Abstract
Gene silencing in budding yeast relies on the binding of the Silent Information Regulator (Sir) complex to chromatin, which is mediated by extensive interactions between the Sir proteins and nucleosomes. Sir3, a divergent member of the AAA+ ATPase-like family, contacts both the histone H4 tail and the nucleosome core. Here, we present the structure and function of the conserved C-terminal domain of Sir3, comprising 138 amino acids. This module adopts a variant winged helix-turn-helix (wH) architecture that exists as a stable homodimer in solution. Mutagenesis shows that the self-association mediated by this domain is essential for holo-Sir3 dimerization. Its loss impairs Sir3 loading onto nucleosomes in vitro and eliminates silencing at telomeres and HM loci in vivo. Replacing the Sir3 wH domain with an unrelated bacterial dimerization motif restores both HM and telomeric repression in sir3Δ cells. In contrast, related wH domains of archaeal and human members of the Orc1/Sir3 family are monomeric and have DNA binding activity. We speculate that a dimerization function for the wH evolved with Sir3’s ability to facilitate heterochromatin formation.
Keywords: dimerization, heterochromatin, Orc1, Sir3, winged helix-turn-helix
Introduction
Large regions of eukaryotic chromatin assume an epigenetically heritable structure that is refractory to gene expression, called heterochromatin or, in budding yeast, silent chromatin. From yeast to man, gene silencing is thought to stem from a more compact folding of the chromatin fibre that sterically restricts DNA accessibility (Gottschling, 1992; Singh and Klar, 1992; Loo and Rine, 1994). Covalent modifications of histone proteins play a major role in determining chromatin compaction, and hypoacetylated histones are characteristic of silent chromatin in budding yeast (Braunstein et al, 1993; Suka et al, 2001). In addition, the binding of non-histone complexes along the nucleosomal fibre can repress productive transcription. In budding yeast, this is achieved by the binding of a heterotrimeric complex of Silent Information Regulator (Sir) proteins Sir2, Sir3 and Sir4, each of which is essential for gene silencing (Rine and Herskowitz, 1987).
The Sir proteins themselves do not recognize specific DNA sequences, but are recruited to discrete loci through protein–protein interactions with multifunctional factors like Rap1, ORC and Abf1, which recognize specific DNA motifs at telomeres and in silencer elements that flank the silent homothallic mating type (HM) loci. At HM loci, an intermediary protein called Sir1 bridges between these factors and the Sir complex (reviewed in Rusche et al, 2003). From these initial nucleation sites, the Sir complex spreads for 3–20 kb along the chromatin fibre (Hecht et al, 1996; Strahl-Bolsinger et al, 1997).
The establishment of silent chromatin requires the NAD-dependent deacetylase activity of Sir2 (Tanny et al, 1999; Imai et al, 2000; Smith et al, 2000; Yang and Kirchmaier, 2006), while Sir3 and Sir4 are thought to play structural roles in the silent chromatin formation (reviewed in Gasser and Cockell, 2001; Rusche et al, 2003; and Moazed et al, 2004). Sir4 forms a tight complex with Sir2, and stimulates Sir2 activity in vitro (Ghidelli et al, 2001; Hoppe et al, 2002; Tanny et al, 2004; Cubizolles et al, 2006). The C-terminal coiled-coil domain of Sir4 dimerizes and interacts with Sir3; this interaction is essential for silencing, because point mutations in this Sir3–Sir4 interface abolish assembly of the trimeric Sir complex (Chang et al, 2003; Rudner et al, 2005; Ehrentraut et al, 2011).
The spread of Sir-mediated repression is limited both by histone modifications that lower affinity of Sir3 for chromatin and by the limited concentration of Sir proteins in the nucleus (Maillet et al, 1996; Marcand et al, 1996; van Leeuwen et al, 2002; Martino et al, 2009; Oppikofer et al, 2011). The overexpression of Sir3, or a balanced overexpression of Sir4 and Sir3, leads to the extension of the silent chromatin domains at telomeres or enhanced repression at silencer-flanked reporter genes (Renauld et al, 1993; Gotta and Gasser, 1996; Hecht et al, 1996; Maillet et al, 1996; Strahl-Bolsinger et al, 1997), suggesting that Sir3, and its dosage, plays major roles in Sir complex spreading and transcriptional repression.
SIR3 arose from the duplication of the ORC1 gene (Kellis et al, 2004), and both encode a highly conserved N-terminal BAH domain, a C-terminal AAA+ ATPase-like domain, plus an extreme C-terminal domain (Gaudier et al, 2007). A chimeric protein formed by exchanging the BAH domain of Sir3 with that of Orc1 can restore mating in a sir3Δ strain, but no other Orc1 subdomain is able to support silencing when integrated into Sir3 (Bell et al, 1995). Overexpression of a N-terminal domain of Sir3 reinforces telomere-proximal silencing in a SIR+ strain and partially restores mating at HM loci in a sir3Δ background if Sir1 is overexpressed (Gotta et al, 1998; Connelly et al, 2006). Consistently, biochemical and structural analyses show that the Sir3 BAH domain interacts with the nucleosome (Onishi et al, 2007; Buchberger et al, 2008; Sampath et al, 2009; Armache et al, 2011). Both H3K79 methylation by Dot1 and H4K16 acetylation by Sas2 reduce the association of Sir3 BAH with chromatin (Onishi et al, 2007), or of holo-Sir3 with nucleosomes in vitro (Martino et al, 2009; Oppikofer et al, 2011), indicating that the BAH domain helps to restrict Sir3-mediated silencing to unmodified nucleosomes.
The AAA+ ATPase-like (AAA) domain of Sir3 (aa 530–845) has lost its ATPase activity, but has gained specific contacts with both Sir4 and unmodified nucleosomes (Ehrentraut et al, 2011). Mutagenesis confirmed that both interactions are essential for silencing. A third structural subdomain of Sir3 occupies its extreme C-terminus (aa 843–978). When fused to lexA and targeted to a subtelomeric reporter, this fragment can recruit Sir proteins and nucleate transcriptional repression (Liaw and Lustig, 2006). It was also reported to self-associate in a yeast-two-hybrid assay (Liaw and Lustig, 2006). Pull-down experiments argued that a slightly longer Sir3 C-terminal fragment (aa 832–978) might bind a central domain of Sir3 (aa 464–728), which contains the N-terminal portion of the AAA+ ATPase module (King et al, 2006). While these results were suggestive of homo- or hetero-dimerization activities, the importance of such interactions for silencing or their redundancy with other potential dimerization domains in Sir3 was never rigorously tested. Intriguingly, this C-terminal domain is conserved among Orc1 homologues and corresponds to the domain III of ORC/CDC6, which in A. pernix and other archaea binds DNA (Bell et al, 1995; Liu et al, 2000; De Felice et al, 2004; Singleton et al, 2004). X-ray structure analysis showed that the archaeal C-terminal domain adopts a winged helix-turn-helix fold (Dueber et al, 2007; Gaudier et al, 2007). However, the sequence identity between the C-termini of archaeal Orc1 and yeast Sir3 is very low (13%), rendering cross-species structure-function predictions unreliable.
To sort out the role of this extreme C-terminal domain of Sir3, we first solved its X-ray crystal structure to 2.7 Å resolution. These 138 amino acids form a winged helix-turn-helix variant (Sir3 wH) that is reminiscent of domain III of Orc1/Cdc6, yet the Sir3 wH has acquired a strong homodimerization function and fails to bind DNA. We show that deletion of the domain (Sir3ΔwH), or loss of its dimerization potential, ablates silencing, which can be restored by attaching a bacterial dimerization motif to Sir3ΔwH. With a combination of structural, biochemical and genetic analyses we demonstrate that the last 138 aa of Sir3 evolved as the crucial homodimerization motif within Sir3, that is essential for the assembly of Sir-dependent silent chromatin in yeast. Distinct features of the wH structure distinguish human and archaeal Orc1 from the silencing-competent S. cerevisiae Sir3 paralogue.
