Abstract
Small-cell lung cancer (SCLC) is a highly malignant carcinoma with poor long-term survival. Effective treatment remains highly demanded. In the present study, we demonstrated that External Qi of Yan Xin Qigong (YXQ-EQ) exerted potent cytotoxic effect towards SCLC cell line NCI-H82 via induction of apoptosis. Global gene expression profiling identified 39 genes whose expression was altered by YXQ-EQ in NCI-82 cells. Among them, semi-quantitative RT-PCR and real-time qPCR analyses confirmed that the gene expression levels of apoptotic proteins death-associated protein kinase 2 and cell death-inducing DFFA-like effector b were upregulated, whereas that of oncoproteins DEK and MYCL1, cell migration-promoting proteins CD24 and integrin-alpha 9, and glycolytic enzyme aldolase A were downregulated. These findings suggest that YXQ-EQ may exert anticancer effect through modulating gene expression in a way that facilitates cancer cell apoptosis while represses proliferation, metastasis, and glucose metabolism.
Keywords: Small-cell lung cancer, Gene expression, Cell death, Anti-cancer effect
Introduction
Lung cancer is the leading cause of cancer deaths. It is divided into small-cell lung cancer (SCLC) and non-SCLC (NSCLC). SCLC disseminates rapidly throughout the body via the blood and lymphatic system, and cancerous cells have already metastasized in most SCLC patients upon diagnosis [1, 2]. Over-expression of oncogenes and deletion and inactivation of tumor suppressor genes lead to deregulation of cell cycle control and promotion of cell survival and proliferation while inhibiting apoptosis [1]. It is not surprising that a microarray analysis in two dozens of SCLC cell lines showed strong evidence for the notion that genetic alterations leading to changed apoptotic balance and pro-cell survival play a critical role in the pathogenesis of SCLC [3].
The current standard treatment for SCLC is chemotherapy and radiation therapy, and sometimes combined with surgery. Although SCLC cancerous cells are generally very sensitive to chemotherapy and radiation therapy, almost all patients relapse [1, 2]. It is the most aggressive subtype of lung cancer. The median survival in SCLC is only 5–12 weeks without treatment. With current treatments, the median survival for limited SCLC is 14–20 months, and 7–10 months for extensive SCLC [2]. Metastasis and relapse after treatment are the two biggest challenges for SCLC treatment. Overall speaking, little progress has been made since the introduction of combination chemotherapy. New methods for treatment of SCLC are imminently needed [1, 2].
The concept External Qi (of qigong) refers to the technology and ability of “Qi deployment” therapy and health preservation of traditional Chinese medicine [4–6]. External Qi therapy of Chinese medicine has long been one of the medical practices in China and is under management by the Chinese health authorities [7]. Long-term clinical observations and ongoing studies have shown that patients with cancer and other medical conditions received significant beneficial effects from exposure to External Qi of Yan Xin Qigong (YXQ-EQ) without noticeable side effects [8, 9]. A body of substantial experimental studies has been conducted to show the effects of YXQ-EQ at molecular, cellular, and microorganism levels [7, 10–14]. Most recently, it was reported that YXQ-EQ exhibited potent cytotoxic effects on BxPC3 pancreatic cancer cells [4], PC3 prostate cancer cells [5] and MDA-MB-231 breast cancer cells [6], respectively, by effectively inhibiting ERK1/2 and/or Akt pathways in these cancer cells, without harming normal cells.
In this study, we used SCLC cell line NCI-H82 as a model system to further examine the anticancer property of YXQ-EQ and its potential underlying mechanisms through microarray analysis of gene expression. Our data showed that YXQ-EQ effectively induced NCI-H82 cell death and induced a gene expression pattern that may promote apoptosis while inhibiting proliferation, migration, and glucose metabolism in SCLC cancer cells.
Materials and methods
SCLC cell culture
NCI-H82 cell line was derived by Gazdar in 1978 from the pleural fluid of a 40-year-old patient with SCLC. This cell line has been well characterized and used as an in vitro model for studying lung cancer [15, 16]. Cells were cultured in 24-well plates with Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal calf serum and 1% Pen/Strep in a 37°C incubator with 5% CO2.
YXQ-EQ treatment
NCI-H82 cells were treated with YXQ-EQ as described previously [13]. Briefly, pre-prepared cultures were randomly assigned to groups of YXQ-EQ treated or non-treated. Pre-prepared cultures of the YXQ-EQ treated group were treated with YXQ-EQ for 10 min each session. The number of treatment sessions and schedules varied in each set of the experiments, which will be specified in later sections. Non-YXQ-EQ treated groups had undergone a sham-operated procedure.
MTT assay
The MTT cell viability assay was performed as described previously [11, 17, 18]. Briefly, MTT (3,(4,5-dimethylthiazol- 2-yl)2,5-diphenyl-tetrazolium bromide) (Sigma, St. Louis, MO, USA) was dissolved in PBS at a concentration of 5 mg/ml. Lysing buffer was prepared as described in the following: sodium dodecyl sulfate (SDS, 20% w/v) (Sigma) was dissolved at 37°C in a solution of 50% of DMF (N,N-dimethyl formamide) (Sigma) and deionized water. The pH was adjusted to 4.7. Twenty-five microliters of the 5 mg/ml stock solution of MTT was added to each well and, after 2 h of incubation at 37°C, 100 μl of the lysing buffer was added. After an overnight incubation at 37°C, absorbance of the samples was read at 562 nm using a microtiter plate ELISA reader.
Flow cytometric cell death detection
Cultured NCI-H82 cells were collected 0, 6, 12, 24, and 48 h after 10 min of YXQ-EQ treatment with trypsin digestion. Apoptosiswas determined by annexin Vstaining according to a protocol outlined in the annexinV-FITC apoptosis detection kit (Beckman Coulter Inc, Fullerton, CA, USA). Briefly, cells washed once with PBS and resuspended in binding buffer were stained with annexin V-FITC and propidium iodide (PI) for 10 min at room temperature in the dark. After staining, 30,000 events for each sample were collected and analyzed using a FACScan flow cytometer (Beckman Coulter Inc).
Microarray analysis
RNA isolation from NCI-H82 cells was carried out with Trizol reagent according to the manufacture’s manual as described previously [17, 18]. The human v2.0 Qiagen Operon oligonucleotide library was printed at the Microarray Research Facility of Oklahoma Medical Research Foundation as described previously [17, 18]. Data normalization, robust regression, and selection of hyper-variable genes were conducted as described previously [19, 20].
