Background: Several components of Drosophila flight muscles are not characterized.
Results: The Z disc protein Z(210) is an adult isoform of Zasp52, and Zasp52 is required for sarcomere structure in adult muscles.
Conclusion: Flight muscles utilize a novel Zasp52 isoform.
Significance: Drosophila Zasp52 is homologous to human ZASP, which is a disease gene.
Keywords: Drosophila, Genetics, Muscle, Myogenesis, Skeletal Muscle, Z-disc, Myofibril, Sarcomere
Abstract
The Z-disc is a critical anchoring point for thin filaments as they slide during muscle contraction. Therefore, identifying components of the Z-disc is critical for fully comprehending how myofibrils assemble and function. In the adult Drosophila musculature, the fibrillar indirect flight muscles accumulate a >200 kDa Z-disc protein termed Z(210), the identity of which has to date been unknown. Here, we use mass spectrometry and gene specific knockdown studies, to identify Z(210) as an adult isoform of the Z-disc protein Zasp52. The Zasp52 primary transcript is extensively alternatively spliced, and we describe its splicing pattern in the flight muscles, identifying a new Zasp52 isoform, which is the one recognized by the Z(210) antibody. We also demonstrate that Zasp52 is required for the association of α-actinin with the flight muscle Z-disc, and for normal sarcomere structure. These studies expand our knowledge of Zasp isoforms and their functions in muscle. Given the role of Zasp proteins in mammalian muscle development and disease, our results have relevance to mammalian muscle biology.
Introduction
The identification and characterization of components of the skeletal muscle myofibril are essential to understanding its normal development and function and are critical to determining how muscle structure is sustained over extended time periods in the animal. A wealth of studies over the past 50 years have served to develop our current understanding of the functional contractile unit of muscle, the sarcomere, in which anchored sliding filaments mediate the process of muscle contraction. The mammalian sarcomere is now known to comprise a large number of different polypeptides, which functionally collaborate to enable efficient posture and locomotion in the animal (reviewed in Ref. 1). Sarcomeres of striated muscles are held together through multi-protein complexes termed Z-discs. These electron dense structures at the borders of each sarcomere integrate filament systems, transmit tension during contraction, and maintain structure and function of the myofibril (2, 3). Recent research has also identified the Z-disc as a nodal point in muscle cell signaling and maintenance, participating in the sensing of mechanical stress, and the transmission of signals to the nucleus (reviewed in Ref. 4).
It is now apparent that mutations affecting several structural components of muscle can cause morbidity and mortality in humans. Mutations affecting components of the thick or the thin filaments can cause a variety of muscular dystrophies and myopathies (5). In addition, mutations affecting the anchoring point of the thin filaments, the Z-discs, can result in muscle disease. Notably, mutations in the myotilin gene cause limb-girdle muscular dystrophy type 1A (6); and a form of myofibrillar myopathy is caused by mutations in the Z-disc alternatively spliced PDZ motif-containing protein (ZASP,2 also known as LIM domain binding 3, or LDB3) (7). In addition, point mutations in the genes encoding a number of Z-disc proteins are associated with dilated cardiomyopathy (reviewed in Ref. 4). These findings underline the critical role that the Z-disc plays in muscle biology, and also reflect the complexity of this structure.
Much interest has recently surrounded the organization and function of Zasp genes, given their roles in mammalian muscle disease, their demonstrated interaction with α-actinin within the Z-disc (8, 9), and their potential involvement in signaling through interaction with other Z-disc components such as calsarcin (29). ZASP proteins contain a PDZ domain, a “ZASP-like motif” (ZM) and one or a few LIM domains. The PDZ domain and ZM motif mediate interactions with α-actinin (10, 11, 12), whereas LIM domains are thought to be critical for additional protein-protein interactions. How these proteins are organized, and how they stabilize other myofibrillar components, is still a significant field of study, and there are clearly several aspects of Z-disc organization and biology that have yet to be uncovered.
Classical approaches to understanding Z-disc composition have utilized biochemical methods to isolate the Z-discs, and subsequently analyze their protein components. The Drosophila system has been used in this venture, and Saide et al. (13) identified a number of Z-disc components. Specifically, these authors demonstrated that Drosophila flight muscle Z-discs comprise α-actinin, and a number of large molecular weight proteins similar to vertebrate titin. In addition, these and subsequent authors also identified a ∼210 kDa component of Z-discs named Z(210), whose accumulation was restricted to the adult muscles, but for which the identity has never been determined (14).
To solve the identity of Z(210) in the context of the fully annotated Drosophila genome, we set out to identify and characterize this protein. Here, we demonstrate that Z(210) is a novel isoform of the Drosophila ZASP gene, Zasp52, and we demonstrate that the exon encoding the Z(210) epitope is adult-specific. When we knock down expression of Zasp52 in the flight muscles, we observe defects in myofibril organization, and a loss of α-actinin localization in the muscles. Our studies help to characterize the myofibrillar components of this important model muscle, and uncover a critical role for Zasp52 in adult muscle structure and function.
EXPERIMENTAL PROCEDURES
Drosophila Methods
y w and w1118 flies were used as controls. The Mhc10 mutant line was obtained from Dr. Sanford Bernstein (San Diego State University, CA). RNAi stocks for Zasp52 (lines 106177 and 36563) were obtained from the Vienna Drosophila RNAi Center. Line 106177 expresses RNAi sequence complementary to exon 20, and has no predicted off-targets. Line 36563 contains an inducible UAS-RNAi construct that drives expression of hairpin RNA complementary to exon 12, and has 17 potential off-targets. We tested both lines and found line 106177 to be more effective in Zasp52 knockdown in adults. Moreover, line 106177 targets exon 20, which is utilized in almost all Zasp52 splice variants known, indicating that it affects the accumulation of all Zasp52 protein isoforms. An RNAi line for Strn-MLCK (stock #3189) was obtained from Bloomington Drosophila Stock Center. Line 3189 expresses RNAi sequence complementary to exon 10 of Strn-MLCK, and has no predicted off-targets. For induction of RNAi in flight muscles, we crossed RNAi lines with lines carrying Act88F-gal4 (15) or 1151-gal4 (16) drivers. Knockdown crosses were carried out at 29 °C. The ability of control flies and Zasp52 KD flies to fly was analyzed by releasing the flies from a vial and determining if they flew or fell to the benchtop.