Results
The C-terminal 138 aa of Sir3 form a winged helix-turn-helix variant that homodimerizes
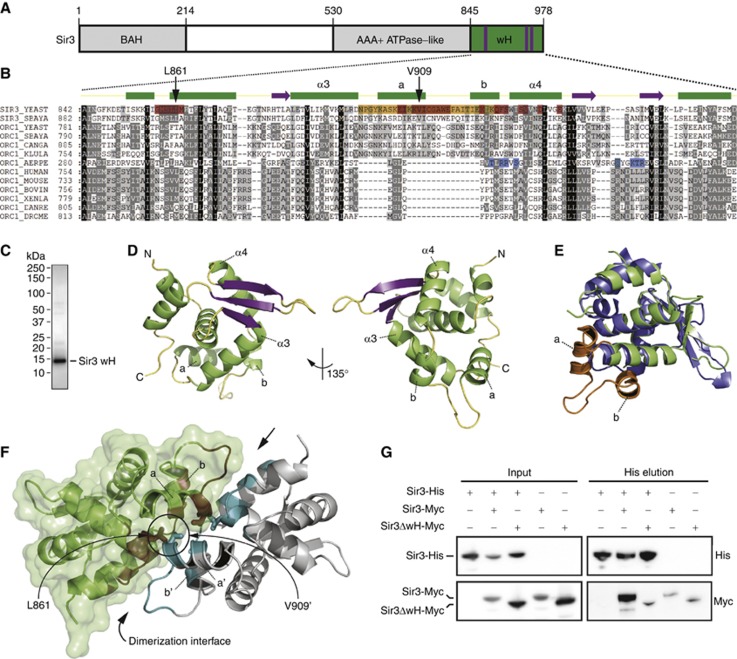
SIR3 arose from the duplication of the ORC1 gene with which it shares a multidomain organization (Figure 1A; Bell et al, 1995; Kellis et al, 2004). While the extreme C-terminal region of archaeal A. pernix Orc1 folds as a winged helix-turn-helix domain that binds DNA (De Felice et al, 2004; Dueber et al, 2007; Gaudier et al, 2007), the function of the homologous domain in ScSir3 was unclear. The multiple sequence alignment of selected archaea, vertebrate, and yeast Orc1/Sir3 wH domains revealed important evolutionary changes (Figure 1B). Notably, yeast Orc1/Sir3 wH domains possess a ∼30 residues insertion (Figure 1B, orange residues) which is missing in archaea and higher eukaryotic Orc1 wH domains. In addition, the specific ApOrc1 residues implicated in DNA binding (Gaudier et al, 2007) are poorly conserved in yeast and higher eukaryotes (Figure 1B, blue residues).
Figure 1.
The C-terminal 138 residues of Sir3 fold into a wH domain and homodimerize. (A) Schematic representation of the Sir3 protein. The C-terminal wH domain in green has been crystallized. Purple bars show the position of the 3-stranded β-sheet wing. (B) Multiple sequence alignment of Orc1/Sir3 wH modules from archaea, budding yeasts and higher eukaryotes. Conserved residues are highlighted in black (100%), dark grey (50%) and light grey (20%). ScSir3 wH residues that contribute to the dimerization interface are in red, the 30-aa insertion is highlighted in orange, and ApOrc1 wH residues that contact DNA are in blue (Gaudier et al, 2007). Above the alignment is the secondary structure of Sir3 wH colour coded as in (D). The L861 and V909 residues mutated in the Sir3-AA construct are highlighted. (C) 2 μg of purified Sir3 wH (aa 840–978) was loaded on an SDS–PAGE and stained with Coomassie blue. (D) Representation of the Sir3 wH structure; α-helices are represented in green, β-sheets in purple and coils in yellow. (E) Superposition of Sir3 and ApOrc1 (PDB 2v1u; Gaudier et al, 2007). The insertion, absent in ApOrc1, forming most of the Sir3 wH homodimerization interface is in orange (see sequence alignment) and displayed as dumbbell-shaped helices (a, b). (F) Representation of a Sir3 wH dimer, the residues involved in the hydrophobic interface are coloured in red and cyan for the green and grey Sir3 wH monomers, respectively. The surface representation of the green Sir3 wH monomer is displayed in light green. The insertion crucial for Sir3 wH dimerization is displayed as dumbbell-shaped helices. (G) His-tagged full-length Sir3 (Sir3-His) was co-expressed in insect cells with Myc-tagged full-length Sir3 (Sir3-Myc) or Sir3 lacking the wH module (Sir3ΔwH-Myc; aa 1–850). Sir3-His was affinity purified, input and His elution were analysed by western blotting using the indicated antibody.
To determine the biochemical and structural features of the predicted Sir3 wH, we expressed the last 138 aa of Sir3 (aa 840–978) in E. coli and purified the protein to ≥95% purity (Figure 1C). Surprisingly, the molecular mass of the purified Sir3 840-978 aa (15.9 KDa) fragment, determined by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) was 33.1 kDa, consistent with recombinant Sir3 840–978 aa being a homodimer in solution (Supplementary Figure S1). The folding of this C-terminal module was determined by solving its X-ray crystal structure.
The extreme C-terminal domain of Sir3 crystallized in space group P3212 with one molecule per asymmetric unit and its structure was solved to 2.7 Å resolution by the multi-wavelength anomalous diffraction (MAD) method using a seleno-methionine derivative (Table I). The final crystal structure displays clear electron density for residues 849–975, that fold into a wH variant featuring a total of 7 α-helices and a 3 stranded β-sheet wing (Figure 1D). A search of the Protein Data Bank (PDB) using DALI (Holm and Rosenstrom, 2010) indicated that the wH domains of archaeal Orc1 (A. pernix, 2V1U) (Gaudier et al, 2007), Orc2 (A. pernix, 1W5T) (Singleton et al, 2004), CDC6P (P. aerophilum, 1FNN) (Liu et al, 2000), and CDCP6 (S. solfataricus, 2QBY) (Dueber et al, 2007) are most similar.
Table 1. Data collection and refinement statistics.
aData collection statistics is reported for unmerged Friedel pairs.
bValues in parentheses refer to the highest resolution shell.
Superposition of Sir3 wH with an archaeal homologue (ApOrc1 wH; Gaudier et al, 2007; Figure 1E) showed that the core helix-turn-helix structures and the β-sheet wing are highly similar (RMSD of 2 Å), but also revealed important structural differences. The most significant differences are due to an additional stretch of 30 aa we identified by sequence alignment (Figure 1B, orange residues), which is absent in the archaeal ApOrc1. This insert is found between the wH core α-helices three (α3, aa 883–896) and four (α4, aa 928–938) and forms two additional α-helices (a, b) that are connected by an elongated turn in coiled conformation (Figure 1B and E; highlighted in orange).
Whereas the asymmetric unit of Sir3 in the crystal contains one Sir3 wH module, the crystal packing reveals two symmetry-related Sir3 wH proteins in the crystal lattice (Figure 1F). The ∼30 aa α-helical insertion contributes most to the buried solvent-accessible surface area of the protein–protein interface covering 1500 Å2, with leucine 861 and valine 909 being located in the hydrophobic core of this contact (Figure 1F; contact surfaces of the two molecules are represented in cyan and dark red). This large contact surface is likely to account for the strong homodimerization observed in solution (Supplementary Figure S1), as predicted by PISA (Protein Interfaces, Surfaces and Assemblies) calculations based on the crystal packing (Krissinel and Henrick, 2007).
In order to test the importance of the wH module for Sir3 dimerization in a competitive assay, we co-expressed in insect cells a His-tagged full-length Sir3 (Sir3-His) with a Myc-tagged full-length Sir3 (Sir3-Myc) or Sir3 lacking the wH module (Sir3ΔwH-Myc; aa 1–850). Affinity purified Sir3-His was tested for interaction with Sir3-Myc and Sir3ΔwH-Myc using an anti-Myc antibody on proteins recovered on Ni beads, which bind the His tag. Strikingly, we observed that the deletion of the wH domain reduced Sir3 self-association to background levels, whereas full-length Sir3-Myc was efficiently recovered with full-length Sir3-His (Figure 1G). Sedimentation analysis by glycerol density gradients showed in addition that Sir3ΔwH-His sedimented more slowly than full-length Sir3-His, consistent with a loss of dimerization (Supplementary Figure S2B). Moreover, we were unable to detect any interaction of Sir3 wH with the Sir3 AAA domain by pull-down assays or isothermal titration calorimetry (Supplementary Figure S3B and C). This suggests that Sir3 wH-mediated homodimerization accounts for most, if not all, Sir3 self-interaction, at least in its soluble form.
The wH fails to interact with DNA or chromatin but promotes Sir3 loading onto nucleosomes
Past work has implicated both the BAH and the AAA domains in dimerization, and in the spreading of Sir3 along the nucleosomal fibre (Connelly et al, 2006; Armache et al, 2011; Ehrentraut et al, 2011). In addition, the Sir3 wH has been proposed to heterodimerize by binding a central Sir3 domain (King et al, 2006; Liaw and Lustig, 2006), while the archaeal ORC/CDC6 wH modules were shown to bind to DNA in concert with the AAA+ ATPase-like domain (De Felice et al, 2004; Singleton et al, 2004; Dueber et al, 2007; Gaudier et al, 2007). Importantly, however, the residues implicated in DNA binding within ApOrc1 (labelled in blue), are not conserved in Sir3 wH (Figures 1B and 2A; Gaudier et al, 2007).
