Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)-encoded immunity in Type I systems relies on the Cascade (CRISPR-associated complex for antiviral defence) ribonucleoprotein complex, which triggers foreign DNA degradation by an accessory Cas3 protein. To establish the mechanism for adaptive immunity provided by the Streptococcus thermophilus CRISPR4-Cas (CRISPR-associated) system (St-CRISPR4-Cas), we isolated an effector complex (St-Cascade) containing 61-nucleotide CRISPR RNA (crRNA). We show that St-Cascade, guided by crRNA, binds in vitro to a matching proto-spacer if a proto-spacer adjacent motif (PAM) is present. Surprisingly, the PAM sequence determined from binding analysis is promiscuous and limited to a single nucleotide (A or T) immediately upstream (−1 position) of the proto-spacer. In the presence of a correct PAM, St-Cascade binding to the target DNA generates an R-loop that serves as a landing site for the Cas3 ATPase/nuclease. We show that Cas3 binding to the displaced strand in the R-loop triggers DNA cleavage, and if ATP is present, Cas3 further degrades DNA in a unidirectional manner. These findings establish a molecular basis for CRISPR immunity in St-CRISPR4-Cas and other Type I systems.
Keywords: DNA interference, DNA-dependent ATPase, nuclease
Introduction
Bacterial viruses (bacteriophages) provide a ubiquitous and often deadly threat to bacterial populations. To survive in hostile environments, bacteria have developed a multitude of antiviral defence systems (Sturino and Klaenhammer, 2006; Labrie et al, 2010). Clustered regularly interspaced short palindromic repeats (CRISPR) together with CRISPR-associated genes (cas) constitute an adaptive immune system, which provides acquired resistance against viruses and plasmids, in bacteria and archaea (Barrangou et al, 2007). The CRISPR-Cas system hijacks short fragments of invasive DNA, integrates them as spacers within the CRISPR array, and subsequently uses them as templates to generate specific small-interfering CRISPR RNA (crRNA) molecules that combine with Cas proteins into effector complexes that trigger degradation of matching foreign nucleic acids, thereby preventing their proliferation and propagation (Al-Attar et al, 2011; Bhaya et al, 2011; Terns and Terns, 2011; Wiedenheft et al, 2012).
CRISPR-Cas systems have been categorized into three main types that differ by the structural organization and function(s) of nucleoprotein complexes involved in crRNA-mediated silencing of foreign nucleic acids (Makarova et al, 2011). In Type I systems (as exemplified by the CRISPR-Cas system of Escherichia coli K12), crRNAs are incorporated into a multisubunit ribonucleoprotein (RNP) complex called Cascade (CRISPR-associated complex for antiviral defence), which binds to matching invasive DNA and triggers degradation by an accessory Cas3 protein (Brouns et al, 2008; Sinkunas et al, 2011; Westra et al, 2012). In Type II systems (as exemplified by the CRISPR1-Cas and CRISPR3-Cas systems of Streptococcus thermophilus), CRISPR-mediated immunity solely relies on the signature Cas9 protein that associates with crRNA to form an effector complex, which specifically cleaves matching target double-stranded DNA (dsDNA) (Garneau et al, 2010; Deltcheva et al, 2011; Sapranauskas et al, 2011; Gasiunas et al, 2012; Jinek et al, 2012). In Type III systems (as exemplified by Sulfolobus solfataricus and Pyrococcus furiosus), Cas RAMP module (Cmr) in association with crRNA recognizes and cleaves RNA in vitro (Hale et al, 2012; Zhang et al, 2012), whereas the CRISPR-Cas system of Staphylococcus epidermidis targets DNA in vivo (Marraffini and Sontheimer, 2010).
The S. thermophilus DGCC7710 model organism (St), for which CRISPR-Cas interference has been demonstrated against phages and plasmids, contains four distinct CRISPR-Cas systems (Horvath and Barrangou, 2010). Direct spacer acquisition and interference activities have been demonstrated for two distinct Type II systems, namely St-CRISPR1-Cas and St-CRISPR3-Cas (Barrangou et al, 2007; Deveau et al, 2008; Garneau et al, 2010; Sapranauskas et al, 2011; Gasiunas et al, 2012). However, until now, neither spacer acquisition nor interference activity has been reported for the St-CRISPR2-Cas or St-CRISPR4-Cas systems, which belong to Type III and Type I systems, respectively. Therefore, we investigated whether the St-CRISPR4-Cas system is functionally active and has the ability to provide immunity against invading DNA.
The St-CRISPR4-Cas system of S. thermophilus DGCC7710 and E. coli CRISPR-Cas system are orthologous (Type I–E) and share genetic structural organization (Horvath and Barrangou, 2010; Sinkunas et al, 2011). In the St-CRISPR4-Cas system, five cas genes are arranged into a cluster (cse1-cse2-cas7-cas5-cas6e) (Figure 1A) analogous to the E. coli cas genes, suggesting that corresponding Cas proteins may assemble into a homologous St-Cascade complex. While in vivo functional activity has not been observed for the St-CRISPR4-Cas system, indirect in vitro evidence suggests that it may be active, at least at the interference step (Young et al, 2012). Indeed, we have previously shown that the Cas3 protein of the St-CRISPR4-Cas system is an active nuclease/helicase, which may play a key role in DNA degradation (Sinkunas et al, 2011).
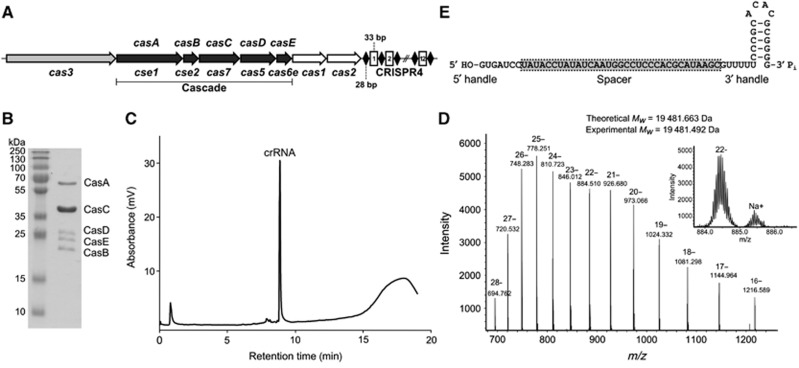
Figure 1.
S. thermophilus CRISPR4-Cas crRNA and Cascade complex. (A) Schematic representation of the CRISPR4-Cas locus containing eight cas genes and twelve repeat-spacer units (conserved 28-bp palindromic repeats 5′-GTTTTTCCCGCACACGCGGGGGTGATCC-3′ are separated from each other by 33-bp spacers of variable sequence). St-Cascade genes homologous to the E. coli Cascade are underlined. Genes names according to Brouns et al (2008) and Makarova et al (2011) are indicated, respectively, above and below corresponding genes. (B) Coomassie blue-stained SDS-polyacrylamide gel of St-Cascade complex proteins isolated using the CasC-His6 protein as bait. (C) IP RP HPLC analysis of mature crRNA. (D) LC ESI-MS analysis of purified S. thermophilus crRNA. Inset shows an enhanced view of the 22-charge state. (E) Architecture of crRNA co-purifying with the St-Cascade protein complex.
Source data for this figure is available on the online supplementary information page.