DNA Methods
All gene names and coordinates are in accordance with the nomenclature adopted by FlyBase (flybase.org), Drosophila genome release 5.48. For RT-PCR, total RNA was isolated from wild type and Zasp52 KD adult thoraces, or from whole wild type third instar larvae, using RNeasy Mini Kit (Qiagen). cDNA synthesis was performed using SMART MMLV-RT (Clontech) according to the manufacturer's protocol. Fragments of Zasp52 were amplified using Advantage 2 Polymerase Mix (Clontech), and alternative splice site selection was confirmed by direct sequencing of the product. Fragment E2-E16 contained exons 2–6, 8–10, 13 and the 5′ part of exon 16; fragment E16-E20 contained exons 16 and 18–20, and fragment E16 contained exon 16 only. To obtain these fragments by PCR, diluted cDNA samples were mixed with the exon-specific primers: E2-E16, 5′-ATGGCCCAACCACAGCTG; 5′-AGACTCCTGTTCCGCCAA; E16-E20, 5′-TCGAGGAGGAGGATTGCTATGAGATGGACA; 5′-GCTAGTCGACTTAGCGCGCGTGATTCTTG; E16, 5′-TCGAGGAGGAGGATTGCTATGAGATGGACA; 5′-TCAAATCGCTCAGGTCTGGCGGAAAGACA.
To create pAFW-Zasp52E16, which expresses Zasp52 exon 16 fused with a FLAG tag, a fragment of exon 16 was amplified using primers: F, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAGGAGGAGGATTGCTATGAGATGGACA;R, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTTCAAATCGCTCAGGTCTGGCGGAAAGACA; and cloned into the Gateway entry vector pDONR221 (Invitrogen). The final expression vector was obtained by recombination between the resulting pDONR221/Zasp52E16 clone and the Gateway destination vector pAFW (Drosophila Genomic Resource Center, DGRC; dgrc.cgb.indiana.edu). A fragment of Myosin heavy chain (Mhc) transcript assayed for loading controls was amplified using the primers: qMhc_F2, 5′-AGTCGCAAATCAGGAGGATG; qMhc_R2, 5′-GATCCAGCAAGACTTCTTCGAG.
Cell Culture
For expression of the FLAG-tagged exon 16 of Zasp52, Drosophila S2 cells were grown in flasks with Schneider's Drosophila Medium (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals), at 24 °C to a density of 2–4 × 106 cells/ml. Cells were then plated into 24-well plates at 5 × 105 cells per well and incubated with DNA and Cellfectin II Reagent (Invitrogen) in serum-free Schneider Drosophila Medium for 18 h at 24 °C. Next, an equal volume of serum-containing culture medium was added, and cells were allowed to grow for an additional 24 h at 24 °C. After this time, growing media were removed from the wells, cells were lysed in 100 μl of Laemmli sample buffer, and heated at 100 °C for 5 min in preparation for SDS-PAGE.
Cryosectioning, Immunofluorescence, and Microscopy
Cryosectioning and immunostaining of adult flies were performed as described before (17). Antibodies to α-actinin, Kettin (also known as Titin and Sallimus (Sls)), and TpnC were obtained from Abcam. Antibody to Z(210) was obtained from Dr. Judith Saide (Boston University Medical Campus, MA) (13), and antibody to Mlp84B was obtained from Kathleen Clark (University of Utah, UT (18)). We used Alexa Fluor antibodies (Molecular Probes) as secondary antibodies in all immunofluorescent stains. Confocal images were generated at the University of New Mexico & Cancer Center Fluorescence Microscopy Shared Resource.
Protein Methods and Western Blotting
Protein samples of adult thoraces were obtained through homogenization of six thoraces per sample in 60 μl of Laemmli sample buffer. For flight muscle samples, muscles of three to six flies were dissected in ice-cold PBS, and homogenized in 20 μl of Laemmli sample buffer. For chemically demembranated (or “skinned”) protein samples, indirect flight muscles were dissected from thoraces of six flies and treated as described by Cripps and Sparrow (19). Briefly, muscles were homogenized in 50 μl of ice-cold solution A containing 10 mm potassium phosphate buffer, pH 7.0, 100 mm NaCl, 2 mm MgCl2, 2 mm EGTA, 1 mm DTT, 0.1 mm PMSF, 0.5% Triton X-100, and Complete Protease inhibitor mixture (Roche). Myofibrils were pelleted at 13,000 × g for 5 min, and washed two times with solution A and two times with solution A without Triton X-100. Washed myofibrils were next pelleted at 13,000 × g for 5 min and resuspended in 60 μl of Laemmli sample buffer. All protein samples in Laemmli buffer were heated at 100 °C for 5 min and then centrifuged at 18,000 × g for 5 min, to remove non-dissolved particles. Protein concentration in samples was measured using the RC DC Protein Assay Kit (Bio-Rad), in order that protein concentrations were normalized between different lanes of the same gel. Proteins were separated in 4–20% acrylamide Mini-Protean TGX gels (Bio-Rad) and either stained with Bio-Safe Coomassie G-250 Stain (Bio-Rad) or electrophoretically transferred onto Amersham Biosciences Hybond ECL membrane (GE Healthcare) for Western blotting. Protein size standards used were Precision Plus Protein Prestained Standards (Bio-Rad). The locations and sizes in kDa of standards are indicated on all stained gels and Western blots. Membranes were immunostained with the same primary antibodies as used for histochemistry. Additionally, we used M2 monoclonal anti-FLAG antibody (Sigma) and rabbit polyclonal antibody to β-actin (#4967, Cell Signaling Technology). Anti-rat, anti-rabbit, and anti-mouse HRP-IgG conjugates (Invitrogen) were used as secondary antibodies, and their chemiluminescent signals was developed using Amersham Biosciences ECL Western blot Analysis System (GE Healthcare). For in-gel protein identification, the band of interest was excised from the Coomassie-stained gel, and shipped to Arizona Proteomics Consortium (Arizona University) for LC-MS/MS analysis.
RESULTS
Identification of Polypeptide Candidates for Z(210)
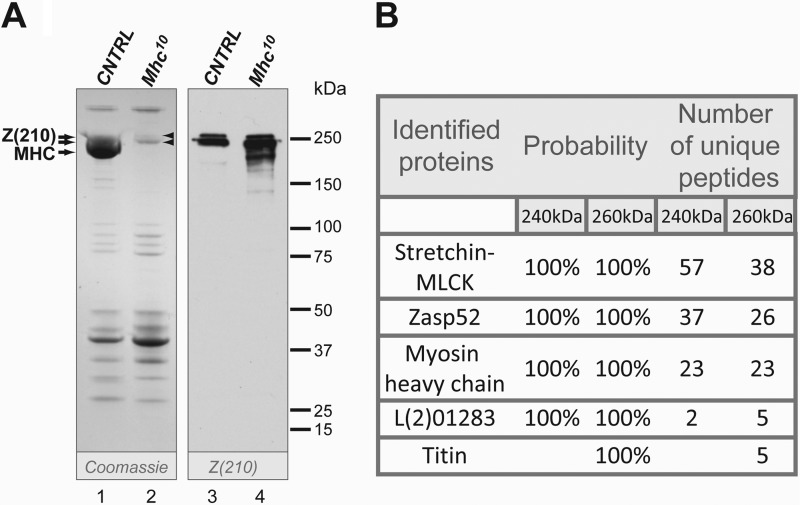
To identify the polypeptide that corresponds to Z(210), we sought to isolate the corresponding protein band from an acrylamide gel, and identify it using mass spectrometry. This approach was technically difficult, since the apparent molecular mass of Z(210) closely matches that of myosin heavy chain (MHC), and the high abundance of MHC might mask the signal arising from rarer polypeptides. To obviate this problem, we sought to identify the band corresponding to Z(210) using protein isolated from an Mhc10 mutant, in which there is no flight muscle myosin produced (20).
