Abstract
In many bacterial pathogens, the second messenger c-di-GMP stimulates the production of an exopolysaccharide (EPS) matrix to shield bacteria from assaults of the immune system. How c-di-GMP induces EPS biogenesis is largely unknown. Here, we show that c-di-GMP allosterically activates the synthesis of poly-β-1,6-N-acetylglucosamine (poly-GlcNAc), a major extracellular matrix component of Escherichia coli biofilms. C-di-GMP binds directly to both PgaC and PgaD, the two inner membrane components of the poly-GlcNAc synthesis machinery to stimulate their glycosyltransferase activity. We demonstrate that the PgaCD machinery is a novel type c-di-GMP receptor, where ligand binding to two proteins stabilizes their interaction and promotes enzyme activity. This is the first example of a c-di-GMP-mediated process that relies on protein–protein interaction. At low c-di-GMP concentrations, PgaD fails to interact with PgaC and is rapidly degraded. Thus, when cells experience a c-di-GMP trough, PgaD turnover facilitates the irreversible inactivation of the Pga machinery, thereby temporarily uncoupling it from c-di-GMP signalling. These data uncover a mechanism of c-di-GMP-mediated EPS control and provide a frame for c-di-GMP signalling specificity in pathogenic bacteria.
Keywords: biofilm, c-di-GMP, glycosyltransferase, poly-GlcNAc, signalling
Introduction
Most bacteria are able to switch from a motile planktonic ‘lifestyle’ to growth in surface-associated multicellular communities known as biofilms. Within these structures, cells are encased in a self-produced extracellular polymeric matrix that is typically composed of proteinaceous adhesin factors, DNA and exopolysaccharides (EPS) (Branda et al, 2005; Flemming and Wingender, 2010). This complex biofilm structure is known to protect bacteria from antimicrobials, physical stresses and the predation by the host immune system. Bacterial biofilms are often associated with chronic infections and infection relapses causing health problems of growing importance (Costerton et al, 1999; Mah and O’Toole, 2001; Davies, 2003; Hall-Stoodley et al, 2004; Fux et al, 2005).
The second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) plays a central role in integrating environmental and cellular cues to control this major bacterial ‘lifestyle’ transition by disfavouring single cell behaviour and by promoting biofilm formation. C-di-GMP is synthesized from GTP by diguanylate cyclases (DGCs) that harbour a conserved GGDEF domain (Paul et al, 2004) and is degraded to the linear dinucleotide pGpG by specific phosphodiesterases (PDEs) that harbour either a conserved EAL (Christen et al, 2005) or HD-GYP domain (Ryan et al, 2006; Hengge, 2009; Schirmer and Jenal, 2009). While DGCs and PDEs have been analysed in detail, both structurally and functionally, little is known about how c-di-GMP acts on downstream targets. Only a few c-di-GMP-specific receptor protein families have been described up to now, for most of which mechanistic details are lacking (Lee et al, 2007; Merighi et al, 2007; Christen et al, 2007; Duerig et al, 2009; Newell et al, 2011; Sondermann et al, 2011).
In Escherichia coli, c-di-GMP regulates several cellular processes including EPS production, the biogenesis of fimbriae, flagellar-based motility and RNA degradation (Pesavento et al, 2008; Monteiro et al, 2009; Boehm et al, 2009; Tagliabue et al, 2010; Boehm et al, 2010; Paul et al, 2010; Fang and Gomelsky, 2010; Tuckerman et al, 2011; Povolotsky and Hengge, 2012). To colonize surfaces, E. coli produces the EPS poly-β-1,6-N-acetylglucosamine (poly-GlcNAc) (Wang et al, 2004). This linear homopolymer was implicated in biofilm formation in a wide variety of pathogenic bacteria including Staphylococcus spp. and Yersinia pestis, where it can promote virulence and contribute to survival in the animal host (Maira-Litrán et al, 2005; O’Gara, 2007; Cerca et al, 2007; Izano et al, 2007, 2008; Bobrov et al, 2008; Choi et al, 2009; Becker et al, 2009; Conover et al, 2010; Pérez-Mendoza et al, 2011; Yakandawala et al, 2011; Bentancor et al, 2012; Skurnik et al, 2012).
In E. coli, poly-GlcNAc is synthesized and secreted by the envelope-spanning Pga machinery (Figure 1A), which is encoded by the pgaABCD operon (Wang et al, 2004). While PgaA and PgaB are required for poly-GlcNAc export, PgaC and PgaD are necessary for poly-GlcNAc synthesis (Figure 1A; Itoh et al, 2008). PgaA is an outer membrane porin that serves to translocate growing poly-GlcNAc chains to the cell surface (Itoh et al, 2008). PgaB is a putative outer membrane lipoprotein that deacetylates about 3% of the GlcNAc residues during poly-GlcNAc export (Wang et al, 2004; Itoh et al, 2008). PgaC is a processive β-glycosyltransferase (GT) of the GT-2 family that is located in the inner membrane and polymerizes poly-GlcNAc from activated UDP-GlcNAc precursor (Saxena and Brown, 1997; Wang et al, 2004; Itoh et al, 2008). The catalytic domain of GT-2 family members is exposed to the cytoplasm (Heldermon et al, 2001; Ciocchini et al, 2006; Bobrov et al, 2008) with sugar transfer through the cytoplasmic membrane being independent of an undecaprenyl phosphate lipid carrier (Gerke et al, 1998). Finally, PgaD is a small protein with two predicted N-terminal transmembrane helices. Its function is unknown and it does not show any obvious similarity to other protein families or domains. However, because PgaD is essential for poly-GlcNAc synthesis (Wang et al, 2004), it was suggested to assist the GT in polymerizing poly-GlcNAc (Itoh et al, 2008).
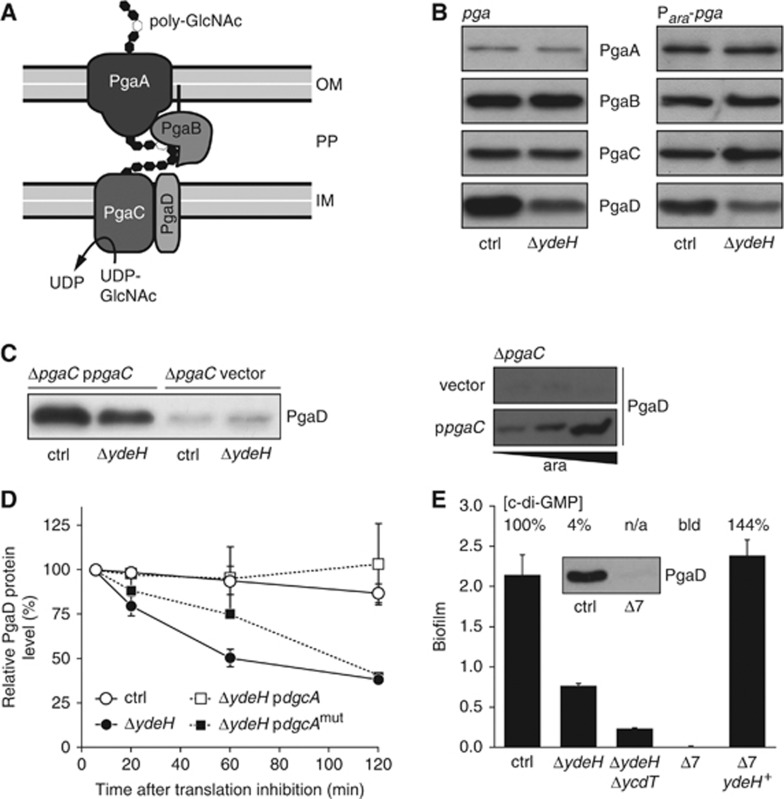
Figure 1.
C-di-GMP controls PgaD stability in a PgaC-dependent manner. (A) Schematic representation of the E. coli Pga machinery. See text for details. IM, inner membrane; PP, periplasm; OM, outer membrane. (B) Immunoblot analysis of 3 × Flag-tagged Pga proteins in the E. coli control strain and ΔydeH mutant. The native pga promoter (left panel) was replaced with the Para promoter (right panel). Expression of the araB-pgaA translational fusion was induced with 0.0002% L-arabinose. (C) PgaD levels depend on PgaC and c-di-GMP. Immunoblots of PgaD–3 × Flag are shown for the indicated mutant strains. Expression of pgaC was induced with 0.0002% L-arabinose (left panel) and with 0, 0.0002 and 0.2% L-arabinose (right panel). (D) Graph showing relative PgaD levels upon blocking protein biosynthesis in exponentially growing cells as an average of two independent experiments with standard deviations. Expression of the heterologous DGC dgcA and its active site mutant dgcAmut (D164N) was not induced (leaky expression). (E) Biofilm formation of strains carrying multiple deletions in genes predicted to encode DGCs. The Δ7 strain carries a total of seven deletions (ΔydeH, ΔycdT, ΔyegE, ΔyfiN, ΔyhjK, ΔydaM, ΔyneF). Error bars are standard deviations of the mean of 3–6 replicas. A representative data set (n=2) of the relative cellular c-di-GMP concentrations of the strains is indicated. n/a, not available; bld, below limit of detection. Inset: Immunoblot of PgaD–3 × Flag in the control strain and the Δ7 mutant. Figure source data can be found in Supplementary data.
The expression of the E. coli pgaABCD operon is tightly regulated on multiple levels. Most importantly, pgaABCD translation is repressed by the action of the RNA binding protein CsrA (carbon storage regulator A) (Wang et al, 2005). This global regulator, whose activity is firmly regulated via a complex signal transduction cascade, antagonistically controls numerous cellular pathways. For example, it promotes motility, glycolysis and virulence, while repressing EPS production and gluconeogenesis (Romeo et al, 1993; Suzuki et al, 2006; Timmermans and Van Melderen, 2010; Romeo et al, 2012). In addition, CsrA inhibits the expression of ydeH and ycdT, two genes encoding DGCs (Jonas et al, 2008). The observation that YdeH stimulates poly-GlcNAc-dependent biofilm formation (Boehm et al, 2009) argued that the expression of this DGC and its target, the Pga machinery, is coupled via CsrA. YdeH and c-di-GMP were shown to control poly-GlcNAc biogenesis on a post-transcriptional level (Boehm et al, 2009), but the mechanism responsible for this induction is unknown.
