containing PGCs and the E11.5–E13.5 genital ridges were fixed in 4% PFA in PBS for 2–4 h on ice, and washed three times with PBS-0.2% Tween20 (PBT) at 4°C. For permeabilization, the tissues were incubated successively in 25, 50, 75, and 90% methanol in PBT for 10 min at 4°C, and were then incubated in 100% methanol. The samples were kept in 100% methanol overnight at −20°C. For hydration, the samples were incubated successively in 90, 75, 50, and 25% methanol in PBT for 10 min at 4°C. After washing with PBT twice, the samples were washed in TBS-0.1% TritonX100 (TBST) for 10 min at 4°C. The samples were blocked with 0.5% BSA-TBST overnight at 4°C and incubated with primary antibodies in the blocking solution for 4 days at 4°C. After washing with TBST eight times, the samples were incubated in secondary antibodies in the blocking solution, washed eight times with TBST, and mounted in Vectashield mounting medium for confocal microscopy.
Using the Spec3 software with which the Olympus FV1000 confocal microscope is equipped, we quantified the H3K27me3 levels in PGCs. We classified the H3K27me3 signal intensities in PGCs from 1 (weakest) to 5 (strongest) and defined the intensities 4 and 5 as high signal intensities. We observed 50 PGCs at each stage.
To explore the relationship between the H3K27me3 levels in PGCs and the cell-cycle state of PGCs, pregnant ICR females mated with Stella–EGFP males were injected intraperitoneally with 1 mg BrdU and embryos were collected after 30 min. The embryonic fragments containing PGCs at E11.5 were dispersed into single cells by incubation with 0.05% trypsin and 0.5 mM EDTA in PBS at 37°C for 5 min. For the staining of BrdU, we used an APC-BrdU Flow Kit (BD Biosciences). The samples were fixed in BD Cytofix/Cytoperm Buffer for 30 min on ice, washed with 1 × BD Perm/Wash Buffer, and incubated in BD Cytoperm Plus Buffer for 10 min on ice. After washing with 1 × BD Perm/Wash Buffer, the samples were re-fixed in BD Cytofix/Cytoperm Buffer for 5 min on ice and washed with 1 × BD Perm/Wash Buffer. The fixed samples were permeabilized in 300 μg/ml DNase solution for 1 h at 37°C, and then incubated in the staining solution (3% BSA, 0.09% sodium azide, 1 × PBS) containing the anti-H3K27me3 and anti-GFP antibodies. After washing with 1 × BD Perm/Wash Buffer, the samples were incubated in the staining solution containing the APC-conjugated anti-BrdU antibody and 10 μg/ml of DAPI and then washed with 1 × BD Perm/Wash Buffer. The stained samples were suspended in 1 × BD Perm/Wash Buffer and spread onto a slide glass with Cyto Spin 4 (Thermo Scientific). Air-dried samples were mounted in Vectashield mounting medium for confocal microscope analysis. We quantified the H3K27me3 levels in PGCs using the Spec3 software as described above.
Western blot analysis
To isolate PGCs for western blot analysis, ICR females were crossed with Stella–EGFP males to obtain Stella–EGFP transgenic embryos. The embryonic fragments containing PGCs of E10.5 and genital ridges of E12.5 embryos were dispersed into single cells as described above. The resulting cells were suspended in 0.1% polyvinyl alcohol (PVA) in PBS and sorted using a BD FACSAria3 Cell Sorter (BD Biosciences). The 2 × 104 GFP-positive and -negative cells were lysed with NuPAGE LDS sample buffer (Invitrogen). Proteins were resolved by NuPAGETM 4–12% Bis-Tris gel (Invitrogen), and electroblotted onto an Immobilon-P transfer membrane (Millipore). The blot was incubated with a blocking solution (10x% skim milk in PBT) for 1 h at room temperature, then incubated in the blocking solution with primary antibodies (anti-UHRF1, 1/200; anti-MVH (Abcam ab13840), 1/500; anti-β-Actin (Cell Signaling #4967), 1/1000) overnight at 4°C. The blot was washed three times with PBT and then incubated in the blocking solution with the secondary antibody (HRP-conjugated anti-rabbit IgG (GE Healthcare), 1/8000) for 1 h at room temperature. After washing three times with PBT, signals were detected by using Chemi-Lumi One Super (Nakarai).