RT-PCR analysis
RT-PCR analysis was performed as described previously [17, 18]. Oligonucleotide primers were designed using the PrimerQuest software (http://biotools.idtdna.com/primerquest/). cDNA products obtained from 25 ng of total RNA were amplified by PCR using the primer pairs: 5′TCAGACC TTCTGAAGCAGCCCATT3′ and 5′AGAGTTACAGGCAC ACGGGCTAAA3′ for DAPK2, 5′GCTAGAGGAGGATG GAACTGC3′ and 5′CCCGTAGAATGTGGCTTTGAC3′ for CIDEB, 5′GCACTACACCTTGCCTTGCTGTTT3′ and 5′AGC AGCTGACGCTGGAGATCTATT3′ for MYCL1, 5′TCAT CGTGGAAGGCAAGAGGGAAA3′ and 5′GCCTGGCCTG TTGTAAAGCAGTTT3′ for DEK, 5′CATGTTGCCCA GGCTGGTTTAGAA3′ and 5′AAGCCTGTAATCCCAGC ACTTTGG3′ for CD24, 5′AGGAATTGCCGTTCAGAG GACTGT3′ and 5′AGAAGAGCTCCCGGGAAACATTGA3′ for ITGA9, 5′AAAGCGAGCCGAGAAGATCTGGAA3′ and 5′GAACAGTTGGCATCAGGCGTTCTT3′ for RCVRN, and 5′TCTGAAGCACCGGAACTTGCTACT3′ and 5′TGCTG GATATTGGTAGGGCATGGT3′ for ALDOA. RT-PCR products were resolved on 2% agarose gels containing 0.5 μg/ml ethidium bromide and images were captured by Quantity One software (Bio-Rad, Hercules, CA, USA).
Real-time qPCR
Real-time qPCR was performed as described previously [17, 18]. First-strand cDNA was synthesized using the same method as in the RT-PCR. Real-time qPCR was carried out in 96-well PCR plates (Applied Biosysystems, Foster City, CA, USA) using a Bio-Rad iCycler. Optimization of primers was verified by one single product of the predicted size with no primer-dimmer bands using both melting curve profile and agarose gel analysis. Quantitative comparison between 0 h control and samples of other time points was calculated using comparative ΔCT: Data were normalized by subtracting the difference of the threshold cycles (CT) between genes of interest and housekeeping gene GAPDH (ΔCT = CT gene of interest – CT GAPDH). The ΔCT from samples at 0 h was then compared to ΔCT of each of the samples at other time points (ΔΔCT = ΔCT 0 h – ΔCT each other time point). The relative change of gene expression between 0 h and other time point was given by this formula: 2ΔΔCT. All PCR reactions were performed at least in triplicate.
Statistical analysis
For YXQ-EQ treatment, percentage of cell viability and apoptosis to the non-treated control were obtained from 5 biologic replicates. Differences between YXQ-EQ treatment and non-treated control were analyzed by two-way analysis of variance, followed by a Bonferroni multiple-comparison post hoc test. t Test was used for comparison of the data from real-time qPCR. P values of less than 0.05 were considered to be statistically significant.
Results
Induction of apoptosis in NCI-H82 cells by YXQ-EQ
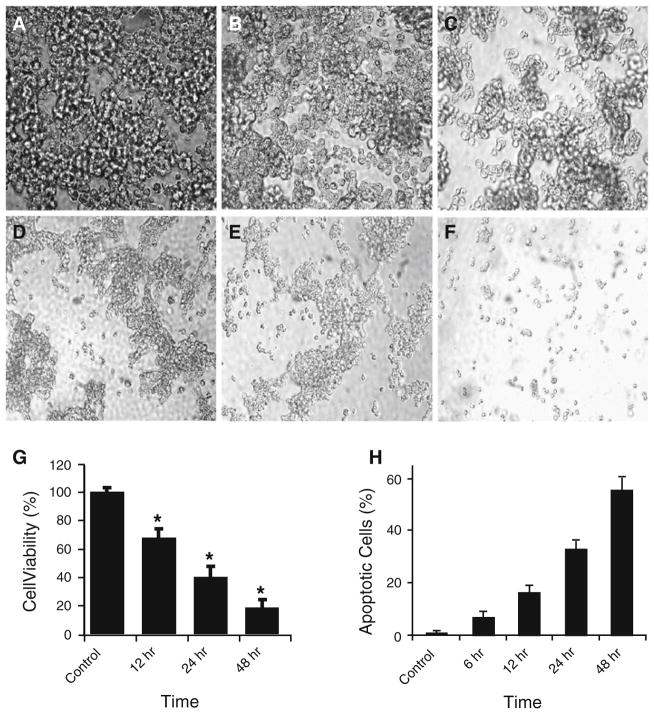
We used SCLC cell line NCI-H82 as an in vitro model to analyze anti-cancer cytotoxicity of YXQ-EQ towards SCLC. This cell line grows as aggregates in suspension and only aggregated cells are viable. Our results showed that while control cells without YXQ-EQ treatment maintained healthy morphology (Fig. 1a), significant cell morphology alteration was observed 12 h after YXQ-EQ treatment (Fig. 1b). By 24 and 48 h after YXQ-EQ treatment, massive cell death could be easily observed under microscope (Fig. 1c, d). On days 5 and 7, many dead cells were disintegrated into debris and disappeared (Fig. 1e, f).
Fig. 1.
YXQ-EQ induced apoptosis in SCLC cells. a–f The morphology of NCI-H82 cells with or without YXQ-EQ treatment. NCI-H82 cells were photographed without YXQ-EQ treatment (a), or 12 h (b), 24 h (c), 48 h (d), 5 days (e), and 7 days (f) after YXQ-EQ treatment, respectively. g Cell viability assessed by MTT assay. NCI-H82 cell viability was monitored without YXQ-EQ treatment (control), or 12, 24, and 48 h after YXQ-EQ treatment. Data presented are mean ± SE of five replicates. Significant difference compared with the untreated cells (P<0.05) is indicated by asterisk. h Induction of apoptosis in NCI-H82 cells by YXQ-EQ. NCI-H82 cells, without YXQ-EQ treatment (control) or 6, 12, 24, and 48 h after YXQ-EQ treatment, respectively, were stained with FITC-conjugated Annexin V and PI. Data presented are mean ± SE of five replicates
We further used MTT assay to quantify the cytotoxicity of YXQ-EQ on NCI-H82 cells by assessing the viability of YXQ-EQ treated or non-treated NCI-H82 cells. Cell viability decreased significantly at 12 h and dropped to less than 20% 48 h after exposure to YXQ-EQ (Fig. 1g). Flow cytometric analysis showed that apoptotic cells appeared as early as 6 h after YXQ-EQ treatment. Apoptotic cells reached 17, 33, and 55% at 12, 24, and 48 h after treatment, respectively (Fig. 1h). In contrast, no significant increase in apoptosis was observed in non-treated cultures at any of these time points. Thus, our findings from microscopic observation, MTT assay, and flow cytometric analysis demonstrated that YXQ-EQ exhibited strong cytotoxicity to SCLC cells. YXQ-EQ induced apoptosis in cancer cells.