In this paper, we unravel a novel allosteric mechanism through which c-di-GMP stimulates poly-GlcNAc-dependent biofilm formation in E. coli. We show that c-di-GMP allosterically activates the PgaCD GT complex. We present genetic and biochemical evidence arguing that c-di-GMP binds to both inner membrane components of the Pga machinery, thereby mediating their productive interaction and the formation of an active GT complex. Finally, we demonstrate that in the absence of c-di-GMP PgaD is rapidly degraded, offering the means to shut-off the Pga machinery in response to a c-di-GMP trough and to temporarily uncouple it from c-di-GMP signalling in the absence of de novo synthesis of Pga components. These studies offer a molecular frame for the widespread c-di-GMP-based activation of bacterial EPS systems and provide the basis for signalling specificity of c-di-GMP-controlled systems.
Results
PgaD in vivo stability depends on c-di-GMP
We have previously shown that PgaD steady-state protein levels are positively controlled by c-di-GMP on a post-transcriptional level (Boehm et al, 2009). This observation was used as an entry point to address the molecular mechanism of c-di-GMP-regulated poly-GlcNAc biogenesis. To mimic the induced state of the Csr regulon, all assays were done in a partial loss-of-function csrA::Tn5 mutant strain background (Romeo et al, 1993), which will be referred to as control strain throughout this work. In order to monitor all Pga complex components individually, 3 × Flag-tagged versions of PgaA, PgaB, PgaC and PgaD were constructed. In the absence of the DGC YdeH, the protein levels of PgaD were reduced, while the levels of the other three Pga proteins remained constant, regardless of whether the pga operon was expressed from its native promoter with the 5′ UTR of pgaA or from the L-arabinose-dependent Para promoter with the 5′ UTR of araB (Figure 1B; Supplementary Figure 1A). Moreover, PgaD levels were strongly reduced in a ΔpgaC mutant, but were restored in a c-di-GMP-dependent manner when pgaC was expressed in trans and were further increased upon overexpression of pgaC (Figure 1C). PgaD levels were still c-di-GMP-dependent in cells expressing a pgaC active site mutant (D256N), arguing that PgaC protein but not PgaC GT activity is required to stabilize PgaD (Supplementary Figure 1B). Finally, expression of the heterologous DGC dgcA (Christen et al, 2006) strongly elevated PgaD levels in a ΔydeH mutant, but only when pgaC was present (Supplementary Figure 1C).
The above data indicated that PgaC and c-di-GMP together control PgaD levels post-translationally. To substantiate this and to demonstrate that the effect is specific for PgaD, pgaD was replaced with yfiR, an unrelated gene from Pseudomonas aeruginosa. The observation that YfiR levels failed to fluctuate in response to c-di-GMP availability excludes the possibility that PgaD levels respond to a c-di-GMP-controlled promoter or to translation initiation control elements within pgaABC (Supplementary Figure 1D). Next, in vivo protein stability of PgaD–3 × Flag was determined under different c-di-GMP concentrations upon blocking de novo protein biosynthesis in exponentially growing cells. While PgaD remained stable over time in strains with normal or increased c-di-GMP levels (control strain and ΔydeH mutant expressing dgcA), the protein was rapidly degraded in strains with low cellular c-di-GMP concentrations (ΔydeH mutant and ΔydeH mutant expressing an active site mutant of dgcA) (Figure 1D; Supplementary Figure 1E).
In summary, these data suggest that c-di-GMP positively modulates PgaD protein stability in a PgaC-dependent manner.
C-di-GMP and PgaD together promote poly-GlcNAc-dependent biofilm formation
The E. coli csrA::Tn5 mutant strain (control strain) forms biofilms under laboratory conditions that fully depend on the EPS adhesin poly-GlcNAc (Wang et al, 2004). To test if c-di-GMP is essential for poly-GlcNAc-dependent biofilm formation, multiple genes coding for potential DGCs (each containing a GGDEF domain) were successively deleted. Concomitant deletions of the two CsrA-controlled genes ydeH and ycdT (Jonas et al, 2008) resulted in a drastic reduction of biofilm formation, while a strain carrying a total of seven deletions (ΔydeH, ΔycdT, ΔyegE, ΔyfiN, ΔyhjK, ΔydaM, ΔyneF) completely lost the ability to form biofilms (Figure 1E). This strain showed a strongly reduced cellular c-di-GMP level in comparison to the control strain (Figure 1E) and will be referred to as Δ7 strain throughout this work. Importantly, both biofilm deficiency and c-di-GMP level could be complemented by reintroducing only ydeH into the bacterial genome (Figure 1E), supporting the idea that YdeH represents the major DGC responsible for poly-GlcNAc induction under these conditions (Boehm et al, 2009). In line with the data described above, PgaD protein was not detectable in the Δ7 mutant (Figure 1E). While c-di-GMP is required for normal PgaD levels under physiological conditions, overexpression of pgaD resulted in a biofilm induction both in the presence and in the absence of YdeH (Supplementary Figure 1F). However, the ΔydeH mutant never reached the same level of biofilm formation as the control strain, arguing that PgaD and c-di-GMP are synergistically needed for optimal biofilm formation.
C-di-GMP enhances PgaC–PgaD interaction
One scenario that could explain PgaC-dependent PgaD stability is a direct interaction of the two membrane proteins. Co-immunoprecipitation experiments using detergent-solubilized membranes revealed that PgaC and PgaD indeed form a stable complex that was resistant to high salt concentrations and up to 2 M urea (Figure 2A). When overexpressed, PgaC and PgaD could be co-purified even from membranes of a Δ7 strain (Figure 2B), arguing that under these conditions c-di-GMP is no longer required for PgaD stability. Together, this suggested that PgaC and PgaD form a stable complex in the cytoplasmic membrane, the formation of which is mediated by c-di-GMP under physiological conditions.
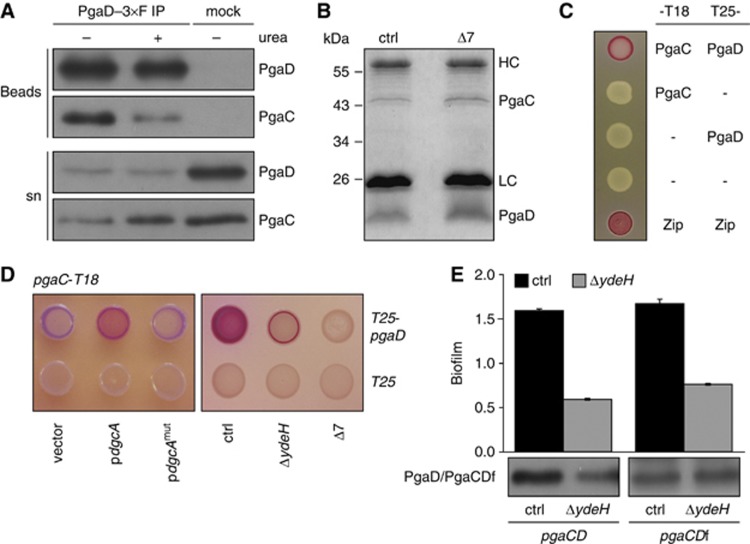
Figure 2.
C-di-GMP enhances PgaC–PgaD interaction. (A) PgaC–6 × His and PgaD–3 × Flag co-immunoprecipitate from detergent-solubilized membranes. Anti-Flag and protein A (mock) IPs were analysed by immunoblots using antibodies against the specific tags. The protein fraction that failed to bind to the beads is indicated (sn, supernatant). 2 M urea was present during the IP procedure as indicated. (B) Co-immunoprecipitation of PgaC and PgaD–3 × Flag from detergent-solubilized membranes of control strain and Δ7 mutant cells overexpressing pgaC and pgaD. IP samples were analysed by Coomassie staining. HC and LC mark heavy and light chains of IgG. (C) Bacterial two-hybrid (BacTH) analysis of PgaC–PgaD interaction. Presence of T18 and T25 fusions is indicated. Zip indicates the leucine zipper positive control. (D) BacTH analysis of c-di-GMP-stimulated PgaC–PgaD interaction. Left panel: Interaction in the presence of a plasmid-borne copy of dgcA or its active site mutant dgcAmut (D164N). Alleles were induced with 0.2% L-arabinose. Right panel: Interaction in strains lacking the DGC YdeH or multiple DGCs (Δ7). See Supplementary Figure 2 for the quantification of interaction strengths. (E) A PgaCD fusion protein is fully functional. Biofilm formation and protein levels of 3 × Flag-tagged PgaD or PgaCD fusion protein (PgaCDf) are indicated for the control strain (black bars) and a ΔydeH mutant (grey bars). Error bars are standard deviations of the mean of 3–6 replicas. Figure source data can be found in Supplementary data.
To test if c-di-GMP is involved in PgaC–PgaD interaction, a bacterial two-hybrid (BacTH) assay was used that is based on the interaction-mediated reconstitution of the split cAMP signalling pathway in E. coli (Karimova et al, 1998). In this assay, full-length PgaC and PgaD showed a robust interaction (Figure 2C), while all truncated variants (e.g., predicted cytosolic parts) were negative (Supplementary Table 2). The interaction was stimulated by the ectopic expression of the heterologous DGC dgcA (Figure 2D; Supplementary Figure 2). Conversely, a step-wise reduction of the cellular c-di-GMP pool gradually lowered the interaction strength. PgaC–PgaD interaction was weakened upon deletion of ydeH and abolished in the Δ7 strain (Figure 2D; Supplementary Figure 2). These data further support the idea that c-di-GMP stimulates PgaC–PgaD interaction or complex stability.
The above results can be interpreted in two different ways. C-di-GMP could regulate poly-GlcNAc production by determining PgaD stability and availability. Alternatively, c-di-GMP could promote PgaC–PgaD interaction with PgaD instability and degradation being a consequence of complex disintegration at low c-di-GMP concentrations. To be able to distinguish between these two possibilities, PgaD was ‘stabilized’ under low c-di-GMP conditions by directly fusing its N-terminus to the C-terminus of PgaC. Surprisingly, the resulting pgaCD fusion construct (pgaCDf) was fully functional and able to complement biofilm formation of a ΔpgaCD mutant in a c-di-GMP-dependent manner (Figure 2E). But in contrast to PgaD, the level of the PgaCD fusion protein (PgaCDf) was unaltered in a strain with lower c-di-GMP concentrations (Figure 2E). These findings reinforce the notion of a direct interplay between PgaC and PgaD and imply that PgaD instability at low c-di-GMP levels is not the cause for Pga control, but may simply result from weak protein interactions under these conditions. These data raise the question why the homologues of PgaC and PgaD exist as two separate proteins in all bacteria harbouring this EPS biogenesis system (see below).