We first confirmed that Z(210) was still present in chemically demembranated flight muscle myofibrils from wild-type and Mhc10 mutants. In the absence of flight muscle MHC, a band of ∼240 kDa was more apparent in Coomassie-stained gels. In the Coomassie-stained gels, we also detected a minor band with molecular mass of ∼260 kDa. This band was hardly visible in control samples, since it was obscured by MHC, but was more clearly visible in the Mhc10 lane (Fig. 1A, left panel, arrowheads). Western blot of the same samples with anti-Z(210) antibody also revealed 2 high molecular weight proteins of 240 and 260 kDa (Fig. 1, right panel), although in subsequent blots (see below) the more minor band was not always detected. Vigoreaux et al. (14) detected minor products of degradation in flight muscle samples stained with anti-Z(210) in addition to main Z(210) band. We also observed lower molecular weight products, but only in flight muscle samples of the myosin mutant (Fig. 1, right panel), that might be the result of higher sensitivity of mutant myofibers to protease cleavage during sample preparation.
FIGURE 1.
Identification of high molecular weight proteins in chemically demembranated flight muscle samples. A, skinned protein samples prepared from w1118 (lanes 1 and 3) and Mhc10 (lanes 2 and 4) flight muscles were resolved in a 4–20% polyacrylamide gel and stained with either Coomassie G-250 (left panel) or anti-Z(210) antibody (right panel). Note the bands of apparent mass ∼240 and ∼260 kDa that are revealed in the absence of MHC (arrowheads). B, results of the LC-MS/MS analysis. The protein bands of ∼240 and 260 kDa (arrowheads in panel A) were excised from the gel and subjected to LC-MS/MS analysis. Probability indicates the level of certainty that the named polypeptide corresponds to a component of the sample analyzed.
The 240 and 260 kDa bands were therefore excised from the gel, and subjected to peptide identification by LC-MS/MS. This analysis identified almost 200 peptides, matching five polypeptides that could correspond to Z(210) (Fig. 1B). Stretchin-MLCK (Strn-MLCK) is a large titin-like polypeptide that was previously identified as a thick filament-associated protein (21, 22). MHC was also identified, and its presence in the lane from a myosin-null mutant probably reflected slight contamination from the adjacent wild-type lane. L(2)01289 was only identified in two peptides for the 240 kDa band or five peptides for the 260 kDa band, and its expression pattern did not match that of a muscle-associated protein (23). Titin, an ortholog of the Drosophila Z-line protein Sallimus (Sls), was identified only in the 260 kDa band. The presence of five Sls peptides in the sample must represent a degraded polypeptide, since the smallest known isoform of Sls is 540 kDa. Finally, Zasp52 was identified in 37 peptides (240 kDa band) and 26 peptides (260 kDa band); the prior identification of this protein as a component of larval muscle sarcomeres (9) indicated that Zasp52 was a strong candidate for Z(210). However, we note that Z(210) is restricted to the adult muscles, whereas Zasp52 is present in embryonic/larval and adult muscles (9, 24, 25).
Identification of Zasp52 as the Protein Corresponding to Z(210)
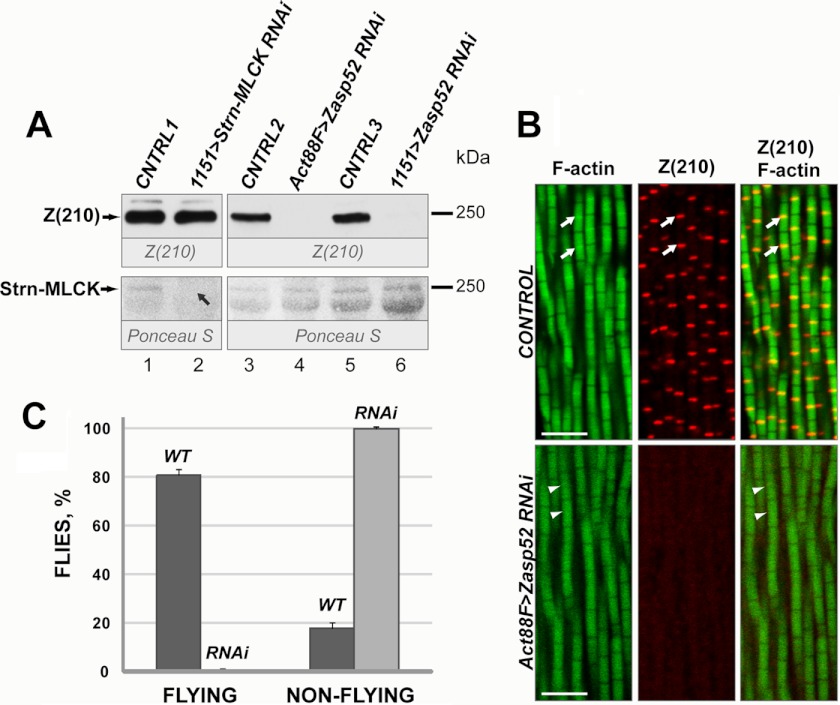
To determine which of the identified polypeptides might correspond to Z(210), we carried out RNAi-mediated knockdowns of the most promising candidates (Strn-MLCK and Zasp52). To achieve this, we crossed the Gal4 drivers 1151-gal4 (16) or Act88F-gal4 (15), to UAS lines that controlled expression of inverted repeats for Strn-MLCK or Zasp52. The accumulation of Z(210) was monitored in each of these samples, alongside either y w controls, or control animals in which the Gal4 drivers were crossed to y w.
The knockdown of Strn-MLCK resulted in the loss of a ∼240 kDa band from the myofibrillar fraction as visualized by Ponceau S staining, but did not affect Z(210) levels based upon immunodetection, for which the signals were equivalent between knockdown and control animals (Fig. 2A, lanes 1 and 2). By contrast, Zasp52 knockdown mediated by either the Act88F or the 1151 driver, resulted in a significant reduction of Z(210) signal relative to control crosses (Fig. 2A, lanes 3–6). These differences did not result from alterations in the total amount of protein loaded, based upon Ponceau S staining (Fig. 2A, lower panel). We conclude that the 240 kDa band, visible by Coomassie staining and Ponceau S staining, represents a mixture of at least two peptides: predominantly Strn-MLCK, with a smaller contribution of Z(210).
FIGURE 2.
