Abstract
Electron transfer proteins play critical roles in determining their reduction potentials by the degree of burial of the redox site within the protein and the degree of permanent polarization of the polypeptide around the redox site. Although continuum electrostatics calculations of protein structures can predict their net effect, quantifying how each contributes separately is difficult. Here, the burial of the redox site is characterized by a dielectric radius Rp, which is a Born-type radius for the protein, the polarization of the polypeptide is characterized by an electret potential φp, which is the average electrostatic potential at the metal atoms, and an electret-dielectric spheres (EDS) model of the entire protein is then defined in terms of Rp and φp. The EDS model shows that for a protein with a redox site of charge Q, the dielectric response free energy is a function of Q2 while the electret energy is a function of Q. In addition, Rp and φp are shown to be characteristics of the fold of a protein and predictive of the most likely redox couple for redox sites that undergo different redox couples.
Keywords: Iron-sulfur proteins, reduction potential, HiPIP, nitrogenase, ferredoxin, Poisson-Boltzmann continuum electrostatics
Introduction
In bioenergetic processes, electrons move over long distances by electron transfer reactions between electron carriers, which are generally redox sites within electron transfer proteins. The reduction potential E° of the redox sites determine the driving force for the reactions, with more favorable electron flow toward the redox site with larger E°. Since nature uses the same redox sites for many different processes, the protein environment surrounding each redox site must play a critical role in determining E°. Thus, understanding how the protein environment affects E° is essential for understanding structure-function relationships in electron transfer proteins.
Computational studies have played an important role in understanding reduction potentials of metalloproteins. Since E° is proportional to the negative of the free energy for the reduction reaction ΔG, the energetics can be divided into the inner sphere contribution ΔGin, which is the energy of adding an electron to the redox site, and the outer sphere contribution ΔGout, which is the change in the interaction energy of the redox site with the protein and solvent upon reduction of the redox site. Many earlier studies of proteins have assumed that ΔGin for a given redox site is independent of the protein, and thus have focused on understanding relative values of E° between different proteins with the same redox site rather than the absolute value of E°[1–6]. However, deviations from experiment can be attributed to the missing inner sphere contribution so the calculations of reduction potentials versus the standard hydrogen electrode (SHE) for [4Fe-4S] proteins by Noodleman, Case, Bashford, and co-workers using density functional theory (DFT) for ΔGin and Poisson-Boltzmann (PB) electrostatics for ΔGout are an important step forward[7,8]. More recent studies[9] indicate that the non-hybrid functional used in those studies gives inner sphere energetics appear somewhat poorer compared to electrospray ionization - photoelectron spectroscopy (EI-PES) detachment energies of [4Fe-4S] cluster analogs measured by Wang and co-workers[10,11] although the geometries were better than some hybrid functionals.
Recently, our group has predicted E° for [4Fe-4S] proteins that are in excellent agreement with experiment (Figure 1) using a simple additive approach we refer to as DFT+PB in which ΔGin is obtained by DFT calculations of gas phase redox site analogs and ΔGout is obtained by PB calculations of the redox site in the protein using partial charges from the DFT calculation[12]. A key factor has been our calibration of the functionals and basis sets for the DFT calculations by comparing the calculated detachment energies of redox site analogs in the gas phase against EI-PES data[9]. Our DFT calculations also confirm that ΔGin for the iron-sulfur clusters is relatively independent of environment[9], although the ligand dihedral conformation may tune it by ~200 mV[13]. The DFT+PB calculations show that the two major factors differentiating ΔGout between proteins are the degree of burial of the redox site in the protein and the degree of permanent polarization of the polypeptide surrounding the redox site[12]. The degree of burial of the redox site determines the dielectric response of the surroundings. According to the Born solvation free energy, the dielectric response for an ion in solution is a function of the charge and radius of the ion and of the dielectric response of the solvent. On the other hand, for the redox site of a protein in aqueous solution, both the low protein dielectric and the high solvent dielectric will contribute; thus, the proximity of the redox site to the surface of the protein and therefore the solvent will affect the dielectric response. Moreover, previous results indicate the proximity to the surface is actually more important than the size of the protein[12]. In addition, the polypeptide in several Fe-S proteins[14] and cytochrome c[2] is polarized around the redox site, creating an electrostatic potential at the redox site that persists even in the limit of zero charge of the redox site[14]. Thus, such a protein is an “electret”, which is a material with quasi-permanent electrical polarization analogous to a permanent magnet[15]. The protein electret is maintained by the hydrogen-bonded, three-dimensional structure of the protein and can easily tune the reduction potential by appropriate mutations.