C-di-GMP acts as an allosteric activator of PgaCD GT activity
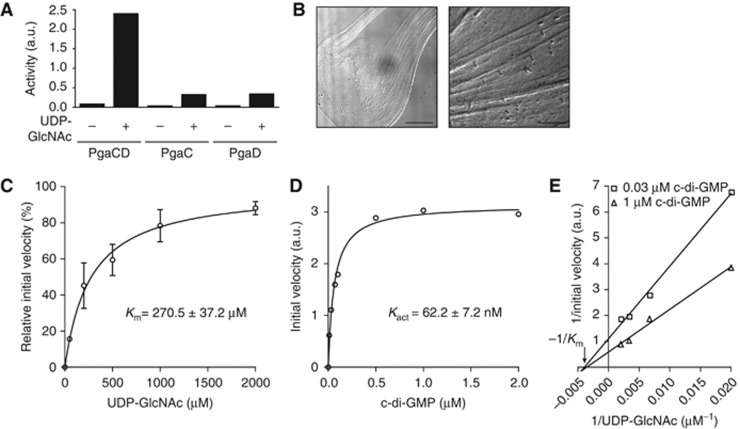
In order to test whether c-di-GMP acts as an allosteric activator for the PgaCD GT complex, an in vitro activity assay was developed with membranes containing PgaCD. GT activity was determined indirectly using a modified enzyme-coupled spectrophotometric assay (Baykov et al, 1988) or directly by measuring UDP-GlcNAc consumption. In agreement with earlier data demonstrating that both PgaC and PgaD are needed for poly-GlcNAc synthesis in vivo (Wang et al, 2004; Itoh et al, 2008), UDP-GlcNAc was only turned over to poly-GlcNAc and UDP by membranes of cells expressing pgaC and pgaD (Figure 3A). Following incubation of active membranes with substrate for several hours, a slimy and viscous reaction product was visualized by light microscopy (Figure 3B). Immunoblot analysis with an anti-poly-GlcNAc antibody confirmed the identity of the reaction product (Supplementary Figure 3A). Experiments to determine the substrate affinity of the PgaCD GT complex revealed a Km for UDP-GlcNAc of 270.5±37.2 μM (Figure 3C). To test if PgaCD GT activity is stimulated by c-di-GMP, initial reaction velocities were measured at varying c-di-GMP concentrations in the presence of a constant UDP-GlcNAc concentration of 50 μM. Under these conditions, c-di-GMP stimulated GT activity >20-fold and curve fitting indicated a c-di-GMP concentration for half-maximal initial velocity (Kact) of 62.2±7.2 nM (Figure 3D). This induction was highly specific as the addition of GTP failed to activate the enzyme and furthermore, the c-di-GMP-mediated activity was fully dependent on the PgaCD machinery (Supplementary Figure 3B). The basal enzymatic GT activity in the absence of exogenously added c-di-GMP correlated with the cellular c-di-GMP concentration of the strain used for pgaCD overexpression and membrane preparation. Almost no basal activity was detected for membranes originating from the Δ7 mutant (Supplementary Figure 3B). A Lineweaver-Burk plot analysis integrating initial reaction velocity data at different UDP-GlcNAc concentrations in the presence of a non-saturating and a saturating c-di-GMP concentration resulted in fitted lines converging close to the x axis, indicating that c-di-GMP affects the Vmax rather than the Km of the enzyme complex (Figure 3E).
Figure 3.
C-di-GMP allosterically stimulates PgaCD glycosyltransferase activity in vitro. (A) GT activity depends on an intact PgaCD complex. Enzyme activities were determined using control strain membranes containing PgaC, PgaD or both proteins in the presence (2 mM) or absence of the substrate UDP-GlcNAc. A representative data set of two independent experiments is shown. (B) Microscopic analysis of the viscous poly-GlcNAc reaction product. Membranes were incubated with 30 mM UDP-GlcNAc for 5 h at 30°C. Scale bars are indicated: 15 μm. (C) Determination of the PgaCD Km for UDP-GlcNAc. Membranes of a Δ7 mutant containing PgaC and PgaD were incubated with increasing concentrations of UDP-GlcNAc in the presence of 1 μM c-di-GMP. Data represent an average of two independent experiments with standard deviations. (D) Stimulatory effect of c-di-GMP on PgaCD GT activity (Kact). Membranes of a Δ7 mutant containing PgaC and PgaD were incubated with increasing concentrations of c-di-GMP in the presence of 50 μM UDP-GlcNAc. A representative data set of two independent experiments is shown. (E) Lineweaver-Burk plot analysis of PgaCD GT activity. Membranes of a Δ7 mutant containing PgaC and PgaD were incubated with increasing concentrations of UDP-GlcNAc in the presence of a non-saturating (0.03 μM) and a saturating (1 μM) c-di-GMP concentration. Negative reciprocal Km is indicated. A representative data set of two independent experiments is shown. GraphPad Prism was used for curve fitting and linear regression. a.u., arbitrary unit.
In summary, these data strongly suggest that c-di-GMP acts as a direct allosteric activator of the PgaCD GT complex.
Concomitant binding of c-di-GMP to both PgaC and PgaD
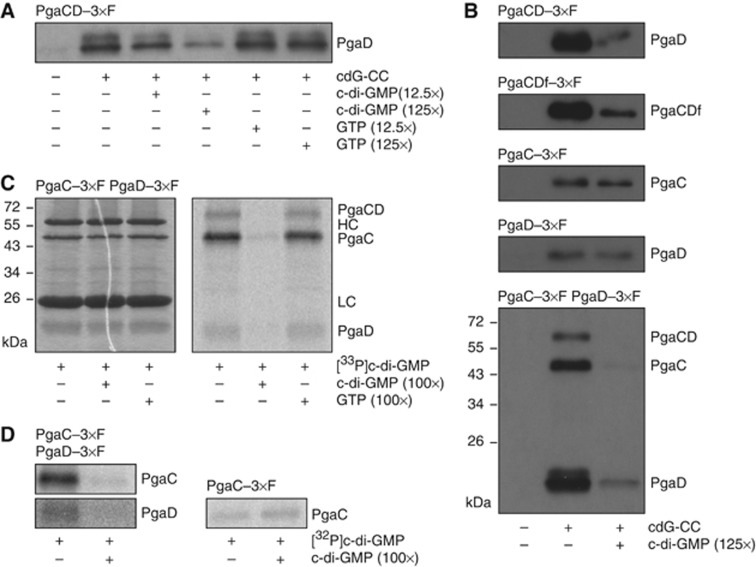
The above in vitro assays argued for a direct role of c-di-GMP as an allosteric activator of PgaCD GT activity. To corroborate these findings, c-di-GMP binding to the PgaCD complex was tested by using a c-di-GMP capture compound (cdG-CC). This molecule consists of a c-di-GMP moiety that is asymmetrically modified at the 2′ hydroxyl of one ribose with a linker connecting to a photo-reactive and a biotin sorting group (Nesper et al, 2012). The PgaCD complex was specifically and competitively captured by the cdG-CC from membrane preparations (Figure 4A). An excess of c-di-GMP, but not GTP, gradually competed with cdG-CC binding. While the PgaCD complex and the PgaCD fusion protein were specifically pulled down, no specific binding was observed when membranes were used that only contained PgaC or PgaD (Figure 4B). Although some residual binding to the cdG-CC was observed under these conditions, the addition of an excess of c-di-GMP failed to compete with this interaction (Figure 4B). When membranes were used that contained 3 × Flag-tagged variants of both PgaC and PgaD, both proteins showed specific cdG-CC binding. A fraction of the PgaC–PgaD heterodimers withstood boiling in SDS sample buffer and appeared as a distinct band on the immunoblot, emphasizing the remarkable stability of these complexes (Figure 4B). Probing cdG-CC samples with an antibody against the biotin moiety of the capture compound revealed that the cdG-CC was covalently crosslinked to both PgaC and PgaD in a competitive way, suggesting that c-di-GMP is able to directly interact with both components of the complex (Supplementary Figure 4A and B).
Figure 4.
Specific binding of c-di-GMP requires PgaC and PgaD. (A) Immunoblot of PgaD captured from membranes containing PgaC and PgaD–3 × Flag. Presence of cdG-CC and competing nucleotides is indicated. (B) Immunoblots of PgaC, PgaD and PgaCD fusion protein (PgaCDf) captured from membranes containing PgaC and PgaD–3 × Flag (first panel), PgaCDf–3 × Flag (second panel), PgaC–3 × Flag (third panel), PgaD–3 × Flag (fourth panel) or PgaC–3 × Flag and PgaD–3 × Flag (fifth panel). Presence of cdG-CC and competing nucleotides is indicated. SDS-resistant heterodimeric PgaCD complexes are indicated (PgaCD). (C) Specific labelling of PgaC and PgaD with [33P]c-di-GMP. Membranes containing PgaC–3 × Flag and PgaD–3 × Flag were UV crosslinked in the presence of [33P]c-di-GMP and competing nucleotides as indicated. Coomassie staining (left panel) and autoradiography (right panel) are shown. HC and LC mark heavy and light chains of IgG. SDS-resistant heterodimeric PgaCD complexes are indicated (PgaCD). (D) Absence of PgaD abolishes c-di-GMP binding. Membranes containing PgaC–3 × Flag and PgaD–3 × Flag (left panels) or PgaC–3 × Flag (right panel) were UV crosslinked in the presence of [32P]c-di-GMP and competing nucleotides as indicated. Only autoradiographies are shown. Figure source data can be found in Supplementary data.
To corroborate these findings, UV light-induced crosslinking experiments with radiolabelled c-di-GMP were performed (Christen et al, 2006). In good agreement with the data obtained with the capture compound, PgaC and PgaD were specifically and competitively labelled with [33P]c-di-GMP when both proteins were present in the membrane fraction (Figure 4C). An excess of c-di-GMP, but not GTP, efficiently outcompeted the [33P]c-di-GMP crosslink to both proteins. It is interesting to note that PgaC labelling was generally much stronger than PgaD labelling. Again, specific c-di-GMP binding and radiolabelling was only observed in membranes containing both proteins, but was lost for PgaC when PgaD was not present (Figure 4D). Interestingly, the presence of the substrate UDP-GlcNAc increased the specific binding of c-di-GMP, indicating some form of communication between the GT active site and the allosteric c-di-GMP binding pocket within the PgaCD complex (Supplementary Figure 4C and D).