Here, we report the isolation and biochemical characterization of the St-Cascade complex, which consists of CasABCDE proteins (Cse1, Cse2, Cas7, Cas5, and Cas6e, respectively) and a 61-nt crRNA. We further demonstrate that crRNA bound to the St-Cascade complex serves as the guide sequence, which specifically recognizes a matching sequence (proto-spacer) in the target DNA. We show that similarly to other Type I systems, St-Cascade binding to the proto-spacer in the invading DNA requires an additional DNA sequence element, a specific proto-spacer adjacent motif (PAM). However, in contrast with other CRISPR systems, the PAM for St-CRISPR4-Cas is promiscuous and limited to a single nucleotide. Furthermore, we show that St-Cascade and St-Cas3 form a functional effector complex, which cleaves target DNA in vitro.
Results
Cloning, expression, and isolation of St-Cascade
In E. coli, CasABCDE proteins (Cse1, Cse2, Cas7, Cas5, and Cas6e, respectively) (Makarova et al, 2011), and crRNA form a Cascade complex (Ec-Cascade) (Brouns et al, 2008) which, together with Cas3, provide interference against invading foreign DNA. We tested the hypothesis that homologous S. thermophilus Cas proteins (Figure 1A) may assemble into a similar St-Cascade complex, and designed the following strategy for complex isolation. First, three compatible heterologous plasmids containing, respectively, a casABCDE cassette, the C-terminal His-tagged variant of casC (casC-His), and six copies of the repeat-spacer-1 unit (6 × SP1) of the S. thermophilus CRISPR4 region, were engineered. Next, all three plasmids were co-expressed in E. coli BL21 (DE3) strain and the St-Cascade complex was purified by subsequent Ni-chelating, size exclusion, and heparin affinity chromatography steps. Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis (Figure 1B) of the isolated complex revealed five bands that matched to individual Cas proteins, suggesting that CasABCDE proteins assemble into a St-Cascade complex similar to that of E. coli. The identity of all Cas proteins in St-Cascade was confirmed by mass spectrometry analysis (Supplementary Table S1). The stoichiometry of the protein complex was not directly determined; however, the band intensity in the SDS–PAGE (Figure 1B) in conjunction with the mass spectrometry analysis of the St-Cascade tryptic digest suggests that the CasC protein is the most abundant protein present in St-Cascade similar to the Ec-Cascade (Jore et al, 2011). Denaturing PAGE analysis revealed that RNA co-purifies with the St-Cascade complex (Supplementary Figure S1).
Characterization of S. thermophilus CRISPR4-Cas crRNA
Next, we used denaturing RNA chromatography in conjunction with electrospray ionization mass spectrometry (ESI-MS) to characterize the mature crRNAs isolated directly from the St-Cascade complex. Denaturing ion pair reverse phase chromatography was used to purify the crRNA directly from the St-Cascade complex (Dickman and Hornby, 2006; Waghmare et al, 2009). The RNA isolated from this complex consisted of a single mature crRNA with a retention time consistent with an approximate length of 60 nt (Figure 1C). Purified mature crRNA was further analysed using ESI-MS to obtain the accurate intact mass. A molecular weight of 19 482 Da was obtained (Figure 1D). In addition, ESI-MS/MS was used to analyse the oligoribonucleotide fragments generated from RNase T1 and RNase A digestion of the mature crRNA (Supplementary Figures S2 and S3). In conjunction with the intact mass analysis and denaturing PAGE (Supplementary Figure S1), these indicate that processing of St-CRISPR4-Cas crRNAs is similar to that of E. coli CRISPR-Cas crRNAs, generating a 61-nt crRNA (consisting of a 7-nt 5′ handle, a 33-nt spacer, and a 21-nt 3′ handle) with 5′-OH and 3′-Pi (MW 19 481.5 Da) (Figure 1E). Further verification of the 3′-Pi termini was obtained upon acid treatment of the crRNA where no change in mass was observed using ESI-MS.
PAM sequence analysis of the S. thermophilus CRISPR4-Cas system
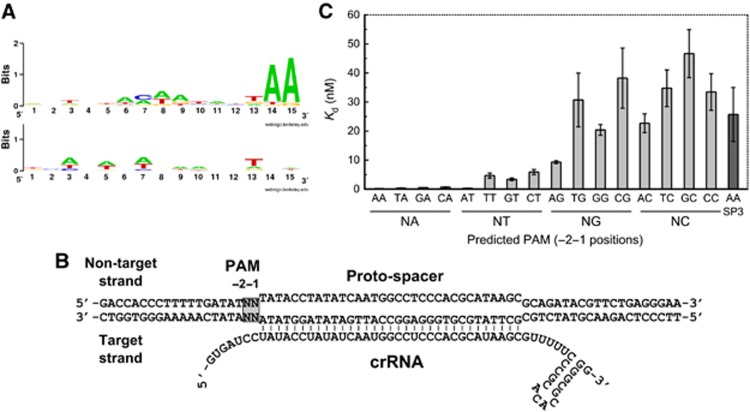
The PAM located in the vicinity of a proto-spacer is absolutely required for silencing of invading DNA by Type I and Type II CRISPR-Cas systems (Deveau et al, 2008; Horvath et al, 2008; Sapranauskas et al, 2011; Semenova et al, 2011). In the E. coli Type I-E system, the PAM corresponds to the 5′-AWG-3′ sequence located immediately upstream of a proto-spacer (Mojica et al, 2009) and is essential for Ec-Cascade binding and subsequent DNA interference (Semenova et al, 2011). On the other hand, experimental analysis of CRISPR repeat boundaries in E. coli suggests a dinucleotide 5′-AW-3′ as PAM (Goren et al, 2012). To determine the putative PAM sequence of the CRISPR4-Cas system, we analysed all currently available CRISPR4 spacer sequences found in S. thermophilus strains. A CRISPR4 locus is present in DGCC7710 (Horvath and Barrangou, 2010) and three other strains from the DuPont culture collection. In DGCC7710, the CRISPR4 locus contains 12 unique spacers, and 26 more unique spacers were identified in the three other CRISPR4-positive strains. Sequence similarity searches, both in public and proprietary sequence databases, showed that most (26 out of 38) of these CRISPR4 spacer sequences have matches (proto-spacers) in S. thermophilus phage sequences. Only perfect matches (100% identity over the complete spacer sequence) between spacer and proto-spacer were considered, providing a set of 106 matching sequences. The sequences located immediately upstream and downstream of these proto-spacers were examined for the presence of a possible PAM. After removal of redundant alleles, a Weblogo representation (Crooks et al, 2004) was used to depict sequence conservation over a 15-nt segment of 28 (upstream) and 21 (downstream) unique sequences (Figure 2A). A 2-base pair (bp) conserved motif 5′-AA-3′ could be identified immediately upstream of the proto-spacers.
Figure 2.
PAM-dependent St-Cascade binding. (A) Predicted PAM for the St-CRISPR4-Cas system. Weblogo representation (Crooks et al, 2004) of 15-nt sequences found immediately upstream (top) and downstream (bottom) of phage proto-spacers that match known CRISPR4 spacers. A conserved, 2-nt PAM (5′-AA-3′) is located immediately upstream of the proto-spacers. (B) A schematic representation of a putative R-loop structure resulting from the St-Cascade binding to the 73-bp oligodeoxynucleotide. Nucleotides NN at −1 and −2 positions of predicted PAM were varied. In the R-loop structure, a target strand bound to the crRNA is engaged into a heteroduplex while the non-target strand is displaced as a single-stranded DNA. (C) PAM sequence dependence of a proto-spacer-1 binding by St-Cascade. Bar diagram shows dissociation constant Kd values obtained by EMSA. Error bars represent standard deviations of average Kd value determined in three separate experiments. Oligoduplex containing a non-matching proto-spacer-3 sequence was used as a non-specific DNA control.