Figure 1.

Comparison of experimental versus calculated reduction potentials for the 2-/3- (below dashed line) and 1-/2- (above dashed line) couples. From left to right, Fe4S4(SCH2CH3)4 (purple), Rg/RtHiPIP (red), Cv, Pr, Pa, and CaFds (blue), Av-ADP, Av, and CpFePs (green), Eh, Ev, Tt, Rt, Rf, and CvHiPIPs (red), and Cp/CvFd (blue).
Since both the burial of the redox site and the electret of the protein are important in determining ΔGout and thus E°, quantifying each is useful for comparing different proteins; however, both are difficult to quantify. For instance, the solvent accessible surface area (SASA)[16] of the redox site can distinguish between partially exposed sites but not completely buried sites, which is important since the distance from the surface affects the dielectric response free energy even for a completely buried site. Also, while the number of hydrogen bonds to the redox site indicates the polarization of the backbone, the contribution of each hydrogen bond is highly dependent on the orientation and distance[17]. Here, using continuum electrostatics calculations of crystal structures, the dielectric radius of the protein Rp is defined to measure the degree of burial of the redox site within the protein and the protein electret potential φp is defined to measure the degree of permanent polarization of the protein surrounding the redox site. Next, the electret-dielectric sphere model is developed in terms of Rp and φp and used to explore how the two factors contribute to the redox properties of differing proteins. Specifically, Rp and φp are shown to be characteristic of the fold of a protein and, for a redox site that can undergo different redox couples, only certain values of Rp and φp are shown to be consistent with reduction potentials that are within the physiological range for each couple. The proteins examined here are homologous proteins from three families of [4Fe-4S]-proteins: HiPIP, ferredoxin (Fd), and nitrogenase iron protein (FeP), plus an example from each of three more families of redox proteins: the [1Fe]-protein rubredoxin (Rd), the blue-copper protein plastocyanin (Pc), and the heme-protein cytochrome c (cyt c) (Figure 2).
Figure 2.
The folds of TtHiPIP, CaFd, and AvFeP (top, left to right) and PfRd, TaCyt c, and PlPc.(bottom, left to right).
Methods
In the DFT+PB approach,[12] the total E° versus the standard hydrogen electrode (SHE) is decomposed as
| (1) |
where n is the number of electrons transferred, F is the Faraday constant, and ΔGSHE/F = 4.43 eV is the absolute electrode potential for the SHE[18]. Although values of 4.28 eV[19] and 4.34 eV[20,21] based on cluster-ion solvation data[22], this value is close to the IUPAC recommended value[23] and the differences will not significantly affect results[24]. ΔGin is calculated using DFT as the difference in free energy between an oxidized and reduced redox site analog in the gas phase and ΔGout is calculated using continuum electrostatics as the change in the interaction energy of the redox site with the surrounding protein and solvent upon reduction of the redox site. In the continuum electrostatics approach, the permittivity of the redox site is εc, permittivity of the protein is εp, and the permittivity of water is εw.
Calculations of Rp and φp
Two physical quantities are defined here to characterize a metalloprotein, utilizing continuum electrostatics calculations of a protein in a dielectric continuum solvent. Although partial charges of the redox sites have been calculated by us for the cases studied here, partial charges for redox sites are not always available. Thus, instead of using the partial charges of the redox site, a system of test charges equal to Q/M are distributed over the M metals of the redox site, where Q is equal to −1, so that the only parameters required available in standard protein force fields. Moreover, while it is possible to “fit” the model for each different charge state, or each different redox couple, the net charge Q is chosen as −1, again for generality in case the net charge of the redox site is not known and to limit the number of definitions. First, the dielectric radius of the protein Rp is defined using the Born solvation free energy equation as
| (2) |
where Q = −1, εin = εp, and εout = εw. ΔsolG = ΔsolGtest is calculated using the PB equation for the system of test charges in place of the partial charges of the redox site in the protein cavity with permittivity εp and the solvent with permittivity εw (Figure 3, F). Next, the protein electret potential, φp, is defined as the average of the electrostatic potential at each metal atom in the redox site. It is calculated using the PB equation for a system consisting of the redox site cavity with no test or partial charges and permittivity εc, the protein cavity with the protein partial charges and permittivity εp, and the solvent with permittivity εw (Figure 3, G). The backbone contribution to the electret potential, φbb, is defined similarly except the side chain partial charges for each residue are set to zero. In the case of ferredoxin, partial charges for the other redox site in the 2- state are included in the calculation of φ.