YXQ-EQ altered gene expression profile of NCI-H82 cells
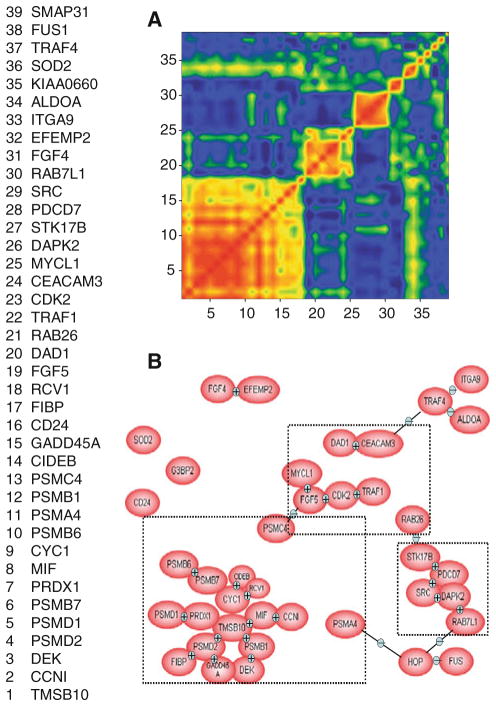
To identify genes that were involved in the anti-cancer effects of YXQ-EQ, cultured NCI-H82 cells were exposed to YXQ-EQ for 10 min. Total RNA was isolated from NCI-H82 cells 12, 24, 36, and 48 h after YXQ-EQ treatment, respectively. Total RNA was also isolated from non-treated NCI-H82 cells as control. Changes in gene expression were analyzed by microarray analysis. Using a selection procedure established previously [20], genes that responded to YXQ-EQ treatment were selected as “hypervariable” genes. Among the 21,329 genes on the array, 39 genes were identified as “hyper-variable genes”. Correlation coefficients have been calculated for the clusters and were represented in graphical outputs as mosaics that could be identified by visual inspection. Correlation mosaics of these 39 genes with hyper-variable expression between YXQ-EQ treated and non-treated cells showed three different major correlation groups (Fig. 2a). Biological association pathways of these 39 genes were modeled using the PathwayAssist (Ariadne Genomics, Rockville, MD). This package extracts functional information on specific genes from the ResNet database using a natural language processing algorithm called MedScan. Data analyzed through this technique can then be resolved into cogent models of the specific biological pathways activated under the experimental conditions used in the array analyses [21]. Three major linked groups were also identified using this analysis (Fig. 2b). Those genes had known annotation information that belonged to various categories, including oncogenes, cell migration-related genes, apoptotic genes and lung cancer-associated genes. Eight of the 39 hypervariable genes were selected for further confirmation based on their known biological function and relevance with cancer development and treatment (Table 1). The changes of their expression levels were evaluated by semi-quantitative RT-PCR and quantitative real-time qPCR and described in the following sections.
Fig. 2.
Differential gene expression induced by YXQ-EQ. Genes with differential expression in control and treated NCI-H82 cells were identified by microarray analysis and selection of hyper-variable genes. a The correlation mosaics for 39 genes highly variable in response to YXQ-EQ treatment. b Biological association pathways of 39 hyper-variable genes were modeled using the PathwayAssist (Ariadne Genomics, Rockville, MD) showing three linked groups
Table 1.
Quantification of representative genes affected by XYQ-EQ by real-time quantitative PCR and semi-quantitative RT-PCR
| # | Accession # | Name | Product size (nt) | Ave. fold change |
|---|---|---|---|---|
| 1 | NM_014326 | Death-associated protein kinase 2 (DAPK2) | 142 | +2.5 (48 h) |
| 2 | NM_014430 | Cell death-inducing DFFA-like effector b (CIDEB) | 247 | +2.3 (48 h) |
| 3 | BC011864 | Lung myelocytomatosis viral oncogene homolog 1 (MYCL1) | 85 | −3.1 (24 h) |
| 4 | NM_003472 | DEK oncogene (DEK) | 200 | −2.3 (24 h) |
| 5 | NM_013230 | CD24 | 83 | −2.1 (48 h) |
| 6 | NM_002207 | Integrin, alpha 9 (ITGA9) | 196 | −2.8 (48 h) |
| 7 | NM_002903 | Recoverin (RCVRN) | 169 | −2.6 (48 h) |
| 8 | NM_000034 | Fructose-1,6-bisphosphate aldolase A (ALDOA) | 54 | −2.3 (36 h) |
The sizes of PCR products and average fold changes measured by real-time qPCR after YXQ-EQ treatment (time point as indicated in the table) for the genes are listed
Up-regulation of apoptosis-promoting genes DAPK2 and CIDE-B
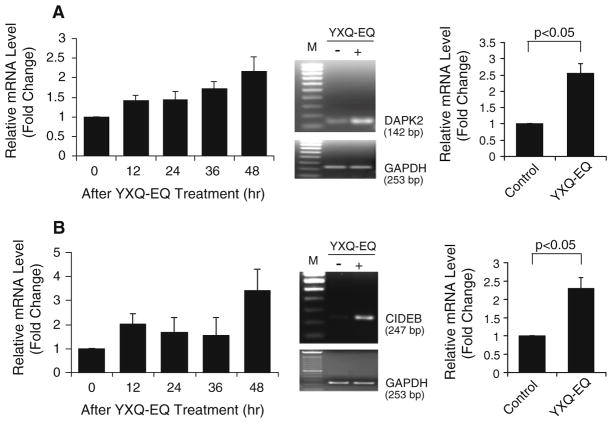
DAPK2 (death-associated protein kinase 2) is a member of the serine/threonine protein kinase family with significant similarity to DAPK1, a positive regulator of apoptosis. Microarray data showed an initial up-regulation of DAPK2 12 h after YXQ-EQ treatment. This increase persisted up to 48 h after the treatment. The time point of 48 h after YXQ-EQ treatment was selected for further confirmation by semiquantitative RT-PCR and real-time qPCR. More than two-fold increase in DAPK2 gene expression in response to YXQ-EQ treatment was detected by real-time qPCR (Fig. 3a; Table 1).