Bisulphite sequence analysis
To isolate PGCs for bisulphite sequence analysis, the Stella–EGFP females were crossed with JF1 males to obtain F1 hybrid embryos. The embryonic fragments containing PGCs of E10.5–13.5 hybrid embryos were dispersed into single cells and sorted using a BD FACSAria3 Cell Sorter as described above. Bisulphite treatment of the samples was performed with Epitect Plus Bisulfite Kits (Qiagen). After bisulphite treatment, the samples were collected using a microcolumn and desulphonated with 0.3 M NaOH. After the desulphonation, DNA was eluted with 15 μl of the elution buffer. We used bisulphite-treated DNA from >500 PGCs for each PCR amplification. Amplification was performed using ExTaq (TaKaRa) under the following conditions: 40 cycles of 96°C for 30 s, 60°C for 1 min (for IAP/LTR (long terminal repeats of IAPs), 53°C for 1 min), and 72°C for 1 min.
The PCR primers used are as follows:
5′-aatggttgaattttagtttttgtttttatggtt-3′ and 5′-accaatacaatcccacatactttatcataaaa-3′ for H19, 5′-gttgggaaggattatgtagagaaaagtatatt-3′ and 5′-ccaaaaacatactcatctttaac-3′ for Kcnq1ot1, 5′-gttaaggaaatttatgggtatagagaataaag-3′ and 5′-ctaaaaaaatcttaacctaactatctacataa-3′ for Peg3, 5′-gtaaagtgattggttttgtatttttaagtg-3′ and 5′-ttaattactctcctacaactttccaaatt-3′ for Peg10, 5′-gttttggttaaataggatgtatttttattattag-3′ and 5′-aacaattatatccattattccaaattaacaata-3′ for Snrpn (Kobayashi et al, 2006), 5′-gttttattttgaaattagaagatagtagttataa-3′ and 5′-cacacccaaacctacaaattctac-3′ for Nnat (Lee et al, 2002), and 5′-ttgtgttttaagtggtaaataaataatttg-3′ and 5′-aaaacaccacaaaccaaaatcttctac-3′ for a mass population of essentially all IAP/LTR (Hajkova et al, 2002). Amplified fragments were cloned into plasmids and sequenced. We used the QUMA web-based tool for the visualization and quantification of the bisulphite sequence data (Kumaki et al, 2008). Polymorphisms between C57BL/6 and JF1 were detected as single-nucleotide polymorphisms (Lee et al, 2002; Kobayashi et al, 2006).
Microarray analysis of gene expression in PGCs
To isolate PGCs for microarray analysis, BDF1 females were crossed with Stella–EGFP males. The embryonic fragments containing PGCs at E10.5–E13.5 were dispersed into single cells and sorted using a BD FACSAria3 Cell Sorter as described above. Total RNAs were extracted from the PGCs and purified using an RNeasy Micro Kit (Qiagen), then subjected to cDNA synthesis and amplification by the methods described previously (Kurimoto et al, 2006, 2007), with a minor modification (12 PCR cycles were used for the initial amplification). Biotin-labelled cRNAs were synthesized and hybridized to the Mouse Genome 430.2 GeneChip microarray (Affymetrix) according to the manufacturer’s instruction. The CEL data were then normalized using the dChip software (Li and Wong, 2001) with the invariant set mode, together with the data for E5.75 epiblast, E9.5 PGCs, ESCs, day 2 male EpiLCs, and day 6 male PGCLCs from a previous study (Hayashi et al, 2011) (Gene Expression Omnibus (GEO) database accession number GSE30056), and the Model-Based Expression Indices (i.e., expression levels) were calculated. Two biological replicates were analysed for each developmental stage and embryonic sex. The microarray data set has been deposited to NCBI GEO with accession number GSE40412.