Figure 2.
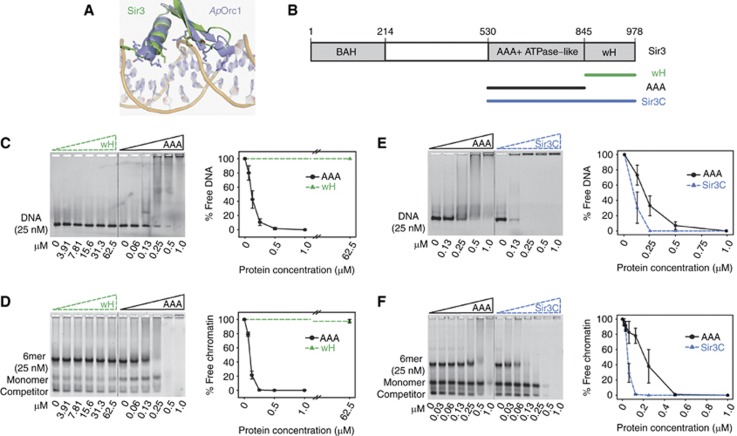
The association of the Sir3 C-terminus to DNA and chromatin is promoted by wH homodimerization. (A) Details on the residues of ApOrc1 wH (blue) interacting with DNA and superposition with the corresponding region of Sir3 (green). (B) Scheme of the modular organization of the Sir3 protein. Constructs used are highlighted. The indicated Sir3 domains were titrated onto a constant amount of Cy3-labelled 147 bp DNA (C, E) or a 6-mer of nucleosomes (D, F). Samples were separated on a native agarose gel, the nucleosomal template was stained with SYBR® safe and the amount of unbound probe was quantified by the intensity of the fluorescent signal. The images are representatives for three or more experiments; quantification represents mean values and s.e.m. of the % of free probe normalized to the input.
To test various models of DNA binding, domain interaction and cooperation in chromatin association, we purified two additional Sir3 C-terminal fragments: the AAA+ ATPase-like (AAA) domain alone (aa 530–845; Ehrentraut et al, 2011) and the entire Sir3 C-terminal half, which includes both AAA and wH domains (Sir3C; aa 527–978; Figure 2B; Supplementary Figure S3A). We first incubated increasing amounts of the Sir3 wH and the AAA module with a constant amount of a Cy3-labelled 147 bp DNA (Huynh et al, 2005; Martino et al, 2009), and monitored interaction by gel shift assay. Scoring the loss of unbound DNA template, we found that Sir3 AAA readily binds DNA, while Sir3 wH does not, even at 2500-fold molar excess (Figure 2C). To see if the Sir3 wH instead binds a chromatin template, we repeated the titration into a constant amount of hexameric (6-mer) nucleosomal arrays, reconstituted from bacterially expressed histones and the Widom 601 repeat (Martino et al, 2009). The AAA domain efficiently interacted with this array (see also Ehrentraut et al, 2011), while Sir3 wH was unable to bind chromatin, even at 2500-fold molar excess (Figure 2D).
In order to test whether the dimerization capacity of the wH domain contributes to the DNA and chromatin binding properties of Sir3 AAA, we compared the binding of the AAA domain with the larger Sir3C fragment, which includes the wH domain (Figure 2B). Strikingly, the presence of the wH, which does not interact with DNA per se, increased Sir3C-DNA binding affinity by approximately 3- to 4-fold over that of the AAA module alone (based on apparent IC50 from gel shift quantitation, Figure 2E). Similarly, when we compared the interaction of the AAA and the larger Sir3C domains (containing the wH domain) with the 6-mer array, the loading of Sir3C onto chromatin was strikingly more efficient (Figure 2F). We found no evidence for heterodimerization between the Sir3 wH and AAA domains by pull-down and isothermal titration calorimetry experiments (Figure 1G; Supplementary Figures S2 and S3). Thus, while we cannot exclude that wH-AAA interactions might occur after the Sir complex is loaded onto chromatin, our data argue that homodimerization of the wH module within the full Sir3C domain stabilizes the association of Sir3C molecules with chromatin, indirectly increasing affinity for the 6-mer array. Contacts with nucleosomes and DNA, on the other hand, are mediated exclusively by the AAA domain (Figure 2, see also Ehrentraut et al, 2011).
Sir3 wH is essential for silencing in vivo, and can be substituted by a bacterial dimerization motif
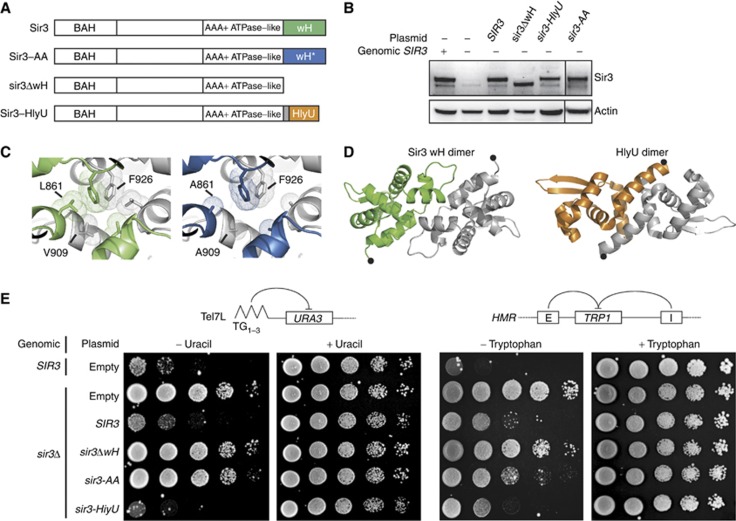
To test whether Sir3 dimerization through its wH module is important to form silent chromatin in vivo, we examined the restoration of silencing in a sir3Δ strain complemented with various Sir3 constructs expressed from a plasmid (Figure 3A). The constructs were cloned together with 1 kb of Sir3 5′- and 3′-UTR to ensure endogenous expression levels, which were checked routinely by western blot (Figure 3B). As expected, expression of full-length SIR3 fully restored silencing of reporter genes inserted at telomere 7L and to a large extent at the HMR locus (Figure 3E). We then tested the complementation efficiency of a construct lacking the wH module (Sir3ΔwH; aa 1–850) which fails to dimerize in vitro (Supplementary Figure S2B). Importantly, we found that Sir3ΔwH, which was expressed at wild-type Sir3 levels, failed to restore silencing at telomere 7L and HMR (Figure 3B and E).
Figure 3.
Sir3 wH homodimerization is essential for telomeric and HMR silencing. (A) Schematic representation of the Sir3 constructs used. Sir3 wH wild-type (green), the L861A-V909A mutant (blue) and the HlyU module from Vibrio vulnificus (orange) are highlighted. (B) Western blot analysis of alkaline yeast extracts (Ruault et al, 2011) of the strains used in (E) show wild-type levels of Sir3 expression, using a polyclonal antibody raised against full-length Sir3 (a kind gift from L Pillus). (C) Modelled representation of the impact L861A and V909A mutations on the extent of the hydrophobic Sir3 wH dimerization interface. (D) Representation of the Sir3 wH or the HlyU dimer. The most C-terminal residues are highlighted by a black dot. (E) The indicated strain was grown overnight in selective media and 10-fold dilutions were dropped on the indicated media. Ability to repress a URA3 gene inserted at the telomere 7L was tested by growth on synthetic media lacking uracil while ability to repress a TRP1 gene inserted at the HMR locus was tested by growth on synthetic media lacking tryptophan. Synthetic complete media (only selecting for plasmids) was used to control for equal deposition.
We next examined whether Sir3 wH homodimerization per se was necessary for silencing. Because PISA analysis suggested that leucine 861 (L861) and valine 909 (V909) contribute strongly to the wH-wH hydrophobic interface, we mutated these two key residues to alanines (Sir3-AA; L861A-V909A). V909 is located at the C-terminal end of the α-helix ‘a’, which is part of the yeast-specific insert described above. It contacts L861 of the other Sir3 wH unit within the crystallized homodimer (Figures 1B, F and 3C). Like Sir3ΔwH, the Sir3-AA mutant had impaired dimerization capacity and sedimented as a monomer in a glycerol gradient (Supplementary Figure S2A and B). Importantly, the Sir3-AA double mutant, which was again expressed at levels comparable to wild-type Sir3 (Figure 3B), led to a complete loss of silencing of the telomere 7L reporter gene, mimicking a full deletion of SIR3 (Figure 3E; Supplementary Figure S4A and B). Sir3-AA also weakly decreased silencing at HMR compared to wild-type Sir3 (Figure 3E). Several other mutations, such as the triple alanine substitution mutant, L861A-V909A-F926A, or mutation of L861 and V909, singly or in combination, into tryptophan (W), arginine (R) or aspartic acid (D) residues, as well as swapping the residues 900–914 with a short glycine-serine-glycine (GSG) linker, instead reduced Sir3 protein levels, although they were expressed from the same promoters. These mutations, which rendered Sir3 unstable, of course, also failed to repress at telomere 7L (hereafter, TPE for Telomere position effect; Gottschling et al, 1990; Supplementary Figure S4A and B).