Figure 3.

The solvation energy of A) the standard atomistic description of a protein by PB methods consists of the solvation energies of B) point charges at metal atoms within the redox site cavity, C) point charges at metal atoms within the protein cavity, and D) point charges at the protein atoms within both cavities. In the EDS model, B, C, and D, are represented by E) a point charge in a sphere of radius Rc representing the redox site cavity, F) a point charge in a sphere of radius Rc representing the protein cavity, and G) an electret potential, φp, due to the polarization of the protein environment.
Electret-Dielectric Spheres Model
The EDS model is a means of interpreting ΔGout (such as from a full PB calculation) based on a physical model in terms of the two quantities Rp and φp, by modeling a redox-active protein as a point charge inside two concentric dielectric spheres immersed in a dielectric continuum solvent (Figure 3). The point charge inside the inner sphere represents the redox site while the outer sphere plus an associate electret represents the protein. The solvation free energy for this model is
| (3) |
where Q is the net charge of the redox site including both the metals and ligands, Rc is the radius of the redox site, Rp is radius of the protein, and φp is the electret potential. Since Rc is sensitive to the exact distribution of the partial charges, it is calculated separately for each couple according to
| (4) |
The first two terms are an extension of the Born solvation free energy for a point charge Q inside two concentric dielectric spheres in a dielectric continuum, which has been used by Warshel to give a physical picture of the protein environment,[25] while the last term arises from the interaction of Q with φp, the permanent electrostatic potential due to the protein electret. Thus, the outer sphere contribution to E° in the EDS model is ΔGEDS = ΔsolGEDS(Qred) − ΔsolGEDS(Qoxd), where Qoxd and Qred are the net charge of the redox site in the oxidized and reduced states, respectively.
All solvation energies and electrostatic potentials were calculated using APBS[26], a program for solving the Poisson-Boltzmann equation, as described fully elsewhere[24] and summarized briefly here. Radii for Connolly surfaces[16] of the proteins and redox sites and partial charges for the protein were from the CHARMM22 parameters[27]. Partial charges from our previous DFT calculations were used for the [4Fe-4S],[24] [1Fe],[28] and blue-copper[29] redox sites while CHARMM22 parameters were used for the heme, where the total charge of each redox site is an integer. The dielectric permittivity were chosen as εc=1, εp=4, and εw=78 and the ionic concentration was set to zero. A 51.2 Å × 51.2 Å × 51.2 Å box with 257 grid points in each direction was used for HiPIP, Fd, Rd, Pc, and cyt c while a 76.8 Å × 76.8 Å × 76.8 Å box with 385 grid points in each direction was used for FeP, at a constant 0.2 Å spacing for all proteins. Electrostatic potentials were calculated as the average over the potential at the eight grid points surrounding the atom of interest.
Crystal structures of the proteins were obtained from the Protein Data Bank (PDB)[30] and are listed in Table S1. Missing heavy atoms in the crystal structure were added from CHARMM22 parameters and hydrogen atom positions were added with HBUILD in CHARMM[31] version 35b1. For crystal structures containing more than one protein in the asymmetric unit or multiple side chain conformations, the calculated reduction potentials were averaged over all structures except for Eh HiPIP, where one structure was not used due to unusual Fe-S bond lengths. The redox site was defined to include all metal ligands up to the Cβ for side chains of the protein. Thus, [Fe4S4(SCH2)4] was used for the [4Fe-4S] proteins, [Fe(SCH2)4] for Pf Rd, the copper, His39, His92, Cys89, and Met97 for Pl Pc, and the heme, His18, and Met80 for Ta cyt c. Note that while the calculation of ΔGin terminates the ligand with a methyl group, the protein has a methylene group and the partial charges of the methyl group are redistributed accordingly for the calculation of ΔGout[24].