Analysis of Strn-MLCK and Zasp52 proteins in knockdown flies. A, Western blot of protein samples prepared from control and knockdown adult thoraces (upper panel). For controls, protein samples were prepared from thoraces of y w (CNTRL1), or of offspring from y w crossed with Act88F-gal4 (CNTRL2), or of offspring from y w crossed with 1151-gal4 (CNTRL3). For Strn-MLCK knockdown, Strn-MLCK RNAi flies were crossed with 1151-GAL4, and offspring were assayed (lane 2). For Zasp52 knockdown, Zasp52 RNAi flies were crossed with either Act88F-gal4 (lane 4) or 1151-gal4 (lane 6), and offspring were assayed. Blots were first stained with Ponceau S to assess equivalent loading of samples (lower panel), followed by staining with anti-Z(210) antibody. A 240 kDa band disappeared on the Ponceau S-stained blot in Strn-MLCK knockdown samples (lower panel, lane 2, black arrow), whereas Z(210) protein accumulation appeared to be unchanged in the same samples (upper panel, lane 2). In Zasp52 knockdown, Z(210) protein disappeared (upper panel, lane 4), or was significantly decreased (upper panel, lane 6), while the intensity of the 240 kDa band on Ponceau S staining was unaltered (lower panel). B, control and Zasp52 KD pharate adults were cryosectioned and stained with phalloidin (left panels) or anti-Z(210) antibody (middle panels). Right panels represent merged anti-Z(210) and phalloidin images. For control, y w were crossed with Act88F-gal4 flies and the offspring were assayed. White arrows point to Z-lines. White arrowheads show absence of Z-lines on phalloidin images of Zasp52 KD flies. Scale bars, 5 μm. C, flight ability of control and Zasp52KD flies. Dark gray bars, WT, offspring from y w crossed with Act88F-gal4; light gray bars, RNAi, Zasp52 knockdown, Zasp52 RNAi flies were crossed with Act88F-gal4. Error bars indicate standard deviation for results from three independent tests.
To determine if the loss of Z(210) immunoreactivity in the Zasp52 knockdowns was reflected in sectioned tissues, we analyzed Z(210) accumulation in the flight muscles of control and knockdown animals. In wild-type, anti-Z(210) labeled very specifically the Z-discs of the IFM myofibrils, consistent with earlier studies (Fig. 2B, arrows) (13, 14). In the Zasp52 knockdown animals, anti-Z(210) labeling was absent (Fig. 2B). We also noted that, in the knockdown animals, IFM myofibril structure was aberrant: the myofibrils showed variations in thickness compared with wild-type, and the Z-discs were indistinct based upon phalloidin staining (Fig. 2B, arrowheads). These observations were consistent with a flightless phenotype of the knockdown flies (Fig. 2C): all tested knockdown females were flightless, compared with 81.1% of control female flies that were able to fly.
Taken together, these data indicate that the most likely identity for Z(210) is an isoform of the Zasp52 gene. Moreover, the function of this gene in the indirect flight muscles is required for normal myofibril structure and function.
Characterization of the Large Flight Muscle Isoform of Zasp52
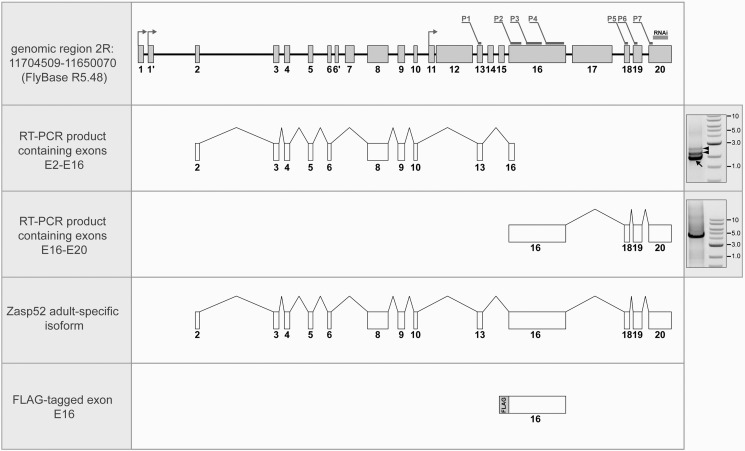
Previous studies of the Zasp52 gene have uncovered significant complexity in its genomic organization and pattern of splicing (24, 25). The current annotation at Flybase.org lists over 20 exons (Fig. 3, top diagram), and a similar number of alternatively-spliced products. Moreover, the gene is expressed at high levels in embryonic and larval stages, in addition to the pupal and adult stages, yet Z(210) has been shown to be adult-specific in its accumulation (13). We therefore sought to identify the exon arrangement of the isoform of Zasp52 corresponding to Z(210), to determine if it corresponds to a known or novel isoform.
FIGURE 3.
A molecular model for the Zasp52 high molecular weight isoform. Main panels, top: genomic region of Zasp52 with genomic coordinates. Exon organization is according to Katzemich et al. (25). The location of the sequence targeted by the 106177 RNAi line is indicated above exon 20. Coding sequences corresponding to the 37 peptides identified by LC-MS/MS are distributed between seven groups, P1-P7, and indicated above the exons. Groups P1 and P5 contain 2 peptides in each; group P2: 3 peptides; P3: 10 peptides; P4: 18 peptides; and groups P6 and P7: one peptide in each. Arrows indicate positions of transcription start sites. Below, from top to bottom: RT-PCR fragment E2-E16 containing exons 2–6, 8–10, 13, and 5′ part of exon 16; RT-PCR fragment E16-E20 containing exons 16 and 18–20; model for high molecular weight isoform of Zasp52 that corresponds to Z(210); exon 16 with the in-frame N-terminal FLAG-tag. Right panels: images of agarose gels containing RT-PCR fragments E2-E16 (upper) and E16-E20 (lower). Arrow points to the product corresponding to E2-E16, arrowheads show RT-PCR products representing partially spliced transcripts. Numbers represent band sizes in kb.
A striking characteristic of the Zasp52 gene structure is the large exon 16, that alone is calculated to contribute 160kDa to the molecular mass of an encoded protein. A splice isoform of Zasp52 that includes most of exons 2–10, plus exon 16 and exons 18–20, would generate a polypeptide whose size is similar to that reported for immunologically detected Z(210) (14). Our proteomic analysis provided some support for this hypothesis, since 30 of the 37 Zasp52 peptides identified from Mhc10 flight muscle myofibrils corresponded to fragments within exon 16 (these peptides are indicated on Fig. 3).
On the other hand, the recent characterization of Zasp52 isoforms by Katzemich et al. (25) and FlyBase (flybase.org) suggested that most exon 16-containing transcripts either have a 5′-truncated exon 16, or utilize an internal promoter, and could only generate polypeptides with relative masses 200 kDa or less.
To resolve this issue, we analyzed Zasp52 transcripts, using RT-PCR of RNA isolated from adult thoraces. In the reaction with primers to exons 2 and 16 we amplified three products in the size range 1.8–2.8 kb. The larger two products contained intron sequences and multiple internal stop codons, and thus could not be translated into long polypeptides (Fig. 3, upper right panel, arrowheads). We believe these two bands simply represent incompletely spliced transcripts. The third product contained exons 2–6, 8–10, 13 and 16 (Fig. 3, upper right panel, arrow), and contained a continuous open reading frame. In the reaction with primers to exons 16 and 20, we amplified one ∼4.5 kb product containing exons 16 and 18–20 (Fig. 3, lower right panel).