We reasoned that if Sir3 wH homodimerization was the only essential function of the wH in gene repression, a chimeric protein consisting of the Sir3ΔwH construct fused to an ectopic dimerization domain might be able to restore silencing. To test this, we made use of the well-characterized transcription factor HlyU from Vibrio vulnificus, which dimerizes readily and is similar in overall size to Sir3 wH, although it shares no sequence identity (Nishi et al, 2010; Liu et al, 2011; Figure 3D). This chimeric construct (sir3-HlyU), like the Sir3-AA mutant, was expressed at wild-type Sir3 levels (Figure 3B) in a sir3Δ strain. Using reporters at both telomere 7L and at the HMR locus, we found that the hybrid protein Sir3-HlyU was able to restore silencing completely, with an efficiency indistinguishable from that of full-length Sir3 protein (Figure 3E). The fact that an ectopic bacterial dimerization module can functionally replace the wH domain of Sir3, conferring silencing function and protein stability, strongly suggests that dimerization is the only essential function of the Sir3 wH domain for Sir-mediated repression in vivo.
Overexpression of Sir3 wH competes with endogenous Sir3 dimerization to relieve TPE
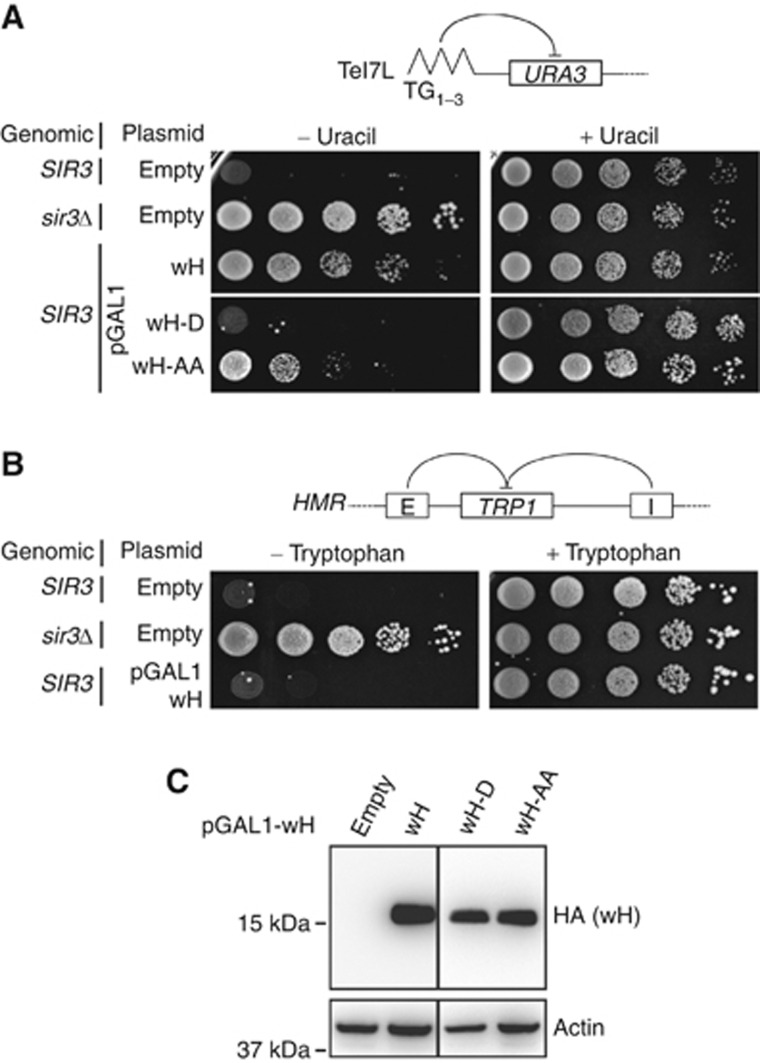
Given the above findings, and past results showing that SIR3 gene dosage affects repression (Renauld et al, 1993; Hecht et al, 1996; Maillet et al, 1996; Strahl-Bolsinger et al, 1997), we reasoned that an excess of Sir3 wH might compete for endogenous full-length Sir3 dimerization and impair silencing. To test this hypothesis, we cloned the wH module under the control of the GAL1 promoter, which allows overexpression in galactose-containing media (Mumberg et al, 1994). We introduced this construct in a SIR3+ strain and monitored TPE at telomere 7L and 5R, as well as silencing at the HMR locus on plates containing galactose to induce Sir3 wH expression. Indeed, overexpression of Sir3 wH abrogated TPE at the two telomeres scored (Figure 4A; Supplementary Figure S5). On the other hand, Sir3 wH overexpression did not affect repression at HMR, where redundant silencers ensure more stable repression (Figure 4B; Maillet et al, 1996; Marcand et al, 1996).
Figure 4.
Overexpression of Sir3 wH, but not of dimerization mutants, derepresses silencing. (A) The indicated strain was grown overnight in selective media and 10-fold dilutions were dropped on the indicated media containing galactose as carbon source, to induce pGAL1promoter-SIR3wH construct overexpression. Ability to repress a URA3 gene at telomere 7L was tested as in Figure 3E. (B) The ability to repress TRP1 inserted at the HMR locus was tested as in Figure 3E. (C) Western blot analysis from alkaline yeast extracts (Ruault et al, 2011) of strains showed in Figure 4A, grown on galactose, to confirm domain expression. wH-D and wH-AA refer to the overexpression of the Sir3 wH domain carrying the single V909D or the double L861A-V909A mutations, respectively.
To confirm that the Sir3 wH-provoked derepression stems from its ability to dimerize and thereby disrupt essential Sir3–Sir3 interactions, we introduced structure-based point mutations in the dimerization interface of Sir3 wH. As detailed in Figure 3, by introducing the same L861A and V909A mutations as we did in full-length Sir3 (Sir3-AA) in the overexpressed wH module alone (wH-AA) we reduced the extent of the wH-wH hydrophobic dimerization interface (Figure 3C). We also replaced V909 by an aspartic acid (wH-D), destabilizing Sir3 wH dimerization by inserting a negative charge at the hydrophobic interface. Both mutated Sir3 wH domains, wH-AA and wH-D, were expressed at similar levels as the wild-type Sir3 wH domain (Figure 4C). Overexpression of the wH-D mutant failed to impair TPE at telomeres 7L and 5R (Figure 4A; Supplementary Figure S5), and the wH-AA mutant derepressed both sites, albeit less efficiently than the wild-type domain (Figure 4A; Supplementary Figure S5). This is consistent with the notion that overexpression of the Sir3 wH domain has a dominant-negative effect on TPE by interfering with endogenous Sir3 dimerization, and again confirms the importance of the large dimerization interface within Sir3 wH domains for Sir-mediated repression.
Sir3 dimerization is dispensable for Sir complex formation, but is essential for Sir complex assembly on chromatin in vivo
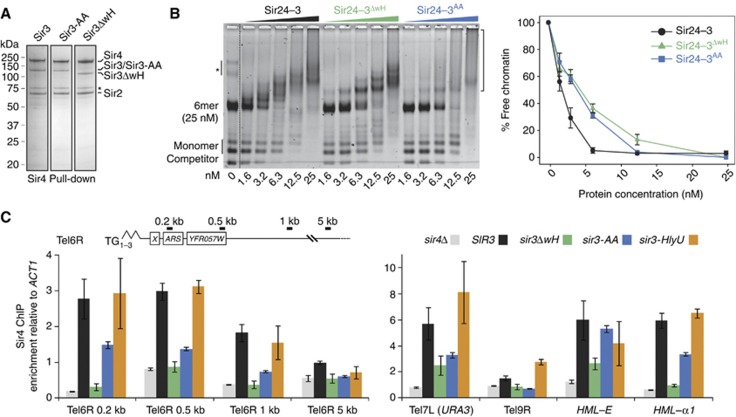
Our in vitro assays revealed that the Sir3 wH domain promotes the loading of the Sir3 C-terminal region onto a reconstituted chromatin template (Figure 2). Given the multiplicity of contacts between the Sir complex and chromatin, we next examined whether Sir3 wH homodimerization also promotes the binding of the holo-Sir complex to chromatin in vitro. We first expressed Sir3-AA, Sir3ΔwH or wild-type Sir3 in insect cells, together with Sir2 and Sir4, and found that all generate stable complexes with Sir2-4 that can be purified from cell extracts (Figure 5A), indicating that the Sir3–Sir4 interaction does not depend on Sir3 dimerization capacity. We then incubated purified Sir2–4 complexes and the corresponding equimolar amount of Sir3, Sir3-AA or Sir3ΔwH (Supplementary Figure S2A) with the 6-mer of reconstituted nucleosomes. Strikingly, disruption of Sir3 dimerization by deletion (Sir3ΔwH) or mutation (Sir3-AA) of the Sir3 wH domain consistently reduced the binding affinity of the holo-Sir complex to chromatin in vitro (Figure 5B). This is consistent with the loss of TPE in cells expressing only the Sir3ΔwH or Sir3-AA forms of Sir3.