PAM is required for St-Cascade binding to the proto-spacer
To determine whether the predicted PAM sequence is important for proto-spacer recognition, we analysed St-Cascade binding to a set of synthetic 73-bp oligoduplexes containing the spacer-1 sequence and variable nucleotides at positions −2 and −1 (Figure 2B). Oligoduplexes were radiolabelled at the 5′-end of the target strand, and the St-Cascade binding affinity was evaluated by electrophoretic mobility shift assay (EMSA). Binding analysis revealed that oligoduplexes fall into three categories with regards to St-Cascade binding. Oligoduplexes containing N(−2)A(−1) nucleotides in the predicted PAM display high binding affinity with Kd ∼0.2 nM, oligoduplexes containing N(−2)T(−1) nucleotides show binding with Kd <10 nM, while all other oligoduplexes except A(−2)G(−1) bind with the same affinity as the non-specific oligoduplex containing spacer-3 instead of spacer-1. Thus, these results suggest that a single nucleotide PAM, A or T (W) at the −1 position upstream of the proto-spacer is required for the St-CRISPR4-Cas system. The G and C nucleotides are not tolerated at this position except for the A(−2)G(−1) dinucleotide (Figure 2C; Supplementary Table S2).
To test whether a non-conserved nucleotide at the −3 position in the vicinity of the predicted PAM is important for spacer recognition, we analysed St-Cascade binding to a set of oligoduplexes containing a conserved A(−2)A(−1) dinucleotide and any nucleotide at the −3 position (SP1-TAA, SP1-AAA, SP1-GAA, and SP1-CAA, respectively) (Supplementary Figure S4A). EMSA analysis revealed that St-Cascade bound all oligoduplexes with a variable N(−3) nt with the same affinity (Supplementary Figure S4B), confirming that the −3 position is not important for St-Cascade binding.
In Type I CRISPR systems, as exemplified by E. coli and P. aeruginosa, target recognition is governed by the crRNA seed sequence located at the 5′-end of the spacer region (Semenova et al, 2011; Wiedenheft et al, 2011) and results in the formation of an R-loop where the target strand of the proto-spacer is engaged into a heteroduplex, while the non-target strand is displaced as single-stranded DNA (ssDNA). To demonstrate the formation of the R-loop upon St-Cascade binding to a proto-spacer, we used the P1 nuclease that specifically cleaves ssDNA regions (Jore et al, 2011; Supplementary Figure S5). In the oligoduplexes SP1-AA and SP1-AG that contain correct PAMs, the non-target strand is susceptible to endonuclease P1 cleavage, while the target strand is resistant to P1 nuclease treatment. On the other hand, in the oligoduplex SP1-CC, which lacks a correct PAM, or in the oligoduplex SP3-AA which contains a PAM but lacks a matching proto-spacer sequence, both DNA strands were resistant to nuclease P1 cleavage. Thus, nuclease P1 assay confirms that an R loop is formed only when both the correct PAM and a matching proto-spacer sequence are present in the oligoduplex.
St-Cascade binding to the proto-spacer triggers St-Cas3 ATPase activity
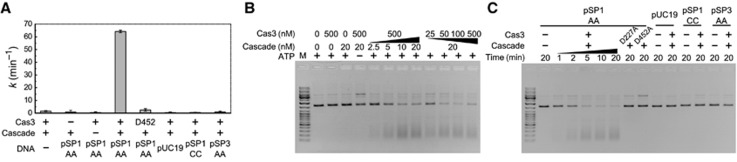
St-Cas3 is a metal-dependent nuclease that possesses a ssDNA stimulated ATPase activity coupled to unwinding of DNA/DNA and RNA/DNA duplexes (Sinkunas et al, 2011). St-Cascade complex binding to the proto-spacer creates an R-loop (Figure 2B) where a non-target strand is displaced as ssDNA and may function as a docking site for Cas3. We used a colorimetric assay to monitor St-Cas3 ATPase activity in the presence of St-Cascade and DNA. The St-Cas3 protein was mixed with the St-Cascade complex and pUC19 plasmid variants that either contain or lack proto-spacer-1 in the context of the correct or mutated PAM (Supplementary Table S3), and ATPase reactions were initiated by addition of ATP and Mg2+ ions. The assay revealed that ATPase activity of St-Cas3 is triggered only in the presence of the pSP1-AA plasmid, containing a matching proto-spacer-1 and a correct PAM (Figure 3A). Plasmids lacking proto-spacer-1 and PAM or containing a different proto-spacer (pSP3-AA) or incorrect CC PAM (pSP1-CC) did not stimulate St-Cas3 ATPase activity in the presence of St-Cascade complex. No ATPase activity was detected for the St-Cas3 ATPase-deficient mutant D452A with an impaired Walker B motif (Sinkunas et al, 2011). Taken together, these results suggest a link between the ATPase activity and Cas3 docking on ssDNA formed upon Cascade binding to the matching proto-spacer flanked by a correct PAM.
Figure 3.
ATPase and nuclease activities of St-Cas3 induced by St-Cascade binding to the proto-spacer dsDNA. (A) ATP hydrolysis rates. Malachite green assay was used to measure ATP hydrolysis through the detection of free phosphate liberated from ATP. Reaction rate constant k values (1/min) were calculated from linear slopes of time courses of phosphate liberation per St-Cas3 amount added. ATPase reactions were conducted at 37°C in the ATPase reaction buffer supplemented with 3 nM supercoiled plasmids, 12 nM St-Cascade and 300 nM St-Cas3 or the ATPase-deficient mutant D452A. Error bars indicate the ±standard deviation for the rate constant k value determined in three separate experiments. (B) dsDNA degradation requires St-Cas3, St-Cascade, and ATP. Nuclease reactions were conducted at 37°C for 10 min in a Nuclease buffer supplemented with 5 nM pSP1-AA and indicated amounts of St-Cas3 and St-Cascade. (C) PAM and a proto-spacer are essential for DNA degradation. Nuclease reactions were conducted at 37°C for indicated time intervals in Nuclease buffer supplemented with 100 nM St-Cas3, 20 nM St-Cascade, and 5 nM of respective supercoiled plasmids.
Source data for this figure is available on the online supplementary information page.
St-Cascade binding to the proto-spacer triggers St-Cas3-mediated plasmid degradation
Cas3 of S. thermophilus is a metal-dependent nuclease that degrades ssDNA using an active site located in the N-terminal HD domain (Sinkunas et al, 2011). dsDNA is resistant to St-Cas3 cleavage (Sinkunas et al, 2011). Consistent with published data on the Thermus thermophilus Cas3 protein (Mulepati and Bailey, 2011), St-Cas3 degraded ss M13mp18 DNA more rapidly in the presence of Ni2+ compared to Mg2+ ions (Supplementary Figure S6A). Since the ATPase activity of St-Cas3 is not supported by Ni2+ ions (Sinkunas et al, 2011), a mixture of Mg2+ and Ni2+ ions that supports both ATPase/helicase and nuclease activities of St-Cas3 was used in further experiments (Supplementary Figure S6B).