Altogether, these data suggest that the PgaCD GT complex represents a novel type c-di-GMP receptor, where ligand binding to two individual proteins promotes their stable interaction and subsequent activation.
Constitutive mutations in pgaD uncouple PgaCD activity from c-di-GMP
To more closely define the c-di-GMP binding site in PgaD, variants with C-terminal truncations were analysed for their ability to stimulate biofilm formation. Although biofilm formation gradually decreased with deletions extending towards the second transmembrane helix, c-di-GMP stimulation was sustained in truncations extending to amino acid R78 (Figure 5A and B). This argued that c-di-GMP binds to a region within the first 78 amino acids of PgaD consisting of only two transmembrane helices with short flanking regions in the cytoplasm, thus suggesting that c-di-GMP modulates the interaction of PgaC and PgaD in the vicinity of the cytoplasmic membrane. To test this hypothesis, we set up a genetic screen to isolate mutations in pgaC and pgaD that facilitate biofilm formation in the absence of c-di-GMP. Error-prone PCR mutagenesis and screening for biofilm-forming colonies in the Δ7 strain using Congo Red agar plates led to the isolation of several constitutive mutants (Supplementary Table 3). With one exception, all mutations in PgaD clustered within a short conserved region between the second transmembrane helix and residue R78 (Figure 5A). Two of the activating pgaD alleles (N75D,K76E and L73Q,K76E,R78C) firmly locked biofilm formation at an intermediate level independently of the availability of c-di-GMP (Figure 5C). In both cases, this constitutive phenotype required the presence of multiple mutations with single changes showing no or little effect (Figure 5C). While the N75D,K76E mutant completely failed to respond to c-di-GMP, the L73Q,K76E,R78C allele retained some residual induction upon ectopic expression of a heterologous DGC (Supplementary Figure 5A). Interestingly, protein levels of both constitutive PgaD mutants were increased in the Δ7 strain, but in contrast to wild-type PgaD they showed no significant response to changes in cellular c-di-GMP concentration (Figure 5D). The stability of these mutant forms was still dependent on the presence of PgaC (data not shown).
Figure 5.
Mutations in PgaD render the PgaCD complex constitutively active and independent of c-di-GMP. (A) Predicted topology of PgaD. Positions of c-di-GMP-independent (orange) and loss-of-function mutations (red) within the most conserved region of PgaD (grey) are indicated. Sites of C-terminal PgaD truncations are marked by triangles. IM, inner membrane; PP, periplasm. Transmembrane helices were predicted using the TMHMM server (Sonnhammer et al, 1998). (B) Biofilm formation of strains expressing C-terminally truncated pgaD alleles as a function of cellular c-di-GMP concentrations. The last residue of each mutant is indicated (A). Δ7 strains harbouring individual pgaD alleles contained plasmids with an IPTG-inducible copy of the heterologous DGC wspR (pwspR) or control plasmids (vector). Expression of plasmid-borne pgaD alleles was induced with 0.2% (left graph) and 0.02% L-arabinose (right graph). Error bars are standard deviations of the mean of 3–6 replicas. (C) Contribution of pgaD mutants to biofilm formation is shown in the control strain (black bars) and the Δ7 mutant (grey bars). Isolated constitutive alleles are underlined. Error bars are standard deviations of the mean of 3–6 replicas. (D) Immunoblot analysis of steady-state levels of wild-type and mutant forms of PgaD–3 × Flag in the control strain and the Δ7 mutant. (E) C-di-GMP-dependent PgaCD GT activity. Membranes of a Δ7 mutant containing either PgaD wild-type or mutant forms were incubated with (black bars) or without c-di-GMP (grey bars) in the presence of 300 μM UDP-GlcNAc. PgaD mutant variants were expressed as PgaCD fusion proteins. A representative data set of two independent experiments is shown with standard errors. a.u., arbitrary unit. (F) The PgaD mutant N75D,K76E is strongly impaired in c-di-GMP binding. Relative amounts of PgaD wild-type or mutant forms captured in the presence (black bars) or absence of excess c-di-GMP (grey bars) are shown as an average of two independent experiments with standard deviations. PgaD variants were expressed as PgaCD fusion proteins. Figure source data can be found in Supplementary data.
Next, the behaviour of the PgaD mutant forms was assayed in the in vitro GT activity assay. To avoid possible stoichiometry problems arising from different overall levels of PgaD, assays were performed with normalized protein levels of the PgaCD fusion protein (Supplementary Figure 5B). Both mutant proteins showed a >3-fold increased basal GT activity in the absence of exogenously added c-di-GMP and could not be stimulated further by the addition of 100 nM c-di-GMP, a concentration that causes approximately half-maximal activation of the wild-type enzyme (Figures 3D and 5E). These data suggested that constitutive PgaD mutants are able to interact with and stimulate PgaC in the absence of c-di-GMP, thereby uncoupling the PgaCD complex from c-di-GMP signalling. To test if these mutants still bind the allosteric ligand in vitro, cdG-CC experiments were performed in the context of the PgaCD fusion protein. Consistent with the data described above, the N75D,K76E mutant almost completely failed to bind the cdG-CC, while the pull-down of the L73Q,K76E,R78C mutant was severely reduced (Figure 5F). These experiments demonstrate that specific mutations in the conserved region of PgaD abolish c-di-GMP binding and at the same time mimic a c-di-GMP-bound state that activates the PgaCD GT complex.
Two conserved residues of PgaD located within the same region, W71 and Y74, were previously shown to be important for the function of the PgaD homologue of Y. pestis (Forman et al, 2006). While the Y74A mutation did not affect E. coli PgaD function, the W71A mutation resulted in an almost complete loss of biofilm formation (Figure 5C). Importantly, while W71 was not required for cdG-CC binding (Figure 5F), the W71A mutation was dominant over the constitutive allele N75D,K76E (Supplementary Figure 5C). This argues that W71 resides downstream of the c-di-GMP-mediated activation in the PgaD signal transduction process.
Constitutive mutations in pgaC influence PgaD protein levels
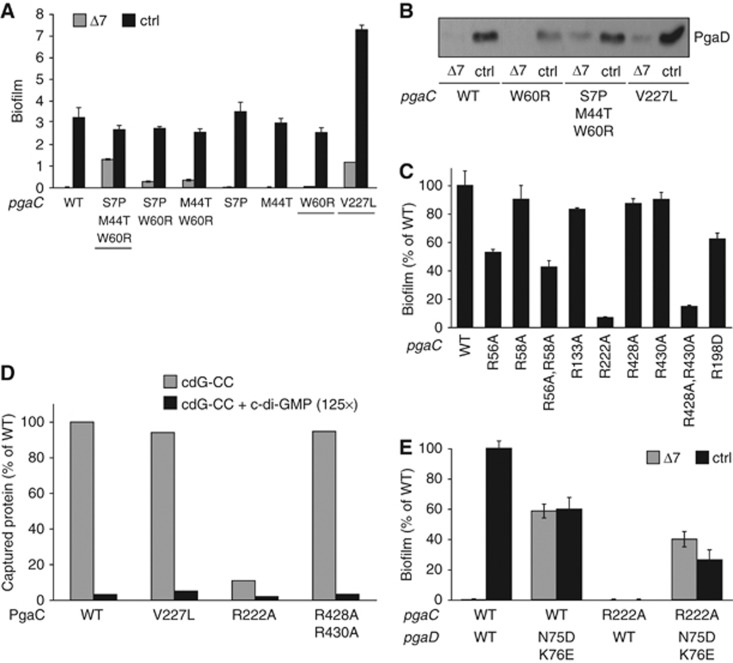
In contrast to the constitutive pgaD mutants, all activating mutations isolated in pgaC retained some level of c-di-GMP stimulation (Figure 6A; Supplementary Table 3). Moreover, they all still depended on the presence of PgaD for biofilm formation (data not shown). In case of the pgaC S7P,M44T,W60R allele, the combination of three mutations contributes to the high level of biofilm formation in a Δ7 strain (Figure 6A). In contrast, the single mutation V227L strongly upregulated biofilm formation both in the Δ7 strain and in a strain expressing diguanylate cyclases (Figure 6A). Co-expression of the pgaC V227L allele with the constitutive pgaD N75D,K76E mutant increased biofilm formation up to the fully induced level observed for the V227L single mutant, even when c-di-GMP was absent (Supplementary Figure 6A). These data indicate that PgaC V227L partially uncouples PgaCD GT activity from c-di-GMP, while the PgaD mutant N75D,K76E has a strong dominant effect that fully releases the PgaCD complex from its c-di-GMP dependency. Because PgaC and c-di-GMP are required for PgaD stability in vivo (see above), we hypothesized that constitutive pgaC mutants should lead to enhanced PgaD levels in the absence of c-di-GMP. As shown in Figure 6B, PgaD was markedly stabilized in Δ7 strains expressing either the triple pgaC mutant S7P,M44T,W60R or the single V227L allele. This further substantiates the idea that c-di-GMP stimulation primarily affects PgaC–PgaD interaction, while PgaD stability is merely a consequence of the allosteric control of the Pga machinery.
Figure 6.
A constitutive PgaD mutant rescues a PgaC mutant unable to bind c-di-GMP. (A) Constitutive pgaC mutants show partial c-di-GMP independence. Contribution of pgaC mutants to biofilm formation is shown in the control strain (black bars) and the Δ7 mutant (grey bars). Isolated constitutive alleles are underlined. Error bars are standard deviations of the mean of 3–6 replicas. (B) Immunoblot analysis of PgaD–3 × Flag in the control strain and the Δ7 mutant expressing different pgaC alleles. (C) Mutational analysis of conserved arginine residues of PgaC (see text and Supplementary Figure 6D). Biofilm formation was determined for strains expressing the respective PgaC variants as PgaCD fusion proteins. R198D was included as a control as this arginine is also conserved in organisms that lack c-di-GMP. Error bars are standard deviations of the mean of 3–6 replicas. (D) The PgaC R222A mutant is strongly impaired in c-di-GMP binding. A representative data set (n=2) of the relative amounts of PgaC wild-type or mutant forms captured in the presence (black bars) or absence of excess c-di-GMP (grey bars) is shown. PgaC variants were expressed as PgaCD fusion proteins. (E) The constitutive PgaD N75D,K76E mutant rescues the PgaC R222A mutant deficient in c-di-GMP binding. Biofilm formation was determined for strains expressing different pgaC and/or pgaD alleles in the control strain (black bars) and the Δ7 mutant (grey bars). Error bars are standard deviations of the mean of 3–6 replicas. Figure source data can be found in Supplementary data.