Figure 5.
Sir3 wH homodimerization is critical for loading or stable binding of Sir complex onto chromatin in vivo. (A) Hi5 insect cells were co-infected with Sir2, Sir4 and the indicated Sir3 expressing virus. Sir4 was affinity purified as described previously (Martino et al, 2009) and a sample elution was loaded on an SDS–PAGE and stained with Coomassie blue. The asterisk indicates a common Sir4 degradation product. (B) The holo-Sir complex or mutant complexes containing either Sir3ΔwH (aa 1–850) or Sir3-AA (L861A and V909A) were titrated into a constant amount of 6-mer of nucleosomes. The images are representative of at least three independent experiments; quantifications show the mean value ±s.e.m. of the % of unbound chromatin compared to the input. The asterisk indicates a contaminant DNA, and the bracket on the right indicates Sir/chromatin complexes. (C) Sir4 ChIP analysis of the same strains used for silencing assay in Figure 3E, the amount of precipitated DNA was measured by qPCR. Enrichment over the ACT1 locus was normalized to the input.
Given the fact that loss of Sir3 wH homodimerization weakened Sir complex loading in vitro (Figure 5B) and compromised TPE in vivo (Figure 3), we asked whether Sir3 wH-mediated dimerization was also necessary for the stable binding and spread of Sir complexes along subtelomeric chromatin in vivo. Alternatively, the mutant Sir complexes might bind chromatin, but simply fail to repress. To address this, we performed chromatin immunoprecipitation (ChIP) analysis using an antibody against Sir4 in strains that express the Sir3 constructs described above (Figure 3), driven by the SIR3 promoter and flanked by SIR3 terminator sequences. All constructs are expressed at levels comparable to wild-type Sir3 (Figure 3B). Using qPCR on chromatin sheared to obtain a resolution of roughly 500 bp, we monitored Sir4 binding to the subtelomeric URA3 reporter gene that monitors silencing at telomere 7L, along with other genomic locations that support Sir-mediated repression.
We found that Sir3ΔwH failed to support Sir4 loading at native telomeres 6R and 9R, and at the HML-α1 gene, while there was reduced Sir4 binding at telomere 7L and at the HML-E silencer (Figure 5C). In addition, the dimerization mutant sir3-AA reduced Sir4 loading at all telomeres tested (Tel6R, 7L, 9R) as well as at HML-α1, while it did not affect binding of Sir4 to the HML-E silencer. Consistent with its ability to restore repression, expression of Sir3-HlyU restored binding of the Sir4 protein at all loci tested (Figure 5C). This confirms that Sir3 dimerization through the wH domain is essential for Sir complex spreading in vivo, although Sir4-silencer binding is largely independent of Sir3 dimerization activity. Our ChIP results correlate perfectly with the silencing phenotypes described in Figure 3, and reveal that Sir3 wH-mediated dimerization is critical for holo-Sir complex loading or stable association on chromatin in vivo. Since the bacterial dimerization domain (HlyU) restores holo-Sir binding, our data argue that dimerization is the essential silencing-specific function of Sir3 wH.
The S. cerevisiae Orc1/Sir3 wH domain evolved to self-associate prior to gene duplication
As discussed above, the Orc1/Sir3 family is conserved throughout evolution. The archaeal A. pernix Orc1 wH was shown to bind DNA (Gaudier et al, 2007), and based on this observation, a similar function was predicted for other Orc1 homologues, including the Sir3 wH domain (Norris and Boeke, 2010; Hickman et al, 2011). Having shown here that Sir3 wH does not bind DNA, and evolved instead into a dimerization module that is crucial for Sir-mediated silencing, we were curious whether the S. cerevisiae and H. sapiens Orc1 wH domains would be more similar to Sir3 or to the archaeal protein.
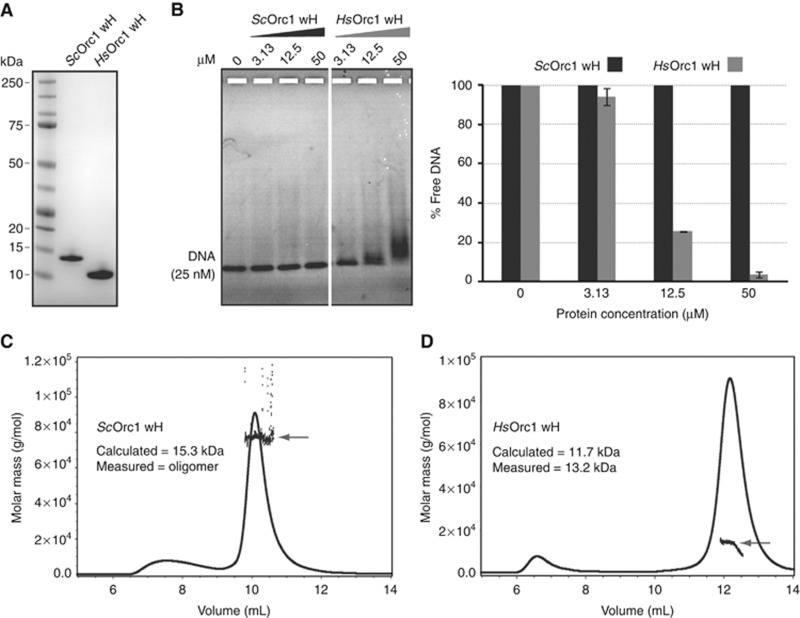
To shed light on the functional conservation of these wH modules, we purified the equivalent domains of ScOrc1 and HsOrc1 after expression in E. coli (Figure 6A). To test whether the ScOrc1 or HsOrc1 wH domains interact with DNA, we titrated an increasing amount of each purified protein into a constant amount of Cy3-labelled 147-bp DNA, and monitored interaction by gel shift assay. We found that the human Orc1 wH domain bound DNA readily, while the yeast Orc1 wH did not (Figure 6B). Next, we investigated the oligomeric states of ScOrc1 and HsOrc1 by SEC-MALS. We found that ScOrc1 wH oligomerizes in solution and, in the conditions used here, forms a range of oligomers, primarily consistent with 5–7 molecules, but ranging in one experiment up to 27 (Figure 6C). This may reflect a need for other subunits of ORC for proper oligomerization. In contrast, the HsOrc1 wH, purified under the same conditions and with homologous boundaries as ScOrc1 and ScSir3, showed no self-association and retained a stable monomeric state in solution (Figure 6D). We conclude that there is a fundamental difference between ScOrc1/Sir3 and both the archaeal and human Orc1 proteins, in that the ScOrc1/Sir3 wH evolved to promote protein–protein interactions, losing DNA binding ability. This most likely occurred before an ancient yeast underwent the genome duplication that generated the two related SIR3 and ORC1 genes.
Figure 6.
Evidence for a functional conservation of Sir3 wH in Saccharomycetales species. (A) 2 μg of the indicated domains was run on an SDS–PAGE and stained with Coomassie blue. (B) The indicated domain was titrated onto a constant amount of a 147-bp Cy3-DNA probe; binding was scored as in Figure 2. Purified ScOrc1 wH, 3 mg/ml (C) or HsOrc1 wH, 7.8 mg/ml (D) was analysed by SEC-MALS as described in Materials and methods. The measured molar mass is represented by black dots (grey arrow) and superposed to the UV (280 nm) trace. The ScOrc1 wH domain was injected five times yielding the following results: 78, 81, 108, 123 and 417 g/mol.
Discussion
The formation of yeast silent chromatin depends on the recruitment of the Sir proteins by factors that bind specific DNA elements, followed by the establishment of Sir–chromatin and Sir–Sir contacts. These layers of interaction contribute to a stable chromatin structure that impairs transcription and protects the linker DNA from nuclease attack. Whereas Sir protein recruitment by Rap1, ORC, Abf1 and Yku has been studied extensively, as have interactions between Sir domains and chromatin, only a few studies have examined the importance of Sir protein dimerization for repression.