To determine whether St-Cas3 docking on the ssDNA formed upon St-Cascade binding triggers nuclease activity, we analysed Cas3-mediated cleavage of plasmid DNA. pSP1-AA plasmid was pre-incubated with St-Cascade and an ATP solution containing Mg2+ and Ni2+ ions, followed by addition of St-Cas3. Under these conditions, the pSP1-AA plasmid was degraded in a St-Cascade and St-Cas3 concentration- and time-dependent manner (Figure 3B and C). On the other hand, the pUC19 plasmid, or plasmids containing a non-matching proto-spacer (pSP3-AA) or a defective PAM (pSP1-CC), were resistant to St-Cas3 cleavage (Figure 3C). ATP hydrolysis was required for pSP1-AA plasmid degradation. In the absence of ATP, the supercoiled pSP1-AA plasmid was converted into a nicked form but not degraded (Figure 3B). The identical cleavage pattern was observed for the St-Cas3 ATPase-deficient mutant D452A in the presence of ATP (Figure 3C). In contrast, the D227A replacement in the nuclease active site (Sinkunas et al, 2011) abolished DNA cleavage activity. Taken together, these data suggest that both ATPase/helicase and nuclease activities of St-Cas3 are required for pSP1-AA plasmid degradation in the presence of St-Cascade. It has been recently reported that Ec-Cascade preferentially binds a negatively supercoiled DNA and that a linear or nicked plasmid is not degraded by Ec-Cascade-Cas3 (Westra et al, 2012). To find out whether DNA supercoiling affects DNA cleavage rate by the St-Cascade-Cas3 system, we monitored degradation rates of supercoiled (pSP1-AA) and linearized (pSP1-AA-BamHI) DNA forms by St-Cascade-Cas3 (Supplementary Figure S12). In contrast to the E. coli CRISPR system, St-Cascade-Cas3 degraded both supercoiled and linear DNA substrates at similar rates (kSC=0.54±0.06/min and klinear=0.43±0.01/min, respectively).
PAM is required for St-Cas3-mediated plasmid degradation
DNA binding studies revealed that St-Cascade binding to a matching proto-spacer sequence requires a correct PAM sequence (Figure 2C). To check whether plasmid DNA cleavage in the in vitro reconstituted interference system follows the same dependence on PAM, we engineered plasmid substrates containing all possible combinations of base pairs at the −2 and −1 positions relative to the proto-spacer (within the predicted PAM), and monitored St-Cas3-mediated cleavage in the presence of St-Cascade and ATP (Supplementary Figure S7). Consistent with previous binding assays, we found that plasmids containing A(−1), T(−1), or A(−2)G(−1) nucleotides upstream of proto-spacer-1 were efficiently degraded, while plasmids with B(−2)G(−1) (where B=T or C or G) or C(−1) sequences were resistant to cleavage. Altogether, DNA binding and cleavage experiments demonstrate that proto-spacer recognition by St-Cascade is PAM dependent, and that subsequent R-loop formation triggers dsDNA degradation by St-Cas3.
St-Cas3 cleaves DNA within the proto-spacer and upstream of the PAM
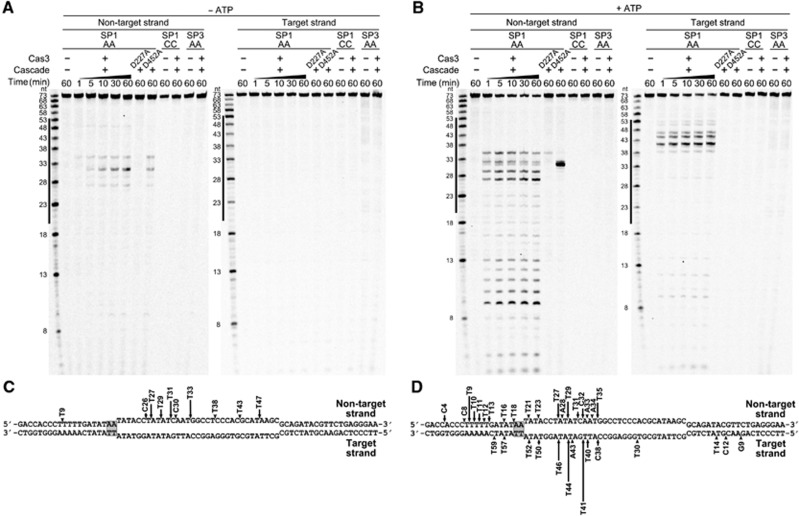
To map St-Cas3 cleavage sites, oligoduplexes SP1-AA, SP1-CC, and SP3-AA were 33P-5′-end-labelled either on the target or non-target strand, and St-Cas3-induced cleavage was assessed on each strand of each duplex in the absence or presence of ATP. In the absence of ATP, only a non-target DNA strand of the SP1-AA oligoduplex is cut in the proto-spacer region, while the target DNA strand is resistant to cleavage (Figure 4A and C). In contrast, in the presence of ATP, both target and non-target strands of the SP1-AA oligoduplex are cleaved at multiple positions (Figure 4B). The non-target strand is extensively cut within the proto-spacer and upstream of the PAM at the 5′-end proximal region. The target strand is extensively cleaved within the proto-spacer with minor cuts occurring at both the 5′- and 3′-proximal termini (Figure 4D). Consistent with plasmid DNA cleavage data, no St-Cas3-mediated cleavage was observed for the oligoduplex lacking a proto-spacer (SP3-AA) or with a mutated PAM sequence (SP1-CC), neither in the presence or absence of ATP. Furthermore, the nuclease-deficient mutant D227A did not cleave the SP1-AA oligoduplex. In contrast, ATPase-deficient D452A mutant cleaved only the non-target strand, in both the absence and presence of ATP (Figure 4). The cleavage pattern of the SP1-AA oligoduplex explains why the nicked DNA form is a major product during plasmid DNA cleavage by the D452 mutant or wild-type (WT) St-Cas3 in the absence of ATP. Interestingly, almost all cleavage sites are located at the 3′-end of pyrimidine (T and C) bases (Supplementary Figure S8). Preference for pyrimidine bases is also characteristic for the St-Cas3 cleavage of single-stranded oligodeoxynucleotides (Supplementary Figure S9). The non-target strand cleavage at the 5′-end proximal side of the oligoduplex suggests subsequent St-Cas3 translocation in the 3′→5′ direction, followed by DNA degradation.
Figure 4.
St-Cas3 cleavage of St-Cascade bound to target dsDNA. Oligoduplexes 33P-labelled in either the non-target or target strand were pre-incubated with St-Cascade without (A) or with (B) ATP and reaction products analysed in denaturating polyacrylamide gels and mapped on the SP1-AA oligoduplex sequence (C, D), respectively. Cleavage reactions were conducted at 37°C in a Nuclease buffer containing 0 mM (A) or 2 mM (B) ATP, 8 nM St-Cascade and 100 nM (B) or 500 nM (A) St-Cas3 or D227A and D452A mutants supplemented with 2 nM SP1-AA, SP1-CC, or SP3-AA 33P-labelled oligoduplexes. Solid lines designate proto-spacer boundaries. Arrows indicate cleavage positions, height of the arrow correlates with a relative amount of cleavage product after 10 min incubation.
Source data for this figure is available on the online supplementary information page.