R222 of PgaC plays an essential role in c-di-GMP-dependent PgaCD activation
In order to identify regions of PgaC involved in c-di-GMP binding, we focussed on arginines as they were shown to play a critical role in c-di-GMP binding (Benach et al, 2007; Habazettl et al, 2011). To identify conserved arginines potentially involved in c-di-GMP binding, PgaC sequences from Gram-negative bacteria harbouring genes encoding GGDEF and EAL domain proteins were compared to PgaC sequences from Gram-negative organisms lacking c-di-GMP (no GGDEF domain proteins) (Supplementary Figure 6D). Based on this analysis, the following six residues, which are only conserved in species with GGDEF domains, were selected and changed to alanines individually or in combination: R56, R58, R133, R222, R428 and R430. Two alleles, R222A and R428A,R430A, were identified that produced normal protein levels in vivo (Supplementary Figure 6B), but almost completely failed to support biofilm formation (Figure 6C). The R222A but not the R428A,R430A mutant also showed a strong binding defect for the cdG-CC (Figure 6D). In agreement with a specific role for R222 in c-di-GMP binding, cells expressing the pgaC R222A allele were unable to stabilize PgaD. In contrast, PgaD was stabilized by the PgaC GT active site mutant (D256N) in a c-di-GMP-dependent manner (Supplementary Figure 6C). Most importantly, when co-expressed with the constitutive pgaD allele N75D,K76E, the PgaC R222A function was restored (Figure 6E). This underscores the tight interplay between PgaC and PgaD and demonstrates that the R222A mutation does not cause a general loss of PgaC activity, but rather specifically affects c-di-GMP binding and GT activation.
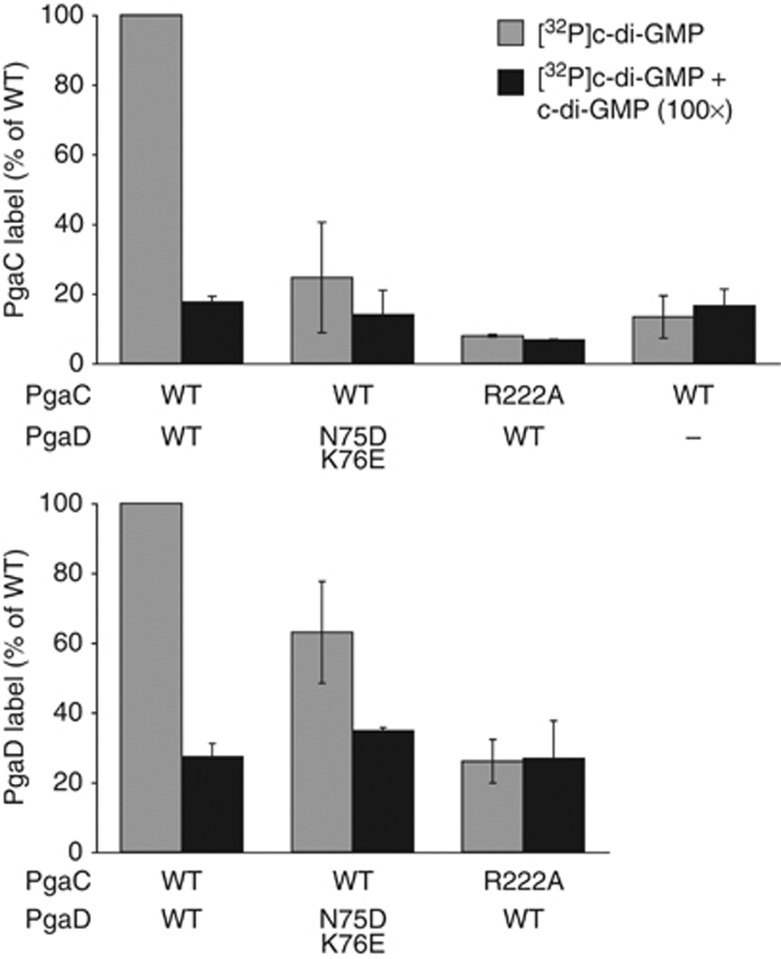
Together, these data suggested a critical role for R222 of PgaC in the c-di-GMP-dependent activation of the PgaCD GT complex and implied that R222 is directly involved in c-di-GMP binding. To test this, UV light-induced crosslinking experiments with radiolabelled c-di-GMP were performed. As shown in Figure 7, both PgaC and PgaD specifically and competitively incorporated radiolabelled c-di-GMP when present in wild-type GT complexes. In contrast, GT complexes containing either PgaD N75D,K76E or PgaC R222A were strongly impaired in c-di-GMP binding. In both cases, the total amount of crosslinked [32P]c-di-GMP was reduced in PgaC as well as in PgaD (Figure 7; Supplementary Figure 7), arguing that individual binding mutations in PgaC or PgaD affect the overall binding of the complex. This is consistent with the observation that cdG-CC binding to the PgaCD complex was strongly reduced for the PgaD N75D,K76E and the PgaC R222A mutant (Figures 5F and 6D). Altogether, these data strongly support the idea that c-di-GMP binds to both PgaC and PgaD, resulting in the tight interaction and activation of the PgaCD GT complex.
Figure 7.
C-di-GMP directly binds to both PgaC and PgaD. Membranes containing wild-type and mutant forms of PgaC–3 × Flag and/or PgaD–3 × Flag were UV crosslinked in the presence of [32P]c-di-GMP and with (black bars) or without excess c-di-GMP (grey bars). Relative PgaC (upper graph) and PgaD (lower graph) autoradiography band intensities are shown as an average of two independent experiments with standard deviations.
Discussion
To transit from a planktonic, single cell to a biofilm-associated community ‘lifestyle’ bacteria undergo a complex and highly regulated process that is globally coordinated by the ubiquitous bacterial second messenger c-di-GMP (Schirmer and Jenal, 2009; Hengge, 2009). One of the key cellular processes directly stimulated by c-di-GMP is the production and secretion of EPS that serve as protective biofilm matrix. Recently, several c-di-GMP receptor proteins were identified that regulate EPS production (Amikam and Galperin, 2006; Merighi et al, 2007; Lee et al, 2007; Whitney et al, 2012). However, their mode of action has remained elusive. To address the molecular principles of c-di-GMP-induced EPS production, we have chosen the E. coli Pga system primarily for reasons of its relatively simple architecture. The secretion of poly-GlcNAc by the Pga machinery was linked to c-di-GMP signalling earlier (Kirillina et al, 2004; Boehm et al, 2009; Tagliabue et al, 2010; Pérez-Mendoza et al, 2011). However, the molecular mechanisms involved remained unclear and, despite of obvious analogies to other EPS secretion systems, none of the canonical c-di-GMP receptor domains is part of the Pga system.
We showed previously that the Pga system is regulated by c-di-GMP on the post-transcriptional level (Boehm et al, 2009). In this study, we close the gap by demonstrating that c-di-GMP allosterically regulates the PgaCD GT complex in the inner membrane. The PgaCD complex represents a novel type c-di-GMP receptor, in which both membrane-integral proteins contribute to ligand binding, thereby mediating robust interaction, PgaD stabilization and activation of the two partners. This is the first example of a c-di-GMP receptor that relies on protein–protein interaction. Several lines of evidence support these findings. Only a PgaCD complex, but not PgaC or PgaD alone, showed specific and competitive ligand binding. Moreover, UV crosslinking of radiolabelled c-di-GMP consistently and specifically labelled both PgaC and PgaD. Because of the close proximity that is needed for covalent zero-length crosslink formation, this strongly implies that amino acid residues from both proteins participate in the formation of the ligand-binding pocket. The observation that PgaC was incorporating more radioactivity than PgaD could reflect the nature of the c-di-GMP binding pocket, since not all amino acid residues show the same propensity for covalent crosslinking to a nucleotide ligand upon UV light irradiation (Meisenheimer and Koch, 1997). These results strongly argue against the possibility that the c-di-GMP binding pocket is entirely contained within PgaC with PgaD triggering the binding-competent conformation of its partner. Concomitant binding of c-di-GMP to PgaC and PgaD is further supported by genetic evidence. We isolated pgaD alleles that uncoupled the PgaCD complex from c-di-GMP signalling in terms of c-di-GMP binding, allosteric GT activation and biofilm formation. These constitutive mutations cluster within a short, positively charged region proximal to the second membrane-spanning domain of PgaD that likely contributes to c-di-GMP binding. In contrast, none of the activating pgaC alleles showed a completely c-di-GMP-‘blind’ phenotype, emphasizing the important role of PgaD in c-di-GMP-mediated GT activation.