The Sir3 C-terminal wH domain is necessary and sufficient for Sir3 dimerization
Sir4 homodimerizes through a C-terminal coiled-coil domain, which also interacts with Sir3, Yku70 and Rap1 (Moretti et al, 1994; Tsukamoto et al, 1997; Chang et al, 2003; Murphy et al, 2003; Rudner et al, 2005). Sir4 dimerization has been proposed to be necessary for silencing by providing a binding site for Sir3 (Chang et al, 2003; Murphy et al, 2003). The importance of Sir3 dimerization has never been tested directly, even though Sir3 has the ability to spread along chromatin with substoichiometric amounts of Sir2 and Sir4 (Hecht et al, 1996; Strahl-Bolsinger et al, 1997). This implied that Sir3–Sir3 interaction should be important for its function in vivo.
Over the years, various Sir3 domains have been proposed to mediate homodimerization or heterodimerization. Both the AAA domain and the BAH domain made suggestive contacts in the crystal packing when crystallized alone (Connelly et al, 2006; Ehrentraut et al, 2011) or together with a nucleosome (Armache et al, 2011). However, the relevance of this finding outside of the crystal context is unclear. Dimerization of the Sir3 BAH could not be detected by co-IP (Buchberger et al, 2008) and only a very weak interaction was observed by analytical ultracentrifugation (Armache et al, 2011). A Sir3 C-terminal fragment that contained both the AAA and the wH domains appeared to dimerize in solution (Chang et al, 2003), while the Sir3 wH and a fragment containing a large N-terminal portion of the AAA domain showed some interaction upon bacterial co-overexpression (King et al, 2006). Indeed, no study to date had carefully mapped the dimerization interface between Sir3 molecules, nor mutated important residues to monitor both a loss of dimerization and of silencing function in vivo. Here, we demonstrate unequivocally that the Sir3 C-terminal 138 residues form a wH module that is necessary and sufficient for dimerization, and that Sir3 homodimerization is crucial for Sir complex loading along nucleosomes in vivo. Genetic, biochemical and structural data support this conclusion rigorously.
The crystal structure of the wH domain presented here reveals a large hydrophobic dimerization interface (1500 Å2), which we show is biologically relevant in contrast to the contact surfaces observed for the BAH and AAA domains (Connelly et al, 2006; Ehrentraut et al, 2011). We confirmed Sir3 wH dimerization in solution by SEC-MALS. Importantly, Sir3 dimerization in insect cell extracts was reduced to background levels when full-length Sir3 was co-expressed with Sir3 lacking the wH domain (Figure 1G). Finally, while holo-Sir3 was shown to migrate as a dimer in a glycerol gradient, Sir3ΔwH or Sir3-AA shifted the Sir3 protein towards a monomeric state. Together, these studies indicate that dimerization of Sir3 is mostly, if not entirely, mediated by the wH domain. In contrast, we found that the AAA domain does not stably dimerizes (Ehrentraut et al, 2011), nor does it interact with the wH domain by either pull-down or isothermal titration calorimetry (this study). While we cannot rule out that novel dimerization interfaces are revealed in Sir3 upon assembly into the holo-Sir complex or upon binding to chromatin, it is made less likely by the fact that either deletion or mutation of the Sir3 wH domain reduces the binding affinity of the holo-Sir complex to chromatin in vitro.
The Sir3 wH does not bind DNA or chromatin, but stabilizes the Sir complex on chromatin
Due to its homology with the crystallized archaeal ApOrc1 C-terminus (Gaudier et al, 2007), the Sir3 wH domain was predicted to bind DNA. However, Sir3 wH neither binds DNA nor chromatin in vitro. Instead it forms a homodimer that reinforces the loading of the Sir3 C-terminal domain onto both templates in binding assays (Figure 2). Previous work using full-length Sir3 showed that holo-Sir3 can bind DNA and nucleosomal templates, forming oligomeric structures in vitro (Georgel et al, 2001; McBryant et al, 2006, 2008). We attribute these chromatin interactions to contacts demonstrated for the BAH and AAA domains (Connelly et al, 2006; Armache et al, 2011; Ehrentraut et al, 2011), while Sir3 wH specifically mediates dimerization.
Our biochemical data suggest that the dimerization of Sir3 wH promotes Sir3 and holo-Sir complex loading onto chromatin in vitro, while ChIP data show that it also promotes the propagation and/or stable binding of the holo-Sir complex along the chromatin fibre in vivo. Given that the Sir4 C-terminal domain has been reported to dimerize as well (Chang et al, 2003; Murphy et al, 2003), it is tempting to speculate that Sir3 and Sir4 dimerization capacities jointly promote the spread of the Sir complex along chromatin, possibly by creating Sir complex dimers (i.e., hexamers) and then interactions between them along the chromatin fibre. While the precise role of Sir4 dimerization in establishing Sir-repressed chromatin remains to be investigated, reducing the Sir3 wH dimerization interface by targeted mutagenesis (Sir3-AA) or deletion of the wH domain, significantly decreased the loading of Sir4 along subtelomeric nucleosomes at telomeres 6R, 7L, 9R and across the HML locus in vivo. Consistently, the Sir3-AA or Sir3ΔwH mutants failed to silence subtelomeric reporter genes. Remarkably, repression could be completely restored to the wH-deficient Sir3 mutant by attaching a bacterial dimerization motif that has no homology with the native Sir3 domain. Reinforcing our conclusion that dimerization of Sir3 is crucial for repression, it was reported that expression of the BAH domain could support measurable HM silencing only when fused to the dimerization-competent lexA module (Connelly et al, 2006). Taken together, we conclude that Sir3 dimerization capacity is a general requirement for gene repression, even when it is mediated by an alternative mechanism that involves only a subdomain of Sir3. In summary, our data show unequivocally that Sir3 wH-mediated homodimerization is essential for loading of Sir proteins onto chromatin and for the silencing of underlying genes.
Consistent with the loss- and gain-of function constructs described above, we also demonstrate that overexpression of the Sir3 wH domain ablates TPE, while a dimerization deficient mutant does not. This refines previous data showing that overexpression of a large Sir3 C-terminal construct (aa 437–978) derepresses TPE, while if it includes the Sir3 wH domain (Enomoto et al, 2000). Although TPE has been shown to be more easily disrupted than HMR silencing in overexpression studies, we note that the deletion of the wH domain fully derepressed both HMR and telomeric reporters, demonstrating the general need of Sir3 wH dimerization for silencing.
A previous study showed that a few amino-acid substitutions that do not affect Sir3 wH dimerization in yeast-2-hybrid assays, interfered with a tethered repression assay (Y900A, Y964A and K973A; Liaw and Lustig, 2006). Our structural studies reveal that these residues are not involved in dimerization, and thus their effects may stem instead from altered intramolecular secondary structure within Sir3, or Sir3 interfaces with other proteins. However, the fact that the HlyU dimerization motif can replace wH to restore TPE, argues against other silencing-specific functions in this C-terminal domain.
Evolutionarily distinct wH domains among Orc1/Sir3 family members
The basic wH fold that we present here for the Sir3 C-terminal domain is conserved from archaea to man, and generally mediates macromolecular interactions (Aravind et al, 2005). The yeast Sir3 wH domain, however, binds neither chromatin nor DNA, in striking contrast to the wH domain of archaeal (De Felice et al, 2004; Dueber et al, 2007; Gaudier et al, 2007) or human Orc1 (this study). Indeed, we show here that a recombinant wH domain of HsOrc1, like the archaeal Orc1 wH, forms a stable monomer in solution and readily binds DNA under the conditions tested here.
Sequence analysis predicts that only budding yeast lineage (Saccharomycetales) homologues, that is, those bearing an insertion in the wH domain, will have a sufficiently large hydrophobic surface to mediate dimerization (Figure 1B). Consistently, the ScOrc1 wH, unlike HsOrc1 wH, self-associates in solution, and fails to bind DNA. On the basis of these functional and sequence similarities between ScOrc1 and ScSir3, and their dissimilarity with Orc1 homologues in other species, we propose that yeast Orc1 wH is closer to Sir3 than to the human Orc1 wH. In fact, the human Orc1 wH more closely resembles that of archaea, at least with respect to macro-molecular interactions. However, given that the critical residues for ApOrc1–DNA interaction are not conserved in man (Figure 1B), the two may bind DNA in different ways.