DNA degradation by Cas3 in the St-CRISPR4-Cas system is unidirectional
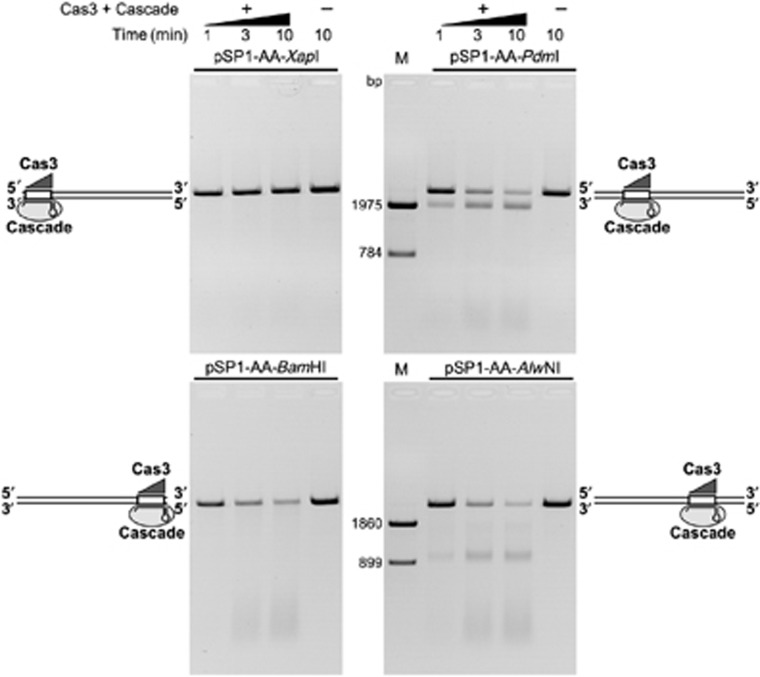
To determine whether the St-Cas3-mediated DNA cleavage is directional, we linearized pSP1-AA plasmid using four different restriction endonucleases (XapI, BamHI, PdmI, AlwNI) to generate a set of linear dsDNA molecules of identical length that have a proto-spacer sequence located at different distances with respect to DNA termini (Supplementary Figure S10). In the pSP1-AA-XapI DNA, the proto-spacer is located almost at the 5′-end, while in the pSP1-AA-PdmI DNA it is located ∼800 bp away from the 5′-terminus of the non-target strand. In two other linear DNA substrates, pSP1-AA-BamHI and pSP1-AA-AlwNI, the proto-spacer sequence is located at the 3′-end or ∼900 bp away from the 3′-end, respectively. Analysis of reaction products resulting from St-Cas3 cleavage in the in vitro reconstituted interference system revealed that the linear DNA pSP1-AA-XapI remained nearly intact, while the pSP1-AA-BamHI substrate was degraded in a similar fashion to the circular plasmid DNA. Furthermore, St-Cas3 acting on the pSP1-AA-PdmI and pSP1-AA-AlwNI substrates produced defined ∼1.9- and ∼0.9-kb products, respectively, while the remaining DNA fragments were degraded into smaller fragments (Figure 5). Interestingly, the reduced mobility of the ∼0.9-kb product resulting from the pSP1-AA-AlwNI cleavage (Supplementary Figure S11) suggests that the St-Cascade (St-Cacade-Cas3) complex remains bound to the cleaved DNA. Taken together, these data are consistent with a model in which Cas3 first makes a double-stranded break in a proto-spacer region (or in its immediate vicinity), and subsequently degrades DNA upstream of the proto-spacer in the 3′→5′ direction in respect to the non-target strand. Therefore, DNA downstream of the proto-spacer remains intact.
Figure 5.
St-Cas3-mediated DNA degradation is unidirectional. Four linear 2759, bp DNA fragments pSP1-AA-XapI, pSP1-AA-PdmI, pSP1-AA-BamHI or pSP1-AA-AlwNI that contain a proto-spacer sequence located at different distance in respect to DNA ends were generated by a restriction endonuclease cleavage of pSP1-AA plasmid. Nuclease reactions were initiated by addition of 20 nM St-Cascade and 100 nM St-Cas3 into a Nuclease buffer containing 5 nM of respective DNA. Linear DNA molecules with St-Cas3 and St-Cascade bound to a proto-spacer are schematically depicted at respective gel picture. M—DNA markers to map cleavage products were obtained by cleaving pSP1-AA with PdmI, XapI and AlwNI, XapI restriction endonucleases, respectively.
Source data for this figure is available on the online supplementary information page.
Discussion
CRISPR-Cas systems have been categorized into three main types that differ by the structural organization and function of RNP complexes involved in crRNA-mediated silencing of foreign nucleic acids. In the E. coli CRISPR-Cas system, a multisubunit RNP complex called Cascade binds to the target DNA and triggers degradation by an accessory Cas3 protein (Brouns et al, 2008). The CRISPR4-Cas system of S. thermophilus DGCC7710 (Horvath and Barrangou, 2010) displays a similar structural organization to that of E. coli (Figure 1A). To show that orthologous S. thermophilus Cas proteins assemble into a similar St-Cascade complex, we have isolated the St-Cascade and demonstrate that it recruits St-Cas3 to form a functional effector complex which cleaves target DNA in vitro. Moreover, we show that mechanistically the process of DNA interference provided by the St-CRISPR4-Cas system can be dissected into target site recognition and cleavage stages, which are executed by St-Cascade and St-Cas3, respectively.
Target DNA recognition by St-Cascade
St-Cascade isolated from the heterologous E. coli host carries a 61-nt crRNA with a 5′-OH and 3′-phosphate, which guides St-Cascade binding to the proto-spacer sequence in the target DNA. In the St-CRISPR4 array, the first 28 nt of the repeat are strictly conserved while the 29th nt is degenerated (C or T). Therefore, we postulate that in the St-CRISPR4 array the repeat and spacer sequences have a length of 28 and 33 nt, respectively. In the orthologous E. coli CRISPR-Cas system, 29-nt repeat and 32-nt spacer sequences were initially proposed (Ishino et al, 1987; Jansen et al, 2002). However, the analysis of sequences of newly inserted repeats in an E. coli CRISPR array in vivo showed that a base previously thought to belong to the repeat is actually derived from the proto-spacer (Goren et al, 2012; Swarts et al, 2012). Therefore, the conserved repeat sequence (‘duplicon’, Goren et al, 2012) in the E. coli CRISPR array was proposed to be 28 nt, delimiting 33-nt spacers. Both E. coli and S. thermophilus processed crRNAs (Figure 1) are composed of 61 nt, suggesting that in the precursor crRNA the cleavage position by Cas6e endoribonucleases is conserved and located at the 21st nt within the repeat sequence. In this case, the mature crRNAs are made of a 7-nt 5′ handle, a 33-nt spacer, and a 21-nt 3′ handle.
Similarly to other Type I systems, St-Cascade binding to the oligoduplexes containing a matching proto-spacer sequence requires a PAM sequence located in the vicinity of the proto-spacer. In the S. thermophilus CRISPR4 system, the PAM predicted by in silico analysis of the matching proto-spacer sequences in S. thermophilus phages is an AA dinucleotide located immediately upstream of the proto-spacer (Figure 2A). Surprisingly, according to EMSA experiments, the PAM required for St-Cascade binding to the proto-spacer sequence is limited to a single A(−1) or T(−1) nucleotide. Nucleotide replacement at the −1 position with a G or C abrogates St-Cascade binding to the proto-spacer, with the only exception of an A(−2)G(−1) dinucleotide that still functions as a PAM and promotes St-Cascade binding, albeit less efficiently than A(−1) or T(−1) variants. In the presence of the correct PAM, St-Cascade binds to the DNA containing a proto-spacer sequence with a sub-nanomolar Kd. It has been shown that in E. coli the L1 loop of CasA protein in Ec-Cascade is involved in the PAM recognition (Sashital et al, 2012). Sequence comparison suggests that a similar loop may be present in the CasA of St-Cascade, however, we were unable to locate a putative PAM recognition motif in the predicted L1 loop of S. thermophilus CasA protein. We suggest that PAM recognition is a key step that triggers subsequent St-crRNA binding to the matching DNA strand, presumably through the seed sequence in the immediate vicinity of the PAM (Semenova et al, 2011; Wiedenheft et al, 2011). Importantly, plasmid DNA cleavage in vitro by the St-Cascade-Cas3 complex shows the same promiscuous PAM pattern identified from the oligoduplex binding studies.