Diagrams for the two main products are shown on Fig. 3, and we conclude that they each correspond to part of a larger isoform, that spans exons 2–20. We were not able to identify a single product from exons 2–20, presumably because the expected size of this product (∼6.16 kb) was prohibitive to RT-PCR. Nevertheless, a transcript with this composition would encode a polypeptide of predicted mass 240 kDa, equal to the detected size of Z(210). Our analysis importantly identified a new isoform of Zasp52 based upon its exon composition.
In our RT-PCR reactions we did not identify longer transcripts with alternatively spliced exons that might correspond to the minor 260 kDa isoform of Zasp52, that sometimes appeared on our protein blots. Based on its molecular weight we can predict that this isoform also contains exon 16, and that it might include exons in addition to the isoform indicated in Fig. 3.
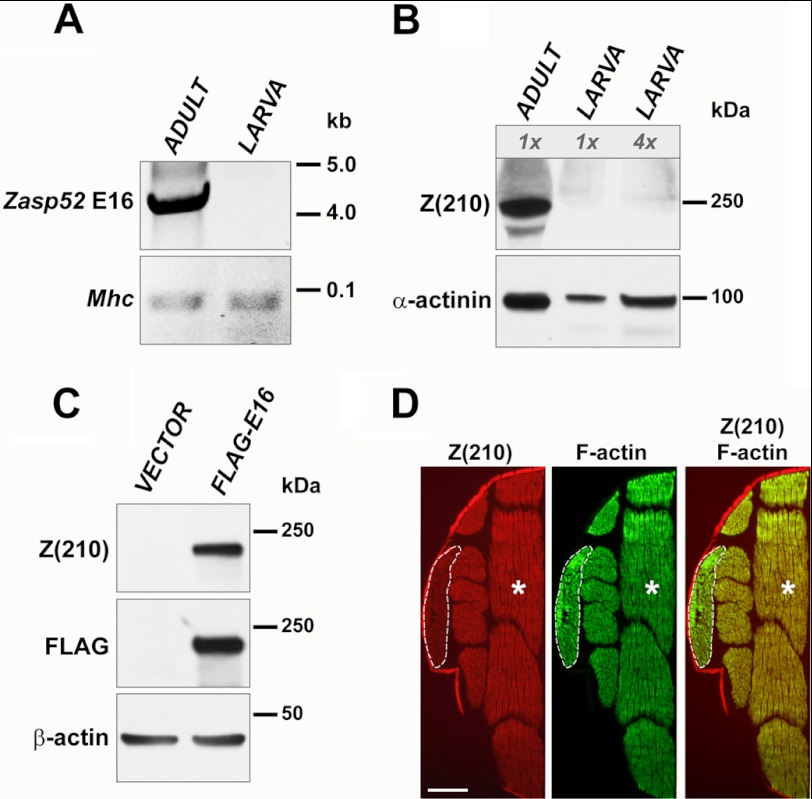
Why does the Z(210) antibody only recognize epitopes in adult tissue? Analysis of high-throughput developmental RNA-sequencing data (23), available at Flybase for Zasp52, suggested that the only exon that might show adult-specific inclusion in transcripts was exon 8. However, when we expressed in S2 cells a truncated FLAG-tagged isoform of Zasp52 containing the spliced exons 2–6, 8–10, and 13, this fragment was not recognized by anti-Z(210) on Western blots, despite being recognized by the FLAG antibody (data not shown).
In parallel, we carried out RT-PCR analysis of larval and adult samples, and found that the large exon 16 was detectable in adults, but not in larvae (Fig. 4A). This finding was at odds with the high-throughput sequencing, yet was confirmed in a number of separate RT-PCR analyses. We also confirmed the absence of Z(210) immunoreactivity in larval tissues by Western blotting: Protein samples prepared from whole larvae did not show Z(210) immunoreactivity, even when the samples contained four times more total protein then the sample from whole thoraces (Fig. 4B, lane 3). The amount of muscle protein in the larval samples was similar to adult thoracic samples, based upon anti-α-actinin staining. This result was consistent with previous finding by Vigoreaux et al. (14) that Z(210) is absent in larval body wall muscle.
FIGURE 4.
The high molecular weight isoform of Zasp52 is adult-specific. A, RT-PCR amplification of cDNA obtained from wild type adult thoraces or from whole larvae. Primers used were designed to amplify exon 16 of Zasp52 (upper panel), or control Mhc transcripts (lower panel). See “Experimental Procedures” for primer sequences. Note that the RT-PCR product was only detected in adult samples. B, Western blot of proteins from thoraces of wild type adults or from whole larvae. The blot was stained with anti-α-actinin antibody to assess equivalence of muscle protein loading (lower panel), and with anti-Z(210) antibody (upper panel). Note absence of Z(210) in larval samples. C, Western blot of proteins expressed in S2 cell culture. S2 cells were transfected either with empty expression vector alone, or with vector expressing FLAG-tagged Zasp52 exon 16. Blots were stained with anti-β-actin antibody, to assess equivalence of protein loading (lower panel), and with anti-Z(210) or anti-FLAG antibody (upper and middle panels, respectively). D, wild type adult flies were sectioned and stained with anti-Z(210) antibody (left panel) or with phalloidin (middle panel). Right panel represents merged anti-Z(210) and phalloidin images. Dashed lines outline TDT muscles, asterisks show flight muscles. Scale bar, 0.5 mm.
Given the apparent adult-specificity of exon 16 as revealed by our RT-PCR analysis, it suggested that this exon might encode the epitope recognized by anti-Z(210). To test this notion, we created an expression construct encoding only exon 16 of Zasp52, fused in-frame to a FLAG tag (Fig. 3). This construct was expressed in Drosophila S2 cells, alongside a control transfection using empty expression vector. No immunoreaction was observed on Western blots of vector-only control transfections; for cells transfected with the FLAG-tagged exon 16 construct, both anti-FLAG and anti-Z(210) recognized a band corresponding to the predicted size of the exon 16 construct in these cell lysates (Fig. 4C).
To confirm the expression pattern of the exon 16-containing isoform, we immunostained with anti-Z(210) antibody cryosections of whole thoraces (Fig. 4D). As expected, both IFMs and TDTs detected very specific Z(210) staining (13, 14). However, in our experiments the jump muscles reacted with the antibody more weakly and, dissimilar to the earlier study (14), showed expression of Z(210) both in small and large cells.
Taken together, our studies describe a previously unknown gene model for Z(210) protein, and characterize it as an isoform of Zasp 52. Moreover, our data significantly expand our understanding of the complexity of Zasp52 expression in Drosophila muscle. We have identified a novel isoform of Zasp52, we have demonstrated it to be adult-specific in its expression pattern, and we have identified an adult-specific exon of this gene. We note that other isoforms of Zasp52 might accumulate in the flight muscles, but Z(210) is the only one that includes the entire exon 16.
