Abstract
EMBO J (2013) 32 3, 354–368 doi:; DOI: 10.1038/emboj.2012.315; published online September 30 2012
The ubiquitous bacterial second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) promotes biofilm formation by an astonishing variety of molecular mechanisms. Steiner et al (2012) now report that c-di-GMP directly stimulates interaction and enzymatic activity of two subunits of the membrane-integrated machinery that produces and secretes the poly-β-1,6-N-acetylglucosamine (PGA) exopolysaccharide in Escherichia coli.
Second messenger signalling has recently been pushed back into the focus of molecular microbiology when c-di-GMP emerged as a molecule that almost ubiquitously promotes the formation of bacterial biofilms. These are surface-attached multicellular communities with cells embedded in a self-produced matrix consisting of adhesins, amyloid proteins, exopolysaccharides and exo-DNA (Karatan and Watnick, 2009). Due to their resistance against antibiotics, desinfectants and the attacks of the immune system, biofilms are involved in chronic infections and in general cause medical harm and technical damage. The synthesis of c-di-GMP seems to represent a common principle in the formation of otherwise highly diverse and species-specific biofilms, and therefore holds promise as a target for general anti-biofilm drugs.
In addition, c-di-GMP signalling is fascinating due to a striking complexity not previously observed for any other bacterial second messenger. On the one hand, the enzymes that ‘make and break’ this cyclic di-nucleotide—diguanylate cyclases (DGC) characterized by GGDEF domains and specific phosphodiesterases (PDE) which carry either EAL or HD-GYP domains—occur in multiples, with sometimes up to 100 or even more GGDEF/EAL/HD-GYP domain proteins in single species. Based on this multiplicity and the observation that many of these proteins seem to be part of larger protein complexes, sequestration of c-di-GMP control modules and local signalling has been proposed (Jenal and Malone, 2006; Hengge, 2009). On the other hand, c-di-GMP acts by a plethora of different molecular mechanisms (summarized in Hengge (2010)). In some species, c-di-GMP binds to untranslated 5′ regions of mRNAs, which act as riboswitches that control transcriptional elongation, transcript processing or translation. Perhaps more common, also proteins of different classes serve as c-di-GMP-binding effectors, and act as transcription factors, as regulatory subunits or domains of enzymes as seen for cellulose synthase or as components that directly target and control the activity of larger cellular structures such as the basal body of the flagellum (Boehm et al, 2010).
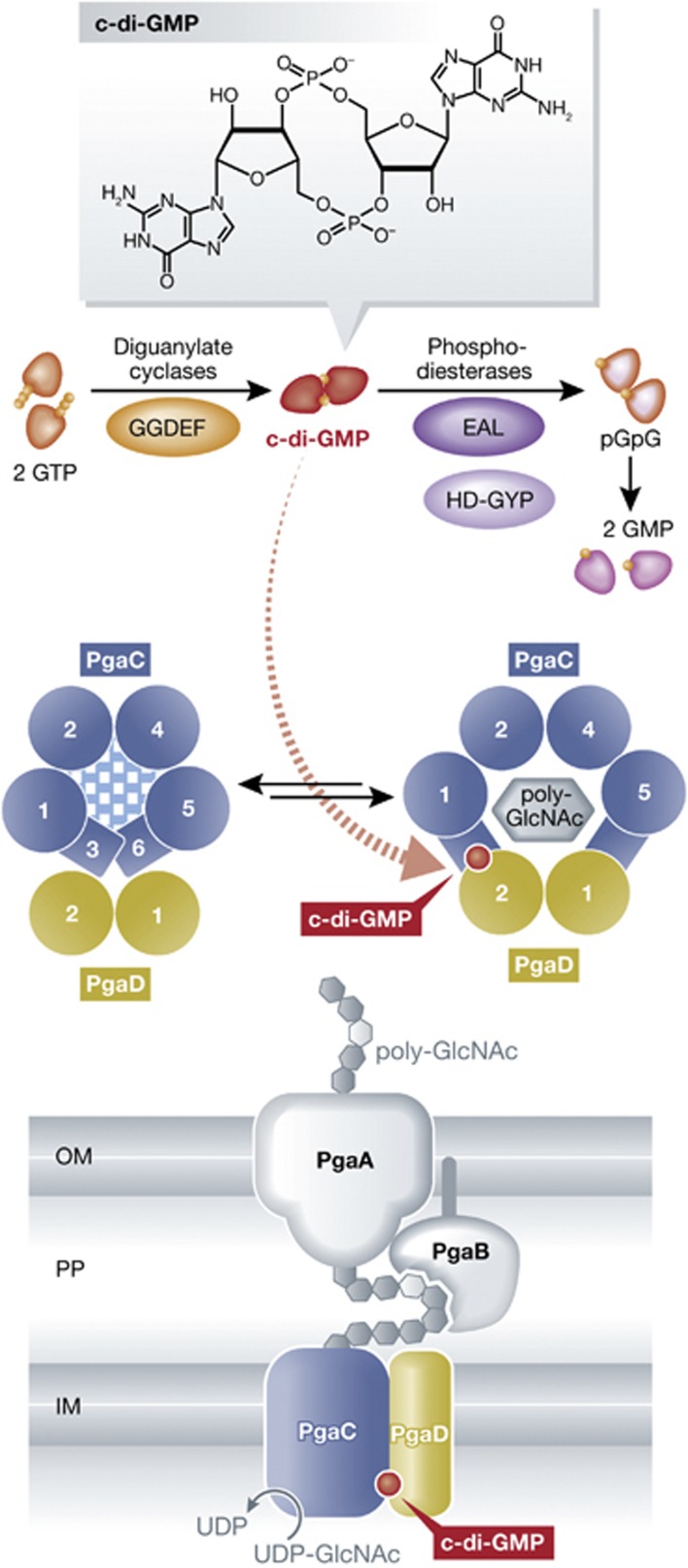
In this issue of EMBO Journal, Steiner et al (2012) now report another novel and so far unique molecular mechanism of c-di-GMP action. The production and secretion of the biofilm-associated exopolysaccharide PGA in E. coli is mediated by a cell envelope-spanning machinery that includes the β-glycosyltransferase (GT) PgaC and the small but crucial protein PgaD in the cytoplasmic membrane, as well as the lipoprotein PgaB and the porin PgaA in the outer membrane (all encoded in the single operon pgaABCD). Based on earlier observations that PGA synthesis and secretion is somehow post-transcriptionally activated by c-di-GMP and in fact in particular by the DGC YdeH (Boehm et al, 2009), this new paper now presents genetic and biochemical evidence that c-di-GMP binds concomitantly to the inner-membrane components PgaC and PgaD and stimulates their direct interaction in a manner that activates GT activity. These findings as well as structural predictions and the analysis of point mutations in PgaD and PgaC, which reduce c-di-GMP binding and/or render PgaCD activity independent of c-di-GMP, allow the authors to come up with a speculative but precise model: c-di-GMP binding stabilizes a heteromeric PgaCD complex, in which the two predicted transmembrane alpha helices of PgaD may team up with the predicted four transmembrane and two membrane-associated alpha helices of PgaC in a conformation that not only stimulates GT activity but also provides a pore large enough for PGA secretion into the periplasm (see Figure 1).
Figure 1.