The discrepancy between the predicted and experimentally determined PAM may be due to several reasons. First, because only a small part of available phage sequence space is explored, it is possible that not all PAM variants were identified in the shallow subset of investigated phage genomes. Alternatively, the requirements for the PAM stringency may differ for the spacer acquisition and interference steps (Swarts et al, 2012). To escape CRISPR interference, bacteriophages often mutate PAM or proto-spacer sequences (Deveau et al, 2008), therefore a short and promiscuous PAM (such as A(−1) or T(−1)) identified for the St-CRISPR4-Cas system may be advantageous for interference. The PAM identified by in silico analysis of the matching proto-spacer sequences in S. thermophilus phages may reflect the more stringent PAM requirement for the spacer acquisition step, putatively executed by Cas1 and Cas2 proteins.
In Type III systems, self versus non-self DNA discrimination, which is essential for CRISPR-mediated immunity, is achieved through base-pairing interactions at −4, −3, and −2 positions upstream of the proto-spacer (Marraffini and Sontheimer, 2010). The complementarity at these positions between the crRNA and the matching bases in the repeat region in the host genome prevents host DNA cleavage. In the St-CRISPR4 repeat sequence, the C nucleotide is conserved at the −1 position and crRNA binding to the complementary spacer sequence will extend the base pairing into the repeat region. Importantly, the DNA containing the C(−1) base is neither bound nor cleaved by the St-Cascade-Cas complex, suggesting that complementary interactions at the −1 position may be important for self versus non-self DNA discrimination in the St-CRISPR4-Cas system.
DNA cleavage by St-Cas3 in the St-Cascade-target DNA complex
Nuclease P1 mapping of the St-Cascade-DNA complex suggests the formation of an R-loop where crRNA and the complementary target DNA strand are engaged into a heteroduplex, and the non-target strand is displaced as a ssDNA. The PAM sequence is critical for R-loop formation. Indeed, if G or C nucleotides are present at the −1 position, no specific St-Cascade binding and concomitant R-loop formation is detected. The R-loop formation is a key pre-requisite for the binding of St-Cas3 protein, which is a ssDNA nuclease that displays an ATPase/helicase activity stimulated by ssDNA (Sinkunas et al, 2011). St-Cas3 alone does not interact with a dsDNA and therefore first requires St-Cascade binding to the dsDNA to generate the R loop where the non-target strand of the proto-spacer is displaced as ssDNA and serves as a platform for the St-Cas3 loading. Indeed, the St-Cas3 ATPase activity is triggered only when the R loop is formed by St-Cascade binding. Our data are consistent with a mechanism proposed recently for the E. coli system where the Cse1(CasA) subunit of Ec-Cascade recruits Cas3, which subsequently catalyses nicking of target DNA through its HD-nuclease domain (Westra et al, 2012). In the absence of ATP, the oligoduplex substrate is only weakly cleaved by St-Cas3 and cleavage is limited to the non-target strand which is displaced as a ssDNA. Consistent with the oligoduplex cleavage pattern without ATP, plasmid DNA under these conditions is converted into a nicked form. In the presence of ATP, the cleavage pattern of both the oligoduplex and plasmid DNA is radically changed. St-Cas3 extensively cuts both DNA strands in the proto-spacer region of the oligoduplex and upstream of the PAM. The plasmid DNA in the presence of ATP is subsequently degraded by St-Cas3. In the Ec-CRISPR-Cas system, a role for supercoiling in the cleavage reaction efficiency has been recently demonstrated (Westra et al, 2012). Our data show that degradation rates of supercoiled and linear DNA forms by St-Cascade-Cas3 are very similar (Supplementary Figure S12). These results do not rule out supercoiling involvement in St-Cascade and target DNA interactions but rather indicate that supercoiling does not limit the cleavage reaction rate.
Our results also reveal that DNA degradation by the St-Cas3 nuclease is directional. After cleaving both DNA strands in the proto-spacer region, St-Cas3 further degrades DNA upstream of the proto-spacer in the 3′→5′ direction in respect to the non-target strand, while DNA downstream of the proto-spacer remains intact. St-Cas3 cleavage of the non-target strand in the 3′→5′ direction generates stretches of ssDNA on the target strand that can serve as a loading platform for the same or other St-Cas3 molecules, promoting further degradation.
It is tempting to speculate that the Cas3 cleavage directionality may contribute to the mechanism of adaptive spacer acquisition proposed recently for the E. coli K12 CRISPR system (Datsenko et al, 2012). According to this mechanism the DNA strand from which new spacers are selected is largely determined by the priming proto-spacer orientation. We show here that Cascade-crRNA binding to the matching proto-spacer sequence determines which strand will be extruded into the R-loop and subjected to degradation in the 3′→5′ direction. In this way, the unidirectional DNA degradation by Cas3 may contribute to the selection of a specific DNA strand from which new spacers are subsequently acquired (Datsenko et al, 2012).
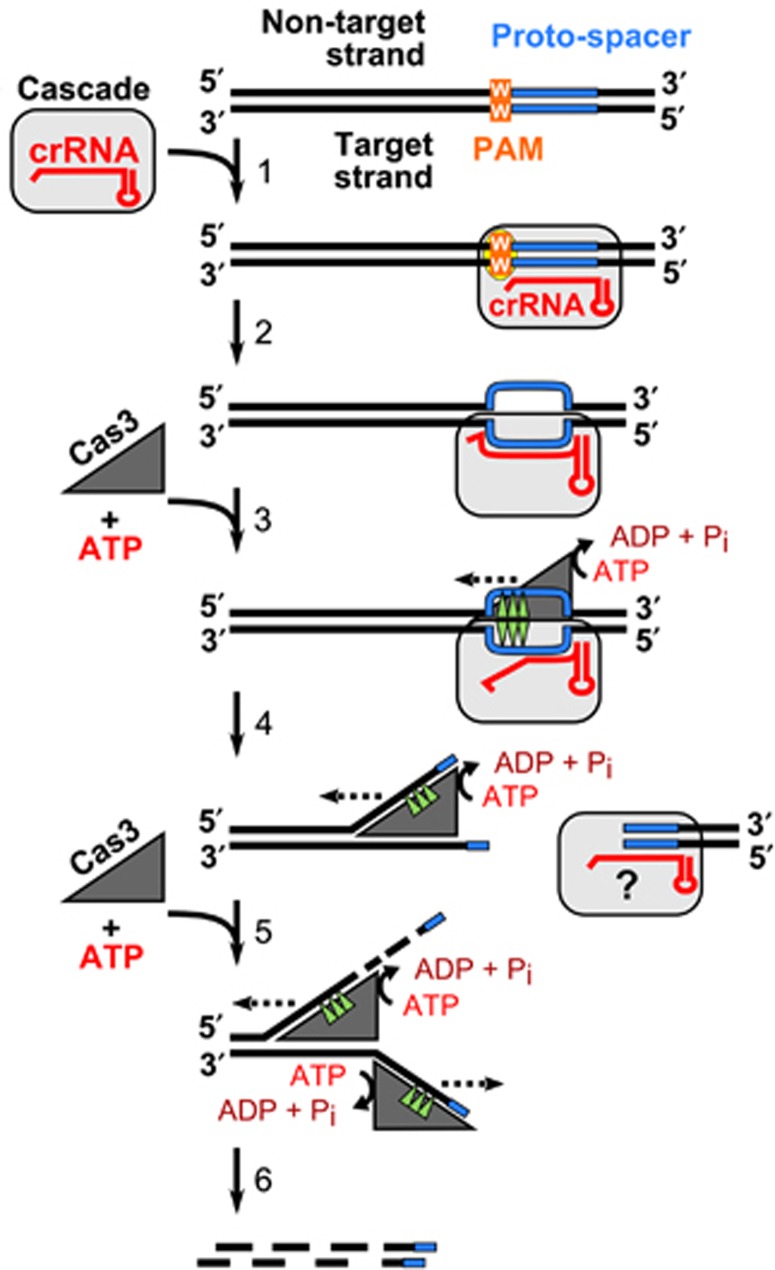
In summary, we have shown that St-Cascade recruits St-Cas3 to form a functional effector complex, which degrades target DNA in vitro. This establishes a molecular basis for CRISPR-based immunity in St-CRISPR4-Cas and other Type I systems (Figure 6). St-Cascade guided by the crRNA locates the target DNA site and, if the correct PAM sequence is present, binds to the matching DNA strand, creating an R-loop that serves as a loading site for the St-Cas3. St-Cas3 binding to the ssDNA triggers ATPase/helicase activity that presumably contributes to Cascade remodelling, making both DNA strands in the proto-spacer region available for Cas3 cleavage. After cleaving both DNA strands within the proto-spacer, Cas3 translocates on the non-target strand in the 3′→5′ direction in an ATP-dependent manner and cleaves the translocating strand using its HD-nuclease domain. This sets the stage for molecular exploitation of the Cas machinery for interference and DNA cleavage.