Requirement of Zasp52 for Maintenance of Z-disc Components in Flight Muscle
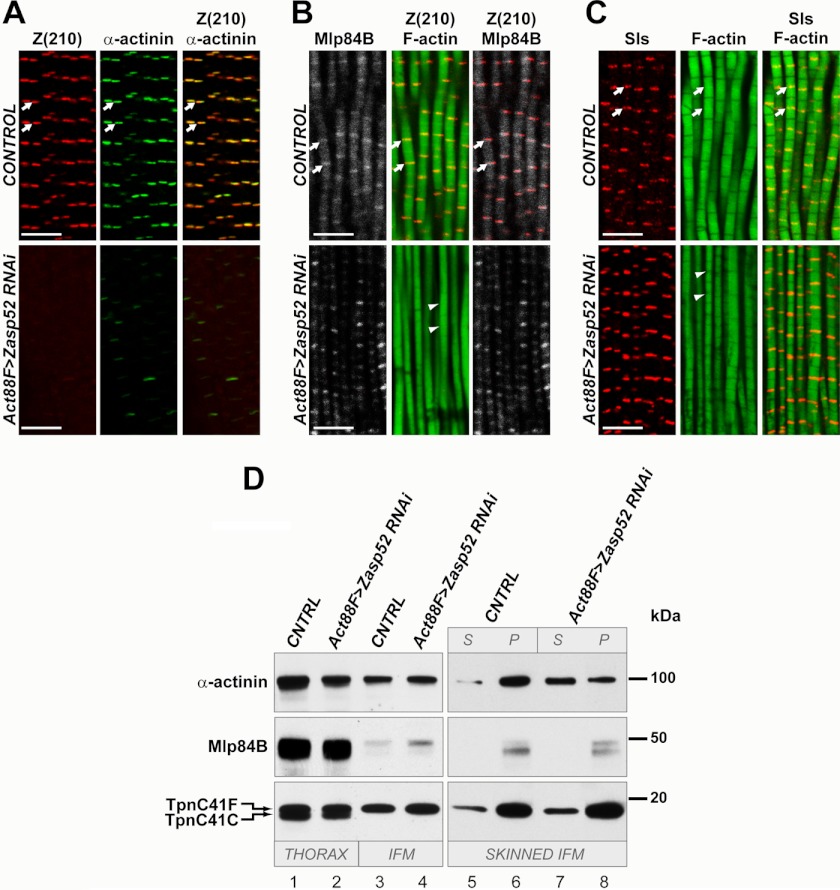
Previous studies of Zasp52 have assessed its requirement in larval body wall muscle for normal muscle structure and function. Zasp52 knockdown studies in embryos showed that this protein is not required for initial sarcomere assembly, but rather is important for Z-line maintenance (27). In larvae, Zasp52 knockdown resulted in a failure of normal myofibril assembly, and mis-localization of α-actinin in the larval muscles (9, 27). Since the myofibrils of adult flight muscles differ significantly in their ultrastructure compared with embryonic and larval muscles, and since the adult muscles accumulate a novel Zasp52 isoform, we sought to define the requirement for Zasp52 in the formation of flight muscle myofibrils. These studies enable us to compare the requirement for Zasp52 in adult muscles, to that in larval muscles. To analyze the function of Zasp52 in the flight muscles, we studied the Act88F>Zasp52 RNAi background that we had used in Fig. 2, and that rendered Z(210) protein levels undetectable and the flies flightless. Knockdown adults were cryosectioned, and analyzed for myofibrillar protein accumulation and localization, using immunofluorescence followed by confocal microscopy. Samples of knockdown animals were also analyzed by Western blotting for the presence of selected myofibrillar proteins.
First, we were interested in the distribution of the crucial Z-disc component α-actinin. In earlier studies, Jani and Schock demonstrated a physical interaction between α-actinin and Zasp52 in larvae (9). The ZASP motif of human ZASP has been shown to be responsible for co-localization with α-actinin (12). This motif is encoded by the constitutive exon 5 of the Drosophila Zasp52 gene, and based upon the structure of the new isoform that we have identified, this adult-specific isoform should also interact with α-actinin. Therefore, we hypothesized that there would be misplacement of α-actinin in Zasp52 knockdown adults. In fact, as with the larval muscles, reduction in Zasp52 levels resulted in a failure of normal localization of α-actinin in cryosectioned tissues (Fig. 5A), indicating a requirement of Zasp52 for α-actinin localization.
FIGURE 5.
Zasp52 is required for proper myofibril organization in adult flies. A, immunostaining of horizontal frozen sections of thoraces from control flies (upper panels) and from Zasp52KD flies (lower panels), using anti-Z(210) (left panels) or anti-α-actinin (middle panels) antibodies. Right panels represent merged anti-Z(210) and anti-α-actinin images. For control, y w were crossed with Act88F-gal4 flies, and offspring were assayed. Note the failure of α-actinin to accumulate at the Z-disks in the knockdown animals. B, immunostaining of frozen sections of control flies (upper panels) and Zasp52KD flies (lower panels), using anti-Z(210), anti-Mlp84B, and phalloidin. For control, y w were crossed with Act88F-gal4, and their offspring were assayed. The middle and right panels represent merged anti-Z(210) and phalloidin, or merged anti-Z(210) and anti-Mlp84B images, respectively. Note that Mlp84B accumulation remains normal in the Zasp52 knockdown muscles. C, immunostaining of frozen sections from control (upper panels) and Zasp52KD (lower panels) adult thoraces, using anti-Sls antibody and phalloidin. For control, y w were crossed with Act88F-gal4 flies, and the offspring were assayed. Note that Sls accumulation and localization is normal in Zasp52 knockdown muscles. A–C: white arrows point to Z-lines. White arrowheads show absence of distinct Z-lines on phalloidin images of Zasp52KD flies. Scale bars 5 μm. D, Western blot of proteins from control or Zasp52KD flies. Proteins from whole thoraces (lanes 1, 2), dissected flight muscles only (lanes 3, 4), or skinned flight muscles (lanes 5–8) were reacted with anti-α-actinin or anti-Mlp84B antibodies. Anti-troponin C (TpnC) staining was used as a protein-loading control. This antibody recognizes two different adult TpnC isoforms, with TpnC41F being expressed in the flight muscles. For control (CNTRL), y w were crossed with Act88F-gal4, and adult offspring were assayed. Note that despite the loss of α-actinin from the Z-discs, apparently normal levels of α-actinin are detected in Zasp52KD muscles. Note that flight muscle levels of Mlp84B are significantly lower than those in whole thoraces. Skinned IFM, lanes 5–8: muscle samples were treated as described under “Experimental Procedures.” Aliquots of supernatant (S) and pellet (P) fractions from the first centrifugation step were analyzed to determine the subcellular locations of the proteins under investigation. Note that Mlp84B and TpnC are predominantly in the pellet (i.e. myofibril fraction) in CNTRL and KD animals. By contrast, α-actinin is present in the pellet in CNTRL animals, but strongly enriched in the supernatant in KD samples.