Fig. 3.
YXQ-EQ upregulated the expression of cell death-promoting genes DAPK2 (a) and CIDE-B (b). Left panel microarray analysis. Middle panel semi-quantitative RT-PCR analysis at 48 h after YXQ-EQ treatment. GADPH was used as loading control. M: 100-bp DNA ladder marker. Right panel real-time qPCR analysis at 48 h after YXQ-EQ treatment. Data presented are mean ± SE of the relative mRNA expression levels (fold changes) of three replicates
Cell death-inducing DFFA-like effector b (CIDE-B), as its name indicates, is an apoptosis-inducing protein. It induces cell death in a caspase-dependent manner through cytochrome c release from mitochondria [22]. As shown in Fig. 3b, 12 h after YXQ-EQ treatment, CIDE-B gene expression was increased and the increase sustained up to 36 h. This up-regulation reached a peak at 48 h after treatment. The upregulation of CIDE-B at the time point of 48 h after YXQ-EQ treatment was confirmed by semiquantitative RT-PCR and real-time qPCR, which had more than twofold increase (Fig. 3b; Table 1).
Down-regulation of oncogenes MYCL1 and DEK
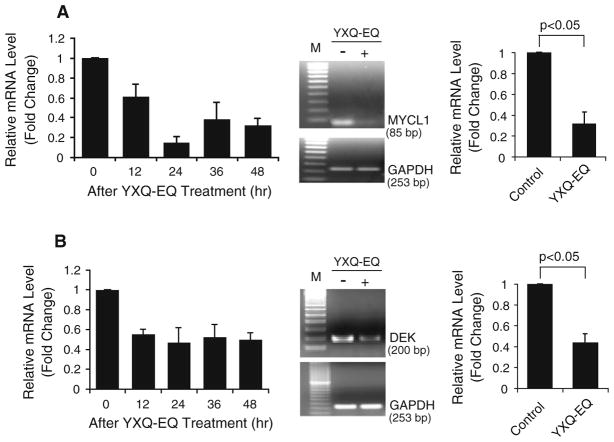
SCLC cell lines, such as NCI-H82, NCI-H209 etc., express MYCL1 [23]. It was recently reported that Myc inhibition in mice elicits regression of established lung tumors [24]. YXQ-EQ treatment of NCI-H82 dramatically reduced the amount of MYCL1 transcripts (Fig. 4a). Microarray data showed an initial down-regulation of MYCL1 at 12 h after YXQ-EQ treatment. This reduction reached a trough at 24 h and remained low at 48 h after the treatment (Fig. 4a). The reduction of MYCL1 expression at 24 h after XYQ-EQ treatment was verified by semi-quantitative RT-PCR and real-time qPCR. There was more than threefold decrease in MYCL1 gene expression in response to YXQ-EQ treatment (Fig. 4a; Table 1).
Fig. 4.
YXQ-EQ downregulated the expression of oncogenes MYCL1 (a) and DEK (b). Left panel microarray analysis. Middle panel semi-quantitative RT-PCR analysis at 24 h after YXQ-EQ treatment, M: 100-bp DNA ladder marker. Right panel real-time qPCR analysis at 24 h after YXQ-EQ treatment. Data presented are mean ± SE of the relative mRNA expression levels (fold changes) of three replicates
The expression of human DEK proto-oncogene in YXQ-EQ- treated NCI-H82 cells was significantly reduced compared with untreated control (Fig. 4b). This decrease was evidenced at 12 h after YXQ-EQ treatment and remained low at 48 h after the treatment. The reduction at 24 h after treatment was confirmed by semi-quantitative RT-PCR. Similar level of decrease was also confirmed by real-time qPCR showing more than twofold down-regulation of this gene by YXQ-EQ treatment at 24-h time point (Fig. 4b; Table 1).
Down-regulation of cell migration regulating genes CD24 and ITGA9
Small-cell lung carcinoma cluster 4 antigen, known as CD24, is a mucin-like adhesion molecule [25]. CD24 is highly expressed in a large variety of human cancers. CD24 was found overexpressed in SCLC [26]. The microarray data showed that the expression of CD24 in YXQ-EQ-treated NCI-H82 cells was significantly reduced compared with untreated control. This decrease was evidenced at 12 h after YXQ-EQ treatment and maintained low through 48 h after the treatment (Fig. 5a). The decrease at the time point of 48 h after the treatment was confirmed by semi-quantitative RT-PCR and real-time qPCR, which had more than twofold reduction (Fig. 5a; Table 1).
Fig. 5.
YXQ-EQ downregulated transcription of cell migration-regulating genes CD24 (a) and ITGA9 (b). Left panel microarray analysis. Middle panel semi-quantitative RT-PCR analysis at 48 h after YXQ-EQ treatment, M: 100-bp DNA ladder marker. Right panel real-time qPCR analysis at 48 h after YXQ-EQ treatment. Data presented are mean ± SE of the relative mRNA expression levels (fold changes) of three replicates
Integrin alpha 9 (ITGA9) was recently reported to mediate tumor cell adhesion and migration [27]. Aberrant upregulation of ITGA9 was found in SCLC [28]. Microarray data showed an initial down-regulation of ITGA9 at 12 h after YXQ-EQ treatment. This decrease reached a trough at 24 h after the treatment and remained low at 48 h after treatment (Fig. 5b). The time point of 48 h after YXQ-EQ was selected for further confirmation by semi-quantitative RT-PCR and real-time qPCR. A more than twofold reduction of ITGA9 gene expression was detected in response to YXQ-EQ treatment (Fig. 5b; Table 1).
Down-regulation of the expression of cancer associated antigens recoverin and Aldolase A
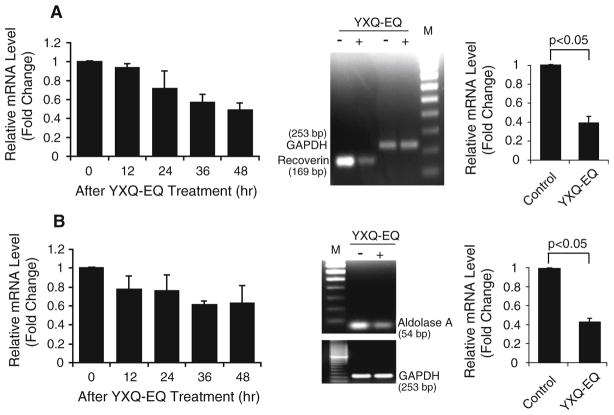
Recoverin (also known as cancer-associated retinopathy protein) is a retina-specific Ca2+-binding protein normally only expressed in neurons in the eye. However, the recoverin protein was detected in 70% of 31 cancer cell lines derived from various cancers including small-cell lung carcinoma [29]. Clinical studies also showed that recoverin was expressed in about 70% of SCLC patients and 85% NSCLC patients, respectively [30, 31]. The aberrant expression of recoverin in cancer cells and presence of recoverin-specific autoantibodies potentially lead to cancer-associated retinopathy (CAR). The microarray data showed that the expression of recoverin in YXQ-EQ-treated NCI-H82 cells decreased significantly compared with untreated control in a time-dependent manner (Fig. 6a). The reduction detected by microarray was confirmed by semi-quantitative RT-PCRand real-time qPCR. The expression of recoverin was down-regulated more than twofold at 48 h after YXQ-EQ treatment (Fig. 6a; Table 1).