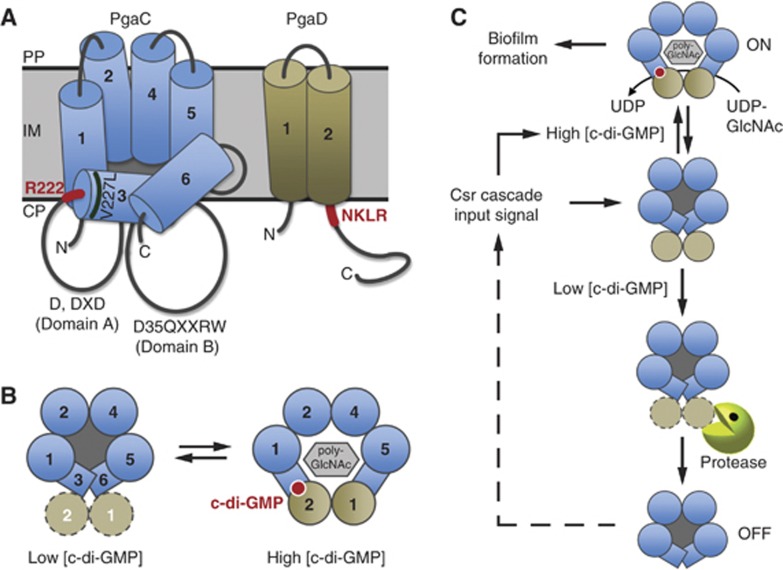
Both in vivo and in vitro data suggest that c-di-GMP is absolutely essential for PgaCD GT activity and poly-GlcNAc-dependent biofilm formation. Our data indicate that c-di-GMP binds to the PgaCD complex with high affinity (Kact=62 nM). Interestingly, c-di-GMP increased the velocity (Vmax) of the GT complex, but not the affinity for its substrate UDP-GlcNAc. This is similar to the findings with cellulose synthase (Aloni et al, 1983; Ross et al, 1987) and implies that UDP-activated sugar molecules are not limiting under conditions that favour EPS synthesis and secretion. This, in turn, is in good agreement with the fact that the Km of PgaCD (270 μM) lies well within the range of reported cellular UDP-GlcNAc concentrations in E. coli (Mengin-Lecreulx et al, 1989; Namboori and Graham, 2008). The strong effect of c-di-GMP on PgaCD activity raises the question of how the second messenger stimulates this enzyme complex. PgaC is a processive β-GT of the GT-2 family, which are thought to function as monomers making use of two active site-containing domains, A and B, for the sugar polymerization reaction (Saxena and Brown, 1997; Tlapak-Simmons et al, 1998; Ciocchini et al, 2006). But how would a growing polysaccharide chain be efficiently transferred across the hydrophobic membrane lipid barrier with as little as four transmembrane domains (TMDs) (Bobrov et al, 2008)? For the Streptococcus hyaluronan synthase, a structural homologue of PgaC, the interaction with cardiolipin molecules was suggested as a solution to this ‘transfer dilemma’ (Tlapak-Simmons et al, 1999). Based on our findings of c-di-GMP-mediated PgaCD complex activation, we propose a central role for PgaD in converting the PgaC GT into a secretion-competent conformation. In our model, c-di-GMP binding to both PgaC and PgaD induces a conformational change that causes the integration of the two transmembrane helices of PgaD into the core of TMDs formed by PgaC. This would convert the loosely associated GT complex into a stable, active and secretion-competent heterodimeric complex by opening up a pore for poly-GlcNAc translocation across the cytoplasmic membrane (Figure 8A and B). The presence of the two membrane-associated domains (MADs) 3 and 6 in the PgaC architecture of our model is based on the membrane topology model determined for the Streptococcus hyaluronan synthase, a homologous protein (Heldermon et al, 2001). In line with this, bioinformatic predictions indicate an increased probability for membrane association of regions 3 and 6 of PgaC. It is thus possible that the c-di-GMP-stimulated interaction between PgaC and PgaD recruits MADs 3 and 6 of PgaC into a secretion-competent transmembrane pore (Figure 8B). The regions in PgaC (R222) and PgaD (NKLR) proposed to be involved in the formation of the c-di-GMP binding site are well positioned to bring together PgaC MAD3 and PgaD TMD2 (Figure 8A and B). Such an arrangement would also explain the strong constitutive effect of the PgaC mutant V227L, as this mutation is located at the N-terminal face of MAD3, in the immediate vicinity of the proposed c-di-GMP binding site (Figure 8A). The formation of a membrane-integral heterodimeric complex as a functional secretion unit is the simplest model to concur with our findings that a PgaCD fusion protein is fully functional, that both proteins are absolutely required for poly-GlcNAc synthesis in vivo and in vitro and that the two TMDs are the critical functional determinants of PgaD. Moreover, the observation that PgaD is strictly required for poly-GlcNAc secretion is in line with a structural requirement for this protein. The association of the GT with a second inner membrane protein essential for its activity seems to be a general phenomenon of homopolymeric EPS secretion systems (Keiski et al, 2010).
Figure 8.
Model for the allosteric activation of the PgaCD glycosyltransferase complex by c-di-GMP. (A) Topology models for PgaC and PgaD in the inner membrane. Orientations of PgaC transmembrane domains (TMDs) are based on this study, on TMHMM server predictions (Sonnhammer et al, 1998), on the proposed topology of the PgaC homologue from Y. pestis (Bobrov et al, 2008) and on a model proposed for the hyaluronan synthase from Streptococcus pyogenes (Heldermon et al, 2001; Weigel and DeAngelis, 2007). TMDs 1, 2, 4 and 5 are true transmembrane domains, while 3 and 6 are membrane-associated domains (MADs). Catalytic domains A and B with the active site of processive GT-2 β-glycosyltransferases are indicated (Saxena and Brown, 1997; Saxena et al, 2001). Regions proposed to be involved in c-di-GMP binding are highlighted in red. The position of the constitutive PgaC mutation V227L is indicated. CP, cytoplasm; IM, inner membrane; PP, periplasm. (B) C-di-GMP binding to the PgaCD complex stabilizes a heterodimeric complex to induce a secretion-competent conformation. Left: Top view of the inactive transient state with loosely associated, highly unstable PgaD. Right: C-di-GMP binding induces a poly-GlcNAc secretion-competent state. (C) Model for the irreversible inactivation of PgaCD upon drop of cellular c-di-GMP levels. Signalling through the Csr cascade induces the synthesis of Pga components and the DGCs YdeH and YcdT. At low c-di-GMP concentrations (e.g., inactive DGCs or highly active PDEs) PgaD is rapidly removed by proteolysis, uncoupling the Pga machinery temporarily from c-di-GMP signalling. Only continuous or renewed input through the Csr signalling cascade will allow cells to reactivate poly-GlcNAc synthesis and secretion, thus providing the Csr cascade with signalling dominance over the enzymes directly regulating the cellular c-di-GMP level.
Among the organisms harbouring a Pga-like poly-GlcNAc secretion system two subfamilies of PgaD proteins exist. All Gram-negative bacteria that are devoid of c-di-GMP signalling harbour a Staphylococcus epidermidis IcaD-like (Gerke et al, 1998) homologue, while the presence of GGDEF domains strongly correlates with PgaD-like proteins (and the presence of R222 in PgaC). This is striking since evidence is accumulating that Staphylococcus spp. are unable to synthesize c-di-GMP (Holland et al, 2008). It can thus be speculated that PgaD-like partners of the PgaC GT family interlink the activity of this EPS system with the cell’s c-di-GMP circuitry. The observation that a PgaCD fusion protein is fully functional and responsive to c-di-GMP raised the question why nature has split this functional unit into two individual polypeptides. We would like to propose that the answer to this question is linked to the observed instability of PgaD when cellular levels of c-di-GMP are low. Rapid removal of PgaD under these conditions would irreversibly shut-off the Pga machinery and temporarily uncouple poly-GlcNAc synthesis and secretion from cellular c-di-GMP levels (Figure 8C). Reinstating poly-GlcNAc production in cells that went through a trough of c-di-GMP would require a derepressed Csr pathway allowing the resynthesis of all Pga components. Such a mechanism would thus elegantly equip the global Csr pathway (Timmermans and Van Melderen, 2010; Romeo et al, 2012) with a clear dominance over short-term fluctuations of c-di-GMP resulting from signal input into different DGCs and PDEs, and by that providing the basis for signalling specificity of c-di-GMP-controlled systems (Figure 8C).
In conclusion, this work shows that in E. coli, poly-GlcNAc-dependent biofilm formation is allosterically controlled through c-di-GMP binding to the membrane-anchored PgaCD complex. Since two proteins have to interact in order to form a ligand-binding pocket, the PgaCD complex represents a novel type c-di-GMP receptor. The elucidation of the details of the specific interaction between the allosteric ligand and the PgaCD complex will require careful biochemical and structural analysis.
Materials and methods
More detailed descriptions of Materials and methods are provided in Supplementary data.
Strains and plasmids used in this study are listed in Supplementary Table 1.
Biofilm assay
Biofilm assays were essentially performed as previously described (Boehm et al, 2009). Briefly, overnight cultures were diluted 1:40 into 200 μl LB medium in 96-well polystyrene microtiter plates (BD Falcon). Plates were incubated for 24 h at 30°C without shaking and optical cell density was recorded, before non-attached cells were discarded and plates were vigorously washed with deionized water from a hose. Attached biomass was stained with a 0.1% crystal violet solution (H2O:isopropanol:methanol 96.7:1.66:1.66, v/v). Following the removal of excess dye, retained crystal violet was dissolved in 200 μl 20% acetic acid and quantified by measuring absorbance at 600 nm. Normalized biofilm formation values (biofilm) are ratios of the optical density of dissolved crystal violet (corresponding to the attached biomass) divided by the optical cell density (measured before discarding non-attached cells). Background values were subtracted. Generally, a single data point represents the average of 3–6 wells per strain and condition. Error bars are standard deviations. Experiments were repeated multiple times.
Immunoblots
For the determination of in vivo steady-state protein levels, cells were grown as for biofilm assays, total cells of 4–6 wells were pooled and adjusted to the same optical density. Proteins were generally separated on 7.5–15% SDS–PAGE gels and blotted onto PVDF membranes (Millipore) according to standard protocols (Laemmli, 1970; Towbin et al, 1979).
For anti-poly-GlcNAc immunoblots, reaction mixtures containing membranes from strain AB1638 harbouring pCD-3xF or pins1 were set up as described for the modified enzyme-coupled spectrophotometric GT activity assay and incubated in the presence of different c-di-GMP concentrations for 1 h at 30°C. In all, 1 μl per sample was spotted onto a nitrocellulose membrane (Hybond-C Extra, Amersham Biosciences).
After overnight incubation in blocking solution (1 × PBS pH 7.4, 0.1% Tween-20, 5% milk powder), immunodetection of proteins was carried out using mouse monoclonal α-Flag M2 (1:10 000; Sigma) or rabbit polyclonal α-His (1:3000; GenScript) antibody. The biotin moiety of the cdG-CC was detected with goat polyclonal α-biotin (1:3000; antibodies-online, Germany) antibody or HRP-coupled Streptavidin (1:4000; SouthernBiotech, USA). Poly-GlcNAc was detected using a rabbit anti-poly-GlcNAc antiserum (1:2000; kind gift of Knut Ohlsen and Wilma Ziebuhr). HRP-conjugated rabbit α-mouse, swine α-rabbit (1:10 000; DakoCytomation, Denmark) or rabbit α-goat (1:10 000; Invitrogen) secondary antibodies were used. Blots were developed with the ECL chemiluminescent substrate (Perkin-Elmer, USA) and photographic films (Fujifilm, Japan). If needed, then band intensities were quantified from scanned X-ray films using the ImageJ software.