By contrast, loss of Zasp52 in the flight muscle myofibrils did not affect the accumulation and localization of the Z-disc-associated protein, Mlp84B (Fig. 5, B and D), despite there being noticeable abnormalities in the structure of the myofibrils (Figs. 2B and 5B). Such myofibrils were missing Z-lines, abnormal in shape, and variable in thickness, indicating that Zasp52 is required for maintaining normal myofibril dimensions in the flight muscles.
In the larval muscles, loss of α-actinin resulted in a failure of Sallimus (Sls; a Drosophila titin) accumulation in Z-discs (9). We therefore also studied Sls protein in the flight muscles of Zasp52 knockdowns. Interestingly, in this experiment, Sls localization did not change in response to Zasp52 removal (Fig. 5C). This indicated that, in the flight muscles, Sls organization may not depend upon α-actinin, as it does in the larval muscles.
Interestingly, by Western analysis, there was still significant accumulation of α-actinin protein in the muscles (Fig. 5D, lanes 1–4), despite a failure to detect α-actinin in Z-disc structures through immunostaining. To understand this apparent contradiction, we studied the subcellular distribution of α-actinin in control and knockdown animals, by analyzing soluble and insoluble fractions of Triton-treated flight muscles (Fig. 5D, lanes 5–8). We found that in wild type, α-actinin was predominantly restricted to the Triton-insoluble fraction, containing myofibrils (Fig. 5D, lanes 5 and 6). In contrast, in Zasp52 knockdown animals, the α-actinin was mostly cytosolic, with a smaller portion of the protein being associated with the myofibrils (Fig. 5D, lines 7 and 8). This finding indicates that when not associated with the myofibrils via interaction with Zasp52, α-actinin protein is stable and not subject to immediate proteosomal degradation. In summary, our data identify the Z-disc protein Z(210) as a novel adult-specific isoform of Zasp52, which is required in flight muscle myofibrils for α-actinin localization and normal sarcomere structure.
DISCUSSION
Understanding how myofibrillar components collaborate in the formation of the mature structure is a central challenge in the field, and is critical to understanding the causes and effects of human muscle diseases. A critical component of the sarcomere is the Z-disc. Here, actin filaments are cross-linked by α-actinin to enable transmission of force along the length of the myofibril, and the Z-disc is a central component of muscle cell signaling from the extracellular environment to the nucleus.
Z(210) was identified by Saide et al. (13) as a Z-disc component, and further shown to be predominantly restricted in its accumulation to the adult flight muscles and the adult jump muscles (14). Here, we identify Z(210) as a novel isoform of Zasp52, which has been identified separately in both Drosophila and vertebrates as a critical component of the Z-disc. ZASP proteins are required for normal myofibril maintenance in mice and zebrafish (11, 28), and Zasp mutations in humans are associated with skeletal and cardiac muscle diseases (reviewed in Ref. 2). Characterized mutations of Zasp52 in Drosophila embryos cause myofibrillar defects in embryonic/larval muscles, including irregular formation of Z-disc structures, and muscle attachment defects (9, 24). We show here that the requirement for Zasp52 in normal muscle structure extends to the adult indirect flight muscles, where myofibrils lacking Zasp52 show defects in their shape and diameter.
What is the molecular basis for these defects? In the embryos, at the stage of myofibril formation, loss of Zasp52 does not change myofibril striation pattern and distribution of α-actinin (27). However, later in embryogenesis muscle striations in Zasp52 mutants disappear, suggesting that Zasp52 is required rather for maintenance of Z-lines than for sarcomere assembly (2, 9). In contrast, in the larva, knockdown of Zasp52 results in mis-localization of α-actinin and absence of Z-lines (9). α-Actinin is a well-known Z-disc protein, where it has a fundamental structural role in sarcomere formation (reviewed in Ref. 3). Nevertheless, during Drosophila development, it plays a different role in distribution of another important structural Z-line protein, Sls (a Drosophila ortholog of titin). In the α-actinin mutant embryos, localization of Sls was unaffected, whereas it was lost from the Z-discs of mutant larvae (9, 27). Here, we document a loss of α-actinin from the sarcomeres in adult Zasp52 knockdown flight muscles, and in this case Sls localization is not affected. Clearly, there are structural and organizational differences between embryonic, larval, and adult muscles that account for this differential sensitivity of Sls to the absence of α-actinin and/or Zasp52.
Our data also show that Z-discs of adult flight muscles have a Zasp52 isoform that is not present in larval sarcomeres. It is not known which isoforms of Zasp52 protein are used for embryonic or larval Z-discs assembly, but we can propose that during Drosophila development different isoforms of the same protein can be involved in sarcomere organization versus maintenance, and different isoforms might have distinct interactions with the other Z-disc proteins.
The large size of Z(210) relative to many other Zasp52 isoforms arises from the adult-specific inclusion of the large exon 16 in the mature mRNA. This exon does not contain known functional domains, thus its role in Zasp activity remains poorly understood. It is possible that inclusion of this exon acts to create additional space between the N-terminal PDZ and ZASP domains and the C-terminal LIM domains, to accommodate a specific structural feature of the adult muscles. How this might be reconciled with the presence of Z(210) in the flight and jump muscles, two muscles with vastly different ultrastructures (26), is not clear.
Interestingly, across species as distant as Drosophila and humans, Zasp genes retain their characteristic feature of producing multiple, alternatively spliced isoforms (see online genomic database at www.ensembl.org). From this and other studies (10, 25), it is evident that Zasp splice-isoforms are often specific only for a subset of muscles. Thus, the evolutionarily preserved diversity of Zasp may be required by specialized somatic muscles to better fit their specific functional demands. We speculate that Zasp splice-isoforms might do this via organization of alternative Z-disc structures, adapted for best performance within specialized muscles. Overall, our studies identify and characterize a critical component of the muscle Z-disc, and demonstrate its requirement for normal myofibril maintenance in a specialized skeletal muscle, through the stabilization of α-actinin.
Acknowledgments
We thank Dr. Judith Saide for the anti-Z(210) antibody and Dr. Kathleen Clark for anti-Mlp84B. Confocal images were generated in the University of New Mexico & Cancer Center Fluorescence Microscopy Shared Resource, funded as detailed on hsc.unm.edu/crtc/microscopy/acknowledgment.shtml. We acknowledge technical support from the Dept. of Biology's Molecular Biology Facility, supported by National Institutes of Health Grant Number 1P20RR18754 from the Institute Development Award (IDeA) Program of the National Center for Research Resources.
This work was supported, in whole or in part, by Grant GM061738 (to R. M. C.) from the National Institutes of Health.
- ZASP
- Z-disc alternatively spliced PDZ motif-containing protein
- ZM
- ZASP-like motif
- Mhc
- myosin heavy chain
- Sls
- Sallimus.