Fig. 6.
YXQ-EQ downregulated recoverin (a) and aldolase A (b) gene expression. Left panel microarray analysis. Middle panel semi-quantitative RT-PCR analysis. M: 100-bp DNA ladder marker. Right panel real-time qPCR analysis. In both middle and right panel, for the expression of recoverin, analyses were performed at 48 h after YXQ-EQ treatment, and 36 h for the expression of aldolase A. Data presented are mean ± SE of the relative mRNA expression levels (fold changes) of three replicates
Aldolase A is a glycolytic enzyme, which is overexpressed in human lung cancer [32]. It has been reported as an antigenic retinal protein in melanoma-associated retinopathy (MAR) [33]. Microarray data showed a trend of down-regulation of Aldolase A starting at 12 h after YXQ-EQ treatment. This decrease reached a trough at 36 h and remained low at 48 h after treatment (Fig. 6b). The expression reduction at 36 h after treatment was confirmed by semi-quantitative RT-PCR and real-time qPCR. More than two-fold reduction of Aldolase A gene expression in response to YXQ-EQ treatment was detected (Fig. 6b; Table 1).
Discussion
In the present study we demonstrated that YXQ-EQ effectively induced apoptosis in SCLC cells, which is in accordance with our previous reports that YXQ-EQ induces apoptosis in pancreatic, prostate and breast cancer cells [4–6]. In order to understand the underlying molecular mechanisms of the cytotoxicity of YXQ-EQ on cancer cells, we examined the changes of gene expression profile in SCLC cell line NCI-H82 induced by YXQ-EQ through microarray analysis and identified 39 genes whose expression was altered by YXQ-EQ. Among them, the gene expression levels of oncogenes MYCL1 and DEK were reduced and the levels of apoptotic genes DAPK2 and CIDE-B were increased, which both could inhibit cancer cell proliferation and lead to cell death and play a role in YXQ-EQ-induced cell death/apoptosis. Meanwhile, the expression of cell migration-promoting adhesion molecules CD24 and ITGA9 was found to be downregulated after YXQ-EQ treatment, suggesting YXQ-EQ may have an inhibitory effect on SCLC cell migration. In support of this notion, we have previously reported that YXQ-EQ suppresses the migration and invasion of metastatic breast cancer cell line MDA-MB-231 in vitro [6]. YXQ-EQ also downregulated the expression of glycolytic enzyme Aldolase A. Thus, our findings indicate that YXQ-EQ may exert anti-lung cancer effect through induction of apoptosis and inhibition of proliferation, migration, and glucose metabolism in cancer cells via simultaneously interfering with the expression of multiple genes involved in various biological pathways and functions.
DAPK2 is a recently identified calcium/calmodulindependent protein kinase that has an intrinsic kinase activity. Increase of intracellular Ca2+ can activate DAPK2 and thereby facilitate apoptosis. Overexpression of DAPK2 induces cell death [34], and restoration of downregulated DAPK2 expression in Hodgkin lymphoma induces apoptosis [35]. Aberrant DNA methylation associated with repressing/silencing DAPK2 gene expression was shown in non-small cell lung cancers, nasopharyngeal carcinomas, and colorectal and gastric cancers [36, 37]. It is not known whether the promoter of DAPK2 gene in SCLC cells is aberrantly methylated to inhibit or silence its expression, but it is reasonable to conjecture that the enhanced expression of DAPK2 in NCI-H82 cells had contributed to the induction of apoptosis in NCI-H82 cells after YXQ-EQ treatment. CIDE-B protein is located in mitochondria and dimerized with other CIDE family members [22]. It induces apoptosis through cytochrome c release. Like DAPK2, overexpression of CIDE-B induces apoptosis in transfected cells [38]. Moreover, inactivation of CIDE-B can inhibit CIDE-B-induced apoptosis [38]. Conceivably, overexpression of CIDE-B in cancer cells would promote cancer cell apoptosis. Elevated expression of DAPK2 and CIDE-B in YXQ-EQ-treated NCI-H82 cells that underwent apoptosis suggests that activating apoptotic pathway(s) which involves DAPK2 and CIDE-B is one of the mechanisms utilized by YXQ-EQ to manifest its cytotoxicity towards cancerous cells.
Besides inducing apoptosis, the other common approach to inhibit cancer cell growth is to downregulate oncogene expression. As transcription factors, Myc family proteins transcriptionally activate several hundred target genes that are involved in diverse biological process [39]. They increase cell proliferation and cause cell-cycle progression and play a pivotal role in tumorigenesis in numerous human cancers of diverse origin [1, 39]. Deregulated expressions of c-Myc, N-Myc, and MYCL1 (L-Myc) are evident in many human cancers and shown to promote cancerous cell proliferating [40]. As a potent oncoprotein that plays a pivotal role as a regulator of tumorigenesis in human cancers, Myc is a very attractive target for cancer treatment. Small-cell lung cancer cell lines, such as NCI-H82, NCI-H209, etc., express MYCL1 [23]. Specific targeting of MYCL1 by antisense DNA in SCLC cell lines effectively inhibits cell proliferation. The growth inhibition by the antisense DNA is correlated with the level of downregulation of MYCL1 expression [41]. Retinoic acidinduced growth arrest and apoptosis in NCI-H82 and NCI-H209 cells is associated with substantial reduction of both c-Myc and MYCL1 expression [23]. It is consistent with the observation that reduction of Myc expression in YXQ-EQ- treated lung cancer cells is correlated with the inhibition of cell proliferation and apoptosis.