Results and Discussion
Calculations of Rp and φp
The dielectric radii Rp (Eqn. 2) and the protein electret potentials φp were calculated for the six types of proteins (Table 1). The values of Rp follow the order Cyt c > HiPIPs > Fd ≈ Pc > Rd > FeP. While Rp, which reflects the burial of the redox site (Figure 4) but is also relatively independent of the size of the protein as measured by the molecular weight or number of residues. The values of φp follow a different order Pc < HiPIP < Cyt c < Rd ≈ Fd < FeP and the values of φbb, the contribution of the backbone to φp, have yet another order Pc Cyt c < HiPIP < Rd < Fd ≈ FeP. The backbone is the major contributor to φp, and interestingly, there is little correlation with the net charge of the proteins. The range in Rp, φp, and φbb for homologous proteins is relatively small while the difference between proteins with different folds is larger especially for φbb, indicating that Rp, φp, and φbb are characteristics of the protein folds. However, within a set of homologous proteins, the size of the protein appears to have more of an effect on Rp and the sequence has more of an effect on φp.
Table 1.
Properties of proteins and the calculated dielectric radii Rp and electret potentials at the redox site due to the full protein φp and the backbone φbb
| Protein | Molecular Weight (D) | Number of Residues | Net Charge (e) | Rp (Å) | φp (eV) | φbb (eV) |
|---|---|---|---|---|---|---|
| HiPIP | 8290 ± 900 | 75 ± 9 | −2 ± 6 | 10.3 ± 0.4 | 0.39±0.15 | 0.36±0.04 |
| Fd | 8019 ± 2000 | 69 ± 16 | −13 ± 4 | 8.9 ± 0.2 | 0.59±0.13 | 0.79±0.03 |
| FeP | 63988 ±4400 | 558 ± 24 | −26 ± 9 | 6.4 ± 0.3 | 0.64±0.13 | 0.80±0.07 |
| Pf Rd | 5901 | 53 | −8 | 6.7 | 0.59 | 0.68 |
| Pl Pc | 11466 | 105 | −4 | 8.9 | 0.05 | 0.12 |
| Ta Cyt c | 11416 | 104 | +9 | 11.2 | 0.47 | 0.21 |
Figure 4.
Spheres of radius Rp (red) located at the center of the metal sites and the Connolly surfaces (white) of TtHiPIP, CaFd, and AvFeP (top row, left to right) and Ta cyt c, Pl Pc, and Pf Rd (bottom row, left to right).
Electret-Dielectric Spheres Model
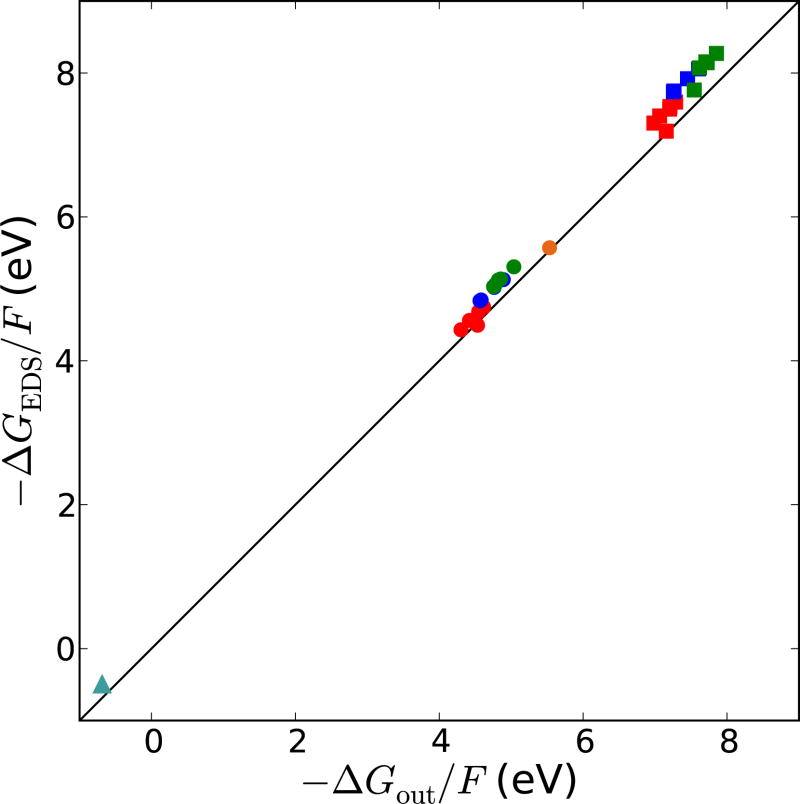
The electret-dielectric spheres model was used to calculate the redox energetics of the six proteins using Rp and φp For the calculations, Rc are 4.34 Å for [Fe4S4(SCH2)4] for the 1-/2-couple, (4.38 Å for [Fe4S4(SCH2)4] for the 2-/3- couple so the value for the 1-/2- is used), 3.46 Å for [Fe(SCH2)4] for the 1-/2- couple, and 3.35 Å for the blue copper site for the 1+/0 couple. The ΔGEDS for the 1-/2- and 2-/3- couples in the concentric dielectric spheres of radius Rc and Rp with the appropriate φp are compared to the full PB calculation of ΔGout (Figure 5) to see how well EDS can model the response to a change in the charge of the redox site. Although good agreement is expected since these quantities are derived from PB calculations albeit from a slightly different charge distribution of the redox site, the agreement also demonstrates that the EDS model is a way of decomposing the full PB calculation. Moreover, the important implication of the agreement is that the EDS model breaks down the solvation free energy into a dielectric response contribution, which depends on Q2, and an electret contribution, which depends on Q. Thus, the degree of burial of the redox site will have a greater effect on the E° of redox sites with a larger absolute net charge, while the electret contribution to E° will be independent of the net charge. In addition, when the