Translation block experiment
To assess the in vivo stability of PgaD–3xF, overnight cultures of strains AB1062 (or AB1569 harbouring pins1), AB1063 (or AB1570 harbouring pins1) and AB1063 harbouring pAB551 or pAC551 were diluted 1:100 into fresh LB medium and cultures were grown to exponential phase (OD600 between 0.8 and 1.8) at 30°C, before protein synthesis was inhibited at time point zero by the addition of 100–200 μg/ml chloramphenicol or 300 μg/ml erythromycin. dgcA alleles encoded on pAB551 and pAC551 were not induced (leaky expression). Samples were harvested at indicated times after translation inhibition and PgaD levels were analysed by immunoblots. Band intensities were quantified using the ImageJ software and normalized to levels present 5 min after translation inhibition for each strain.
Co-immunoprecipitation
Membranes containing overexpressed proteins of interest from strain AB2043 harbouring pC-His-D-3xF (experiment A) or from strains AB1638 harbouring pCD-3xF and AB2043 harbouring pCD-3xF (experiment B) were used for anti-Flag co-immunoprecipitation experiments. In experiment A, membrane fraction (∼4 mg/ml total protein) was solubilized in IP Solubilization Buffer A (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 M NaCl, 0.5% DDM) for 4 h at 4°C with end-over-end agitation. Thereafter, sample was ultracentrifuged (100 000 g, 1 h, 4°C), the supernatant was split into three parts and incubated either with 20 μl prewashed anti-Flag M2 affinity gel (Sigma) (with or without 2 M urea) or with the same amount of prewashed protein A agarose (Roche) (mock IP) for 45 min at 4°C with end-over-end agitation. Aliquots of the protein fraction that did not bind to the beads (supernatant) were harvested, before beads were washed multiple times with IP Wash Buffer A (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 M NaCl, 0.1% DDM±2 M urea). Immunoprecipitated proteins and supernatant controls were analysed by immunoblots. In experiment B, membrane fractions (∼3–5 mg/ml total protein) were solubilized in IP Solubilization Buffer B (50 mM Tris–HCl pH 7.5, 1 M NaCl, 5% glycerol, 1 mM DTT, 0.5% DDM) overnight at 4°C with end-over-end agitation. After ultracentrifugation (100 000 g, 1 h, 4°C), supernatants were collected and incubated with 40 μl prewashed anti-Flag M2 affinity gel (Sigma) overnight at 4°C with end-over-end agitation. Beads were washed multiple times with IP Wash Buffer B (50 mM Tris–HCl pH 7.5, 1 M NaCl, 5% glycerol, 0.1% DDM), before immunoprecipitated proteins were analysed by Coomassie staining.
Membrane preparation
Overnight precultures of strains AB1638 or AB2043 harbouring the desired plasmid for protein overexpression (or strains AB1775, AB1776 and AB1777) were diluted 1:100 into 1 l LB medium and cultures were grown at 30°C to OD600 of 0.2, before expression of plasmid-borne genes was induced with 0.2% L-arabinose for 5 h. Cells were harvested by centrifugation, resuspended in 5–10 ml ice-cold French Press Buffer (50 mM HEPES pH 7, 5 mM CaCl2, 1 mM DTT, Complete Mini EDTA-free protease inhibitors (Roche)) and lysed by passage three times through a French pressure cell (Vanderheiden et al, 1970). Lysate was clarified by centrifugation (27 000 g, 70 min, 4°C), before membranes were pelleted by ultracentrifugation (120 000 g, 90 min, 4°C). Membranes were generally resuspended in ∼250 μl French Press Buffer and stored at −80°C.
GT activity assays
Modified enzyme-coupled spectrophotometric assay. PgaCD GT activity was indirectly determined with a modified enzyme-coupled spectrophotometric assay (Baykov et al, 1988). Briefly, 50 μl reaction mixtures containing membranes from strains AB1775, AB1776 or AB1777 (∼10 mg/ml total protein) in GT Activity Buffer (50 mM HEPES pH 7, 5 mM CaCl2, 5 mM MgCl2) were incubated for 5 h at 30°C with or without 2 mM UDP-GlcNAc. The pH of the reactions was increased to 8–8.5 by adding 0.1 M NaOH and taking them up in SAP Buffer (50 mM Tris–HCl pH 9, 10 mM MgCl2), before reactions were incubated with 1.5 μl shrimp alkaline phosphatase (SAP) (Promega) for 80 min at 37°C. Phosphate content (indirect measure for UDP) was determined spectrophotometrically at 630 nm using the colour reagent containing molybdate and malachite green (Baykov et al, 1988). Background value was subtracted.
FPLC anion exchange column assay. Standard 100 μl reaction mixtures contained membranes from strain AB2043 harbouring the desired plasmid for protein overexpression (∼0.3–0.6 mg/ml total protein), varying UDP-GlcNAc concentrations (between 50 μM and 2 mM) and different c-di-GMP concentrations (between 0 μM and 2 μM) in GT Activity Buffer (50 mM HEPES pH 7, 5 mM CaCl2, 5 mM MgCl2). Whenever different mutants were compared, membrane inputs were adjusted with an immunoblot beforehand. Reactions were incubated between 0 and 180 min at 30°C, before they were stopped by boiling for 5 min at 98°C. Samples were cleared by centrifugation (16 100 g, 1 min, 25°C) and supernatants were taken up in 900 μl 1 mM sodium acetate. Nucleotides UDP and UDP-GlcNAc were separated on an anion exchange column (1 ml Resource Q, GE Healthcare) mounted on an ÄKTA Purifier FPLC unit (GE Healthcare) with a linear gradient of sodium acetate from 1 mM to 1 M and monitored with Unicorn software. Initial linear PgaCD GT reaction velocities were determined by plotting integrated peak areas against reaction incubation times using GraphPad Prism.
C-di-GMP capture compound (cdG-CC) binding assay
CdG-CC (Caprotec Bioanalytics, Germany) experiments were carried out in 200 μl 12-tube PCR strips (Thermo Scientific) as previously described (Nesper et al, 2012) with some modifications. In all, 100 μl samples generally contained membranes from strain AB1638 harbouring the desired plasmid (∼3–4 mg/ml total protein) and 20 mM UDP-GlcNAc in Binding Buffer (20 mM HEPES pH 7.5, 50 mM potassium acetate, 10 mM magnesium acetate, 10% glycerol, 5 mM MgCl2, 1.5 mM CaCl2). Whenever different mutants were compared, experiments were performed in the context of the PgaCD fusion protein and membrane inputs were adjusted with an immunoblot beforehand. A 12.5- or 125-fold molar excess of c-di-GMP or GTP was added to competition experiments and strips were preincubated for 30 min at 30°C with end-over-end agitation. After the addition of 0.8 or 8 μM cdG-CC, strips were wrapped in aluminium foil and incubated for 2 h at 30°C with end-over-end agitation. Samples were UV irradiated at 310 nm for 4 min at 4°C using a caproBox (Caprotec Bioanalytics, Germany), before they were taken up in a final volume of 200 μl Capture Solubilization Buffer (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 M NaCl, 0.5% DDM) and solubilized for 4 h at 4°C with end-over-end agitation. After ultracentrifugation (100 000 g, 1 h, 4°C), an aliquot of the supernatants was saved and the rest incubated with 35 μl magnetic streptavidin beads (Dynabeads MyOne Streptavidin C1, Invitrogen) in PCR strips (Thermo Scientific) for 40 min at 4°C with end-over-end agitation. Beads were collected with a magnet (caproMag, Caprotec Bioanalytics, Germany) and washed 9 × with 200 μl Capture Wash Buffer (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 M NaCl, 0.1% DDM), before captured proteins were analysed by immunoblots. If different mutants were compared, then band intensities were quantified using the ImageJ software and band intensities were normalized to the total solubilized protein amount of each sample.
UV crosslinking with [32/33P]c-di-GMP
UV light-induced crosslinking experiments were performed as previously described (Christen et al, 2005, 2006) in conical 96-well plates (Greiner Bio-One). In all, 25 μl samples generally contained membranes from strain AB1638 harbouring p2-3xF or p6a (∼30 mg/ml total protein) and 20 mM UDP-GlcNAc in Binding Buffer (20 mM HEPES pH 7.5, 50 mM potassium acetate, 10 mM magnesium acetate, 10% glycerol, 5 mM MgCl2, 1.5 mM CaCl2). Whenever different mutants were compared, membrane inputs were adjusted with an immunoblot beforehand. For competition experiments, a 100-fold molar excess of c-di-GMP or GTP was added. Plates were preincubated sealed with a foil for 35 min at 30°C on a rocking platform, before the addition of 1 or 2 μM radiolabelled [32/33P]c-di-GMP. After a second incubation for 2 h at 30°C, foils were removed and 96-well plates were UV irradiated at 254 nm for 20 min using a Bio-Link crosslinker (Vilber Lourmat, France). Thereafter, samples were taken up in a final volume of 200 μl Crosslinking Solubilization Buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 5% glycerol, 1 mM DTT, 0.5% DDM) and solubilized overnight at 4°C with end-over-end agitation. After ultracentrifugation (100 000 g, 1 h, 4°C), supernatants were incubated with 40 μl anti-Flag M2 magnetic beads (Sigma) overnight at 4°C with end-over-end agitation. Beads were washed multiple times with IP Wash Buffer B (50 mM Tris–HCl pH 7.5, 1 M NaCl, 5% glycerol, 0.1% DDM) and the help of a magnet, before immunoprecipitated proteins were analysed by Coomassie staining and autoradiography. If needed, then band intensities were quantified using the ImageJ software and autoradiography band intensities were normalized to protein amounts on Coomassie-stained gels.
Supplementary Material
Acknowledgments
We thank Jacob G Malone for assistance with error-prone PCR; Alain Casanova for the construction of strain AB1313 and plasmid pAC551; Knut Ohlsen and Wilma Ziebuhr for the generous gift of antiserum against poly-GlcNAc; and Régis Hallez for helpful advice and discussions. C-di-GMP measurements were provided by Prof. Volkhard Kaever from the Medizinische Hochschule Hannover. This work was supported by Swiss National Science Foundation grant 31003A_130469 to UJ and a fellowship from the Werner Siemens Foundation (Zug) to SS.