Supplementary Material
Acknowledgments
We thank H Ohta, K Hayashi, Y Kumaki, and Y Seki for their discussion of this study. We are grateful to T Shiroishi for the JF1 strain and to H Koseki and J Sharif for the Uhrf1-knockout ESCs. This study was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; by JST-PRESTO/CREST/ERATO; and by the Mitsubishi Foundation.
Author contributions: SK designed and conducted the experiments and wrote the manuscript. KK conducted the microarray experiments. MY and TH assisted with the immunofluorescence experiments. MS conceived the project, designed the experiments, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aoto T, Saitoh N, Sakamoto Y, Watanabe S, Nakao M (2008) Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression. J Biol Chem 283: 18905–18915 [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Chang DH, Cattoretti G, Calame KL (2002) The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev 117: 305–309 [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science 330: 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P, Jiricny J (1996) A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature 383: 735–738 [DOI] [PubMed] [Google Scholar]
- Gaudet F, Rideout WM 3rd, Meissner A, Dausman J, Leonhardt H, Jaenisch R (2004) Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol 24: 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert S, Forne T, Weber M (2012) Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA (2008) Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA (2002) Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA (2010) Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M (2011) Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532 [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H (2008) Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, Capel B (2012) Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet 8: e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H (2006) Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res 113: 130–137 [DOI] [PubMed] [Google Scholar]
- Koide T, Moriwaki K, Uchida K, Mita A, Sagai T, Yonekawa H, Katoh H, Miyashita N, Tsuchiya K, Nielsen TJ, Shiroishi T (1998) A new inbred strain JF1 established from Japanese fancy mouse carrying the classic piebald allele. Mamm Genome 9: 15–19 [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, Okano M (2008) QUMA: quantification tool for methylation analysis. Nucleic Acids Res 36: Web Server issueW170–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M (2006) An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res 34: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M (2007) Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc 2: 739–752 [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M (2008) Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 22: 1617–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W (2003) Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93 [DOI] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F (2002) Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129: 1807–1817 [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC (2000) Dynamics of DNA replication factories in living cells. J Cell Biol 149: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R (1998) Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143: 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor GR, Zambrowicz BP, Soriano P (1995) Tissue non-specific alkaline phosphatase is expressed in both embryonic and extraembryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development 121: 1487–1496 [DOI] [PubMed] [Google Scholar]
- Miura M, Watanabe H, Sasaki T, Tatsumi K, Muto M (2001) Dynamic changes in subnuclear NP95 location during the cell cycle and its spatial relationship with DNA replication foci. Exp Cell Res 263: 202–208 [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK (2004) Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem 279: 52353–52360 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563 [DOI] [PubMed] [Google Scholar]
- O’Keefe RT, Henderson SC, Spector DL (1992) Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol 116: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, Ishino F (2001) A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73: 232–237 [DOI] [PubMed] [Google Scholar]
- Payer B, Chuva de Sousa Lopes SM, Barton SC, Lee C, Saitou M, Surani MA (2006) Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis 44: 75–83 [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA (2002) A molecular programme for the specification of germ cell fate in mice. Nature 418: 293–300 [DOI] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K (2012) Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139: 15–31 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y (2008) Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9: 129–140 [DOI] [PubMed] [Google Scholar]
- Sato M, Kimura T, Kurokawa K, Fujita Y, Abe K, Masuhara M, Yasunaga T, Ryo A, Yamamoto M, Nakano T (2002) Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech Dev 113: 91–94 [DOI] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y (2005) Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278: 440–458 [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M (2007) Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P (2007) Genetic and epigenetic regulators of pluripotency. Cell 128: 747–762 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Snow MH (1981) Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J Embryol Exp Morphol 64: 133–147 [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T (2000) The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 14: 841–853 [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H (2011) Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M (2006) Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod 75: 705–716 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y (2012) Tet1 controls meiosis by regulating meiotic gene expression. Nature (advance online publication 14 November 2012; doi:; DOI: 10.1038/nature11709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M (2008) Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet 40: 1016–1022 [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Seebart C, Tomschik M (2007) Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J 21: 1294–1310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.