Human DEK proto-oncogene encodes a 43-kDa nucleic acid binding phosphoprotein. Though it is ubiquitously expressed in mammalian cell nucleus, the expression level is high in proliferating cells and low in resting and terminally differentiated cells [42]. It is a senescence inhibitor. Overexpression of DEK protein is implicated in human carcinogenesis [42, 43]. Since DEK is a senescence inhibitor, reduction of its expression should also help to induce apoptosis in YXQ-EQ-treated cancer cells. It was recently reported that downregulation of DEK expression through RNA interference in human tumor cells result in cell death [44]. Inhibition of cell proliferation and induction of apoptosis are two interrelated but distinct biological processes. Downregulating the expression of oncogenes MYCL1 and DEK while upregulating the expression of apoptotic DAPK2 and CIDE-B in YXQ-EQ treated cancer cells suggests that YXQ-EQ can inhibit cancer cell growth through suppressing oncogene expression in addition to inducing apoptosis through up-regulation of expression of pro-apoptotic genes.
CD24 is a cell surface molecule used as a B cell lineage differentiation marker. It participates in regulation of both cell proliferation and cell–cell interaction. Overexpression of CD24 in various human cancers facilitates cancer cell metastasis and is strongly associated with a more aggressive course of the disease and poor prognosis. It has been suggested that if the expression of CD24 in cancer cells is downregulated, cancer cell proliferation and metastasis may be inhibited [45]. It was recently reported that downregulation of CD24 expression in colorectal tumors inhibits tumor growth [46]. Therefore, CD24 is considered a potential therapeutic target in cancer biology [45, 47]. ITGA9 is a member of the integrin family. Aberrant upregulation of ITGA9 was found in SCLC [28], although its precise role in cancer development and progression remains obscure. However, integrins connect cells to components of the extracellular matrix or to counter receptors on other cells mediating cell adhesion, migration, and other cell–cell contact. It is very likely the overexpression of ITGA9 in SCLCs is also linked to the high metastatic nature of lung cancer cells, and ITGA9 has recently been reported to promote tumor cell adhesion and migration [48]. Thus, CD24 and ITGA9 are very likely among the factors that make the SCLC cells highly prone to metastasis. Downregulation of CD24 and ITGA9 in NCI-H82 cells would affect the capability of cells to migrate, eventually resulting in inhibition of metastasis. Therefore, down-regulation of the expression of CD24 and ITGA9 by YXQ-EQ suggests that YXQ-EQ may inhibit cancer cell migration and metastasis. In agreement, YXQEQ has been shown to repress breast cancer cell migration and invasion [6].
Recoverin is one of the paraneoplastic antigens, which is exclusively expressed within photoreceptor cells and retinal bipolar cells under normal circumstances. It is not known why recoverin is aberrantly expressed in tumor cells including lung cancers. Aberrantly expressed recoverin is a highly pathogenic protein [30, 31]. It is believed that the aberrant expression of recoverin in malignant tumors localized outside the nervous system triggers the production of autoantibodies by the immune system resulting in CAR, an autoimmune syndrome characterized by sudden, progressive loss of vision in association with circulating anti-retinal autoantibodies. Reduction of recoverin in NCIH-82 by YXQ-EQ treatment indicates YXQ-EQ may lower the incidence of SCLC-associated CAR development. Recoverin protein was also found to be associated with caveolin/G-protein-coupled receptor kinases (GRKs) in transformed and cancer cells [49], which suggest the possible involvement of recoverin in caveolin/GRK-dependent regulation of tumor progression, metastasis, and drug resistance [49]. YXQ-EQ may affect these processes through downregulation of the expression of recoverin.
Aldolase A, as a glycolytic enzyme, is expressed in developing embryo and in many adult tissues its expression is downregulated [50]. However, it is overexpressed in human lung cancer and malignant pleural effusion cells [32, 48]. It has been suggested to be involved in glucose metabolic reprogramming that benefits cancer cells [48]. Down-regulation of Aldolase A in SCLC cells by YXQ-EQ may interfere with glucose metabolism that is important in cancer cell proliferation and metastasis.
In summary, our data indicate that the expression of genes involved in multiple cellular processes, such as apoptosis, proliferation, migration, adhesion, and glucose/ energy metabolism, is modulated by YXQ-EQ when it induces cell death in cancer cells. These findings provide novel insights into the mechanisms underlying the anticancer effect of YXQ-EQ and important references for further studies.
Acknowledgments
This study was supported by the Yan Xin Foundation and Grants P20 RR16478-04, P20 RR020143, P20 RR15577, 01700172 from NIH, and Grant HR04-110F from OCAST.
Abbreviations
- SCLC
Small-cell lung cancer
- TCM
Traditional Chinese medicine
- YXQ
Yan Xin Qigong
- YXQ-EQ
External Qi of Yan Xin Qigong
- FITC
Fluorescein isothiocyanate
- PI
Propidium iodide
- ITGA9
Integrin alpha 9
- VCAM1
Vascular-cell-adhesion molecule 1
- GRKs
G-protein-coupled receptor kinases
- MYCL1 (or L-Myc)
Lung myelocytomatosis viral oncogene homolog 1
- DAPK2
Death-associated protein kinase 2
- CIDE-B
Cell death-inducing DFFA-like effector b
- CAR
Cancer-associated retinopathy
- RCVRN
Recoverin
- ALDOA
Aldolase A
Contributor Information
Xin Yan, The Institute of Chongqing Traditional Chinese Medicine, Chongqing, China. New Medical Science Research Institute, New York, NY 10107, USA.
Feng Li, University of Oklahoma Health Sciences Center, Oklahoma, OK 73104, USA.
Igor Dozmorov, Microarray Research Facility, Oklahoma Medical Research Foundation, Oklahoma, OK 73104, USA.
Mark Barton Frank, Microarray Research Facility, Oklahoma Medical Research Foundation, Oklahoma, OK 73104, USA.
Ming Dao, Department of Materials Science and Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Michael Centola, Microarray Research Facility, Oklahoma Medical Research Foundation, Oklahoma, OK 73104, USA.
Wei Cao, University of Oklahoma Health Sciences Center, Oklahoma, OK 73104, USA.
Dan Hu, Email: dan_hu@rics.bwh.harvard.edu, Center for Neurologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, HIM, Room 730, 77 Avenue Louis Pasteur, Boston, MA 02115, USA.