Author contributions: SS, CL, AB and UJ conceived and designed the experiments. SS and CL performed the experiments. SS, CL, AB and UJ analysed the data. SS and UJ wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aloni Y, Cohen R, Benziman M, Delmer D (1983) Solubilization of the UDP-glucose:1,4-beta-D-Glucan 4-beta-D-Glucosyltransferase (Cellulose Synthase) from Acetobacter xylinum. J Biol Chem 258: 4419–4423 [PubMed] [Google Scholar]
- Amikam D, Galperin MY (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22: 3–6 [DOI] [PubMed] [Google Scholar]
- Baykov AA, Evtushenko OA, Avaeva SM (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171: 266–270 [DOI] [PubMed] [Google Scholar]
- Becker S, Soares C, Porto LM (2009) Computational analysis suggests that virulence of Chromobacterium violaceum might be linked to biofilm formation and poly-NAG biosynthesis. Genet Mol Biol 32: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF (2007) The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J 26: 5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentancor LV, O’Malley JM, Bozkurt-Guzel C, Pier GB, Maira-Litrán T (2012) Poly-N-acetyl-beta-(1-6)-glucosamine is a target for protective immunity against Acinetobacter baumannii infections. Infect Immun 80: 651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD (2008) Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol 10: 1419–1432 [DOI] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U (2010) Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141: 107–116 [DOI] [PubMed] [Google Scholar]
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U (2009) Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 72: 1500–1516 [DOI] [PubMed] [Google Scholar]
- Branda SS, Vik A, Friedman L, Kolter R (2005) Biofilms: the matrix revisited. Trends Microbiol 13: 20–26 [DOI] [PubMed] [Google Scholar]
- Cerca N, Maira-Litrán T, Jefferson KK, Grout M, Goldmann DA, Pier GB (2007) Protection against Escherichia coli infection by antibody to the Staphylococcus aureus poly-N-acetylglucosamine surface polysaccharide. Proc Natl Acad Sci USA 104: 7528–7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litrán T (2009) The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191: 5953–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U (2006) Allosteric control of cyclic di-GMP signaling. J Biol Chem 281: 32015–32024 [DOI] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, Jenal U (2007) DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci USA 104: 4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U (2005) Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280: 30829–30837 [DOI] [PubMed] [Google Scholar]
- Ciocchini AE, Roset MS, Briones G, Iñón de Iannino N, Ugalde RA (2006) Identification of active site residues of the inverting glycosyltransferase Cgs required for the synthesis of cyclic beta-1,2-glucan, a Brucella abortus virulence factor. Glycobiology 16: 679–691 [DOI] [PubMed] [Google Scholar]
- Conover MS, Sloan GP, Love CF, Sukumar N, Deora R (2010) The Bps polysaccharide of Bordetella pertussis promotes colonization and biofilm formation in the nose by functioning as an adhesin. Mol Microbiol 77: 1439–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322 [DOI] [PubMed] [Google Scholar]
- Davies D (2003) Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2: 114–122 [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U (2009) Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23: 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Gomelsky M (2010) A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8: 623–633 [DOI] [PubMed] [Google Scholar]
- Forman S, Bobrov AG, Kirillina O, Craig SK, Abney J, Fetherston JD, Perry RD (2006) Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology 152: 3399–3410 [DOI] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P (2005) Survival strategies of infectious biofilms. Trends Microbiol 13: 34–40 [DOI] [PubMed] [Google Scholar]
- Gerke C, Kraft A, Süssmuth R, Schweitzer O, Götz F (1998) Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273: 18586–18593 [DOI] [PubMed] [Google Scholar]
- Habazettl J, Allan MG, Jenal U, Grzesiek S (2011) Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J Biol Chem 286: 14304–14314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108 [DOI] [PubMed] [Google Scholar]
- Heldermon C, DeAngelis PL, Weigel PH (2001) Topological organization of the hyaluronan synthase from Streptococcus pyogenes. J Biol Chem 276: 2037–2046 [DOI] [PubMed] [Google Scholar]
- Hengge R (2009) Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7: 263–273 [DOI] [PubMed] [Google Scholar]
- Holland LM, O’Donnell ST, Ryjenkov DA, Gomelsky L, Slater SR, Fey PD, Gomelsky M, O’Gara JP (2008) A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J Bacteriol 190: 5178–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, Romeo T (2008) Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol 190: 3670–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Sadovskaya I, Vinogradov E, Mulks MH, Velliyagounder K, Ragunath C, Kher WB, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB (2007) Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb Pathog 43: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Sadovskaya I, Wang H, Vinogradov E, Ragunath C, Ramasubbu N, Jabbouri S, Perry MB, Kaplan JB (2008) Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb Pathog 44: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors O (2008) The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70: 236–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95: 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiski C-L, Harwich M, Jain S, Neculai AM, Yip P, Robinson H, Whitney JC, Riley L, Burrows LL, Ohman DE, Howell PL (2010) AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD (2004) HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol 54: 75–88 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S (2007) A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65: 1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, O’Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9: 34–39 [DOI] [PubMed] [Google Scholar]
- Maira-Litrán T, Kropec A, Goldmann DA, Pier GB (2005) Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated Staphylococcal poly-N-acetyl-beta-(1-6)-glucosamine. Infect Immun 73: 6752–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenheimer KM, Koch TH (1997) Photocross-linking of nucleic acids to associated proteins. Crit Rev Biochem Mol 32: 101–140 [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Siegel E, van Heijenoort J (1989) Variations in UDP-N-acetylglucosamine and UDP-N-acetylmuramyl-pentapeptide pools in Escherichia coli after inhibition of protein synthesis. J Bacteriol 171: 3282–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S (2007) The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65: 876–895 [DOI] [PubMed] [Google Scholar]
- Monteiro C, Saxena I, Wang X, Kader A, Bokranz W, Simm R, Nobles D, Chromek M, Brauner A, Brown RM Jr, Römling U (2009) Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences. Environ Microbiol 11: 1105–1116 [DOI] [PubMed] [Google Scholar]
- Namboori SC, Graham DE (2008) Enzymatic analysis of uridine diphosphate N-acetyl-D-glucosamine. Anal Biochem 381: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U (2012) A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Prot 75: 4874–4878 [DOI] [PubMed] [Google Scholar]
- Newell PD, Boyd CD, Sondermann H, O’Toole GA (2011) A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9: e1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gara JP (2007) ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett 270: 179–188 [DOI] [PubMed] [Google Scholar]
- Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM (2010) The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a ‘backstop brake’ mechanism. Mol Cell 38: 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U (2004) Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18: 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R (2008) Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22: 2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povolotsky TL, Hengge R (2012) ‘Life-style’ control networks in Escherichia coli: Signaling by the second messenger c-di-GMP. J Biotechnol 160: 10–16 [DOI] [PubMed] [Google Scholar]
- Pérez-Mendoza D, Coulthurst SJ, Sanjuán J, Salmond GPC (2011) N-acetyl-glucosamine-dependent biofilm formation in Pectobacterium atrosepticum is cryptic and activated by elevated c-di-GMP levels. Microbiology 157: 3340–3348 [DOI] [PubMed] [Google Scholar]
- Romeo T, Gong M, Liu MY, Brun-Zinkernagel A (1993) Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175: 4744–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T, Vakulskas CA, Babitzke P (2012) Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol (advance online publication, 5 June 2012; doi:10.1111/j.1462-2920.2012.02794.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, Vroom E, de, Van der Marel GA, Van Boom JH, Benziman M (1987) Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325: 279–281 [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He Y-W, Zhang L-H, Heeb S, Cámara M, Williams P, Dow JM (2006) Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA 103: 6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Saxena IM, Brown RM Jr (1997) Identification of cellulose synthase(s) in higher plants: sequence analysis of processive beta-glycosyltransferases with the common motif ‘D, D, D35Q(R,Q)XRW’. Cellulose 4: 33–49 [Google Scholar]
- Saxena IM, Brown RM Jr, Dandekar T (2001) Structure-function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry 57: 1135–1148 [DOI] [PubMed] [Google Scholar]
- Schirmer T, Jenal U (2009) Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7: 724–735 [DOI] [PubMed] [Google Scholar]
- Skurnik D, Davis MR Jr, Benedetti D, Moravec KL, Cywes-Bentley C, Roux D, Traficante DC, Walsh RL, Maria-Litràn T, Cassidy SK, Hermos CR, Martin TR, Thakkallapalli EL, Vargas SO, McAdam AJ, Lieberman TD, Kishony R, LiPuma JJ, Pier GB, Goldberg JB et al. (2012) Targeting pan-resistant bacteria with antibodies to a broadly conserved surface polysaccharide expressed during infection. J Infect Dis 205: 1709–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann H, Shikuma NJ, Yildiz FH (2011) You’ve come a long way: c-di-GMP signaling. Curr Opin Microbiol 15: 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6: 175–182 [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T (2006) Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20: 2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabue L, Antoniani D, Maciag A, Bocci P, Raffaelli N, Landini P (2010) The diguanylate cyclase YddV controls production of the exopolysaccharide poly-N-acetylglucosamine (PNAG) through regulation of the PNAG biosynthetic pgaABCD operon. Microbiology 156: 2901–2911 [DOI] [PubMed] [Google Scholar]
- Timmermans J, Van Melderen L (2010) Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 67: 2897–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlapak-Simmons VL, Baggenstoss BA, Clyne T, Weigel PH (1999) Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and Streptococcus equisimilis. J Biol Chem 274: 4239–4245 [DOI] [PubMed] [Google Scholar]
- Tlapak-Simmons VL, Kempner ES, Baggenstoss BA, Weigel PH (1998) The active Streptococcal hyaluronan synthases (HASs) contain a single HAS monomer and multiple cardiolipin molecules. J Biol Chem 273: 26100–26109 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A (2011) Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol 407: 633–639 [DOI] [PubMed] [Google Scholar]
- Vanderheiden GJ, Fairchild AC, Jago GR (1970) Construction of a laboratory press for use with the French pressure cell. Appl Microbiol 19: 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T (2005) CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56: 1648–1663 [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF, Romeo T (2004) The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186: 2724–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel PH, DeAngelis PL (2007) Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J Biol Chem 282: 36777–36781 [DOI] [PubMed] [Google Scholar]
- Whitney JC, Colvin KM, Marmont LS, Robinson H, Parsek MR, Howell PL (2012) Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J Biol Chem 287: 23582–23593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakandawala N, Gawande PV, LoVetri K, Cardona ST, Romeo T, Nitz M, Madhyastha S (2011) Characterization of the poly-beta-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl Environ Microbiol 77: 8303–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.