protein docks to a redox partner, Rp will increase if the redox site is buried by the redox partner but φp will be relatively unaffected so that only the dielectric response contribution to E° is expected to change significantly. One caveat is that the proteins are assumed to be fairly rigid so that neither the redox site nor the surrounding protein changes much upon reduction. This is thought to be true for the proteins studied here. For instance, even though nucleotide binding to FeP causes a conformational change that decreases the distance the [4Fe-4S]-cluster of FeP and the P-cluster of nitrogenase[32], the environmental change around the [4Fe-4S]-cluster has a small effect on E° (φp is reduced by < 0.05 eV and Rp increases by < 0.2 Å for Av FeP). Other metal sites such as the found in some copper proteins may experience significant changes in the coordination sphere, and other proteins undergo conformational changes. Thus, this approach should be considered a baseline, from which deviations can be studied.
Figure 5.
−ΔGout/F versus − ΔGEDS/F for the 1-/2- (circles) and 2-/3- (squares) redox couples of HiPIP (red), Fd (blue), FeP (green), Pc (cyan), and Rd (orange).
Classifying Redox Properties of Metalloproteins
A plot of Rp versus φbb shows that the burial of the redox site and the polarization of the backbone differ between non-homologous proteins but are relatively consistent within a given fold (Figure 6A), which demonstrates that such a plot can be used to classify protein types. Moreover, the values of Rp and φp are particularly interesting when comparing proteins with the same redox site when the site can achieve multiple redox couples, such as the [4Fe-4S] clusters. Since ΔGin for two redox couples (−0.232 eV for 1-/2- and 3.452 eV for 2-/3-), the range of values of Rp and φp that correspond to a biological range of Eº from −0.6 V to 0.4 V can be found by solving Eqn. (3). When the values of Rp and φp for the [4Fe-4S] proteins are plotted along with the ranges consistent for each couple, the HiPIPs fall in the range corresponding to the 1-/2- couple, the Fd and FeP fall within the range corresponding to the 2-/3- couple (Figure 6B).
Figure 6.

A) Rp versus φbb for cyt c (purple), HiPIP (red), Fd (blue), FeP (green), Pc (cyan), and Rd (orange). B) Rp versus φp for HiPIP (red), Fd (blue), and FeP (green). The shaded area indicates the combination of Rp and φp that results in a E° between 0.4 V and E° = −0.6 V with ΔGin = −0.232 eV (horizontal lines) for the 1-/2- couple or ΔGin = 3.452 eV (vertical lines) for the 2-/3- couple.
Conclusions
The outer sphere contribution to the reduction potential for six types of metalloproteins are examined here in terms of the degree of burial of the redox site inside the protein and the permanent polarization of the protein around the redox site. The dielectric radius of the protein, Rp, is used to measure the degree of redox site burial and the protein electret potential, φp, is used to measure the permanent polarization. Using PB calculations, Rp is shown to be determined mainly by the burial of the redox site within the protein, with only slight dependence on the total size of the protein, while φp is shown to be due mainly to the backbone, although the side chains can alter the values somewhat. Thus, Rp and φp are mainly a function of the fold of the protein, but especially φp can be tuned by the sequence, which is consistent with our previous findings on the effects of fold and sequence[12].