C-di-GMP accumulation promotes complex formation of PgaC and PgaD resulting in synthesis and secretion of the biofilm matrix exopolysaccharide poly-GlcNAc (PGA). Among the diguanylate cyclases (which carry GGDEF domains) in E. coli, mainly YdeH is involved in generating c-di-GMP that binds simultanously to the membrane-inserted proteins PgaC and PgaD. C-di-GMP binding stabilizes the PgaCD complex, activates its glycosyltransferase activity and allows secretion of the product PGA. Without c-di-GMP bound, PgaD is subject to proteolysis. Circles with numbers symbolize the transmembrane alpha helices of PgaC and PgaD as seen perpendicular to the plane of the cytoplasmic membrane. For further details, see text.
In addition, it was observed that PgaD is degraded when the cellular c-di-GMP production is low (in mutants that lack the DGC YdeH alone or in combination with additional DGCs). This suggests a powerful mechanism for the shut-off of PGA synthesis and secretion that can only be reversed by resynthesis of PgaD. This synthesis, however, requires the sustained inhibition of the RNA-binding protein CsrA by two small RNAs, CsrB and CsrC (Suzuki et al, 2002)—otherwise, CsrA would interfere with the translation of pgaABCD mRNA (Wang et al, 2005). CsrA is actually a global regulator that binds to many mRNAs, including that of the DGC YdeH, which is also downregulated (Jonas et al, 2008). In other words, regulation by the RNA-based Csr system combined with PgaD proteolysis provides a fine-tuned and highly dynamic regulatory checkpoint for rapidly and efficiently turning on and off PGA synthesis in response to still unknown environmental and/or stress conditions.
Overall, the study by Steiner et al (2012) demonstrates a novel mechanism of c-di-GMP action that extends the currently known functional repertoire of second messengers and provides a conceptual framework for the future analysis of other and often more complex synthesis machineries for biofilm-related exopolysaccharides. Future challenges with the PGA system include the structural analysis of the c-di-GMP-modulated PgaCD complex to unravel mechanistic aspects in greater detail. Also, the question arises why a distinct DGC, that is, YdeH, seems to have relative priority over other DGCs in activating the Pga system. Although in terms of expression, YdeH is one of the minor DGCs (Sommerfeldt et al, 2009), its expression and possibly its activity are stimulated under the particular conditions where CsrA is inactivated and PGA production is turned on; alternatively, YdeH signalling in PgaCD activation could be more direct. In any case, it may be predicted that bacterial second messengers and, in particular, c-di-GMP will continue to surprise us with much more complex and diverse mechanisms of action than ever anticipated.
Footnotes
The author declares that she has no conflict of interest.
References
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackerman M, Kaever V, Sourjik V, Roth V, Jenal U (2010) Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141: 107–116 [DOI] [PubMed] [Google Scholar]
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackerman M, Schirmer T, Jenal U (2009) Second messenger signaling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 72: 1500–1516 [DOI] [PubMed] [Google Scholar]
- Hengge R (2009) Principles of cyclic-di-GMP signaling. Nat Rev Microbiol 7: 263–273 [DOI] [PubMed] [Google Scholar]
- Hengge R (2010) Cyclic-di-GMP reaches out into the bacterial RNA world. Sci Signal 3: pe44. [DOI] [PubMed] [Google Scholar]
- Jenal U, Malone J (2006) Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40: 385–407 [DOI] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors O (2008) The RNA binding protein CsrA controls c-di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70: 236–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E, Watnick P (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73: 310–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R (2009) Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155: 1318–1331 [DOI] [PubMed] [Google Scholar]
- Steiner S, Lori C, Boehm A, Jenal U (2012) Allosteric acivation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein–protein interaction. EMBO J 32: 354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T (2002) Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol 184: 5130–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T (2005) CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56: 1648–1663 [DOI] [PubMed] [Google Scholar]