Figure 6.
CRISPR-mediated interference mechanism. Cascade binding to a matching proto-spacer in the presence of the correct PAM generates an R-loop where the crRNA and the complementary target DNA strand are engaged into a heteroduplex, while the non-target strand is displaced as a single-stranded DNA providing a platform for the Cas3 loading (1). ssDNA binding stimulates the Cas3 ATPase activity that may trigger Cascade remodelling making both DNA strands in the proto-spacer region available for the Cas3 cleavage (2). After cleaving both DNA strands at the proto-spacer Cas3 translocates on the non-target strand in the 3′→5′ direction (dashed line) in the ATP-dependent manner and chops the translocating strand using the HD-nuclease domain (3). A stretch of single-stranded DNA created on the complementary strand may promote binding of another Cas3 molecule (4) followed by concomitant cleavage resulting in the degradation of both strands of invading DNA (5).
Materials and methods
Cloning, expression, and purification of proteins
Streptococcus thermophilus DGCC7710 genomic DNA was used as a template for polymerase chain reactions (PCRs) to clone the casABCDE gene cassette and casC into pCDF-Duet1 and pBAD24-CHis expression vectors, respectively. A CRISPR locus containing six copies of the repeat-spacer-1 unit (6 × SP1) of the WT S. thermophilus CRISPR4 system was assembled from oligonucleotides and cloned into pACYC-Duet1 vector (Supplementary Table S3). Full sequencing of cloned DNA fragments confirmed perfect matches to the original sequences.
The St-Cascade complex was expressed in Escherichia coli BL21 (DE3) grown in LB broth (BD) supplemented with ampicillin (25 μg/ml), chloramphenicol (17 μg/ml), and streptomycin (25 μg/ml). Cells were grown at 37°C to OD600nm of ∼0.5 and expression was induced with 0.2% (w/v) arabinose and 1 mM IPTG for 3 h. Harvested cells were disrupted by sonication and cell debris removed by centrifugation. The St-Cascade complex was first purified on the Ni2+-charged HiTrap column (GE Healthcare) followed by Superdex 200 (HiLoad 16/60; GE Healthcare) and heparin (GE Healthcare) chromatography steps. The St-Cascade complex was stored at +4°C in a buffer containing 20 mM tris(hydroxymethyl)aminomethane (Tris)–HCl (pH 8) and 750 mM NaCl. The St-Cascade complex was subsequently analysed by SDS–PAGE and the sequence of the CasABCDE proteins was further confirmed by mass spectrometry of tryptic digests. Cascade complex concentration was estimated by Bradford assay (Fermentas) using bovine serum albumin (BSA) as a reference protein. Conversion to molar concentration was performed assuming that the St-Cascade stoichiometry CasA1:B2:C6:D1:E1:crRNA1 is analogous to that of the E. coli Cascade (Jore et al, 2011).
St-Cas3 and its mutants were produced and purified as previously described (Sinkunas et al, 2011).
Extraction of crRNA
Nucleic acids co-purified with St-Cascade were isolated by phenol:chloroform:isoamylalcohol (PCI) (25:24:1, v/v/v) extraction. Purified nucleic acids were incubated with DNase I (Fermentas) supplemented with 2.5 mM MgCl2 or RNase A/T1 (Fermentas) for 30 min at 37 °C. Nucleic acids were separated on a denaturing 15% polyacrylamide gel and visualized by SybrGold (Invitrogen) staining.
HPLC purification of crRNA
All samples were analysed by ion-pair reversed-phased-HPLC on an Agilent 1100 HPLC with UV260nm detector (Agilent) using a DNAsep column 50 mm × 4.6 mm I.D. (Transgenomic, San Jose, CA). The chromatographic analysis was performed using the following buffer conditions: (A) 0.1 M triethylammonium acetate (TEAA) (pH 7.0) (Fluka); (B) buffer A with 25% LC MS grade acetonitrile (v/v) (Fisher). The crRNA was obtained by injecting purified St-Cascade complex at 75°C using a linear gradient starting at 15% buffer B and extending to 60% B in 12.5 min, followed by a linear extension to 100% B over 2 min at a flow rate of 1.0 ml/min. Analysis of the 3′ terminus was performed by incubating the HPLC-purified crRNA in a final concentration of 0.1 M HCl at 4°C for 1 h. The samples were concentrated to 10–20 μl on a vacuum concentrator (Eppendorf) prior to ESI-MS analysis.
ESI-MS analysis of crRNA
ESI-MS was performed in negative mode using an UHR TOF mass spectrometer (maXis) (Bruker Daltonics), coupled to an online capillary liquid chromatography system (Ultimate 3000, Dionex, UK). RNA separations were performed using a monolithic (PS-DVB) capillary column (50 mm × 0.2 mm I.D., Dionex). The chromatography was performed using the following buffer conditions: (C) 0.4 M 1,1,1,3,3,3,-Hexafluoro-2-propanol (HFIP, Sigma-Aldrich) adjusted with triethylamine (TEA) to pH 7.0 and 0.1 mM TEAA, and (D) buffer C with 50% methanol (v/v) (Fisher). RNA analysis was performed at 50°C with 20% buffer D, extending to 40% D in 5 min followed by a linear extension to 60% D over 8 min at a flow rate of 2 μl/min, 250 ng crRNA was digested with 1 U RNase A and RNaseT1 (Applied Biosystems). The reaction was incubated at 37 °C for 4 h. The oligoribonucleotide mixture was separated on a PepMap C-18 RP capillary column (150 mm × 0.3 μm I.D., Dionex) at 50°C using gradient conditions starting at 20% buffer C and extending to 35% D in 3 min, followed by a linear extension to 60% D over 40 min at a flow rate of 2 μl/min. The mass spectrometer was set to select a mass range of 250–2000, m/z and the capillary voltage was kept at −3650 V. Oligoribonucleotides with −2 to −4 charge states were selected for tandem mass spectrometry using collision induced dissociation.