References
- 1.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 2.Thatcher N, Faivre-Finn C, Lorigan P. Management of small-cell lung cancer. Ann Oncol. 2005;16(Suppl 2):235–239. doi: 10.1093/annonc/mdi700. [DOI] [PubMed] [Google Scholar]
- 3.Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R, Minna JD, Pollack JR. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25:130–138. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- 4.Yan X, Shen H, Jiang HJ, Zhang CS, Hu D, Wang J, Wu XQ. External Qi of Yan Xin Qigong differentially regulates the Akt and extracellular signal-regulated kinase pathways and is cytotoxic to cancer cells but not to normal cells. Int J Biochem Cell Biology. 2006;38:2102–2113. doi: 10.1016/j.biocel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Yan X, Shen H, Jiang HJ, Zhang CS, Hu D, Wang J, Wu XQ. External Qi of Yan Xin Qigong induces G2/M arrest and apoptosis of androgen-independent prostate cancer cells by inhibiting Akt and NF-kappa B pathways. Mol Cell Biochem. 2008;310:227–234. doi: 10.1007/s11010-007-9684-2. [DOI] [PubMed] [Google Scholar]
- 6.Yan X, Shen H, Jiang H, Hu D, Zhang C, Wang J, Wu X. External Qi of Yan Xin Qigong Induces apoptosis and inhibits migration and invasion of estrogen-independent breast cancer cells through suppression of Akt/NF-kB signaling. Cell Physiol Biochem. 2010;25:263–270. doi: 10.1159/000276560. [DOI] [PubMed] [Google Scholar]
- 7.Lu Z. Scientific Qigong exploration. Amber Leaf Press; Malvern: 1997. [Google Scholar]
- 8.IYXQA. Yan Xin Qigong collectanea. LOTUS; New York: 1997. [Google Scholar]
- 9.Ming Z. The new frontiers of modern sciences—an introduction to YanXin Qigong. Xinhua Press; Beijing: 1988. [Google Scholar]
- 10.Yan X, Fong YT, Wolf D, Shen H, Zaharia M, Wang J, Wolf G, Li F, Lee GD, Cao W. XY99-5038 promotes long-term survival of cultured retinal neurons. Int J Neurosci. 2002;112:1209–1227. doi: 10.1080/00207450290026166. [DOI] [PubMed] [Google Scholar]
- 11.Yan X, Fong YT, Wolf G, Wolf D, Cao W. Protective effect of XY99-5038 on hydrogen peroxide induced cell death in cultured retinal neurons. Life Sci. 2001;69:289–299. doi: 10.1016/s0024-3205(01)01122-5. [DOI] [PubMed] [Google Scholar]
- 12.Yan X, Lin H, Li HM, Traynor-Kaplan A, Xia ZQ, Lu F, Fang Y, Dao M. Structure and property changes in certain materials influenced by the external qi of qigong. Materials Research Innovations. 1999;2:349–359. [Google Scholar]
- 13.Yan X, Shen H, Zaharia M, Wang J, Wolf D, Li F, Lee GD, Cao W. Involvement of phosphatidylinositol 3-kinase and insulin-like growth factor-I in YXLST-mediated neuroprotection. Brain Res. 2004;1006:198–206. doi: 10.1016/j.brainres.2004.01.068. [DOI] [PubMed] [Google Scholar]
- 14.Yan X, Xia ZQ, Shen H, Traynor-Kaplan A. External Qi of Yan Xin life science technology can revive or suppress enzyme activity of phosphatidylinositol 3-kinase. Bull Sci Technol Soc. 2002;22:403–406. [Google Scholar]
- 15.Xiao L, Lang W. A dominant role for the c-Jun NH2-terminal kinase in oncogenic Ras-induced morphologic transformation of human lung carcinoma cells. Cancer Res. 2000;60:400–408. [PubMed] [Google Scholar]
- 16.Shinoda C, Maruyama M, Fujishita T, Dohkan J, Oda H, Shinoda K, Yamada T, Miyabayashi K, Hayashi R, Kawagishi Y, Fujita T, Matsui S, Sugiyama E, Muraguchi A, Kobayashi M. Doxorubicin induces expression of multidrug resistance-associated protein 1 in human small cell lung cancer cell lines by the c-jun N-terminal kinase pathway. Int J Cancer. 2005;117:21–31. doi: 10.1002/ijc.21094. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Tang Y, Li F, Turner S, Li K, Zhou X, Centola M, Yan X, Cao W. 17beta-estradiol (betaE2) protects human retinal Muller cell against oxidative stress in vitro: evaluation of its effects on gene expression by cDNA microarray. Glia. 2006;53:392–400. doi: 10.1002/glia.20291. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, Tang Y, Li F, Frank MB, Huang H, Dozmorov I, Zhu Y, Centola M, Cao W. Protection against hydrogen peroxide-induced cell death in cultured human retinal pigment epithelial cells by 17beta-estradiol: a differential gene expression profile. Mech Ageing Dev. 2005;126:1135–1145. doi: 10.1016/j.mad.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Dozmorov I, Centola M. An associative analysis of gene expression array data. Bioinformatics. 2003;19:204–211. doi: 10.1093/bioinformatics/19.2.204. [DOI] [PubMed] [Google Scholar]
- 20.Dozmorov I, Knowlton N, Tang Y, Shields A, Pathipvanich P, Jarvis JN, Centola M. Hypervariable genes—experimental error or hidden dynamics. Nucleic Acids Res. 2004;32:e147. doi: 10.1093/nar/gnh146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Frank MB, Dozmorov I, Cao W, Cadwell C, Knowlton N, Centola M, Anderson RE. Identification of mouse retinal genes differentially regulated by dim and bright cyclic light rearing. Exp Eye Res. 2005;80:727–739. doi: 10.1016/j.exer.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZM, Guo K, Toh SY, Zhou ZH, Li P. Mitochondria localization and dimerization are required for CIDE-B to induce apoptosis. J Biol Chem. 2000;275:22619–22622. doi: 10.1074/jbc.C000207200. [DOI] [PubMed] [Google Scholar]
- 23.Weber E, Ravi RK, Knudsen ES, Williams JR, Dillehay LE, Nelkin BD, Kalemkerian GP, Feramisco JR, Mabry M. Retinoic acid-mediated growth inhibition of small cell lung cancer cells is associated with reduced myc and increased p27(Kip1) expression. Int J Cancer. 1999;80:935–943. doi: 10.1002/(sici)1097-0215(19990315)80:6<935::aid-ijc21>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarn JA, Jackson DG, Bell MV, Jones T, Weber E, Sheer D, Waibel R, Stahel RA. The small-cell lung-cancer antigen cluster-4 and the leukocyte antigen Cd24 are allelic isoforms of the same gene (Cd24) on chromosome band 6q21. Cytogenet Cell Genet. 1995;70:119–125. doi: 10.1159/000134075. [DOI] [PubMed] [Google Scholar]