The fact that the wH domains of both yeast Sir3 and Orc1 acquired a ∼30 aa insert that generates a large protein–protein interface, and the ability to self-interact, rather than bind DNA, makes it highly likely that these changes in the wH domain sequence arose prior to the genome duplication event that generated S. cerevisiae and the SIR3/ORC1 gene pair (Kellis et al, 2004). Consistently, the characteristic insertion in the wH domain is also found in the budding yeast Kluyveromyces lactis, even though its genome did not undergo endoduplication (Hickman and Rusche, 2010). Intriguingly, Orc1 from K. lactis acts in conjunction with the deacetylase Sir2 and the histone-binding protein Sir4 to generate telomeric and mating type heterochromatin (Hickman and Rusche, 2010). The fact that ScOrc1 cannot substitute for Sir3 in the budding yeast SIR complex, despite having a similar wH domain, most likely reflects the well-characterized specialization of the Sir3 AAA domain for Sir4 interaction (Ehrentraut et al, 2011).
We still do not know how important ScOrc1 self-association is for its function in either replication or silencing, although we show here that dimerization of the same domain in Sir3, is crucial for silencing in yeast. We note that the changes in the wH domain that promote dimerization, correlate well with the appearance of a divergent ‘silencing’ branch of the Orc1/Sir3 family. While other mechanisms of chromatin-mediated repression, such as those involving Heterochromatin Protein 1 or Polycomb repressive complex 1, also depend on components that have homodimerization potential, it has never been shown that the dimerization domain can be swapped for a dimerization motif from bacteria (Fujisaki et al, 2003; Czypionka et al, 2007; Canzio et al, 2011). This demonstration for yeast Sir3 confirms that its dimerization capacity is essential for efficient Sir complex loading onto chromatin, that is, for both telomeric and mating type silencing. By ensuring the ability of haploid spores to switch mating type and conjugate, mating type silencing is, of course, essential for the survival of the species.
Materials and methods
Multiple sequence alignment
Orc1 wH sequences from higher eukaryotes were aligned with ClustalW (www.ebi.ac.uk) and submitted to HHPRED (Soding et al, 2006) to yield an alignment with the A. pernix Orc1 wH sequence of which the structure is known (Gaudier et al, 2007). A ClustalW alignment of yeast Sir3 and Orc1 wH sequences was then added according to a structural alignment computed with DALI (Holm and Rosenstrom, 2010) using crystal structures of the A. pernix Orc1 and Sir3 wH domains.
Purification of protein domains
The Sir3 wH (aa 840–978), Sir3C (aa 527–978), ScOrc1 wH (aa 783–914) and HsOrc1 wH (aa 760–861) constructs were cloned into a pOPINF vector using the In-Fusion system (Clontech) (Berrow et al, 2007), expressed in the E. coli strain BL21 Rosetta pLysS and affinity purified via a His tag using ProBond Ni-NTA resin (Invitrogen) according to manufacturer’s instructions. Tags were removed by HRV 3C protease digestion (Novagen) and Sir3 domains were further purified by gel filtration on a HiLoad 16/60 Superdex 75 column in 50 mM Tris pH 7.5, 200 mM NaCl, 0.02% NaN3 and 1 mM TCEP. The Sir3 AAA (aa 530–845) was purified as described previously (Ehrentraut et al, 2011). Point mutations were introduced by site directed mutagenesis using Pfu DNA polymerase (Promega). For the chimera Sir3-HlyU, the coding region for aa 851–978 was deleted and replaced with AscI and XbaI sites using site directed mutagenesis. The cDNA of HlyU_Vv was amplified by PCR (without the ATG start codon) inserting AscI and XbaI sites and a SGSG linker. The cDNA of HsOrc1 (IRAK013J04) was obtained through RIKEN BRC, Japan (Ota et al, 2004; Otsuki et al, 2005; Itoh et al, 2006; Kimura et al, 2006). Purity was confirmed by SDS–PAGE and Coomassie blue staining; protein concentration was measured by UV absorbance (280 nm).
Structure determination
Native and selenomethionine substituted Sir3 wH crystals were grown at 20°C by sitting-drop vapour diffusion in 96-well crystallization plates from mixtures containing equal volumes of protein (22 mg/ml in 20 mM Tris pH 8.5, 200 mM NaCl, 0.02% NaN3 and 1 mM TCEP) and reservoir solutions (0.1 M sodium citrate pH 5.6, 0.2 M ammonium acetate and 30% (w/v) PEG-4000) and were flash-frozen in mother liquor made up to 25% (v/v) ethylene glycol. Diffraction data were collected at beam lines X06DA and X10SA at the Swiss Light Source synchrotron in Villigen, Switzerland. Detailed Materials and methods for structure determination and refinement can be found in Supplementary data. Coordinates have been deposited in the PDB database under accession code 3ZCO.
SEC-MALS experiments
Purified domains were concentrated to 2–8 mg/ml and filtered through a 0.1 μM Amicon filter before injection. In all, 100 μl of each protein was separated on a Wyatt SEC 300A 7.8 × 300 mm column equilibrated in 20 mM Tris pH 7.4, 200 mM NaCl, 0.02% NaN3 at a flow rate of 0.5 ml/min at 4°C. Light scattering was recorded on an in-line miniDAWN TREOS 3-angle light scattering detector (Wyatt Technology) and protein concentration detected with an in-line Optilab Trex refractive index detector. Data analysis was done using the Zimm fitting method in Astra V software (Wyatt Technology).
Co-immunoprecipitation from insect cells
Full-length or truncated Sir3-ΔwH was co-infected in Hi5 cells as indicated in Figure 1G. Cells were lysed by gentle sonication in 50 mM HEPES pH 7.5, 150 mM NaCl, 0.1% NP-40, and Sir3-HIS-HA purified in batch using ProBond Ni-NTA resin (Invitrogen). Beads were washed with 50 mM Na2HPO4 pH 8.0, 500 mM NaCl, 50 mM imidazole, 0.05% NP-40 and eluted in 2 × the bead volume with 50 mM Na2HPO4 buffer pH 8.0, 300 mM NaCl, 250 mM imidazole, 0.05% NP-40. Co-purification of Myc-tagged Sir3 constructs was analysed by immunoblotting using HIS-hrp (Abcam, ab1187) or Myc (9E10) antibodies.
Chromatin and DNA interaction assays
In vitro reconstitution of nucleosomal arrays and binding assays were performed as described previously (Oppikofer et al, 2011). See also description in Supplementary data.
Overexpression and complementation assays in yeast
For overexpression assays, the genomic sequence coding for Sir3 wH (aa 840–978) was HA tagged, amplified by PCR and cloned into the pRS425-GAL1 vector (Mumberg et al, 1994) using Pst1 and Sal1 sites. For complementation assays, the SIR3 gene together with 1 kb 5′- and 3′-UTR was amplified by PCR and cloned into pRS415 using BamHI and NotI sites. Deletion and point mutations were introduced by site directed mutagenesis using Pfu DNA polymerase (Promega). Telomeric silencing used GA503 (SIR3+) and GA7055 (sir3Δ) strains, while HMR silencing used GA484 (SIR3+) and GA7292 (sir3Δ) strains. SIR3 deletion was achieved by replacing the endogenous gene together with 1 kb 5′- and 3′-UTR with a kanamycin resistance cassette (Supplementary Table S1). Silencing of the indicated reporter genes (URA3 or TRP1) was monitored as described (Gotta et al, 1998), by growth on synthetic media lacking either uracil or tryptophan.
ChIP and qPCR experiments
ChIP experiments were carried out as previously described (Braunstein et al, 1993), with an antibody raised against GST-Sir4 C-terminus used at 750 ng/20 μl MagSi-proteinA 1.0 beads (MagnaMedics). Precipitated DNA was purified using the AccuPrep PCR purification kit (Bioneer) after overnight incubation at 65°C to reverse the formaldehyde cross-linking. The GA5822 sir4Δ strain (Kueng et al, 2012) was used as control. Quantification of the precipitated DNA was performed by qPCR on a StepOnePlus instrument (Applied Biosystems); enrichments were normalized to input and the ACT1 locus. Primers have been used previously (see Supplementary Table S2). Experiments were performed in biological and technical duplicates.
Supplementary Material
Acknowledgments
We would like to thank the Gasser laboratory for discussion and support. We thank Lorraine Pillus (University of California, San Diego, California) for the Sir3 antibody and Jeong-Sun Kim (Chonnam National University, Gwangju, Republic of Korea) for the HlyU_Vv cDNA. We thank the Research Association for Biotechnology, Dr Yoshihide Hayashizaki (RIKEN OSC) and Dr Sumio Sugano (Tokyo University Graduate School) for the HsOrc1 cDNA (IRAK013J04). The Gasser laboratory is supported by the Novartis Research Foundation and the EU network Nucleosome 4D. SK was supported by an EMBO long-term fellowship, an FWF Schroedinger fellowship and the Swiss SystemsX initiative C-CINA.