The EDS model is a simple model of a redox protein that can be used to break down the total outer sphere response of the reduction potential into the dielectret response energy and the electret energy. Moreover, it shows that the dielectric response energy determined by Rp is dependent on Q2 while the electret energy determined by φp is dependent on Q. The definitions of Rp and φp provide a useful way of quantitating comparisons of how redox site burial and protein polarization contribute to the redox properties of different proteins. Rp and φp, or better yet φbb, are characteristics of the fold and therefore are useful in identifying how an unknown protein may behave, especially in comparison to a known protein with the same redox site but perhaps different fold. In particular, Rp, and φp are shown to be useful for identifying the physiological couple for redox sites with more than one couple.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM045303) and by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute in the Laboratory of Computational Biology (Z99-TW999999-03). The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the U.S. Government. The continuum electrostatic calculations were performed on computers funded through the William G. McGowan Foundation and Georgetown University.
References
- 1.Langen R, Jensen G, Jacob U, Stephens P, Warshel A. J Biol Chem. 1992;267:25625. [PubMed] [Google Scholar]
- 2.Gunner MR, Honig B. Proc Natl Acad Sci U S A. 1991;88:9151. doi: 10.1073/pnas.88.20.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartz P, Beck B, Ichiye T. Biophys J. 1996;71:2958. doi: 10.1016/S0006-3495(96)79533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ergenekan CE, Thomas D, Fischer JT, Tan ML, Eidsness MK, Kang CH, Ichiye T. Biophys J. 2003;85:2818. doi: 10.1016/S0006-3495(03)74705-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck BW, Xie Q, Ichiye T. Biophys J. 2001;81:601. doi: 10.1016/s0006-3495(01)75726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo Y, Ergenekan C, Fischer J, Tan M, Ichiye T. Biophys J. 2010;98:560. doi: 10.1016/j.bpj.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouesca JM, Chen JL, Noodleman L, Bashford D, Case DA. J Am Chem Soc. 1994;116:11898. [Google Scholar]
- 8.Torres RA, Lovell T, Noodleman L, Case DA. J Am Chem Soc. 2003;125:1923. doi: 10.1021/ja0211104. [DOI] [PubMed] [Google Scholar]
- 9.Niu S, Ichiye T. Mol Simul. 2011;37:572. [Google Scholar]
- 10.Niu S, Wang XB, Nichols JA, Wang L-S, Ichiye T. Journal of Physical Chemistry, A. 2003;107:2898. [Google Scholar]
- 11.Wang X-B, Niu S, Yang X, Ibrahim SK, Pickett CJ, Ichiye T, Wang L-S. J Am Chem Soc. 2003;125:14072. doi: 10.1021/ja036831x. [DOI] [PubMed] [Google Scholar]
- 12.Perrin BS, Jr, Ichiye T. Proteins: Struct, Funct, Bioinf. 2010;78:2798. doi: 10.1002/prot.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu S, Ichiye T. J Am Chem Soc. 2009;131:5724. doi: 10.1021/ja900406j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan EA, Yelle RB, Beck BW, Fischer JT, Ichiye T. Biophys J. 2004;86:2030. doi: 10.1016/S0006-3495(04)74264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böttcher CJF. Theory of Electric Polarization: Dielectrics in Static Fields. Vol. 1 Elsevier Scientific Pub. Co; Amsterdam: 1973. [Google Scholar]
- 16.Connolly ML. Science. 1983;221:709. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 17.Sheridan R, Allen L, Carter C. J Biol Chem. 1981;256 [PubMed] [Google Scholar]
- 18.Reiss H, Heller A. The Journal of Physical Chemistry. 1985;89:4207. [Google Scholar]
- 19.Lewis A, Bumpus JA, Truhlar DG, Cramer CJ. J Chem Educ. 2007;84:934. [Google Scholar]