- 26.Jackson D, Waibel R, Weber E, Bell J, Stahel RA. Cd24, a signal-transducing molecule expressed on human b-cells, is a major surface-antigen on small-cell lung carcinomas. Cancer Res. 1992;52:5264–5270. [PubMed] [Google Scholar]
- 27.Lydolph MC, Morgan-Fisher M, Hoye AM, Couchman JR, Wewer UM, Yoneda A. Alpha 9 beta 1 integrin in melanoma cells can signal different adhesion states for migration and anchorage. Exp Cell Res. 2009;315:3312–3324. doi: 10.1016/j.yexcr.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Hibi K, Yamakawa K, Ueda R, Horio Y, Murata Y, Tamari M, Uchida K, Takahashi T, Nakamura Y, Takahashi T. Aberrant up-regulation of a novel integrin alpha-subunit gene at 3p21.3 in small-cell lung-cancer. Oncogene. 1994;9:611–619. [PubMed] [Google Scholar]
- 29.Maeda A, Ohguro H, Maeda T, Wada I, Sato N, Kuroki Y, Nakagawa T. Aberrant expression of photoreceptor-specific calcium-binding protein (recoverin) in cancer cell lines. Cancer Res. 2000;60:1914–1920. [PubMed] [Google Scholar]
- 30.Bazhin AV, Savchenko MS, Belousov EV, Jaques G, Philippov PP. Stimulation of the aberrant expression of a paraneo-plastic antigen, recoverin, in small cell lung cancer cell lines. Lung Cancer. 2004;45:299–305. doi: 10.1016/j.lungcan.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Bazhin AV, Savchenko MS, Shifrina ON, Demoura SA, Chikina SY, Jaques G, Kogan EA, Chuchalin AG, Philippov PP. Recoverin as a paraneoplastic antigen in lung cancer: the occurrence of anti-recoverin autoantibodies in sera and recoverin in tumors. Lung Cancer. 2004;44:193–198. doi: 10.1016/j.lungcan.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Ojika T, Imaizumi M, Abe T, Kato K. Immunochemical and immunohistochemical studies on 3 aldolase isozymes in human lung-cancer. Cancer. 1991;67:2153–2158. doi: 10.1002/1097-0142(19910415)67:8<2153::aid-cncr2820670825>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y, Jia L, He S, Hurley MC, Leys MJ, Jayasundera T, Heckenlively JR. Melanoma-associated retinopathy a paraneoplastic autoimmune complication. Arch Ophthalmol. 2009;127:1572–1580. doi: 10.1001/archophthalmol.2009.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai T, Nomura F, Hoshino K, Copeland NG, Gilbert DJ, Jenkins NA, Akira S. Death-associated protein kinase 2 is a new calcium/calmodulin-dependent protein kinase that signals apoptosis through its catalytic activity. Oncogene. 1999;18:3471–3480. doi: 10.1038/sj.onc.1202701. [DOI] [PubMed] [Google Scholar]
- 35.Tur MK, Neef I, Jost E, Galm O, Jager G, Stocker M, Ribbert M, Osieka R, Klinge U, Barth S. Targeted restoration of down-regulated DAPK2 tumor suppressor activity induces apoptosis in Hodgkin lymphoma cells. Journal of Immunotherapy. 2009;32:431–441. doi: 10.1097/CJI.0b013e31819f1cb6. [DOI] [PubMed] [Google Scholar]
- 36.Wong TS, Chang HW, Tang KC, Wei WI, Kwong DLW, Sham JST, Yuen APW, Kwong YL. High frequency of promoter hypermethylation of the death-associated protein-kinase gene in nasopharyngeal carcinoma and its detection in the peripheral blood of patients. Clin Cancer Res. 2002;8:433–437. [PubMed] [Google Scholar]
- 37.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 38.Erdtmann L, Franck N, Lerat H, Le Seyec J, Gilot D, Cannie I, Gripon P, Hibner U, Guguen-Guillouzo C. The hepatitis C virus NS2 protein is an inhibitor of CIDE-B-induced apoptosis. J Biol Chem. 2003;278:18256–18264. doi: 10.1074/jbc.M209732200. [DOI] [PubMed] [Google Scholar]
- 39.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Opinion—analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 40.Ponzielli R, Katz S, Barsyte-Lovejoy D, Penn LZ. Cancer therapeutics: targeting the dark side of Myc. Eur J Cancer. 2005;41:2485–2501. doi: 10.1016/j.ejca.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 41.Dosakaakita H, Akie K, Hiroumi H, Kinoshita I, Kawakami Y, Murakami A. Inhibition of proliferation by L-Myc antisense DNA for the translational initiation site in human small-cell lung-cancer. Cancer Res. 1995;55:1559–1564. [PubMed] [Google Scholar]
- 42.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein—an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 43.Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Munger K, Wells SI. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA, Cripe TP, Grosveld GC, Wells SI. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol. 2004;35:255–262. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 46.Sagiv E, Starr A, Rozovski U, Khosravi R, Altevogt P, Wang T, Arber N. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res. 2008;68:2803–2812. doi: 10.1158/0008-5472.CAN-07-6463. [DOI] [PubMed] [Google Scholar]
- 47.Pass MK, Quintini G, Zarn JA, Zimmermann SM, Sigrist JA, Stahel RA. The 5′-flanking region of human CD24 gene has cell-type-specific promoter activity in small-cell lung cancer. Int J Cancer. 1998;78:496–502. doi: 10.1002/(sici)1097-0215(19981109)78:4<496::aid-ijc17>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Lin CC, Chen LC, Tseng VS, Yan JJ, Lai WW, Su WP, Lin CH, Huang CYF, Su WC. Malignant pleural effusion cells show aberrant glucose metabolism gene expression. Eur Respir J. 2011;37:1453–1465. doi: 10.1183/09031936.00015710. [DOI] [PubMed] [Google Scholar]
- 49.Miyagawa Y, Ohguro H, Odagiri H, Maruyama I, Maeda T, Maeda A, Sasaki M, Nakazawa M. Aberrantly expressed recoverin is functionally associated with G-protein-coupled receptor kinases in cancer cell lines. Biochem Biophys Res Commun. 2003;300:669–673. doi: 10.1016/s0006-291x(02)02888-7. [DOI] [PubMed] [Google Scholar]
- 50.Kellen JA, Chan A, Caplan B, Malkin A. Aldolase isoenzyme patterns during human ontogeny and in lung, kidney and breast-cancer. Enzyme. 1980;25:228–235. doi: 10.1159/000459257. [DOI] [PubMed] [Google Scholar]