REFERENCES
- 1. Lieber R. (2009) Skeletal Muscle Structure, Function, and Plasticity: The Physiological Basis of Rehabilitation, Lippincott Williams & Wilkins, a Walter Kluwer business, Baltimore, MD [Google Scholar]
- 2. Sheikh F., Bang M. L., Lange S., Chen J. (2007) “Z”eroing in on the role of Cypher in striated muscle function, signaling and human disease Trends Cardiovasc Med. 17, 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luther P. K. (2009) The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 30, 171–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frank D., Frey N. (2011) Cardiac Z-disc signaling network. J. Biol. Chem. 286, 9897–9904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bönnemann C. G., Laing N. G. (2004) Myopathies resulting from mutations in sarcomeric proteins. Curr. Opin. Neurol. 17, 529–537 [DOI] [PubMed] [Google Scholar]
- 6. Hauser M. A., Horrigan S. K., Salmikangas P., Torian U. M., Viles K. D., Dancel R., Tim R. W., Taivainen A., Bartoloni L., Gilchrist J. M., Stajich J. M., Gaskell P. C., Gilbert J. R., Vance J. M., Pericak-Vance M. A., Carpen O., Westbrook C. A., Speer M. C. (2000) Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum. Mol. Genet. 9, 2141–2147 [DOI] [PubMed] [Google Scholar]
- 7. Selcen D., Engel A. G. (2005) Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 57, 269–276 [DOI] [PubMed] [Google Scholar]
- 8. Zhou Q., Ruiz-Lozano P., Martone M. E., Chen J. (1999) Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to α-actinin-2 and protein kinase C. J. Biol. Chem. 274, 19807–19813 [DOI] [PubMed] [Google Scholar]
- 9. Jani K., Schöck F. (2007) Zasp is required for the assembly of functional integrin adhesion sites. J. Cell Biol. 179, 1583–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faulkner G., Pallavicini A., Formentin E., Comelli A., Ievolella C., Trevisan S., Bortoletto G., Scannapieco P., Salamon M., Mouly V., Valle G., Lanfranchi G. (1999) ZASP: a new Z-band alternatively spliced PDZ-motif protein. J. Cell Biol. 146, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou Q., Chu P. H., Huang C., Cheng C. F., Martone M. E., Knoll G., Shelton G. D., Evans S., Chen J. (2001) Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell Biol. 155, 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klaavuniemi T., Ylänne J. (2006) Zasp/Cypher internal ZM-motif containing fragments are sufficient to co-localize with α-actinin–analysis of patient mutations. Exp. Cell Res. 312, 1299–1311 [DOI] [PubMed] [Google Scholar]
- 13. Saide J. D., Chin-Bow S., Hogan-Sheldon J., Busquets-Turner L., Vigoreaux J. O., Valgeirsdottir K., Pardue M. L. (1989) Characterization of components of Z-bands in the fibrillar flight muscle of Drosophila melanogaster. J. Cell Biol. 109, 2157–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vigoreaux J. O., Saide J. D., Pardue M. L. (1991) Structurally different Drosophila striated muscles utilize distinct variants of Z-band-associated proteins. J. Muscle Res. Cell Motil. 12, 340–354 [DOI] [PubMed] [Google Scholar]
- 15. Bryantsev A. L., Baker P. W., Lovato T. L., Jaramillo M. S., Cripps R. M. (2012) Differential requirements for Myocyte Enhancer Factor-2 during adult myogenesis in Drosophila. Dev. Biol. 361, 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anant S., Roy S., VijayRaghavan K. (1998) Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development 125, 1361–1369 [DOI] [PubMed] [Google Scholar]
- 17. Morriss G. R., Bryantsev A. L., Chechenova M., LaBeau E. M., Lovato T. L., Ryan K. M., Cripps R. M. (2012) Analysis of skeletal muscle development in Drosophila. Methods Mol. Biol. 798, 127–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stronach B. E., Renfranz P. J., Lilly B., Beckerle M. C. (1999) Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis. Mol. Biol. Cell 10, 2329–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cripps R. M., Sparrow J. C. (1992) Biochem. Genet. 30, 159–168 [DOI] [PubMed] [Google Scholar]
- 20. Collier V. L., Kronert W. A., O'Donnell P. T., Edwards K. A., Bernstein S. I. (1990) Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 4, 885–895 [DOI] [PubMed] [Google Scholar]
- 21. Champagne M. B., Edwards K. A., Erickson H. P., Kiehart D. P. (2000) Drosophila stretchin-MLCK is a novel member of the Titin/Myosin light chain kinase family. J. Mol. Biol. 300, 759–777 [DOI] [PubMed] [Google Scholar]
- 22. Patel S. R., Saide J. D. (2005) Stretchin-klp, a novel Drosophila indirect flight muscle protein, has both myosin dependent and independent isoforms. J. Muscle Res. Cell Motil. 26, 213–224 [DOI] [PubMed] [Google Scholar]
- 23. Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., Brown J. B., Cherbas L., Davis C. A., Dobin A., Li R., Lin W., Malone J. H., Mattiuzzo N. R., Miller D., Sturgill D., Tuch B. B., Zaleski C., Zhang D., Blanchette M., Dudoit S., Eads B., Green R. E., Hammonds A., Jiang L., Kapranov P., Langton L., Perrimon N., Sandler J. E., Wan K. H., Willingham A., Zhang Y., Zou Y., Andrews J., Bickel P. J., Brenner S. E., Brent M. R., Cherbas P., Gingeras T. R., Hoskins R. A., Kaufman T. C., Oliver B., Celniker S. E. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benna C., Peron S., Rizzo G., Faulkner G., Megighian A., Perini G., Tognon G., Valle G., Reggiani C., Costa R., Zordan M. A. (2009) Post-transcriptional silencing of the Drosophila homolog of human ZASP: a molecular and functional analysis. Cell Tissue Res. 337, 463–476 [DOI] [PubMed] [Google Scholar]
- 25. Katzemich A., Long J. Y., Jani K., Lee B. R., Schöck F. (2011) Muscle type-specific expression of Zasp52 isoforms in Drosophila. Gene Expr. Patterns 11, 484–490 [DOI] [PubMed] [Google Scholar]
- 26. O'Donnell P. T., Bernstein S. I. (1988) Molecular and ultrastructural defects in a Drosophila myosin heavy chain mutant: differential effects on muscle function produced by similar thick filament abnormalities. J. Cell Biol. 107, 2601–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rui Y., Bai J., Perrimon N. (2010) Sarcomere formation occurs by the assembly of multiple latent protein complexes. PLoS Genet. 6, e1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van der Meer D. L., Marques I. J., Leito J. T., Besser J., Bakkers J., Schoonheere E., Bagowski C. P. (2006) Zebrafish cypher is important for somite formation and heart development. Dev. Biol. 299, 356–372 [DOI] [PubMed] [Google Scholar]
- 29. Frey N., Olson E. N. (2002) Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 277, 13998–14004 [DOI] [PubMed] [Google Scholar]