- 20.Lewis A, Bumpus JA, Truhlar DG, Cramer CJ. J Chem Educ. 2004;81:586. [Google Scholar]
- 21.Han W-G, Noodleman L. Inorg Chem. 2011;50:2302. doi: 10.1021/ic1020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tissandier MD, Cowen KA, Feng WY, Gundlach E, Cohen MH, Earhart AD, Coe JV, Thomas R, Tuttle J. Journal of Physical Chemistry A. 1998;102:7787. [Google Scholar]
- 23.Trasatti S. Pure Appl Chem. 1986;58:955. [Google Scholar]
- 24.Perrin B, Jr, Niu S, Ichiye T. J Comput Chem. doi: 10.1002/jcc.23169. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warshel A, Papazyan A, Muegge I. J Biol Inorg Chem. 1997;2:143. [Google Scholar]
- 26.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci U S A. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 28.Luo Y, Niu S, Ichiye T. J Phys Chem A. 2012;116:8918. doi: 10.1021/jp3057509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller CS, Tran K, Niu S, Wang L-S, Ichiye T. Unpublished data. [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187. [Google Scholar]
- 32.Tezcan FA, Kaiser JT, Mustafi D, Walton MY, Howard JB, Rees DC. Science. 2005;309:1377. doi: 10.1126/science.1115653. [DOI] [PubMed] [Google Scholar]
- 33.Parisini E, Capozzi F, Lubini P, Lamzin V, Luchinat C, Sheldrick G. Acta Crystallographica, Section D. 1999;55:1773. doi: 10.1107/s0907444999009129. [DOI] [PubMed] [Google Scholar]
- 34.Noodleman L, Lovell T, Liu T, Himo F, Torres RA. Curr Opin Chem Biol. 2002;6:259. doi: 10.1016/s1367-5931(02)00309-5. [DOI] [PubMed] [Google Scholar]
- 35.Breiter DR, Meyer TE, Rayment I, Holden HM. J Biol Chem. 1991;266:18660. doi: 10.2210/pdb2hip/pdb. [DOI] [PubMed] [Google Scholar]
- 36.Benning MM, Meyer TE, Rayment I, Holden HM. Biochemistry. 1994;33:2476. doi: 10.1021/bi00175a016. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez A, Benini S, Ciurli S. Acta Crystallogr, Sect D: Biol Crystallogr. 2003;59:1582. doi: 10.1107/s0907444903014604. [DOI] [PubMed] [Google Scholar]
- 38.Rayment I, Wesenberg G, Meyer TE, Cusanovich MA, Holden HM. J Mol Biol. 1992;228:672. doi: 10.1016/0022-2836(92)90849-f. [DOI] [PubMed] [Google Scholar]
- 39.Dauter Z, Wilson K, Sieker L, Moulis J, Meyer J-M. Proc Natl Acad Sci U S A. 1996;93:8836. doi: 10.1073/pnas.93.17.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giastas P, Pinotsis N, Efthymiou G, Wilmanns M, Kyritsis P, Moulis J-M, Mavridis IM. J Biol Inorg Chem. 2006;11:445. doi: 10.1007/s00775-006-0094-9. [DOI] [PubMed] [Google Scholar]
- 41.Backes G, Mino Y, Loehr TM, Meyer TE, Cusanovich MA, Sweeney WV, Adman ET, Sanders-Loehr J. J Am Chem Soc. 1991;113:2055. [Google Scholar]
- 42.Dauter Z, Wilson KS, Sieker LC, Meyer J, Moulis J-M. Biochemistry. 1997;36:16065. doi: 10.1021/bi972155y. [DOI] [PubMed] [Google Scholar]
- 43.Schlessman JL, Woo D, Joshua-Tor L, Howard JB, Rees DC. J Mol Biol. 1998;280:669. doi: 10.1006/jmbi.1998.1898. [DOI] [PubMed] [Google Scholar]
- 44.Jang SB, Seefeldt LC, Peters JW. Biochemistry. 2000;39:641. doi: 10.1021/bi991694v. [DOI] [PubMed] [Google Scholar]
- 45.Strop P, Takahara PM, Chiu H-J, Angove HC, Burgess BK, Rees DC. Biochemistry. 2001;40:651. doi: 10.1021/bi0016467. [DOI] [PubMed] [Google Scholar]
- 46.Sarma R, Barney BM, Hamilton TL, Jones A, Seefeldt LC, Peters JW. Biochemistry. 2008;47:13004. doi: 10.1021/bi801058r. [DOI] [PubMed] [Google Scholar]
- 47.Bau R, Rees DC, Kurtz DM, Scott RA, Huang H, Adams MWW, Eidsness MK. J Biol Inorg Chem. 1998;3:484. [Google Scholar]
- 48.Crowley PB, Matias PM, Mi H, Firbank SJ, Banfield MJ, Dennison C. Biochemistry. 2008;47:6583. doi: 10.1021/bi800125h. [DOI] [PubMed] [Google Scholar]
- 49.Takano T, Dickerson R. Proc Natl Acad Sci U S A. 1980;77:6371. doi: 10.1073/pnas.77.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.