Author contributions: MO, SK and SMG designed most experiments and interpreted results. MO performed most experiments with the following exceptions. HG collected X-ray diffraction data, solved the Sir3 wH structure, produced the wH alignment and supervised the structural work. SK performed pull-down experiments from insect cells, and helped with in vivo assays and ChIP. JJK performed SEC-MALS analysis and helped with cloning and purification of proteins from E. coli. MH purified the Sir3 AAA domain and performed ITC analysis under the supervision of AGL. MO, SK, HG, AGL and SMG wrote the manuscript. SMG supervised the work.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 [DOI] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE (2011) Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334: 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B (1995) The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83: 563–568 [DOI] [PubMed] [Google Scholar]
- Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res 35: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev 7: 592–604 [DOI] [PubMed] [Google Scholar]
- Buchberger JR, Onishi M, Li G, Seebacher J, Rudner AD, Gygi SP, Moazed D (2008) Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol 28: 6903–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B (2011) Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell 41: 67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JF, Hall BE, Tanny JC, Moazed D, Filman D, Ellenberger T (2003) Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure 11: 637–649 [DOI] [PubMed] [Google Scholar]
- Connelly JJ, Yuan P, Hsu HC, Li Z, Xu RM, Sternglanz R (2006) Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol Cell Biol 26: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubizolles F, Martino F, Perrod S, Gasser SM (2006) A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol Cell 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Czypionka A, de los Panos OR, Mateu MG, Barrera FN, Hurtado-Gomez E, Gomez J, Vidal M, Neira JL (2007) The isolated C-terminal domain of Ring1B is a dimer made of stable, well-structured monomers. Biochemistry 46: 12764–12776 [DOI] [PubMed] [Google Scholar]
- De Felice M, Esposito L, Pucci B, De Falco M, Manco G, Rossi M, Pisani FM (2004) Modular organization of a Cdc6-like protein from the crenarchaeon Sulfolobus solfataricus. Biochem J 381 (Part 3): 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM (2007) Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Ehrentraut S, Hassler M, Oppikofer M, Kueng S, Weber JM, Mueller JW, Gasser SM, Ladurner AG, Ehrenhofer-Murray AE (2011) Structural basis for the role of the Sir3 AAA+ domain in silencing: interaction with Sir4 and unmethylated histone H3K79. Genes Dev 25: 1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, Johnston SD, Berman J (2000) Identification of a novel allele of SIR3 defective in the maintenance, but not the establishment, of silencing in Saccharomyces cerevisiae. Genetics 155: 523–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki S, Ninomiya Y, Ishihara H, Miyazaki M, Kanno R, Asahara T, Kanno M (2003) Dimerization of the Polycomb-group protein Mel-18 is regulated by PKC phosphorylation. Biochem Biophys Res Commun 300: 135–140 [DOI] [PubMed] [Google Scholar]
- Gasser SM, Cockell MM (2001) The molecular biology of the SIR proteins. Gene 279: 1–16 [DOI] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB (2007) Structural basis of DNA replication origin recognition by an ORC protein. Science 317: 1213–1216 [DOI] [PubMed] [Google Scholar]
- Georgel PT, Palacios DeBeer MA, Pietz G, Fox CA, Hansen JC (2001) Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc Natl Acad Sci USA 98: 8584–8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli S, Donze D, Dhillon N, Kamakaka RT (2001) Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. Embo J 20: 4522–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Gasser SM (1996) Nuclear organization and transcriptional silencing in yeast. Experientia 52: 1136–1147 [DOI] [PubMed] [Google Scholar]
- Gotta M, Palladino F, Gasser SM (1998) Functional characterization of the N terminus of Sir3p. Mol Cell Biol 18: 6110–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE (1992) Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA 89: 4062–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hickman MA, Froyd CA, Rusche LN (2011) Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage. Eukaryot Cell 10: 1183–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Rusche LN (2010) Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc Natl Acad Sci USA 107: 19384–19389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38 (Web Server issue): W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D (2002) Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh VA, Robinson PJ, Rhodes D (2005) A method for the in vitro reconstitution of a defined ‘30 nm’ chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol 345: 957–968 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Itoh M, Yasunishi A, Imamura K, Kanamori-Katayama M, Suzuki H, Suzuki M, Carninci P, Kawai J, Hayashizaki Y (2006) Constructing ORFeome resources with removable termination codons. Biotechniques 41: 44, 46, 48 passim. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T et al. (2006) Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res 16: 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DA, Hall BE, Iwamoto MA, Win KZ, Chang JF, Ellenberger T (2006) Domain structure and protein interactions of the silent information regulator Sir3 revealed by screening a nested deletion library of protein fragments. J Biol Chem 281: 20107–20119 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Kueng S, Tsai-Pflugfelder M, Oppikofer M, Ferreira HC, Roberts E, Tsai C, Roloff TC, Sack R, Gasser SM (2012) Regulating repression: roles for the sir4 N-terminus in linker DNA protection and stabilization of epigenetic States. PLoS Genet 8: e1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw H, Lustig AJ (2006) Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol Cell Biol 26: 7616–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM (2000) Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell 6: 637–648 [DOI] [PubMed] [Google Scholar]
- Liu M, Rose M, Crosa JH (2011) Homodimerization and binding of specific domains to the target DNA are essential requirements for HlyU to regulate expression of the virulence gene rtxA1, encoding the repeat-in-toxin protein in the human pathogen Vibrio vulnificus. J Bacteriol 193: 6895–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Rine J (1994) Silencers and domains of generalized repression. Science 264: 1768–1771 [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Marcand S, Buck SW, Moretti P, Gilson E, Shore D (1996) Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev 10: 1297–1309 [DOI] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM (2009) Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Krause C, Hansen JC (2006) Domain organization and quaternary structure of the Saccharomyces cerevisiae silent information regulator 3 protein, Sir3p. Biochemistry 45: 15941–15948 [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Krause C, Woodcock CL, Hansen JC (2008) The silent information regulator 3 protein, SIR3p, binds to chromatin fibers and assembles a hypercondensed chromatin architecture in the presence of salt. Mol Cell Biol 28: 3563–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Rudner AD, Huang J, Hoppe GJ, Tanny JC (2004) A model for step-wise assembly of heterochromatin in yeast. Novartis Found Symp 259: 48–56 discussion 56–62, 163–169 [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GA, Spedale EJ, Powell ST, Pillus L, Schultz SC, Chen L (2003) The Sir4 C-terminal coiled coil is required for telomeric and mating type silencing in Saccharomyces cerevisiae. J Mol Biol 334: 769–780 [DOI] [PubMed] [Google Scholar]
- Nishi K, Lee HJ, Park SY, Bae SJ, Lee SE, Adams PD, Rhee JH, Kim JS (2010) Crystal structure of the transcriptional activator HlyU from Vibrio vulnificus CMCP6. FEBS Lett 584: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Norris A, Boeke JD (2010) Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev 24: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D (2007) Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell 28: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Oppikofer M, Kueng S, Martino F, Soeroes S, Hancock SM, Chin JW, Fischle W, Gasser SM (2011) A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J 30: 2610–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K et al. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 36: 40–45 [DOI] [PubMed] [Google Scholar]
- Otsuki T, Ota T, Nishikawa T, Hayashi K, Suzuki Y, Yamamoto J, Wakamatsu A, Kimura K, Sakamoto K, Hatano N, Kawai Y, Ishii S, Saito K, Kojima S, Sugiyama T, Ono T, Okano K, Yoshikawa Y, Aotsuka S, Sasaki N et al. (2005) Signal sequence and keyword trap in silico for selection of full-length human cDNAs encoding secretion or membrane proteins from oligo-capped cDNA libraries. DNA Res 12: 117–126 [DOI] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE (1993) Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev 7: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I (1987) Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruault M, De Meyer A, Loiodice I, Taddei A (2011) Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast. J Cell Biol 192: 417–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Hall BE, Ellenberger T, Moazed D (2005) A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol 25: 4514–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sampath V, Yuan P, Wang IX, Prugar E, van Leeuwen F, Sternglanz R (2009) Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol Cell Biol 29: 2532–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Klar AJ (1992) Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev 6: 186–196 [DOI] [PubMed] [Google Scholar]
- Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB (2004) Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J Mol Biol 343: 547–557 [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA 97: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Remmert M, Biegert A, Lupas AN (2006) HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res 34 (Web Server issue): W374–W378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745 [DOI] [PubMed] [Google Scholar]
- Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D (2004) Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol Cell Biol 24: 6931–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H (1997) Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900–903 [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Yang B, Kirchmaier AL (2006) Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Mol Biol Cell 17: 5287–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.