Electrophoretic mobility shift assays
Synthetic oligoduplexes (Metabion) 73 bp in length were used in EMSA experiments (Supplementary Table S3). Each oligoduplex contained a 33-bp proto-spacer sequence corresponding to the first spacer (spacer-1) of the S. thermophilus CRISPR4 locus and various PAM sequences. In control experiments, 73 bp oligoduplex containing a proto-spacer-3 instead of proto-spacer-1 was used. An oligodeoxynucleotide corresponding to the target strand was labelled at the 5′-end using T4 polynucleotide kinase (PNK) (Fermentas) and [γ33P]ATP (Hartmann Analytic) and an oligoduplex assembled by mixing the labelled target and unlabelled non-target strands at a molar ratio of 1:1.5, followed by annealing in 2 mM Tris–HCl buffer (pH 8). Increasing concentrations of St-Cascade were incubated with 0.1 nM of radioactively labelled oligoduplex in the binding buffer (40 mM Tris, 20 mM acetic acid, 1 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0, 150 mM NaCl, 0.1 mg/ml BSA, 10% glycerol) for 20 min at 37°C. The samples were subjected to electrophoresis in 8% (w/v) polyacrylamide gel, and visualized using a FLA-5100 phosphorimager (Fujilm). The Kd values for St-Cascade-DNA complexes were calculated as previously described (Tamulaitis et al, 2006). Kd values represent the average value of three independent experiments.
P1 nuclease footprinting
Oligoduplexes SP1-AA, SP1-AG, SP1-CC, and SP3-AA were 33P-5′-end-labelled at either the target or non-target strand for probing with P1 nuclease. Labelled oligoduplex at 2 nM concentration was incubated with or without 10 nM of Cascade complex at 37°C for 15 min in 20 μl of buffer containing 10 mM Tris–HCl (pH 8), 100 mM NaCl, and 0.1 mg/ml BSA. Then, 0.02 U of P1 nuclease (Sigma) in 20 μl of 30 mM sodium acetate buffer (pH 5.3) was added and incubated at 37°C for 10 min. The reactions were stopped by addition of phenol–chloroform followed by sodium acetate/isopropanol precipitation. The cleavage products were separated on a denaturing 20% polyacrylamide gel and visualized by autoradiography. Products of dideoxy sequencing reactions (‘Cycler Reader DNA Sequencing kit’; Fermentas) of TS132 or TS133 oligonucleotides were used as size markers.
ATPase assay
ATPase reactions were conducted at 37°C in the ATPase reaction buffer (10 mM Tris–HCl (pH 7.5 at 25°C), 75 mM NaCl, 40 mM KCl, 7% (v/v) glycerol, 0.1 mg/ml BSA, 1.5 mM MgCl2, 2 mM ATP) containing 3 nM supercoiled double-stranded plasmid (Supplementary Table S3), 12 nM of Cascade complex, and 300 nM of Cas3 or D452A mutant. Reactions were initiated by adding MgCl2 and ATP to a mixture of the other reaction components. Malachite green assay kit (BioAssay Systems) was used to measure ATP hydrolysis through the detection of liberated-free phosphate as previously described (Sinkunas et al, 2011).
Nuclease assay
Supercoiled or linearized pUC19 (Fermentas) or its derivative plasmids were used as substrates in the DNA cleavage assay. Cleavage reactions were performed at 37°C for indicated time intervals in the Nuclease buffer (10 mM Tris–HCl (pH 7.5), 75 mM NaCl, 40 mM KCl, 7% (v/v) glycerol, 1.5 mM MgCl2, 0.1 mM NiCl2, 2 mM ATP). Supercoiled or linearized plasmid DNA at 5 nM concentration was incubated with 20 nM of Cascade complex and 100 nM of Cas3 or its mutants unless otherwise stated. Reactions were initiated by addition of Cas3 and stopped by mixing with 3 × stop solution (67.5 mM EDTA, 27% (v/v) glycerol, 0.3% (w/v) SDS). Reaction products were analysed by 0.8% (w/v) agarose gels electrophoresis and visualized by ethidium bromide staining.
To monitor oligoduplex (Supplementary Table S3) cleavage, either the target or non-target strands were 33P-5′-end-labelled and 2 nM of labelled oligoduplex was incubated with 4 nM of Cascade complex and 100 or 500 nM of Cas3 in the presence or absence of ATP, respectively. The cleavage products were separated on a denaturing 20% polyacrylamide gel and visualized by autoradiography. Products of dideoxy sequencing reactions (‘Cycler Reader DNA Sequencing kit’; Fermentas) of 33P-5′-end-labelled oligodeoxynucleotides (Supplementary Table S3; section Cleavage markers) were used as size markers.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr Christophe Fremaux and members of the Siksnys laboratory for helpful discussions. This work was funded by the European Social Fund under Global Grant measure. MJD acknowledges support from the Engineering and Physical Sciences Research Council (UK) and the Biotechnology and Biological Sciences Research Council (UK).
Author contributions: TS, GG, and VS designed research; TS performed research; SPW and MJD performed MS analysis; PH and RB contributed new reagents/analytic tools; TS, GG, SPW, MJD, RB, PH, and VS analysed data; and TS, GG, and VS wrote the paper.
Footnotes
PH and RB are employees of DuPont Nutrition and Health. GG, VS, RB, and PH are inventors on patent applications related to CRISPR.
References
- Al-Attar S, Westra ER, van der Oost J, Brouns SJJ (2011) Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem 392: 277–289 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45: 273–297 [DOI] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E (2012) Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3: 945. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman MJ, Hornby DP (2006) Enrichment and analysis of RNA centered on ion pair reverse phase methodology. RNA 12: 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis M-È, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71 [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren MG, Yosef I, Auster O, Qimron U (2012) Experimental Definition of a Clustered Regularly Interspaced Short Palindromic Duplicon in Escherichia coli. J Mol Biol 423: 14–16 [DOI] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV 3rd, Graveley BR, Terns RM, Terns MP (2012) Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell 45: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coûté-Monvoisin A-C, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R (2008) Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190: 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169: 5429–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders APL, Dickman MJ, Doudna JA, Boekema EJ, Heck AJR, van der Oost J, Brouns SJJ (2011) Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18: 529–536 [DOI] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ (2010) Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463: 568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740 [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S (2011) Structural and biochemical analysis of the nuclease domain of the clustered regularly interspaced short palindromic repeat (CRISPR) associated protein 3(CAS3). J Biol Chem 3: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39: 9275–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA (2012) Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46: 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJJ, Severinov K (2011) Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA 108: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturino JM, Klaenhammer TR (2006) Engineered bacteriophage-defence systems in bioprocessing. Nat Rev Microbiol 4: 395–404 [DOI] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, Brouns SJ (2012) CRISPR Interference Directs Strand Specific Spacer Acquisition. PLoS ONE 7: e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Mucke M, Siksnys V (2006) Biochemical and mutational analysis of EcoRII functional domains reveals evolutionary links between restriction enzymes. FEBS Lett 580: 1665–1671 [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns RM (2011) CRISPR-based adaptive immune systems. Curr Opin Microbiol 14: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare SP, Pousinis P, Hornby DP, Dickman MJ (2009) Studying the mechanism of RNA separations using RNA chromatography and its application in the analysis of ribosomal RNA and RNA:RNA interactions. J Chromatogr A 1216: 1377–1382 [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJ, van der Oost J (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338 [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, Doudna JA (2011) RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA 108: 10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, Fremaux C, Horvath P, Barrangou R, Verberkmoes NC (2012) Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS One 7: e38077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, Naismith JH, Spagnolo L, White MF (2012) Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 45: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.